Abstract
Bacillus anthracis is surrounded by an anti-phagocytic capsule that is entirely composed of γ-linked d-glutamic acid (γdPGA). γdPGA is required for virulence and is produced in large quantities following spore germination. We have previously described the isolation of several γdPGA-reactive mAbs. The reagents are effective in both immunoprotection and diagnostic applications. The current work was done to further investigate the specificity of γdPGA-reactive mAbs. The specificity of each mAb was characterized using surface plasmon resonance. Our results indicate that each mAb is stereoselective for binding to d-glutamic acid oligomers, but to varying degrees. In particular, mAb F26G3 is highly selective for γdPGA; alterations in stereochemistry disrupted recognition. These differences in mAb reactivity suggest that binding of γdPGA by mAb F26G3 is more specific than non-directional ionic interactions between a negatively charged antigen and a positively charged antibody.
Keywords: antibody, anthrax, polyglutamic acid capsule, stereoselective, molecular model
1. Introduction
Bacillus anthracis is the causative agent of anthrax and a category A biothreat. Virulent strains are encapsulated by a polymer of γ-linked d-glutamic acid (γdPGA), a structure that is unusual among human pathogens (Hanby and Rydon, 1946; Haurowitz and Bursa, 1949; Avakyan et al., 1965; reviewed in Candela and Fouet, 2006). Capsule formation begins immediately upon spore germination, and presents a major obstacle to the mammalian host response (Zwartouw and Smith, 1956; Maynell and Maynell, 1964; Wang and Lucas, 2004; Drysdale et al., 2005). We previously reported that γdPGA is detectable in serum in both murine and non-human primate models of pulmonary anthrax using a monoclonal antibody (mAb)-based immunoassay (Kozel et al., 2004; Kozel et al. 2007; Boyer et al. 2009). Current diagnosis of anthrax is time-consuming and requires the isolation of bacteria by culture. It is likely that novel targets for immunoassay, such as the bacterial capsule, will allow for a rapid diagnosis and, subsequently, reduce mortality through early treatment (Sweeney et al., 2011).
Specificity is a key requirement for diagnostic assays. With anthrax, the intrinsic properties of the capsule present a unique obstacle. Whereas many targets for immunoassay are globular proteins, γdPGA is flexible, polyvalent, and carries a significant negative charge. Others have demonstrated that antibodies may bind with high specificity to small peptide targets (Landsteiner and van der Scheer, 1929; Hofstetter et al., 1999), however, previous reports found antibody recognition of γdPGA to be more generalized. Studies done by Goodman and colleagues demonstrated that rabbit polyclonal antibody (pAb) generated against whole cells of B. anthracis may additionally react with small peptide antigens that incorporate aspartic acid, alanine, and lysine (Goodman and Nitecki, 1966). Furthermore, Goodman noted that anti-capsular pAb did not distinguish between d- and l-isomers of glutamic acid, or polymers that were linked via the β- or γ-carboxyl moieties. Together, these observations contributed to the hypothesis that antibody recognition of polyglutamic acids relied less on the orientation of the carboxyl moieties, and more on the overall secondary and tertiary structural features of the antigen.
Given the results of previous studies that used pAb, it was of interest to determine the binding specificity of several mAbs that react with the B. anthracis capsular antigen. To accomplish our analysis, we surveyed binding of four capsule-reactive mAbs to polyglutamic acids that were enantiomerically pure (d- or l-homopeptides). All four mAbs preferentially bound γdPGA, however, the results identified a spectrum of mAb specificities, likely due to antigen flexibility and polyvalence. Notably, mAb F26G3 displayed a remarkable preference for γdPGA both in strength of binding and the total number of antigen:antibody complexes that were measurable on a twenty-five residue peptide. Together, these results indicate that antibody interactions with poly-glutamic acids are highly dependent on antigen stereochemistry.
2. Materials and Methods
2.1 mAb production
The Immunization protocols for production and isolation of the murine antibodies F24F2 (IgG3), F24G7 (IgG3), F26G3 (IgG3), and F26G4 (IgG3) have been described (Kozel et al., 2004). Hybridoma cell lines were cloned by limiting dilution. mAb-secreting cell lines were grown in tissue culture in an Integra CL 1000 culture flask (Integra Biosciences, East Dundee, IL), and mAbs were isolated by affinity chromatography on protein A (Pierce, Rockford, IL).
2.2 Poly-glutamic acid
γdPGA and γlPGA polypeptides were synthesized by the Nevada Proteomics Center (University of Nevada, Reno) from 9-fluorenylmethoxy carbonyl-d or l-glutamic acid (O-t-butyl) (Bachem, Peninsula Laboratories, San Carlos, CA) using 9-fluorenylmethoxy carbonyl chemistry. The peptides were purified to approximately 95% using a C8 YMC column on a Thermo Separations (San Jose, CA) P4000 preparative liquid chromatograph.
2.3 Surface plasmon resonance - affinity determination
Binding experiments were performed using surface plasmon resonance (SPR) with a BIAcore ×100 instrument (GE Healthcare, Piscataway, NJ). The running and sample buffer for all experiments was HBS buffer, pH 7.4, containing 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P20 (HBS-EP+). For ligand preparation, 10 mg of γdPGA or γlPGA oligomers (25 residues) were biotinylated by standard amine coupling chemistry (Pierce, Rockford, IL) and purified by size exclusion chromatography (Pierce Rockford, IL). Biotinylated peptides were immobilized onto a SA sensor chip until immobilization levels of 80–90 response units (RU) were reached (GE Healthcare). A flow cell was left unmodified for reference subtraction. To evaluate binding, mAb samples were diluted in HBS-EP+ and analyzed at concentrations of 5–333 nM (γdPGA) and 26–833 nM (γlPGA). At each concentration, mAb was injected over the modified chip surface at 30 µl/min for 180 s. The chip surface was regenerated between runs with a 1 min pulse of 2 M MgCl2. Affinity constants were determined using the 3-parameter Hill equation in Sigma Plot software. A plot of the double reciprocals of binding by all mAbs (1/C versus 1/Rmax) gave a regression that was linear (data not shown).
2.4 Surface plasmon resonance - competition assays
Inhibition assays were performed by incubating each mAb (8.3 nM) with a five-residue isopeptide of γdPGA or γlPGA at 37°C for 1 h. Inhibitor concentrations ranged from 16–500 µM. Following incubation, binding of each sample to a sensor surface immobilized with a twenty five-residue isopeptide of γdPGA was analyzed. The percentage inhibition was calculated from a comparison with binding signal achieved by mAb alone. The concentration of free peptide required for 50% inhibition of mAb binding (IC50) was calculated by a hyperbolic regression analysis using SigmaPlot software. The inhibition constant (KI) was calculated using: KI = IC50/(1 + ([C]/KD)) (Cheng Y and Prusoff WH, 1973).
2.5 Molecular model
2.5.1 Computational methods
The model was constructed de novo taking advantage of the well-defined repetitive domain structure of immunoglobulins (Ig). Each two-chain domain was constructed individually. The template of the structurally conserved region (SCR) of each chain was selected from candidates identified from FUGUE (de Bakker et al., 2001; Shi, Blundell, and Mizugchi, 2001; Williams et al., 2001) ORCHESTRAR, as implemented in SYBYL (Tripos Inernational, St. Louis, MO.) and FASTA (Berman et al., 2000) searches of the Protein Data Bank (www.rsb.org).
The amino acid sequence of the template was converted to that of the target using the spl (SYBYL) script “atomic horror.” Gaps and insertions in the SCR were repaired by the SYBYL loop search utility. Side chains were relaxed. As necessary, the two chains comprising the folding domain were docked using the PDB template files as guides. The structure was refined using molecular dynamics constrained by fixed distance, range distance, and torsion constraints. The search loop algorithm was applied to repair gaps that remained in the loop regions (from insertions and deletions). Constrained molecular dynamics was reapplied. The four folding domains were assembled into Fab and Fc fragments. The initial docks were guided by Ig structures deposited in the PDB and subsequently refined by the docking program HEX (Ritchie and Venkatraman, 2010). The assembled Fab and Fc domains were refined by constrained molecular dynamics (MD). Sequence gaps were repaired by the loop search algorithm and refined by constrained MD. Constrained MD was applied first in continuum solvent followed by extensive MD in explicit solvent (TIP3). Finally, unconstrained MD was applied. The AMBER force field was used throughout the MD simulations.
The quality of the target structure was accessed by a variety of statistical comparisons with experimentally determined Ig structures. These tests include i) configuration of peptide bonds, ii) Ramachandran plots, iii) side chain rotomer probability, iv) surface/buried positional probability, and v) strain energy. Only the Fab fragment was used for this report and only to illustrate properties relevant to experimental results reported in this manuscript.
2.5.2 Conformational landscape
The SYBYL software package was used to determine the conformational landscape of γdPGA. A peptide of three γ-linked d-glutamyl residues (trimer) was constructed. Here, the end groups were included to simulate the effect of adjacent residues in a larger polymer. The AMBER force-field and charge set were used in MDa simulations; however, both were slightly modified to account for the different geometry. A molecular database of rotamers of the single bonds of the backbone (N-Cα, Cα-Cβ, Cβ-Cγ, Cγ-Cδ) of the central residue was constructed through the use of a conformational search algorithm. Using molecular mechanics (steepest descent and Powell minimizers, no simplex) each conformer was minimized in two stages. First the terminal residues were relaxed and then the entire trimer was relaxed.
3. Results
3.1 Affinity measurements by SPR
A panel of mAbs that are reactive with the capsular polypeptide of B. anthracis (γdPGA) has been described (Kozel et al., 2007). The mAbs resulted from immunization of BALB/c mice with soluble capsule that was isolated from Bacillus licheniformis culture supernatant fluids. Unlike B. anthracis, which produces a capsule of enantiomerically pure γdPGA, B. licheniformis incorporates a varied content of d- and l-glutamic acids to its capsular filaments. The relative proportion of d- and l-glutamic acids may be altered through the addition of metal cations (Leonard CG, Housewright RD, and Thorne CB, 1958). The purified capsule of B. licheniformis that was used for the generation of the present mAb library consisted of ~84% d-glutamic acid.
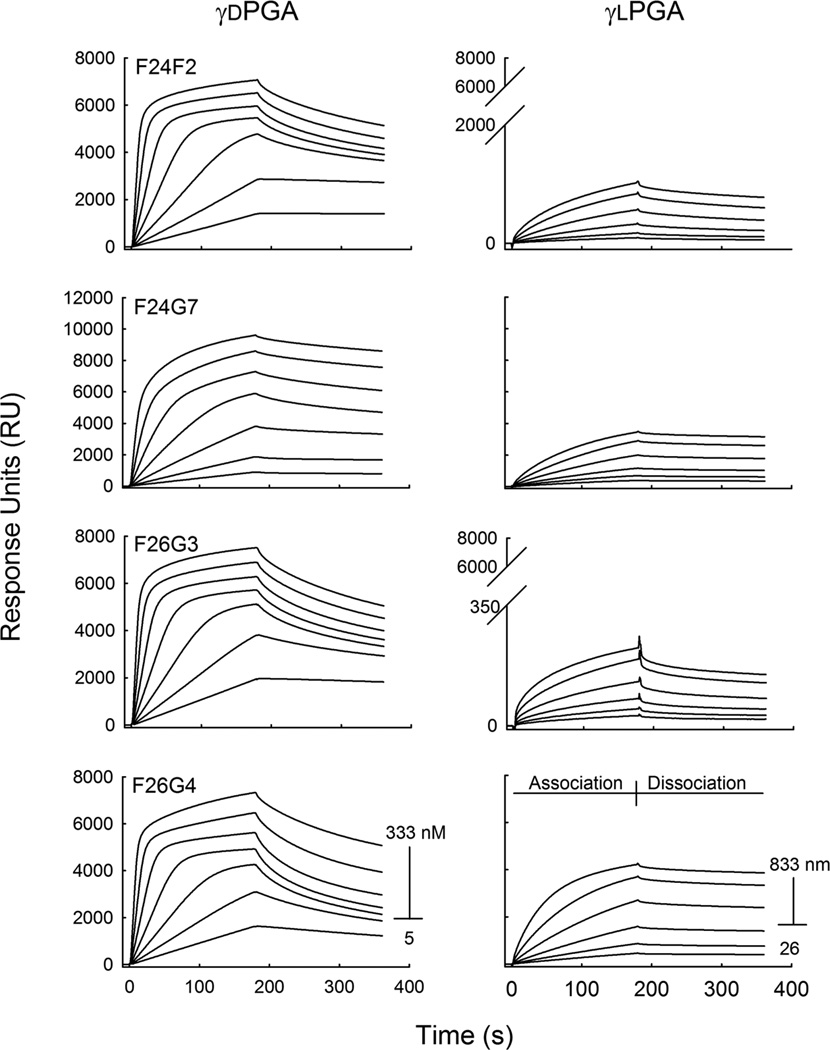
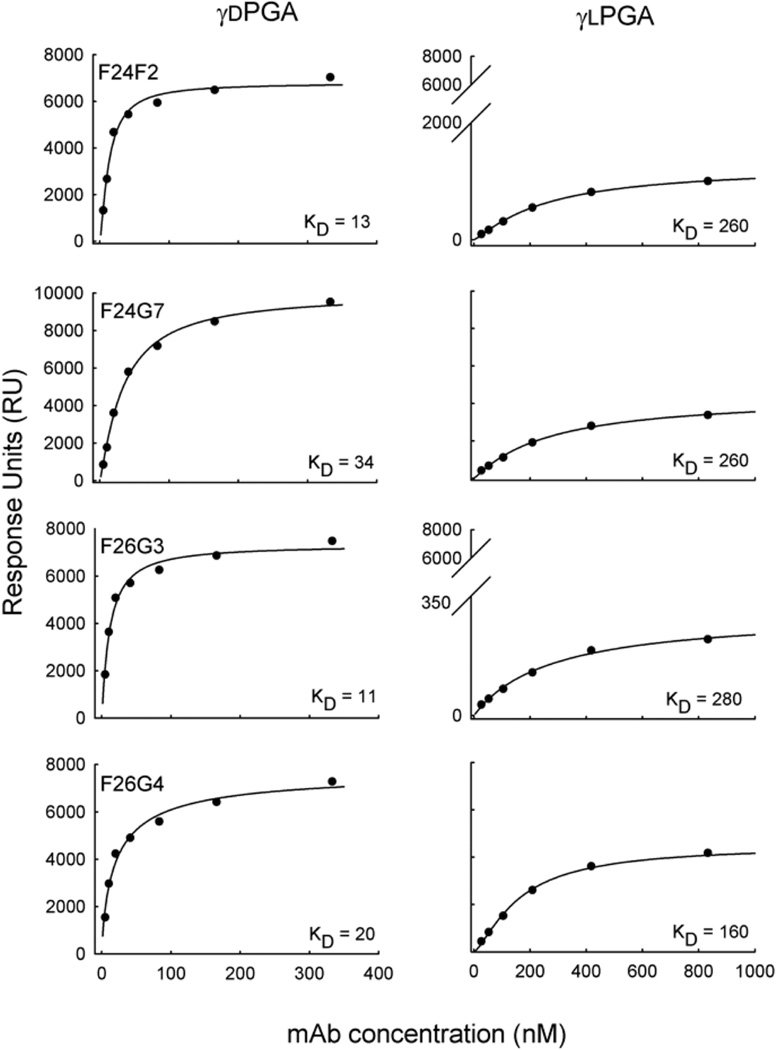
The mixed content of the immunizing agent raises questions as to the true specificity of the library of γdPGA mAbs. To determine the specificity, each mAb was first assayed to establish its binding affinity with oligomers of synthesized, enantiomerically pure γdPGA The analysis was done using SPR. The binding surface was prepared by direct immobilization of biotinylated γdPGA oligomer (25 residues, 25mer) to a streptavidin-coated sensor chip. Total immobilization of γdPGA 25mer was 80 RU. In the SPR platform, binding response is a function of weight located on the sensor surface (1000 RU = 1 ng/mm2). Therefore, it is possible to calculate the total antigen density of the immobilized surface. At 80 RU, the predicted average distance between γdPGA 25mer is 82 Å. This distance is approximately the same as the known Stokes radius of an IgG, which is 75 Å. The distance between γdPGA molecules is large enough to permit binding of at least 1 mAb to each antigen, yet small enough for interactions to occur among bound mAbs. A titratable increase in RU was observed following a 180 s pulse of each mAb concentration (Fig. 1). A regression analysis of RU versus mAb concentration found that all mAbs bound to γdPGA with nano molar affinity (Fig. 2). The calculated binding strengths displayed a hierarchy that was evident but subtle, placing F24F2≈F26G3>F26G4>F24G7. The difference in γdPGA-binding affinity between the strongest (F26G3) and weakest (F24G7) binders was 11:34 nM or ΔG = 700 cal/mol; equivalent to the loss or gain of up to two hydrogen bonds. The observed range in affinity constants was also accompanied by a hierarchy in the total amount of mAb on the sensor chip surface at saturation (RUmax), indicating variability in total mAb:Ag complexes formed on the sensor surface (F24G7>F26G4≈F26G3≈F24F2). These differences were also subtle. The highest ratio of mAb per γdPGA molecule was 2.7 (F24G7); in comparison, the lowest ratio was 1.8 (F24F2) (Table 1).
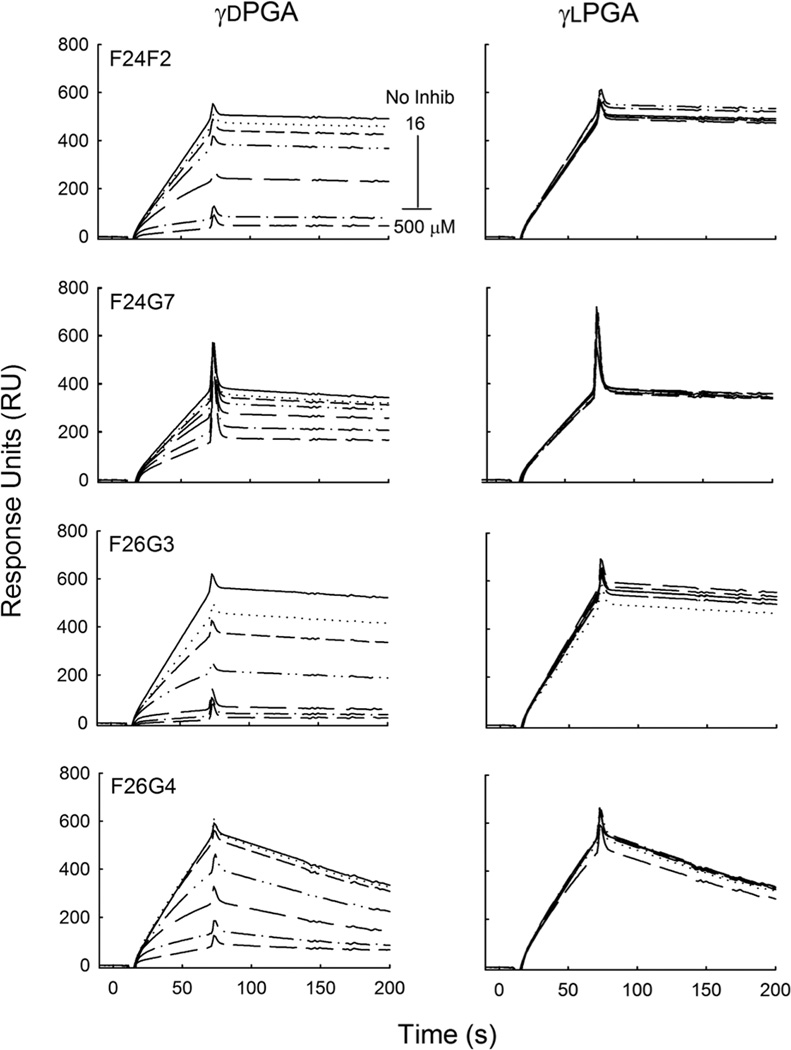
Fig. 1.
Binding specificity of anti-capsular mAbs. Synthetic γd- and γlPGA 25mer were immobilized to separate sensor chips at 80 and 90 RU, respectively. mAb binding was analyzed in a two-fold dilution series at a mAb concentration of 5.5–333 nM to the γdPGA coated surface (left), and at a mAb concentration range of 26–833 nM to the γlPGA coated surface (right). The corresponding sensorgrams are given as a function of response generated over time for each mAb concentration. Note the substantial differences in the magnitude of response for binding of d versus L antigens.
Fig. 2.
Binding affinity of anti-capsular mAbs to γd- and γlPGA-coated surfaces by surface plasmon resonance. The total RU that was achieved on each surface following a 180 s pulse is given as a function of mAb concentration. The 3-parameter Hill equation was used to create the regression line. The dissociation constants of each mAb were determined as: y = axb/(cb+xb), where c equals the apparent dissociation constant (KD).
Table 1.
Binding and immunochemical properties of anti-capsular mAbs
| γdPGA | γlPGA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mAb | Subclass | KDa | KDb | KIc | RUmax | mAb:Ag | KDd | KDb | KIc | RUmax | mAb:Ag |
| F24F2 | IgG3 | 0.5 | 13 | 73 | 6800 | 1.8 | nt | 260 | >500 | 1300 | 0.3 |
| F24G7 | IgG3 | 0.36 | 34 | 340 | 10000 | 2.7 | nt | 260 | >500 | 4000 | 1.0 |
| F26G3 | IgG3 | 0.37 | 11 | 48 | 7300 | 2.0 | nt | 280 | >500 | 300 | 0.07 |
| F26G4 | IgG3 | 0.51 | 20 | 98 | 7800 | 2.1 | nt | 160 | >500 | 4500 | 1.1 |
Intrinsic affinity using fluorescence perturbation (µM), results adapted from Kozel et al., 2007
Functional affinity using SPR (nM)
Inhibition constant using fluid and solid phase antigen in SPR format (µM)
Intrinsic affinity using fluorescence perturbation (µM); nt, not tested
Each mAb was also assayed for reactivity with enantiomerically pure γlPGA 25mer. The amount of γlPGA that was immobilized to a sensor chip was 90 RU, which corresponds to 78 Å between each antigen molecule. Initial studies that used the identical mAb concentrations in the evaluation of binding to the γdPGA 25mer surface displayed limited reactivity (data not shown). A nearly 10-fold increase in mAb concentration, as compared to γdPGA experiments, was required to obtain useful binding data (Fig. 1). Interaction of the four mAbs with immobilized γlPGA was quite different when compared to γdPGA. All mAbs displayed larger dissociation constants and lower RUmax, but at variable magnitudes. MAb F26G4 bound to γlPGA with a dissociation constant of 160 nM. In contrast, mAbs F24F2, F24G7 and F26G3 bound γlPGA with markedly lower affinities of 260, 260, and 280 nM, respectively. When compared to γdPGA binding, these differences accounted for 8–23 fold changes in dissociation constants or ΔG = 1.3–1.9 Kcal/mol. Stark differences were also observed in the saturation densities of mAb on the γlPGA sensor surface, as was evidenced by the lower RUmax values. MAbs F24G7 and F26G4 saturated the chip surface at relatively higher densities of 4400 and 4500 RU, respectively. Lower saturation densities were observed in mAb F24F2 (1300 RU) and, more strikingly, mAb F26G3 (300 RU). The calculated ratio of mAb per γlPGA molecule ranged from 1 (mAbs F24G7 and F26G4) to ~0.1 (mAbs F24F2 and F26G3) (Table 1).
3.2 Competitive assays
Binding inhibition experiments were also performed to further investigate the activity of mAbs in our library. We wanted to determine if binding specificity was influenced by the state of the antigen, i.e. solid versus fluid phase. It is possible that fluid-phase antigen may assume conformations not found in the solid phase, and that such conformations are reactive with the mAb-binding site. Alternatively, it can be hypothesized that epitope accessibility may be hindered by direct immobilization to the sensor surface. To this end, experiments were designed such that mAb binding to a γdPGA-immobilized sensor chip could be inhibited after incubation with a soluble ligand.
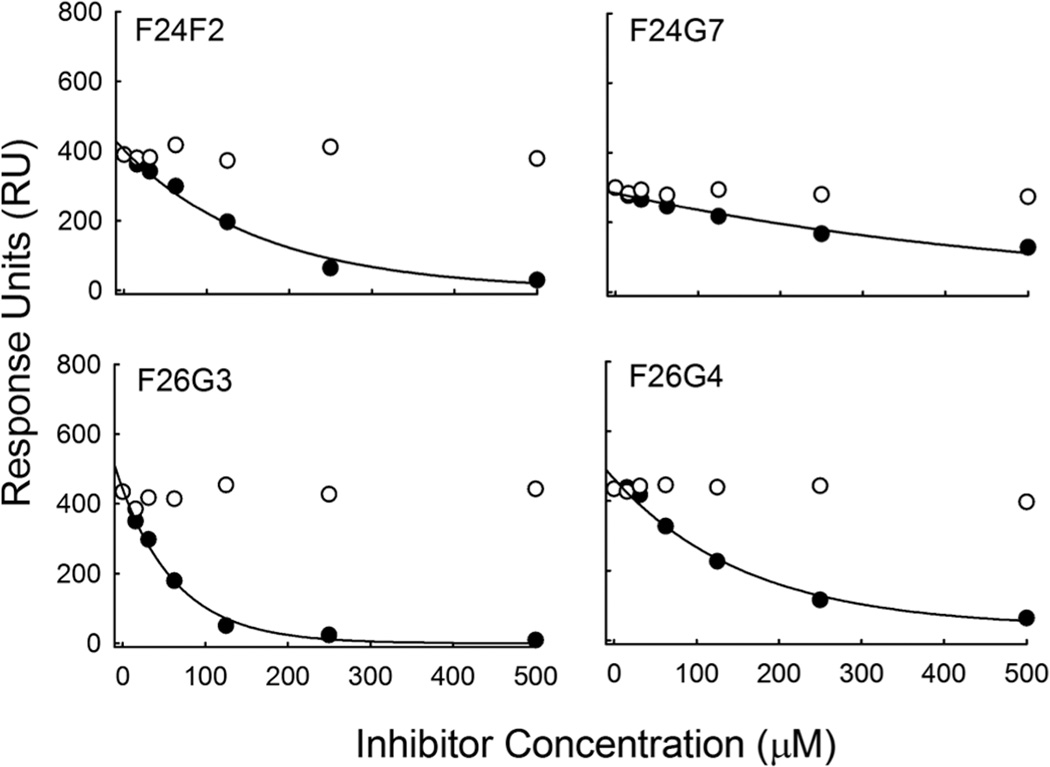
Initially, a fixed concentration of each mAb (8.3 nM) was pre-incubated with free γdPGA 5mer in varying concentration (16–500 µM) before injection over a γdPGA-immobilized sensor surface. The mAb concentration of 8.3 nM was chosen as it was either at or below the calculated KD of all four mAbs. All mAbs were inhibited from binding to solid phase γdPGA 25mer by the presence of soluble γdPGA 5mer but at varying degrees (Fig. 4). MAb F26G3 bound free-γdPGA with an inhibition constant (KI) of 48 µM. In contrast, mAbs F24F2, F24G7, and F26G4 displayed increased KI values of 73, 98, and 340 µM, respectively. These trends were similar to the previous direct-binding experiments, which showed a weaker affinity for mAb F24G7 as compared to the others. The study was repeated by incubating each mAb with free γlPGA 5mer. Analysis showed that binding to the γdPGA-coated sensor surface was not affected by the presence of the soluble γlPGA peptide, even when excess inhibitor concentrations were present (500 µM) (Fig. 4).
Fig. 4.
Determination of inhibition constants. Binding response is given as a function of inhibitor concentration.The closed circles represent γdPGA 5mer, whereas the open circles represent the γlPGA 5mer. A titratable decrease in RU was not observed in any of the capsule-reactive mAbs when a γlPGA inhibitor was present. Hyperbolic fits of the inhibition that was observed with soluble γdPGA were used to calculate the concentration of inhibitor necessary to achieve a fifty percent signal decrease (IC50). IC50 values were then used to derive the KI value for each mAb.
Additional studies with mAb F26G3 were done to assess the effect of polymer length on antibody recognition. A previous study found that five glutamyl residues optimally fill the antibody binding pocket (Kozel et al., 2007). Larger peptides, therefore, may be used to determine if contacts are made outside the binding pocket; such contacts may contribute to specificity. To this end, 10- and 25-residue length γd- or γlPGA peptides were assayed for inhibition activity. Using ten times the KI, a complete loss of surface binding was observed for γdPGA inhibitors. However, identical amounts of γlPGA were insufficient to inhibit binding to surface-associated γdPGA when using either the 10- and 25-residue peptides (data not shown).
3.3 Molecular Modeling
MAb F26G3 was chosen for the creation of a molecular model. The model is intended to illustrate the nature of γdPGA-mAb interactions. Homology modeling was used to create a representation of the F26G3 binding pocket. The VH and VL CDRs were introduced to the structurally conserved regions obtained from template structures deposited in the RCSB PDB. Conformational minimization was performed to obtain a low-energy structure. Molecular dynamics simulations were used to visualize interactions between the mAb and γdPGA. The solvent accessible surface area (SASA) of the binding pocket is shown in Fig. 5, along with the mAb residues that have contact with the antigen at four-angstroms. These residues are present on both the heavy and light chains and include Tyr, Trp, Ala, His, Lys, and Arg. Finally, the model shows that five d-glutamyl residues occupy the binding site, this observation is consistent with previous experimental data (Kozel et al., 2007).
Fig. 5.
The binding pocket of mAb F26G3. The solvent accessible surface of the binding pocket is shown in green. Additionally, a single γdPGA 5mer is shown in space-fill with hydrogen, carbon, nitrogen, and oxygen being represented in white, grey, blue, and red, respectively. Docking simulations show that 5 γ-glutamyl residues optimally fill the binding pocket.
The number of possible conformations in the γdPGA backbone was estimated by exploring the conformational space of a three residue γ-d-glutamyl molecule. The highly flexible propionate side chain connecting the amides of γ-d-glutamate has a theoretical 81 conformers; four single bonds each with three rotational energy minima. A conformational search algorithm was used to generate a database of conformers. These were then minimized using molecular mechanics. For each γ-d-glutamate monomer, 28 conformers were found within 1.4 kcal/mol of the global energy minimum.
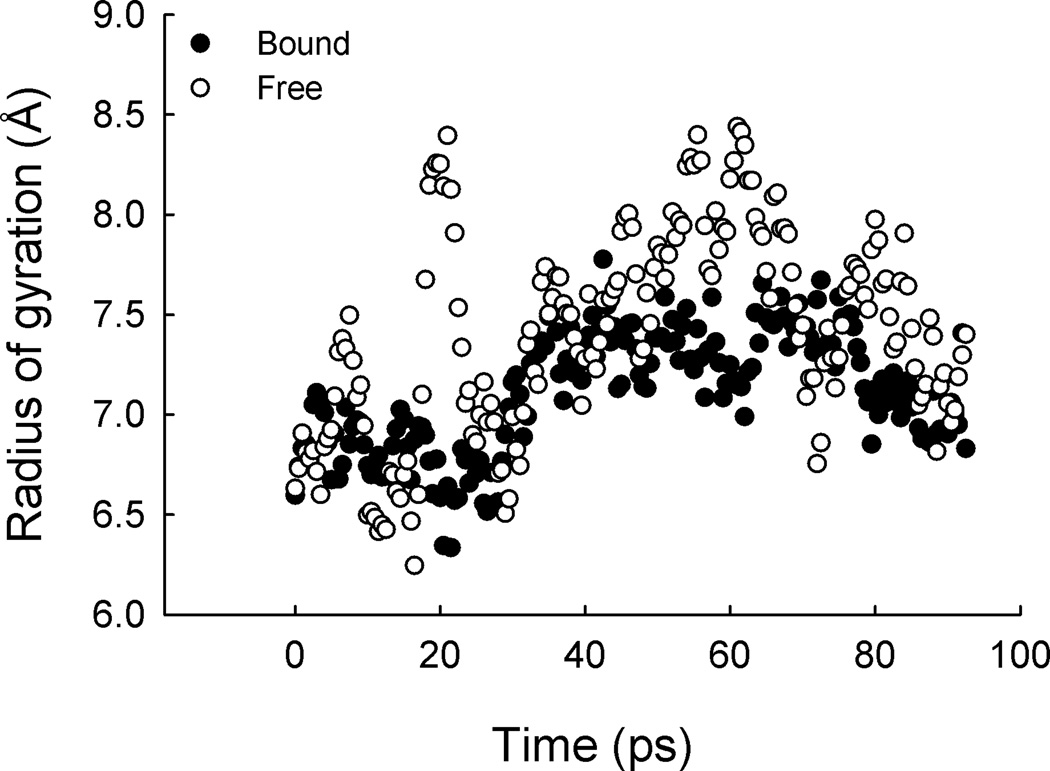
The effect of mAb-binding on antigen mobility was assessed. Antigen mobility was measured as the radius of gyration (ROG) using molecular dynamics simulation. A γdPGA 25mer was docked to mAb F26G3. The ROG was measured for the five γ-d-glutamyl residues within the binding pocket, as well as five unbound γ-d-glutamyl residues on the far side of the polymer. The results indicate bound γdPGA is less mobile, in comparison to unbound γdPGA; <ROG> = 7.1 Å ± 0.3 and <ROG > = 7.4 Å ± 0.5, for bound versus unbound, respectively. Although these values appear similar, qualitative analysis of ROG suggest otherwise (Fig. 6). Here, two conformers, each with a lifetime of 30 ps (<ROG > = 6.8 ± 0.2 and 7.3 ± 0.2) dominate the bound section; in contrast, no stable conformer was detected in the unbound section. Therefore, binding energy is sufficient to restrict the residue motion into identifiable conformers.
Fig. 6.
Mobility of mAb-bound and free γdPGA. Molecular dynamic simulation was used to analyze the difference of antigen mobility in the bound a free states. A γdPGA 25mer was docked to mAb F26G3. The five γd-glutamyl peptides in the binding pockets were measured for lateral mobility using the radius of gyration (closed circles). Alternatively, five γd-glutamyl peptides on the unbound portion of the 25mer were measured for comparison (open circles). Note the restriction in movement.
4. Discussion
Molecular recognition between antibody and antigen involves interactions among surfaces that are continuously in a state of flux. Binding reactions may involve necessary conformational changes by the antigen or antibody before the formation of the antigen-antibody complex. Alternatively, conformational changes may be necessary within the antigen-antibody complex itself. Here, a flexible bivalent antibody interacts with a flexible, multivalent and highly disordered antigen. Interactions between the two surfaces are, therefore, complex. In this study, mAbs reactive with the capsular polypeptide of B. anthracis were assayed for binding activity using surface plasmon resonance, which allows the visualization of antigen-antibody complex formation in realtime, and without chemical reporters. Surface plasmon resonance also facilitates the calculation of the total number of antigen-antibody complexes, as weight is directly proportional to the response signal. Given the length and flexibility of the antigen used in this study (PGA 25mer), it is likely that the antigen-antibody complexes occurred both as a function of inter-antigen (cross-linking between two separate 25mers) and intra-antigen binding.
To estimate the total number binding sites per 25mer filament, the binding response at several mAb concentrations was recorded and extrapolated to predict the theoretical RUmax when an infinite amount of mAb is present. Under these conditions, all second order rate processes become infinitely large and first order reactions, such as prior isomerization and induced fit become important. On a γdPGA-coated sensor surface, an average of 2 (mAbs F24F2, F26G3, F26G4) to 3 (mAbs F24G7) bound mAbs per individual antigen molecule. This observation is consistent with the polyvalent nature of the antigen, and suggests that a high number of overlapping binding sites are present. This knowledge suggests the likelihood that the total maximum binding for each mAb is not limited by the number of possible epitopes on a single antigen, but the point at which the repulsive interactions between mAbs (molecular crowding) are greater than the attractive energy of binding. We assume that the individual, capsule-reactive mAbs differ in the ability to orient themselves on the sensor surface to promote optimal packing. Because each mAb that was used in this study is a murine IgG3, we can assume that structural differences affecting segmental flexibility are not a factor. Thus, the differences in Rmax may rely, at least in part, on epitope abundance, accessibility, and orientation.
Prior sequencing data confirmed the variable region of each mAb in this group is unique; however, the four mAbs tested here show remarkable similarity in binding affinities and maximum surface densities for γdPGA. Preliminarily, these results suggest that all four mAbs have closely similar epitopes; however, binding of each mAb to γlPGA suggests otherwise. Here, for each mAb the density of binding was less and the dissociation constants larger. At saturation; one mAb bound per γlPGA 25mer (F24G7 and F26G4), one mAb bound every one in three γlPGA 25mer (F24F2), and most strikingly, one mAb bound every one in ten γlPGA 25mer (F26G3). We can assume that the conformational flexibility of the γlPGA compensates for the difference in stereochemistry; however, the favorable antigen conformers become less abundant and only permit a low density of antibody binding. Interestingly, mAbs F24G7 and F26G4 have the weakest binding to γdPGA, and the least ability to discriminate between d- and l-isomers. The relatively smaller changes in affinity (~8 fold for γdPGA versus γlPGA) and higher saturation densities supports this hypothesis. Most likely, the reduced differences in the binding of γdPGA versus γlPGA for these mAbs is a consequence of the high flexibility and polyvalent nature of the antigen; such qualities allow for multiple antigen conformers to exist with similar binding energies. Overall, the data suggest that antigen stereochemistry influences both the total amount and energetic favorability of conformers recognized by all mAbs in this library.
Competition experiments offer further insight into antibody binding. Our previous findings indicated that the KD values of all four γdPGA-reactive mAbs were nearly identical (Kozel et al., 2007 and Table 1). These measurements were determined by fluorescence perturbation and relied on alteration of the microenvironment of tryptophan residues found in the binding pocket of each mAb. Such measurements may be used to determine the intrinsic affinity of antibody binding, which refers to the strength of a single antigen-antibody interaction in solution. Using a small γdPGA 5mer, the intrinsic affinity of each mAb was determined to be ~500 nM. In the current study, the functional affinity of each mAb to the surface-bound γdPGA was much higher (~10–50×). The increase in functional versus intrinsic affinity that was observed for each mAb is likely due to restriction of antigen flexibility in three-dimensional space and the ability of the antibody to bind the antigen bivalently. In a competitive environment, the mAbs always favored binding to surface-associated over fluid-phase antigen. The large KI values (48–340 µM) from these experiments support this observation. Interestingly, mAb F24G7 was relatively insensitive to the addition of free γdPGA 5mer. Prior experiments using fluorescence perturbation also demonstrated that mAb F24G7 had 2–3 fold lower levels of maximum quenching when compared to the other anti-capsular mAbs (Kozel et al., 2007).
The observation that antibodies reactive with this capsular polypeptide are stereo-selective rather than stereo-specific is not surprising. The poly-glutamic acid antigen is highly flexible; such antigens present special considerations for molecular recognition. The binding pockets of all mAbs in this study accommodate five-glutamyl peptides. This knowledge was derived experimentally (Kozel et al., 2007) and in silico (mAb F26G3) (Fig. 5). Because the backbone of this γ-linked homopolymer contains four rotatable bonds, each residue can theoretically exist in 81 rotamers. A systematic conformation search using a γdPGA 3mer confirms that each glutamyl residue exists in a large number of conformers; however, the energies of each conformer are sufficiently different that only a few will be populated at 310K. Here, we calculate the total number to be 28. Therefore, the 5-residue antigen may assume 285(or 1.7 × 107) theoretical conformations. It is reasonable, with such numerable conformations possible, that molecular recognition of γd- or γlPGA may not be completely specific. It is almost certain that a sub-population of conformers from each antigen will highly resemble one another.
The antibody binding pocket may also exist is several conformers; however, the amino acids that make up the complementarity determining regions (CDR) of the antibodies are considerably more restricted in movement than the free polyglutamic acid antigen. For example, the CDRs consist of stretches of amino acids that connect to framework regions in the variable domain. The flexibility of these loop structures are inhibited by the end-to-end distance between two framework regions. As a consequence, the CDR may move freely within the confines of excluded volume and electrostatic energy, but is limited from lateral movement. In contrast, the antigen has no such restrictions and is only limited by excluded volume and electrostatic energy. Additionally, the number of rotatable bonds in each CDR-associated amino acid is two, whereas the number of rotatable bonds in each residue of γ-linked PGA is three. Therefore, we assume for this study that the flexibility of the antigen is more influential than the flexibility of the antibody for the bimolecular interaction; however, this observation is certainly not true for all antigens.
Despite the flexibility and charge-density of the antigen, it is likely that mAb-binding occurs following recognition of a particular epitope and not merely non-directional ionic forces between a negatively charged antigen and a positively charged antibody. Molecular dynamics simulations of a 25mer bound to mAb F26G3 support this hypothesis. Here, the conformational difference between bound and free portions of the antigen were evident. When bound, discrete conformations were observed during time-averaged experiments; these conformations had some of the lowest energies of all possible conformers. The unbound portions of the antigen, however, were considerably more mobile. The differences in the radius of gyration for bound versus unbound antigen illustrate this point (Fig. 6). Notably, unbound antigen displayed increased mean, standard deviation, and range values for radius of gyration, when compared to bound antigen; accordingly, no such stable conformers were identified during time-course experiments. These observations suggest mAb-binding leads to a restriction in antigen flexibility, as contacts between specific residues are critical to the binding interaction.
The molecular model of mAb F26G3 found that 12 residues are in 4Å contact with the antigen. Of these 12 residues, there are: 1-Ala, 2-Arg, 1-His, 2-Lys, 1-Trp and 5-Tyr. Previous reports of amino acid usage in the complementarity determining region (CDR) of antibodies have indicated that Tyr is the most common (~40%) (Zemlin et al., 2003). The Tyr content of mAb F26G3 approximates this value (42%). Overall, Tyr is associated with antibodies that share high complementarity with antigen, as hydrogen bonding and Van der Waals forces are the most prevalent. In contrast, Arg, His, and Lys are uncommon to the CDRs (≤1% usage). Here, these 3 amino acids constitute 42% of the 4Å contacts as well. Most likely, the overuse of these basic amino acids is a consequence of the acidic nature of the antigen. Molecular modeling reveals the formation of specific salt-bridges between the antibody and antigen, which supports this hypothesis. Finally, the presence of Arg in the binding pocket is associated with reduction in antigen specificity (Birtalan et al., 2008). This finding supports our observation that the binding pocket is selective, but not specific for γdPGA.
Overall, the current study highlights the ability of mAbs to discern between nearly identical structures. Previous observations concluded that polyclonal antibodies show little selectivity for enantiomers of polyglutamic acids. Evaluation of our mAb library yielded similar results with mAbs F24G7 and F26G4, both exhibiting a subtle preference for γdPGA. In contrast, mAbs F24F2 and, in particular, F26G3 were more selective for γdPGA. In practical terms, this degree of specificity may be advantageous in reagent selection for the construction of immunoassays to detect the capsular antigen of B. anthracis.
Fig. 3.
Competition assays using short peptide inhibitors. A single concentration of each capsule-reactive mAb was incubated with synthetic oligopeptide (5 residue) of either γdPGA (left) or γlPGA (right). Concentrations of the inhibitors ranged from 16–500 µM in a two-fold dilution series. The binding of mAb:inhibitor mixtures were analyzed by a 60 s pulse over a γdPGA-immobilized sensor chip.
Highlights.
We determined the binding affinities and specificities of several murine mAbs that react with the capsule of Bacillus anthracis.
Binding of all mAbs was not solely dependent on non-directional ionic forces.
All mAbs preferentially bound γ-linked D glutamic acid over γ-linked L glutamic acid.
The murine mAb F26G3 was the most selective for γ-linked D glutamic acid.
A molecular model of mAb F26G3 shows five glutamic acid residues optimally fill the binding site and that two conformations have 30 picosecond lifetimes.
Acknowledgments
This work was supported by Public Health Service grants R01AI059348 and R01AI093365 from the National Institutes of Allergy and Infectious Diseases (TRK).
Glossary
- γdPGA
γ-linked d-polyglutamic acid
- γlPGA
γ-linked l-polyglutamic acid
- mAb
monoclonal antibody
- pAb
polyclonal antibody
- SPR
surface plasmon resonance
- KI
inhibition constant
- KD
dissociation constant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AuCoin DP, Sutherland MD, Percival AL, Lyons CR, Lovchik JA, Kozel TR. Rapid detection of the poly-gamma-d-glutamic acid capsular antigen of Bacillus anthracis by latex agglutination. Diagn Microbiol Infect Dis. 2009;64:229–232. doi: 10.1016/j.diagmicrobio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nuc Acid Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sindhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- Boyer AE, Quinn CP, Hoffmaster AR, Kozel TR, Saile E, Marston CK, Percival A, Plikaytis BD, Woolfitt AR, Gallegos M, Sabourin P, McWilliams LG, Pirkle JL, Barr JR. Kinetics of lethal factor and poly-d-glutamic acid antigenemia during inhalational anthrax in rhesus macaques. Infect Immun. 2009;77:3432–3441. doi: 10.1128/IAI.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela T, Fouet A. Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol Microbiol. 2005;57:717–726. doi: 10.1111/j.1365-2958.2005.04718.x. [DOI] [PubMed] [Google Scholar]
- Candela T, Fouet A. Poly-gamma-glutamate in bacteria. Mol Microbiol. 2006;60:1091–1098. doi: 10.1111/j.1365-2958.2006.05179.x. Review. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Bateman A, Burke DF, Miguel RN, Mizuguchi K, Shi J, Shirai H, Blundell TL. HOMSTRAD: adding sequence information to structure-based alignments of homologous protein families. Bioinform. 2001;17:748–749. doi: 10.1093/bioinformatics/17.8.748. [DOI] [PubMed] [Google Scholar]
- Drysdale M, Heninger S, Hutt J, Yahua C, Lyons CR, Koehler TM. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalational anthrax. The EMBO Journal. 2005;24:221–227. doi: 10.1038/sj.emboj.7600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta P, Tipple CA, Lavrik NV, Datskos PG, Hofstetter H, Hofstetter O, Sepaniak MJ. Enantioselective sensors based on antibody-mediated nanomechanics. Anal Chem. 2003;75:2342–2348. doi: 10.1021/ac034031z. [DOI] [PubMed] [Google Scholar]
- Goodman JW, Nitecki DE. Immunochemical studies on the poly-γ-D-glutamyl capsule of Bacillus anthracis. I. Characterization of the polypeptide and of the specificity of its reaction with rabbit antisera. Biochem J. 1966;5:657–665. doi: 10.1021/bi00866a036. [DOI] [PubMed] [Google Scholar]
- Hanby WE, Rydon HN. The capsular substance of Bacillus anthracis . Biochem J. 1946;40:297–309. [PubMed] [Google Scholar]
- Haurowitz F, Bursa F. The linkage of glutamic acid in protein molecules. Biochem J. 1949;40:297–307. doi: 10.1042/bj0440509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter H, Cary JR, Eleniste PP, Hertweck JK, Lindstrom HJ, Ranieri DI, Smith GB, Undesser LP, Zeleke JM, Zeleke TK, Hofstetter O. New developments in the production and use of stereoselective antibodies. Chirality. 2005;17:S9–S18. doi: 10.1002/chir.20099. [DOI] [PubMed] [Google Scholar]
- Hofstetter O, Hofstetter H, Wilchek M, Schurig M, Green BS. Chiral discrimination using an immunosensor. Nat Biotech. 1999;17:371–374. doi: 10.1038/7927. [DOI] [PubMed] [Google Scholar]
- Hofstetter O, Hofstetter H. Antibodies as chiral selectors for the determination of enantioenrichment. Enantiomer. 2001;6:153–158. [PubMed] [Google Scholar]
- Inglesby TV, O’Toole T, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Friedlander AM, Gerberding J, Hauer J, Hughes J, McDade J, Osterholm MT, Parker G, Perl TM, Russell PK, Tonat K. Working group on civilian biodefense Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA. 2002;287:2236–2252. doi: 10.1001/jama.287.17.2236. Review. [DOI] [PubMed] [Google Scholar]
- Janse I, Hamidjaja RA, Bok JM, van Rotterdam BJ. Reliable detection of Bacillus anthracis,Francisella tularensis and Yersinia pestis by using multiplex qPCR including internal controls for nucleic acid extraction and amplification. BMC Microbiol. 2010;10:314. doi: 10.1186/1471-2180-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-dl-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis . J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel TR, Murphy WJ, Brandt S, Blazar BR, Lovchik JA, Thorkildson P, Percival A, Lyons CR. mAbs to Bacillus anthracis capsular antigen for immunoprotection in anthrax and detection of antigenemia. Proc Nat Acad Sci. 2004;101:5042–5047. doi: 10.1073/pnas.0401351101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel TR, Thorkildson P, Brandt S, Welch WH, Lovchik JA, AuCoin DP, Vilai J, Lyons CR. Protective and immunochemical activities of monoclonal antibodies reactive with the Bacillus anthracis polypeptide capsule. Infect Immun. 2007;75:152–163. doi: 10.1128/IAI.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsteiner K, van der Scheer J. Serological differentiation of steric isomers. J Exp Med. 1928;48:315–320. doi: 10.1084/jem.48.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CG, Housewright RD, Thorne CB. Effects of some metallic ions on glutamyl polypeptide synthesis by Bacillus subtilis . J Bacteriol. 1958;76:499–503. doi: 10.1128/jb.76.5.499-503.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Watarai M, Cheun HI, Shirahata T, Uchida I. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J Infect Dis. 2002;186:227–233. doi: 10.1086/341299. [DOI] [PubMed] [Google Scholar]
- Meynell E, Meynell GG. The roles of serum and carbon dioxide in capsule formation by Bacillus anthracis . J Gen Microbiol. 1964;34:153–164. doi: 10.1099/00221287-34-1-153. [DOI] [PubMed] [Google Scholar]
- Ritchie DW, Venkatraman V. Ultra-fast FFT protein docking on graphics processors. Bioinformatics. 2010;26:2398–2405. doi: 10.1093/bioinformatics/btq444. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. (2001) [DOI] [PubMed] [Google Scholar]
- Silvaieh H, Schmid MG, Hofstetter O, Schurig V, Gübitz G. Development of enantioselective chemiluminescence flow- and sequential-injection immunoassays for alpha-amino acids. J Biochem Biophys Methods. 2002;53:1–14. doi: 10.1016/s0165-022x(02)00087-8. [DOI] [PubMed] [Google Scholar]
- Sweeney DA, Hicks CW, Cui X, Li Y, Eichacker PQ. Anthrax infection. Am J Respir Crit Care Med. 2011;184:1333–1341. doi: 10.1164/rccm.201102-0209CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TT, Lucas AH. The capsule of Bacillus anthracis behaves as a thymus-independent type 2 antigen. Infect Immun. 2004;72:5460–5463. doi: 10.1128/IAI.72.9.5460-5463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MG, Shirai H, Shi J, Nagendra HG, Mueller J, Mizuguchi K, Miguel RN, Lovell SC, Innis CA, Deane CM, Chen L, Campillo N, Burke DF, Blundell TL, de Bakker PI. Sequence-structure homology recognition by iterative alignment refinement and comparative modeling. Proteins. 2001;(Suppl. 5):92–97. doi: 10.1002/prot.1169. [DOI] [PubMed] [Google Scholar]
- Zanuy D, Alemán C, Muñoz-Guerra S. On the helical conformation of un-ionized poly(gamma-d-glutamic acid) Int J Biol Macromol. 1998;23:175–184. doi: 10.1016/s0141-8130(98)00047-6. [DOI] [PubMed] [Google Scholar]
- Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Kirkham PM. Expresssed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Zwartouw HT, Smith H. Polyglutamic acid from Bacillus anthracis grown in vivo: structure and agressin activity. Biochem J. 1956;63:437–442. doi: 10.1042/bj0630437. [DOI] [PMC free article] [PubMed] [Google Scholar]