Abstract
Glioma, the common brain tumor, which arises from the glial cells, offers worse prognosis and therapy than any other tumors. Despite the genetic and pathological diversities of malignant gliomas, common signaling pathways that drive cellular proliferation, survival, invasion and angiogenesis have been identified. Very often, various tyrosine kinase receptors are inappropriately activated in human brain tumors and contribute to tumor malignancy. During such tumourous states where multiple pathways are involved, a few of them are responsbile for cell differentiation, proliferation and anti-apoptosis. Computational simulation studies of normal EGFR signaling in glioma together with the mutant EGFR mediated signaling and the MAPK signaling in glioma were carried out. There were no significant cross talks observed between the mutant EGFR and the MAPK pathways and thus from the simulation results, we propose a novel concept of ‘multiple-targeting’ that combines EGFR and Ras targeted therapy thereby providing a better therapeutic value against glioma. Diallyl Disulfide (DADS) that has been commonly used for Ras inhibition in glioma was taken for analyses and the effect of inhibiting the EGFR downstream signaling protein with this DADS was analyzed using the simulation and docking studies.
Keywords: computational simulation, multiple targeting, glioma, EGFR, MAPK signaling
Introduction
Molecularly targeted therapies are transforming the treatment of cancer at various levels.1 Small molecule inhibitors that target the cancer dependent enzymes raise the possibility of rational approaches to cancer therapy. The wealth of molecular information from the recent genomics technologies offer a remarkable opportunity for new target discovery.2,3 Systems level view of perturbed networks or pathways can provide promising therapeutic strategies. Glioma is known as a highly cellular tumor with poorly differentiated, round or pleomorphic cells that are occasionally multinucleated, nuclear atypia, anaplasia, endothelial proliferation and pseudopalisading necrosis. Multiple genetic level changes involve in the development of primary and secondary gliomas. Hence, inhibitions of multiple targets are essential to control the rapidly growing tumour cells.4 There are two major oncogenic pathways such as PI3K pathway and MAPK pathway. The tyrosine kinase activity of growth factor receptors is stimulated upon the binding of their cognate growth factors, which results in the stimulation of multiple downstream signaling cascades. These signaling pathways are important for a wide range of cellular functions including protein synthesis, transcription, angiogenesis, regulation of the cell cycle, cell proliferation and survival. Many components of these intracellular signaling pathways are identified as the potential therapeutic targets for developing new treatments against malignant gliomas,5 which are associated with poor prognosis despite all therapeutic options currently available.4
In PI3K signaling pathway, the EGFR gets activated upon binding to the epidermal growth factor thereby recruiting the PI3K pathway to the cell membrane. This pathway converts phosphatidylinositol-4, 5-bisphosphate (PIP2) to the second-messenger molecule PIP3. This second messenger then activates the downstream effector molecules, such as AKT and mTor, mammalian target of rapamycin, which assist to induce the cellular proliferation and block apoptosis, while PTEN terminates the PIP3 signal. The mutant receptor EGFRvIII is persistently activated in the absence of ligand, owing to an in-frame deletion within the extracellular ligand-binding domain.6 This activation plays an important role in the biological processes like cell growth, metabolism, survival and mitogenesis.
The MAPK (mitogen-activated protein kinase) super-family is composed of three major sets of kinases: the extracellular-receptor kinases (ERKs) and two types of MAPK-related kinases that respond to cellular stress and inflammatory signals. These signals are typically transduced through small GTPases of the p21Ras super family composed of Ras, Rho and Rab families along with a few specific kinase activities. The EGF-receptor and NGF-receptors signal the p38 and ERK MAPKs, through the activation of Ras-GTPase.7 Though, EGFR normally correlates with the downstream signaling of MAPK signaling pathway, recent studies have shown that the mutant EGFR VIII shown distinct signaling response with PI3K pathway which significantly dominates the MAPK and STAT3 pathways. This suggest that, there would certainly be very less cross talk between the mutant EGFR pathway and so course mediated MAPK pathway in glioma.8
These two pathways were modeled and simulated using the parameters obtained from literature and the reaction kinetics databases such as KMedDB (http://sysbio.molgen.mpg.de/KMedDB) and SABIO-RK (http://sabio.villa-bosch.de/SABIORK), a curated database of biochemical reactions and kinetic properties. An investigation of model differences is important while analyzing the dynamics of signal transduction computational models.9 Hence, the normal and mutant EGFR mediated PI3K signaling combined with the MAPK signaling were modeled to analyze the effect of proliferation and thereby recognizing an alternate strategy, ‘multiple-targeting’10 for glioma therapy. This study, thus, predicts the systems behavior by integrating several levels of useful information.
Methodology
All the simulations in this research work were performed using the Cell Designer software (www.celldesigner.org),11 whose networks are able to link with simulation and other analysis packages through the systems biology workbench (SBW) (http://sbw.kgi.edu/).12 The kinetic parameters used for modeling the pathways are summarized in Table 1 and the ODEs of the model are given in Table 2. The initial model was simulated with the concentration of normal EGFR and the model was altered to simulate the mutant EGFR kinetics.
Table 1.
List of Kinetic parameters.
| Proteins | Initial concentration (nM) | Km (nM) | Velocity of the substrate (nM.S−1) | References |
|---|---|---|---|---|
| F | 0.25 | 0.41 | 0.312 | 13 |
| IGFR | 0.25 | 0.41 | 0.481 | 13 |
| PI3K | 6.02 | 1 | 0.0857 | 13 |
| PTEN | 0.03 | 0.125 | 18.3 | 13 |
| PIP3 | 10 | 1 | 0.0040 | 13 |
| PKB | 0.05 | 1 | 0.005 | 13 |
| RAF | 1 | 0.805 | 0.70601 | 13 |
| EGF | 100 | 0.25 | 800000 | 14 |
| TGF | 100 | 0.25 | 800000 | 14 |
| EGFR | 1.11 | 3.3 | 2.6933 | 14 |
| PLC | 0.2 | 0.031 | 25 | 14 |
| grb2 | 0.1 | 0.0005 | 0.003 | 14 |
| SOS | 0.5 | 0.0093 | 0.13 | 14 |
| Ras GDP | 1 | 0.50505 | 0.02 | 15,16 |
| Ras GTP | 0.1 | 0.50505 | 0.02 | 15,16 |
| Raf inactive | 0 | 0.50505 | 0.02 | 15,16 |
| Raf active | 0.5 | 2.5641 | 10 | 15,16 |
| Mek active | 0.6 | 0.15909 | 0.3 | 15,16 |
| Mek inactive | 0 | 0.0463 | 0.3 | 15,16 |
| Erk active | 3.8 | 15.657 | 6 | 15,16 |
| Erk inactive | 0 | 7.8 | 0.25 | 15,16 |
| IP3 | 0.0142 | 0.006 | 9.9821 | 15,16 |
| DAG | 0.16 | 0.15 | 17187.09 | 15,16 |
| Ca | 0.9 | 1.2 | 0.3428 | 15,16 |
| PKC | 80 | 0.62 | 9.482 | 15,16 |
| CAM | 0.1 | 1.2 | 0.0092 | 15,16 |
| CAMK | 0.83 | 0.25 | 7.623 | 15,16 |
| MEK | 3.6 | 0.9 | 0.8888 | 15,16 |
| ERK | 7.5 | 0.085 | 667435.7 | 15,16 |
| MTOR | 0.025 | 1 | 0.2439 | 15,16 |
| RAS | 2 | 0.61 | 0.5363 | 15,16 |
Table 2.
ODEs of the model.
| Raf +ras gtp<->raf.ras gtp->raf* | D[Ras.GTP]/dt=- k11y(11)y(12)+k12y(13)+k13y(13) |
| D[Raf]=-k11y(11)y(12)+k12y(13) | |
| D[RasGTP.Raf]/dt=k11y(11)y(12)-k12y(14)-k13y(14) | |
| D[Raf*]/dt=-k13y(13) | |
| Raf*+mek<->raf*.mek->mek* | D[Raf*]=-k14y(14)y(15)+k15y(16)+k16y(16) |
| D[pp2A]/dt=-k14y(14)y(14)+k15y(16) | |
| D[Raf*.pp2A]/dt=k14y(14)y(15)-k15y(12)-k16y(12) | |
| D[Raf]/dt=k16y(16) | |
| Mek*+erk<->mek*.erk->erk* | D[Raf*]/dt=-k17y(14)y(17)+k18y(18)+k19y(18) |
| D[Mek]/dt=-k17y(14)y(17)+k18y(18) | |
| D[Raf*.Mek]/dt=k17y(14)y(17)-k18y(19)-k19y(19) | |
| D[Mek*]/dt=-k19y(18) | |
| Raf*+PP2A<->raf*.pp2A->raf | D[Mek*]/dt=-k20y(19)y(20)+k21y(21)+k22y(21) |
| D[pp2A]=-k20y(19)y(20)+k21y(21) | |
| D[Mek*.pp2A]/dt=k20y(19)y(20)-k21y(17)-k22y(17) | |
| D[Mek]/dt=k22y(21) | |
| Mek*+pp2A<->mek*.pp2A->mek | D[Mek*]/dt=-k23y(19)y(22)+k24y(23)+k25y(23) |
| D[Erk]/dt=-k23y(19)y(22)+k24y(23) | |
| D[Mek*.Erk]/dt=k23y(19)y(22)-k24y(24)-k25y(24) | |
| D[Erk*]/dt=-k25y(23) | |
| Erk*+mkp<->erk*.mkp->erk | D[Erk*]/dt=-k26y(24)y(25)+k27y(26)+k28y(26) |
| D[Mkp]/dt=-k26y(24)y(25)+k27y(26) | |
| D[Erk*.Mkp]/dt=k26y(24)y(25)-k27y(22)-k28y(22) | |
| D[Erk]/dt=k28y(26) |
Diallyl Disulfide (DADS) is found to be effective in inhibiting tumor growth through H-ras inhibition, that results in the improved neurological and extended life span of the species that received DADS as a pretreatment.17 The structure of this compound was generated using ACD Chemsketch (www.acdlabs.com) and was docked with three different kinases such as initiating EGFR, the downstream signaled PI3K and ERK using Autodock 4.0 (www.autodock.scripps.edu) to examine the effect of DADS against the kinases.
Results and Discussion
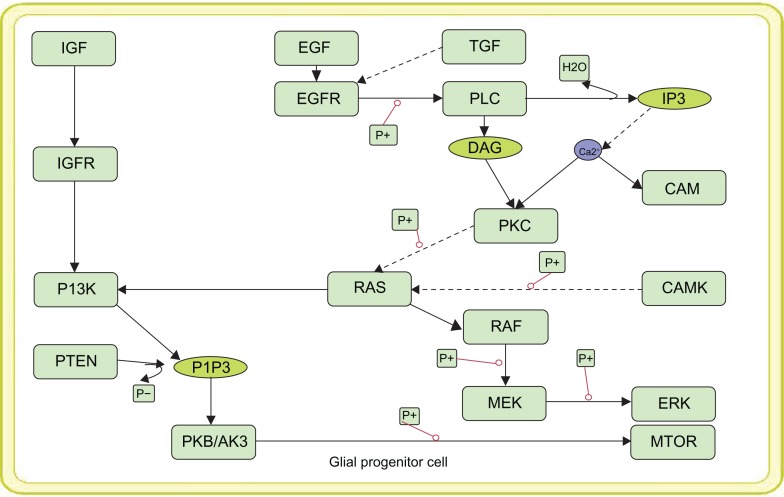
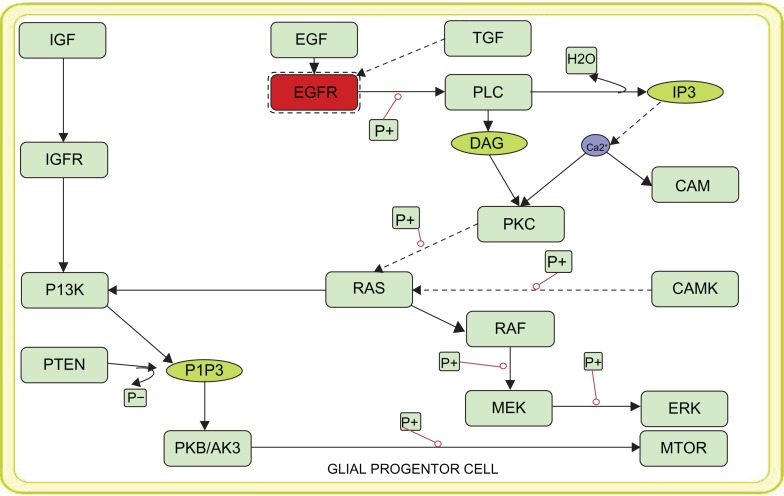
Vibrant modeling of the disease mechanism is often limited to mechanistic details of the entities available within the pathways. Thus using the quantitative information of reaction rates and the molecular concentration of such entities, we have developed a mathematical model of the oncogenic pathways of glioma such as normal EFGR mediated P13 signaling pathway (Fig. 1), mutant EGFR (Fig. 2) mediated downstream signaling of PI3K pathway and the MAPK signaling oncogenic pathway. The kinetic parameters and the chemical equations along with the concentration of the normal and mutant EFGR obtained from the literature were used to simulate the oncogenic pathways. Simulations of these pathways were carried out under different time scales in order to analyze the levels of all the components in the pathway. For the purpose of simulation, the ions present in the model were given a default value of 1.0. All the other parameter values were obtained from the previous works.13–16
Figure 1.
Pictorial view of the constructed model of Normal EGFR mediated PI3K signaling.
Figure 2.
Representation of the constructed model of mutant EGFR (varied parameters from that of normal EGFR) mediated PI3K signaling.
Simulation Analysis
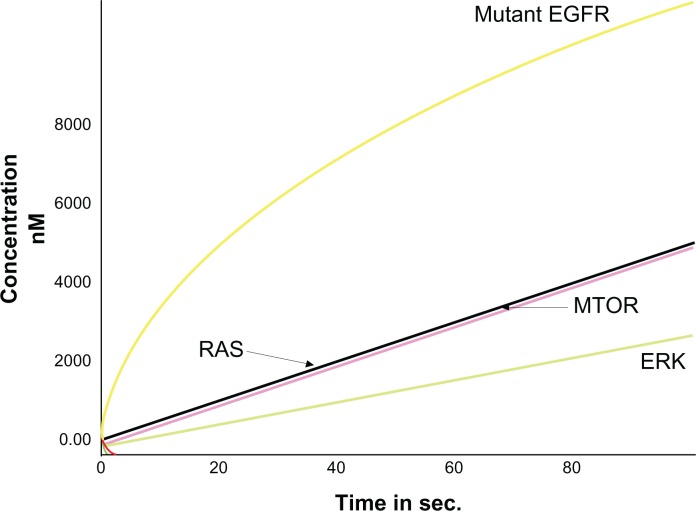
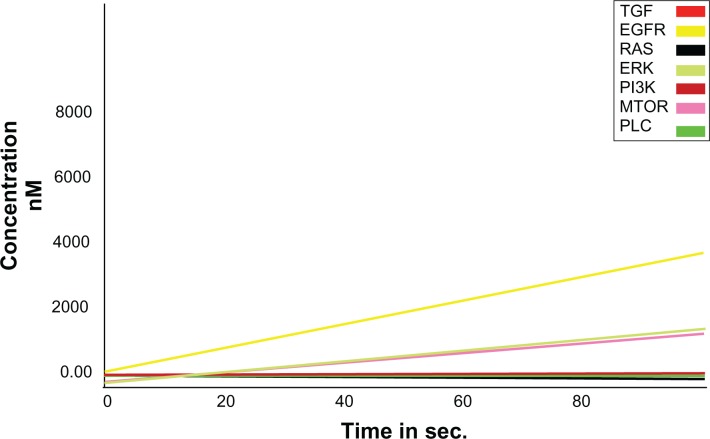
Results of normal EGFR simulations showed that a high level of EGFR expression was achieved by increasing the timescale when compared with other entities within the pathway. Further, it was observed that the expression of mutant EGFR was higher than other proteins involved in the pathway (Fig. 3). This serves as an initiation state and the mutant EGFR increased the expression of other onco-proteins involved within the pathway. The time scale analyses of the models were carried out and the levels of different entities at different time periods were recorded. It was observed that when the DADS was added to inhibit the mutant EGFR, expression of the mutant EGFR reduced followed by the decrease in the levels of downstream signaling proteins (Fig. 4). Thus it is evidently recognized that cell proliferation could be decreased by inhibiting the mutant EGFR system.
Figure 3.
Expression of mutant EGFR (Shown in Yellow).
Figure 4.
Inhibited Mutant EGFR kinetics (Shown in yellow).
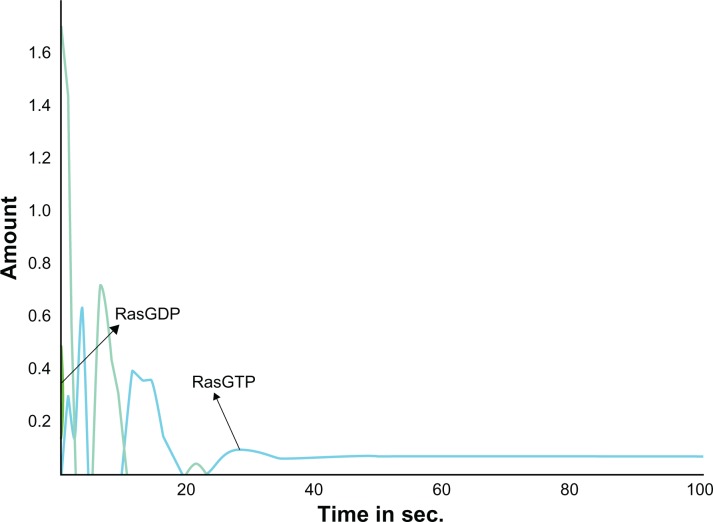
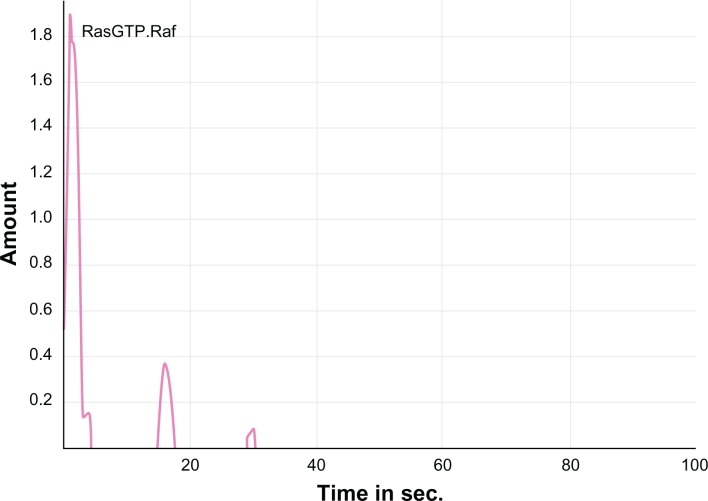
Similarly, the computational simulation of the modeled MAPK pathway resulted in the increased expression of Ras. GTP proteins thereby activating the Ras (Fig. 5) from its unbound Ras. GDP form to a bound Ras. GTP form. This in turn activated the cascading proteins Raf, Mek and Erk which were up-regulated by increasing the expression levels of these proteins upon increased time scales. This activation was achieved from the normal EGFR mediated signaling. The active form of Ras. GTP protein was inhibited with DADS (Fig. 6) and a prolongation in the activation process of the cascading proteins was observed. Hence, the reduced signaling of oncogenes may decrease cell proliferation. Further, the predicted Ki values from the docking analysis were used in the model to examine the inhibition kinetics of the kinases. Targeting PI3K with DADS gave an improved effect of anti-proliferation. Hence, as the DADS inhibition rate is in the order of millimolar, a novel compound may be designed so that, the mutant EGFR which signals the kinases can be inhibited to yield a better therapeutic value against glioma.
Figure 5.
Activation of Ras. GTP by the Normal EGFR signaling.
Figure 6.
Inhibition of activated Ras. GTP. Raf.
From this study, we propose that combined targeting of these two proteins will provide new insights in cancer therapy. Thus there arises a highly demanding need to develop compounds favoring multiple targeting18 that would be a promising therapeutic agent in future.
Docking Analysis
In order to examine the effect of DADS against the kinases, their structures [1I8I (crystal structure of Mutant EGFR VIII), 1E8X (crystal structure of PI3K) and 1TVO (crystal structure of ERK)] obtained from PDB (www.rcsb.org) were docked with the inhibitor, Diallyl Disulfide (DADS) (Fig. 7). The docking calculations were performed using Autodock, an automated docking program that uses a Lamarckian genetic algorithm19 and empirical binding free energy function.20
Figure 7.
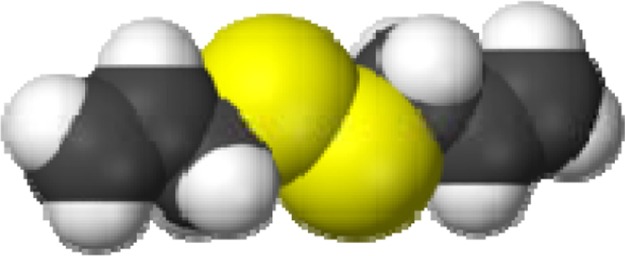
Structure of Diallyl Disulfide (DADS).
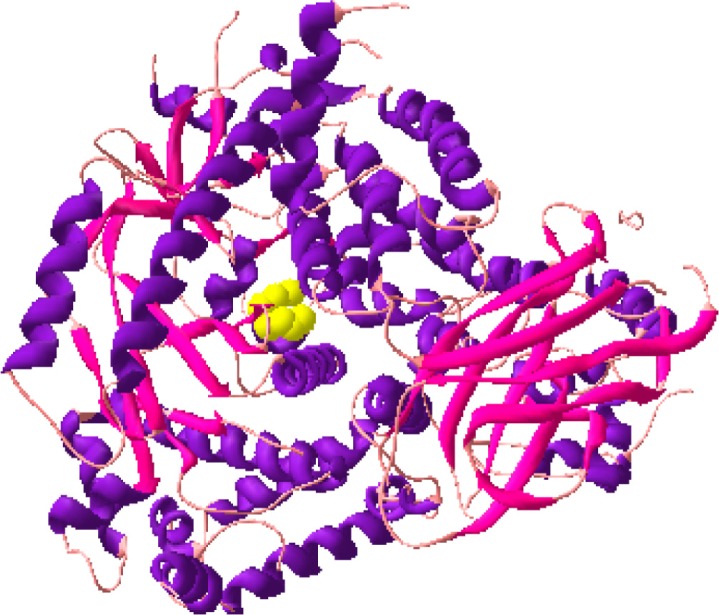
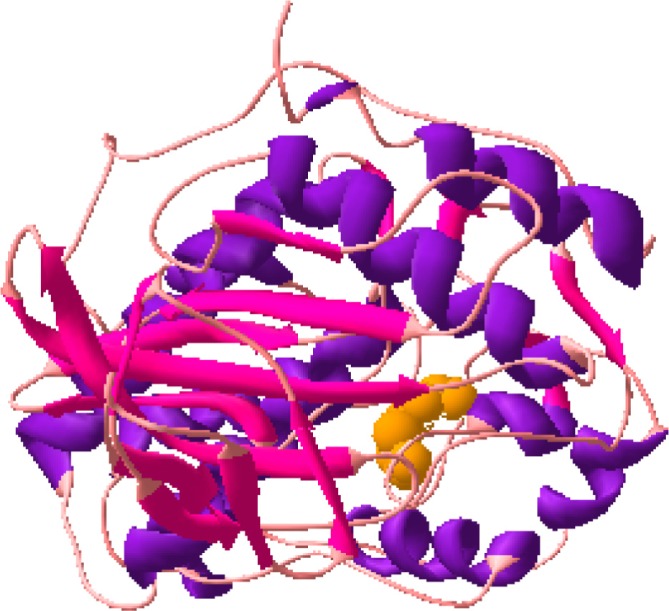
The docked structures of DADS with PI3k (Fig. 8a), DADS with PI3K (Fig. 8b) and DADS with EGFR (Fig. 8c) are shown in Figure 8. Thus the docking calculations gave a new focus into the activity of DADS to inhibit the kinases with Ki at millimolar range. Further, the binding free energies of the DADS-enzyme complexes were also calculated to observe the affinity between them. These calculations were carried out using the equation,
Figure 8a.
Docked structure of PI3K and DADS.
Figure 8b.
Docked structure of ERK and DADS.
Figure 8c.
Docked Structure of Mutant EGFRvIII and DADS.
According to this free energy calculation, DADS possessed higher affinity against the PI3K, with a binding score of −3.85 kcal/mol. Although the inhibitor does not possess a higher inhibition rate (1.50 mM) at a temperature of 298.15 K, a potential compound with the ability to pass the blood brain barrier and produce a enhanced therapeutic effect can be designed.
Conclusion
In this work, we have carried out computational modeling and simulation of the normal EGFR, mutant EGFR and MAPK signaling pathways in glioma and the expression levels of proteins in the pathways were observed. The mutant EGFR was identified to possess higher levels of expression. Further, the signaling onco-proteins, PI3K, ERK and mutant EGFRvIII were docked with DADS, a well known glioma inhibitor, to analyze the inhibition effect of DADS against the mutant EGFR and the downstream signaling proteins. The results show that both mutant EGFR and Ras. GTP can be potentially inhibited with a single inhibitor to obtain sound therpeutic effects against glioma.
Thus we propose that the ‘multiple-targeting’ or ‘combined-targeting’ drug therapy could yield an improved therapeutic value against diseases like glioma. Also, we put forward a novel computational method for drug designing that involves both kinetic modeling and docking calculations to identify suitable target and the target combinations to obtain more powerful therapeutic effects. This mechanism based drug design strategy would provide promising outcome and help the scientific community to understand the disease mechanisms more clearly and thereby design appropriate drug candidates that will eventually become a drug.
Abbreviations
- EGFR
epidermal growth factor receptor;
- DADS
diallyl disulfide;
- MAPK
mitogen-activated protein kinase;
- ERK
extracellular signal-regulated kinase;
- PI3K
phosphoinositide 3 kinase;
- STAT3
Signal transducer and activator of transcription 3;
- PTEN
Phosphatase and tensin homolog.
Footnotes
Conflict of Interest
The authors report no conflicts of interest.
References
- 1.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–7. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 2.Liu ET, Karuturi KR. Microarrays and Clinical Investigations. N Engl J Med. 2004;350:1595–7. doi: 10.1056/NEJMp048050. [DOI] [PubMed] [Google Scholar]
- 3.Ebert BL, Golub TR. Genomic approaches to hematologic malignancies. Blood. 2004;104:923–32. doi: 10.1182/blood-2004-01-0274. [DOI] [PubMed] [Google Scholar]
- 4.Reardon DA, Wen PY. Therapeutic Advances in the Treatment of Glioblastoma: Rationale and Potential Role of Targeted Agents. Oncologist. 2006;11:152–64. doi: 10.1634/theoncologist.11-2-152. [DOI] [PubMed] [Google Scholar]
- 5.Frieboes HB, Lowengrub JS, Wise S, et al. Computer simulation of glioma growth and morphology. NeuroImage. 2007;37:S59–70. doi: 10.1016/j.neuroimage.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular Determinants of the Response of Glioblastomas to EGFR Kinase Inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 7.Yoon S, Seger R. The extracellular signal-regulated kinase: Multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 8.Paul H, Huang, Mukasa Akitake, et al. White, Quantitative analysis of EGFRvIII cellular signalling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proceedings of the National academy of Sciences of the United States of America. 2007;104:12867–72. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada S, Taketomi T, Yoshimura A. Model analysis of difference between EGF pathway and FGF pathway. Biochemical and Biophysical Research Communications. 2004;314:1113–20. doi: 10.1016/j.bbrc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, Xiao J, Sun Q, et al. A Single Decoy Oligodeoxynucleotides Targeting Multiple Oncoproteins Produces Strong Anticancer Effects. Mol Pharmacol. 2006;70:1621–9. doi: 10.1124/mol.106.024273. [DOI] [PubMed] [Google Scholar]
- 11.Funahashi A, Matsuoka Y, Jouraku A, Kitano H, Kikuchi N. CellDesigner: a modeling tool for biochemical networks. Proceedings of the 38th conference on Winter simulation, Winter Simulation Conference; Monterey, California. 2006. [Google Scholar]
- 12.Herbert MH, Sauro M, Finney Andrew, et al. Next Generation Simulation Tools: The Systems Biology Workbench and BioSPICE Integration. OMICS. 2003;7(4):355–72. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 13.Gise VA, Lorenz P, Wellbrock C, Hemmings B, Berberich-Siebelt F, Rapp UR. Troppmair Apoptosis suppression by Raf-1 and MEK1 requires MEK- and phosphatidylinositol 3-kinase-dependent signals. J Mol Cell Biol. 2001;21:2324–36. doi: 10.1128/MCB.21.7.2324-2336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatakeyama M, Kimura S, Naka T, et al. A computational model on the modulation of MAPK and Akt pathways in heregulin-induced ErbB signaling. Biochem J. 2003;373 doi: 10.1042/BJ20021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivaska J, Nissinen L, Immonen N, Eriksson JE, Kähäri VM, Heino J. Integrin 2β1 Promotes Activation of Protein Phosphatase 2A and Dephosphorylation of Akt and Glycogen Synthase Kinase 3β. Mol Cell Biol. 2002;22:1352–9. doi: 10.1128/mcb.22.5.1352-1359.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalisch BE, Demeris CS, Ishak M, Rylett RJ. Modulation of nerve growth factor-induced activation of MAP kinase in PC12 cells by inhibitors of nitric oxide synthase. J Neurochem. 2003;87:1321–32. doi: 10.1111/j.1471-4159.2003.02057.x. [DOI] [PubMed] [Google Scholar]
- 17.Perkins E, Calvert J, Lancon JA, Parent AD, Zhang J. Inhibition of H-ras as a treatment for experimental brain C6 glioma. Molecular Brain Research. 2003;111:42–51. doi: 10.1016/s0169-328x(02)00668-x. [DOI] [PubMed] [Google Scholar]
- 18.de Philippe M, Gilles F, Marc P. Multiple Targeting by the Antitumor Drug Tamoxifen: A Structure-Activity Study. Current Medicinal Chemistry. Anti-Cancer Agents. 2004;4:491–508. doi: 10.2174/1568011043352696. [DOI] [PubMed] [Google Scholar]
- 19.Garrett M, Morris David S, et al. Belew, Olson Arthur J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19:1639–62. [Google Scholar]
- 20.Huey R, Morris GM, Olson AJ, Goodsell DS. A Semiempirical Free Energy Force Field with Charge-Based Desolvation. J Computational Chemistry. 2007;28:1145–52. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]