Abstract
The enzyme human paraoxonase 1 (huPON1) has demonstrated significant potential for use as a bioscavenger for treatment of exposure to organophosphorus (OP) nerve agents. Herein we report the development of protein models for the human isoform derived from a crystal structure of a chimeric version of the protein (pdb ID: 1V04) and a homology model derived from the related enzyme diisopropylfluorophosphatase (pdb ID: 1XHR). From these structural models, binding modes for OP substrates are predicted, and these poses are found to orient substrates in proximity to residues known to modulate specificity of the enzyme. Predictions are made with regard to the role that residues play in altering substrate binding and turnover, in particular with regard to the stereoselectivity of the enzyme, and the known differences in activity related to a natural polymorphism in the enzyme. Potential mechanisms of action of the protein for catalytic hydrolysis of OP substrates are also evaluated in light of the proposed binding modes.
Introduction
The human enzyme paraoxonase 1 (huPON1) possesses the ability to hydrolyze a variety of substrates, including organophosphorus (OP) nerve agents, in a catalytic manner – a capability for which it has received considerable attention as a potential bioscavenger of chemical warfare agents.i Predictions on the basis of the rate of inhibition of acetylcholinesterase in vivo (in animal models) suggest that the wild-type natural form of huPON1 (WT-huPON1) does not have sufficient catalytic activity to act as a bioscavenger. However, variants of huPON1 with enhanced kinetic activity for deactivation of OP agents could provide protection against nerve agents in vivo. It has been estimated that an in vivo concentration of a nerve agent of 0.1 μM is capable of eliciting a toxic response.ii This estimate suggests that a decrease in KM of two orders of magnitude (e.g., a KM of 1.0 to 10 μM), coupled with a 100 fold increase in kcat of WT-huPON1 may be required to achieve the levels of catalytic activity needed for an efficient bioscavenger. Given this dual challenge of a need to alter both KM and kcat simultaneously, in silico design can provide an efficient approach to identifying potential variants with the necessary properties. Attempts to achieve such an enhancement using state-of-the-art protein engineering techniques are in progress; however, this work is complicated by the dearth of information regarding the nature of the active site of the human enzyme, as only a single crystal structure has been reported, featuring an apo-version (with no inhibitor or substrate bound) of a recombinant, gene-shuffled variant of the enzyme. The putative active site has been identified primarily through mutagenesis studies which have demonstrated altered reaction selectivity.iii Using such methods to identify the catalytic mechanism, however, is complicated by the presence of two bound calcium ions for the enzyme; many mutations in the calcium binding regions result in loss-of-function, which can be either due to a change in the protein folding or due to disruptions of the reaction mechanism.
The human form of the protein is extremely difficult to purify to homogeneity while maintaining functionality, and attempts to obtain structural information of the wild-type (WT) protein by crystallographic methods have been unsuccessful to date. The sole crystal structure published at present is that of a recombinant, gene-shuffled variant derived from the human, rabbit, rat, and mouse paraoxonases; this variant, known as G2E6, shares 86% sequence homology with the WT human protein (PDB entry 1V04).iv The crystal structure of G2E6 has been resolved at 2.2 Å resolution, with two fragments remaining unresolved: the N-terminal 15 residues of the protein as well as a flexible loop from residues 72–79, both of which are in the putative high-density lipoprotein (HDL) binding domain. The center of the β-propeller contains two bound calcium ions, which have been designated ‘catalytic’ and ‘structural’ on the basis of mutagenesis studies. In the crystal structure, a single phosphate ion is coordinated to the ‘catalytic’ calcium ion (see Figure 1).
Figure 1.
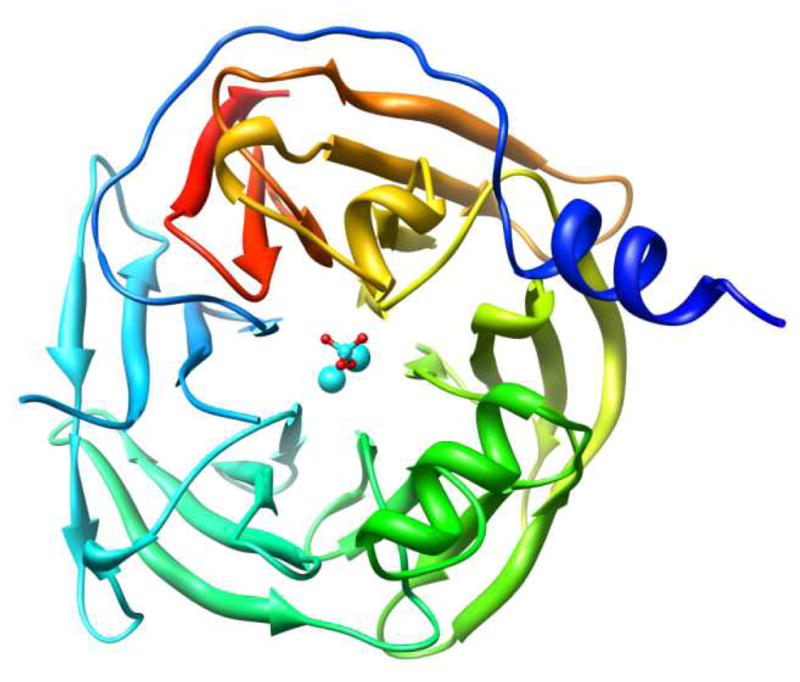
X-ray crystallographic structure of the G2E6 variant of human paraoxonase 1 (huPON1), resolved at 2.2 Å resolution (pdb ID 1V04).iv
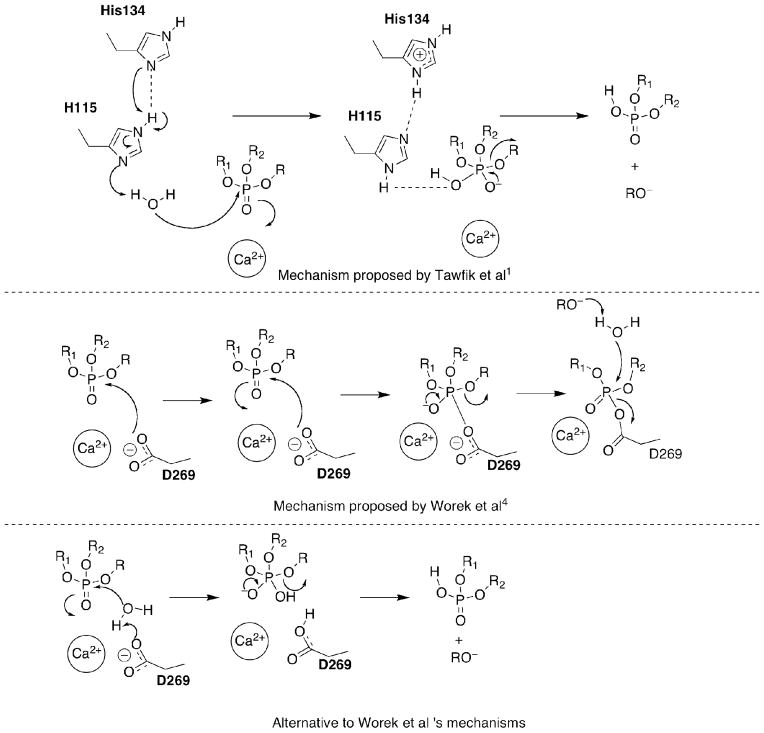
Due to this lack of a crystal structure of a complete form of the human enzyme, the mechanism of action of huPON1 for hydrolysis of organophosphorus compounds remains in doubt. Postulated mechanisms include general base catalysis of substrate by activated water molecules (the source of which is also debated) or direct hydrolysis by a nucleophilic residue in the active site pocket. Possible sources for the generation of hydroxide include either the H115/H134 and D269/H285 dyads, or direct activation of water by coordination to the ‘catalytic’ calcium ion (Figure 2).1,v
Figure 2.
With regard to hydrolysis of organophosphorus compounds, there have been results from mutagenesis studies which cast doubt onto several of the postulated sources of the nucleophile. For example, mutations at H115 have been identified which preserve OPase activity, including H115W, suggesting that a catalytic dyad of these residues is not operative in the mechanism.3 A recent publicationvi of a neutron-scattering structure of DFPase, an enzyme highly similar in structure to PON1, shows the coordination of an intact water molecule to the calcium ion. This result suggests that coordination to the calcium ion alone may be insufficient to generate hydroxide in the active site. The authors postulated that the reaction mechanism for PON1 involved the direct hydrolysis of substrate by D269, which is likely to be in proximity to a substrate coordinated via the phosphoryl oxygen to calcium. We believe, however, that the coordination of D269 to calcium reduces its nucleophilicity, and further, that this carboxylate residue is too distant from the phosphorus center to participate in hydrolysis. The lack of demonstrated ‘aging’vii in PON1, or the isolation of any covalent intermediates of the enzyme with either substrates or inhibitors, further discounts a direct hydrolysis mechanism by an active-site residue; in the cholinesterases, following the formation of the OP-enzyme adduct, the secondary expulsion of alkyl groups occurs as an internally catalyzed process known as aging.viii In contrast, in carboxylesterases, a covalent conjugate with OPs is formed without a subsequent aging step, but the outcome is a stable complex of nerve agent-adducted enzyme.ix Accordingly, we postulate that the most likely mechanism of the protein for OPase activity is general base catalysis, with hydroxide generated in proximity to E53, D269, and/or H285.
The use of computational modeling to study enzymes is a rapidly growing field, although the majority of work being done in the field is in the area of small molecule design and optimization. However, small molecules are not the only possible drug leads, and the emerging field of protein engineering also benefits substantially from computational approaches, in particular for the optimization of substrate binding and catalysis of protein-based biological drugs. Computational techniques can be used to model and predict, or rationalize after the fact, the effects of mutations in proteins, and can evaluate postulated enzymatic reaction mechanisms.x However, for these methods to work, there must first be an accurate computational model of the enzyme of interest, which can be difficult, particularly for membrane-bound proteins or ones which are difficult to crystallize. In addition, the validation of computational models via measurable experimental properties is a constant requirement. In this article, we describe in detail the methods involved in the preparation of a structural model of huPON1 and our attempt to understand the dynamics of the structure and ligand binding using molecular dynamics simulations, molecular docking, and estimation of free energies of binding using several methodologies.
Materials and Methods
Model Preparation
Human paraoxonase 1 (huPON1) is a 355 residue, monomeric protein, with a 6-bladed β-propeller structure (Figure 1), which in serum is associated with high density lipoprotein (HDL).xi The HDL association is believed to be limited to the loops on the top portion of the protein.iii,xi
Preparation of a computational model of the G2E6 variant of huPON1 was performed using the AMBER 9 suite of programs. Protonation states of titratable residues were assigned using the program pdb2pqr,xii with a solvent pH of 7.0. The FF03xiii force field was employed for modeling of the protein; simulations of ligands were performed using parameters assigned using the GAFFxiv force field. Parameters for the calcium ions were derived from the published literature, as implemented in the AMBER FF03 force field; these parameters have been calibrated to reproduce the experimental free energy of solvation of calcium ions.xv To generate structures for huPON1, the final structure of G2E6 following 20 ns of MD simulations was used as a starting point for additional MD simulations on a model with restored human WT residues using in silico mutagenesis.
MD Simulations
The total number of residues in the G2E6 protein model is 340, along with two calcium ions; the N-terminal 15 residues were unresolved in the crystal structure as described above, and were not reconstructed prior to MD. The missing residues from 72 to 79 were reconstructed by comparison with a published homology model of PON1 designed by comparison with the related enzyme DFPase (pdb ID: 1XHR), and the reconstructed surface loop was incorporated into the available crystal structure of PON1 prior to extensive MD simulations.iii,xvi A total of 24074 TIP3Pxvii waters were added in a rectangular periodic box of 91 × 105 × 96 Å3. The protein contains a single disulfide bond, between cysteines 42 and 353, which was included in modeling studies. Energy minimizations and molecular dynamics simulations were performed using the sander module within AMBER 9.xviii A five-step equilibration procedure was employed in preparing the model for production MD simulations. First, a 1000-step energy minimization was performed. This was followed by a four-step warming regime with constant volume, in which the protein was heated to 300K over 4 ps of MD simulations, with a 2 fs time step in each case. No restraints were employed on the protein structure during this heating regime, and no instabilities were observed from analyses of the subsequent MD trajectories. These equilibration steps were followed by production MD (NPT) simulations. The temperature and pressure of the system were regulated using the Berendsenxix scheme of heat bath coupling, and a coupling time of 1.0 ps. While the Berendsen temperature bath has been demonstrated to result in instabilities in certain types of simulations,xx the use of explicit solvation and coupling to the temperature bath largely remove these concerns, and the method has been successfully used in the study of various proteins.xxi The SHAKExxii algorithm was employed to constrain bonds involving hydrogen atoms, and the particle-mesh Ewald (PME) methodxxiii was used with a 10.0 Å cutoff for non-bonded interactions. Periodic boundary conditions were employed in all simulations.
Docking Simulations
Molecular docking simulations were performed using Autodock 4.0.xxiv A total of 8 ‘snapshots’ were taken from the MD trajectories at 0.5 ns intervals over the last 4 ns of the 20 ns of production MD, and these 8 snapshots were used for the docking simulations. Automated receptor preparation was performed, including merging of non-polar hydrogens, and solvent molecules were removed. Autodock 4.0 allows for limited receptor side-chain flexibility, and we chose residues identified as the largest hits in initial rigid docking to be flexible: K70, H115, F222, I291, F292, and V346. The applied scoring grid (see Supporting Information) had a 0.375 Å grid spacing, with dimensions of 18.75 × 15.00 × 15.00 Å, centered above the catalytic calcium ion, to ensure the treatment of all possible binding poses. To generate charges for the OP ligands, each structure was optimized at the B3LYP/6-31+G(d,p) level of theory, and then ChelpGxxv nuclear-centered atomic charges were obtained at the B3LYP/6-311+G(d,p)//B3LYP/6-31+G(d,p) level of theory using Gaussian03.xxvi,xxvii,xxviii,xxix Each of these calculations used the standard basis sets available in Gaussian. The combination of the B3LYP functional and augmented basis sets is generally considered adequate for the accurate prediction of atomic charges using the ChelpG method.xxx For the docking protocol, non-polar hydrogens were merged into adjacent heavy atoms. Additional details on the docking protocol are included in the Supporting Information.
After analysis of the various docking poses for different OP ligands bound into the active-site snapshots, subsequent MD simulations were performed on select calcium-bound receptor-ligand complexes obtained from the docking simulations. A similar minimization protocol was utilized as described above for the initial G2E6 model; the only difference was the inclusion of a moderate, flattened parabolic restraint on the calcium-phosphoryl oxygen bond coordinate, from 2.5 to 4.0 Å, to allow for enhanced relaxation of the ligand-receptor contacts prior to the possible dissociation of substrate. This was found to improve on the sub-optimal treatment of receptor flexibility in the docking protocol, and resulted in a reduction in the number of dissociative poses. A total of 4 ns of unrestrained MD simulations were performed on each ligand-receptor complex. A series of coordinate snapshots was extracted from the production MD trajectory from the terminal 1.5 ns of simulations, at 10 ps intervals, from which individual trajectories were generated for the unbound ligand, free receptor, and complex. Poisson-Boltzmann (MM-PBSA)xxxi and Generalized Born (MM-GBSA)xxxii simulations were performed on these snapshots, using the sander module to calculate individual components of the free energy for each component as in eq 1. From the individual results, the overall free energy of binding was calculated using eq 2.
| (1) |
| (2) |
The non-polar (SA) terms were estimated using the MSMS algorithmxxxiii using the equation GSA = γ SASA + β, with γ and β set to 0.00542 kcal/(mol Å−2) and 0.92 kcal/mol, respectively, and using a probe radius of 1.4 Å for estimating the solvent accessible surface area. For the polar (Gpolar) energy terms, the Generalized Born and Poisson-Boltzmann methods were both utilized as implemented in the AMBER software package. In the GB calculations, dielectric constants of 1 and 78.5 were utilized with AMBER mbondi2 radii. The TSsolute term represents temperature and solute entropy, and in these calculations this term was omitted. As the binding energies were only compared within ligand families, the effect of entropy changes was estimated to be minimal. Such methods have been employed in estimating binding energies for organic molecules with good agreement with experimental data.xxxiv,xxxv
Umbrella sampling was employed to study the potential energy surface for removal of the substrates from coordination to the calcium ions. A 30 kcal/mol-Å2 force constant was placed on the calcium-phosphoryl oxygen bond distance, which was restrained at 0.2 Å intervals from 2.0 to 7.0 Å; thus, a total of 26 parallel trajectories were simulated over 100 ps for each ligand-bound orientation. The Weighted Histogram Analysis Method (WHAM)xxxvi was used to analyze the output of the parallel, restrained trajectories, from which the free energy as a function of distance was predicted.
Results
Preparation of the G2E6 Model
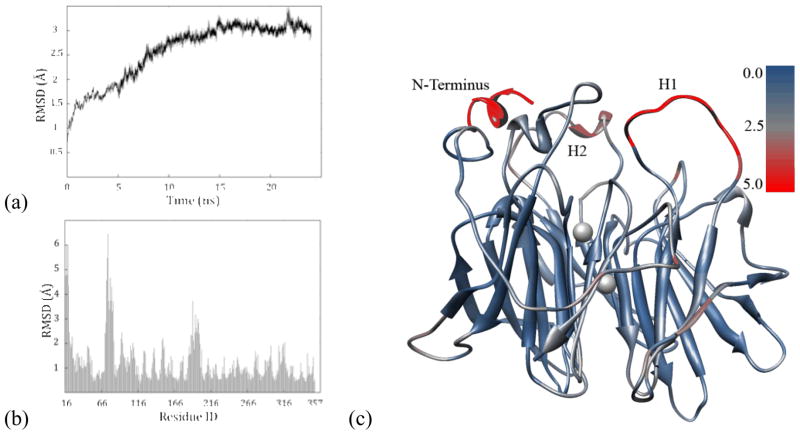
Generation of the G2E6-based model of huPON1 required reconstruction of the loop from residues 72–79. This was accomplished by adaptation of the loop structure contained within a homology model of PON1 previously developed from the related protein diisopropylfluorophosphatase (DFPase),iii,xvi which has a similar six-bladed, β-propeller structure. A total of 20 ns of MD simulations were performed, following the procedure described above, on the G2E6 (chimeric) model system. To measure the stability of the model, the structural and energetic properties were monitored over the course of the MD simulations (see Supporting Information). The root-mean-squared deviation (RMSD) from the starting coordinates was monitored for all atoms as a function of time (Figure 3a), the results of which suggested that the initial portion of the simulation resulted in fairly substantial structural changes, but after ~10 ns, the structure of the protein was largely stable for the remainder of the MD simulation. An additional analysis of the α-carbon RMSD of each individual residue relative to the starting structure was performed to provide insight into the location of the most mobile residues in the protein (Figure 3b).
Figure 3.
(a) All atom RMSD of the chimeric G2E6 model over 20 ns of MD. (b) RMSD of α carbons in the G2E6 model after 20 ns of MD simulations, relative to the x-ray crystal structure of the protein, in angstroms (Å). (c) G2E6 protein model following 20 ns of MD with a colored scale illustrating the RMSD relative to the x-ray crystal structure, in Å (pdb ID 1V04).
As can be seen in Figure 3, the regions with the largest changes relative to the crystal structure of the chimeric protein are the loops on the ‘top’ of the protein, corresponding to the putative HDL binding domain, as well as the more flexible turns units on the periphery of the β-propeller. The missing loop in the crystal structure, from residues 72–79 (Loop H1), was found to display the highest amounts of flexibility, along with the N-terminal domain (neglecting the unresolved 15 N-terminal residues) and an additional flexible surface loop (Loop H2). The lack of resolution of these loops in the available crystallographic structure of G2E6 also supports their inherent flexibility.
Calcium Ion Coordination and the Active Site
The ‘catalytic’ calcium ion for G2E6 in the published structure for the chimeric isoformiv is coordinated by 5 basic residues (E53, N168, N224, D269, N270), as well as one crystallographic water molecule, and with a phosphate from the buffer coordinated to the metal ion. In preparing the computational model system, the phosphate anion was removed prior to the MD simulations. Following 20 ns of MD, some changes were observed in the coordination environment of the calcium ion. The calcium ion itself was observed to move < 1 Å toward the D269 portion of the pocket, and, while the coordination sphere was preserved, some movement of the basic residues around the calcium was observed (Figure 4). In particular, the backbone carbonyl of N224 was found to swing in and coordinate to the calcium ion, which was coupled with movement of the side-chain amide upward toward the now-vacant coordination site that the phosphate anion had occupied. A similar movement was observed for the carboxylates of both E53 and D269 as well. Overall movement of the α-carbons of the calcium coordinating residues was approximately 1 Å relative to the G2E6 crystal structure.
Figure 4.
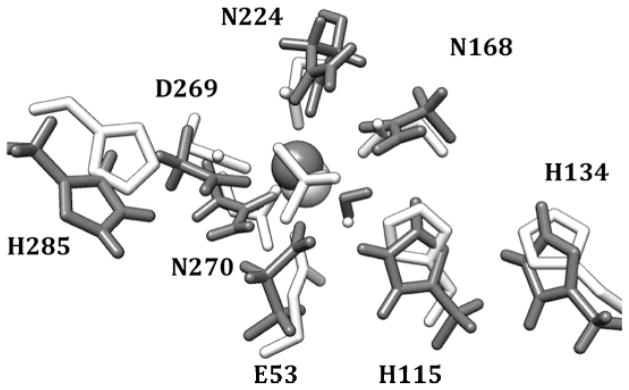
Putative active site of the G2E6 variant of huPON1 following 20 ns of MD simulations (gray) and the orientation of these residues in the x-ray crystal structure (white).
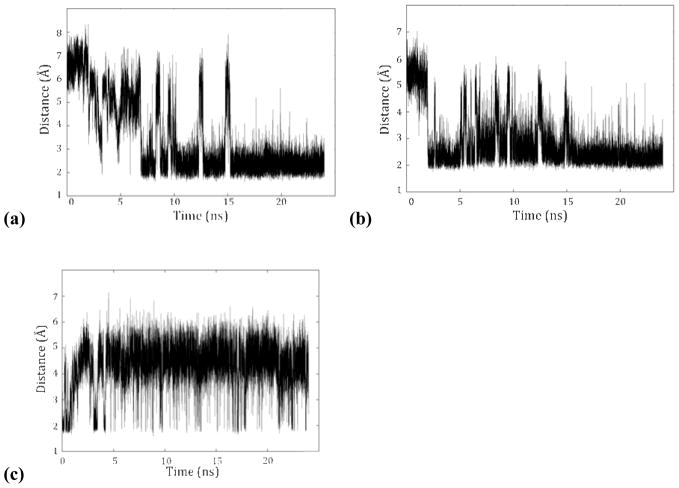
In proximity to the catalytic calcium ion of PON1 are two residue pairs within hydrogen bond distance: the D269/H285 and H115/H134 dyads, both of which have been suggested as possible bases for activation of water for the hydrolysis of substrate (Figures 1 and 4). Over the course of the MD simulations, the D269/H285 dyad demonstrates a dynamic hydrogen bond between the protonated ε nitrogen of H285 and the carboxylate of D269, with the fluxional motion primarily in the histidine side-chain; η-1 coordination of the catalytic calcium ion by the other carboxylate oxygen of D269 is preserved throughout the simulation (Figure 5c). In the other dyad, hydrogen bonding between the δ nitrogen of H115 and the ε hydrogen of H134 is conserved over the simulation, with a second hydrogen bond between E53 and the ε hydrogen of H115 also formed early in the trajectory (Figure 5a,b). The latter interaction is not observed in the crystal structure, with a distance between the two residues of 3.7 Å.
Figure 5.
(a) E53–H115, (b) H115–H134, and (c) D269–H285 hydrogen-bonding distances over 20 ns of molecular dynamics simulations. The H115 hydrogen bonds have over 90% occupancy over the terminal 10 ns, while the D269–H285 bond is more fluxional.
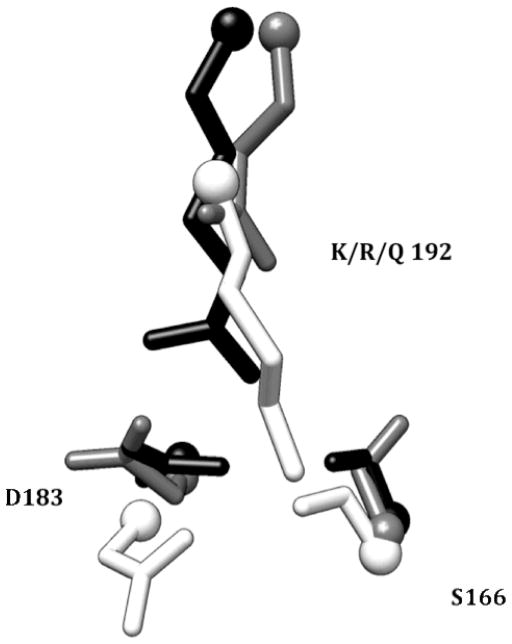
In the G2E6 variant of human paraoxonase, the vast majority of the substitutions relative to human WT protein are on the exterior of the protein, distal from the active site itself. These substitutions tend to be non-polar-to-polar mutations which presumably play a role in improving folding and solubility.iv Within the active site periphery, only two residues are modified in the variant, at positions 166 and 192. In the human WT protein, residue 192 is polymorphic, with isoforms having either arginine or glutamine, with detectible differences in sensitivity to organophosphorus compounds resulting from the polymorphism.xxxvii In G2E6, residue 192 has been modified to lysine, with a second modification at 166 from N to S. Interestingly, in the x-ray crystal structure of G2E6,iv the residues are only 2.9 Å apart, suggesting that the N166S mutation might be compensatory. In the MD trajectory for the G2E6 model, K192 is observed to hydrogen-bond with both S166 (as observed in the MD simulation) as well as D183; the latter is a residue in closer proximity to the active site cavity. The loop on which K192 resides is flexible enough that these interactions are fluxional over the course of MD, possibly corresponding to active site breathing modes (Figure 6, and Supporting Information).
Figure 6.
Region around the polymorphic residue 192, from the G2E6 x-ray crystal structureiv (192K, white), as well as the 192Q (gray) and 192R (black) model systems.
With no other modifications to the putative active site, residue 192 is highly suggestive for playing a role in the considerable differences in OPase activity between the WT and G2E6 variants of huPON1.xxxviii Accordingly, we performed MD simulations on G2E6 K192R and K192Q mutants, to examine the effects on the hydrogen bonding and electrostatic interactions in this pocket of the active site.
Examination of the G2E6 K192R mutant model over the MD trajectory suggests a similar hydrogen-bonding interaction to that observed in the G2E6 “WT” (K192) protein model; R192 is observed to hydrogen bond extensively with both D183 and N166, with a high degree of occupancy, and the arginine mutant is also similar to the lysine in G2E6 WT in terms of electrostatic interactions. In contrast, when Q192 was simulated, the distance to D183 was too long for hydrogen bonding to occur, leaving both D183 and S166 available to hydrogen bond with other neighboring residues (Figure 6). Accordingly, we postulate that the differences in activity of huPON1 WT and the G2E6 variant may result from alterations in the electrostatic and hydrogen-bonding interactions available in the region around residues 183 and 192.
Molecular Binding Studies of Nerve Agents
In the G2E6 model of PON1, molecular docking simulations were performed to study the binding of a variety of nerve agent substrates (Figure 7) into the active site of the protein. This included a number of known nerve agents, as well as paraoxon.
Figure 7.
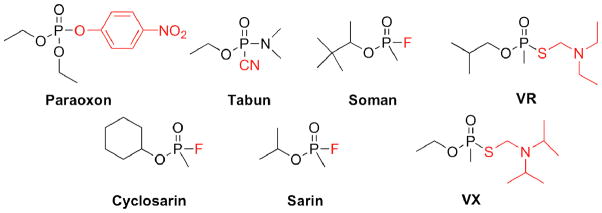
OP compounds including nerve agents employed in binding studies of the huPON1 models. Stereocenters exist at the phosphorus center of all molecules except paraoxon, and an additional chiral center is present in soman. The leaving groups are shown in red.
Receptor flexibility was restricted to six residues bracketing the putative active site region: K70, H115, F222, I291, F292, and V346 (Figure 8). These residues were chosen based on their proximity to the active site and their ability to modulate the active site cavity. To allow for additional sampling of receptor movement, docking simulations were prepared for 8 snapshots of the receptor model spanning 4 ns at 0.5 ns intervals (from the final 4 ns of the simulation); each of these models was used for complete docking simulations with all of the ligands, including stereoisomers and possible protonation states. For the V-series nerve agents, the amino fragment was protonated in accordance with the likely pKa in serum; for tabun, a similar assignment was not made due to the proximity of the amino group to the phosphorus center.
Figure 8.
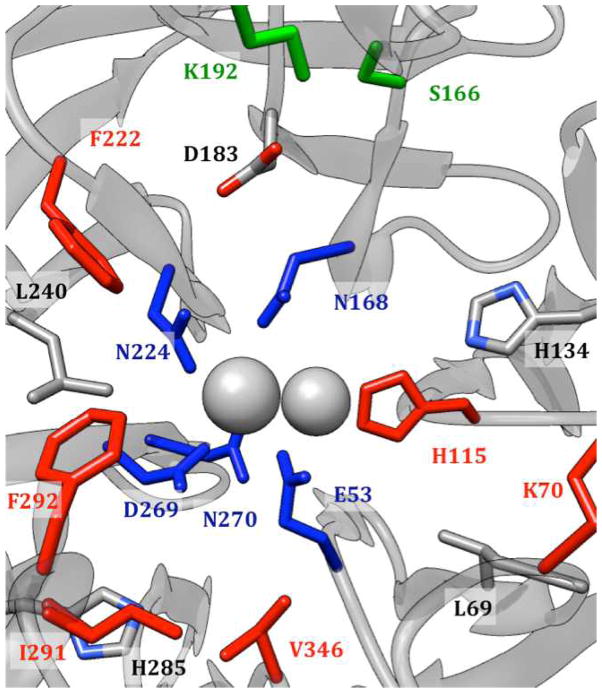
Active site of huPON1, including the 6 flexible residues from docking (in red), calcium-coordinating residues (in blue), residues 166 and 192 (in green) and other potentially important residues.
Evaluation of the bound orientations obtained showed a range of poses spanning the active site region, both coordinated to the catalytic calcium ion and more localized in the HDL binding domain on the upper portion of the protein. A consistent theme in the postulated reaction mechanisms of PON1 is that coordination of the substrate to the ‘catalytic’ calcium ion is a prerequisite for hydrolysis, with polarization of the phosphorus–oxygen bond lowering the reaction barrier, and with the calcium ion acting as an oxyanion holexxxix for the developing negative charge on the phosphoryl oxygen. Moreover, the presence of a phosphate anion coordinated to calcium in the published crystal structure of G2E6 further supports this notion. Accordingly, while we did observe a number of poses lacking such coordination, these were not carried on to subsequent refinement with molecular dynamics techniques. Of the poses that were coordinated to calcium, several consistent binding modes were observed. We chose to group these poses based upon the directionality of the leaving group in the case of the V-series agents, as well as paraoxon, while for the G-agents, the O-alkyl fragment was used.
The most frequently observed poses from molecular docking oriented the largest substituents on phosphorus toward two portions of the active site of the protein: the cavity formed in proximity to H115/H134, and extending across to D183/K192, and the pocket on the opposite side of the active site formed from F222, L240, H285, and V346 (Figure 9). For the V-series nerve agents, orientations placing the leaving group in the D183/K192 pocket resulted in hydrogen-bonding interactions between the protonated amino fragment and D183, a motif which also was observed following MD refinement, as described below. In the other pocket, poses either lacked explicit hydrogen-bond acceptors, or the amino group was hydrogen-bonded with D269 (which also coordinates the active-site calcium). Similar poses were predicted for binding of paraoxon in the active site, with the exception that hydrogen bonding with D183 was not observed; rather, orientation of the p-nitrophenoxide moiety toward that portion of the active site resulted in either hydrogen bonding with K192, or π-π stacking with H134. Of the two principal binding modes, orientation of the thioalkylamino or p-nitrophenoxide leaving group toward D183/K192 also places it anti to the D269/H285 pocket, in a pose suitable for hydrolysis by a nucleophile generated in that vicinity. However, the alternative pose positioning the leaving group in proximity to L240/F222 is not expected to be productive for hydrolysis.
Figure 9.
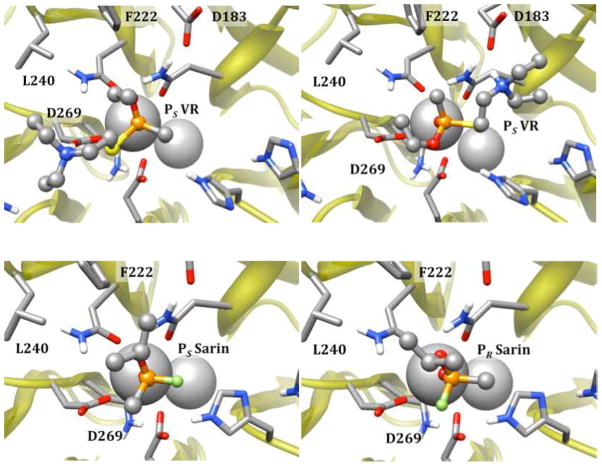
Representative binding modes for V-agents ((PS)-VR, top) and G-agents (PS and PR sarin, bottom) in the G2E6 model active site observed from molecular docking.
For the G-series nerve agents, which lack the large, polar leaving group that paraoxon and the V-series agents possessed, the binding modes predicted with docking were somewhat different. For sarin, the lowest energy docking pose for the PS enantiomer oriented the O-isopropyl fragment in proximity to F222 and L240, with the leaving group fluoride anti to D269. In contrast, for the PR enantiomer, the same orientation of the isopropyl group results in the methyl substituent orienting anti to D269 (Figure 9). To orient the leaving group fluoride anti to D269 for the PS isomer would require placing the O-isopropyl group into the somewhat more congested active site region around V346. Similarly, for cyclosarin, the PR isomer also fails to orient the leaving group anti to calcium in any of the docking poses (see Supporting Information). These findings are consistent with the preference of WT huPON1 for the PS configuration of the G-series nerve agents, and with prior computational studies on fluorogenic OP-analogues docked into the PON1 crystal structure.v
It should be noted that while poses were obtained from docking which were consistent with certain mechanistic hypotheses, the energetic ordering of binding poses was not sufficiently large to distinguish them beyond the margin of error of the docking protocol (see Supporting Information). Energies for docking poses were generally within 2–3 kcal/mol for the 10 lowest energy binding modes predicted for a given substrate molecule, including orientations placing the substrate within the HDL binding domain; furthermore, the energetic ordering for the two modes was not consistent across the ligand library, even for the V-series agents. This is, we suspect, a limitation of the docking protocol when applied to a very open active-site cavity without the possession of a known binding mode from which to prune the output of the calculation.
While the energetic output from the docking simulations was not useful in a quantitative sense as a prediction of binding and turnover of substrate, the conformational sampling of the active site was useful in terms of both identifying general binding modes for substrates, and in suggesting residues for which mutations might alter the substrate specificity of the protein. A tally of the ligand-receptor contacts by distance suggested a number of residues in the active site that are in proximity to the calcium ion, as well as in the peripheral regions, which could be involved in binding specificity for organophosphorus agents.
Interestingly, a number of residues from this list have already been identified as modulating substrate specificity in mutagenesis studies, which further supports the notion that the docked orientations are sampling valid substrate binding modes, despite the coarse nature of the methodology (Table 1). To improve the treatment of receptor flexibility, and to obtain more reliable binding energies for comparison of poses, we employed molecular dynamics simulations to study the ligand-receptor complexes. This methodology has been successfully employed in the modeling of ligand-receptor binding even for molecules with highly similar structural features.xxxiv The inclusion of explicit solvation also allows for improved handling of electrostatic interactions in the receptor and in the complex.
Table 1.
Residues in huPON1 identified as modulating selectivity and activity by mutagenesis studies, and residues indicated by docking ‘hits’ with OP substrates.
| Residue ID | ||
|---|---|---|
| Experiment | Docking Hits | Location in huPON1 |
| 53 | Calcium Binding Site | |
| 69 | 69 | Periphery of active site |
| 70 | Periphery of active site | |
| 71 | Unresolved Loop | |
| 72 | Unresolved Loop | |
| 74 | Unresolved Loop | |
| 75 | Unresolved Loop | |
| 76 | Unresolved Loop | |
| 79 | Unresolved Loop | |
| 115 | 115 | Near to E53 |
| 168 | Calcium Binding Site | |
| 183 | Near to 168/192 | |
| 190 | Loop over Active Site | |
| 192 | Loop over Active Site | |
| 193 | Loop over Active Site | |
| 196 | Loop over Active Site | |
| 222 | 222 | Periphery of Active site |
| 224 | Calcium Binding Site | |
| 240 | 240 | Periphery of Active site |
| 269 | Calcium Binding Site | |
| 285 | Periphery of Active site | |
| 291 | 291 | Periphery of Active site |
| 292 | 292 | Periphery of Active site |
| 293 | Periphery of Active site | |
| 332 | 332 | Calcium Binding Site |
| 346 | 346 | Periphery of Active site |
Molecular Dynamics Refinement of Binding Modes
Binding pose refinement was performed using a similar molecular dynamics protocol to that described above for preparation of the original protein models. An initial restraint on the bond distance between the ‘catalytic’ calcium and the phosphoryl oxygen of the ligand was utilized for the initial minimization and heating steps, in addition to the restraints on the protein α-carbons as described above, to allow for removal of close contacts and other potential instabilities. The employed constraint was a flat–bottomed parabolic restraint (see Supporting Information), with a minimum from 2.5 to 4.0 Å, allowing for limited mobility of the ligand while maintaining contact with the calcium ion. This constraint was removed in latter steps of the MD simulations.
Following MD simulations, it was discovered that some of the docked poses were ultimately not stable in their coordinated orientation, and dissociation of some previously bound ligands from the calcium ion was observed. Of the poses which were ultimately stable following extensive (> 2 ns) MD simulations without restraints on the system, the orientations of the leaving group were generally in the two binding modes first observed from docking: oriented toward either D183 or K192, or between H285 and L240. The interactions in the former tended to include significant electrostatic components arising from hydrogen-bonding interactions with the charged residues in the pocket, or with bridging waters coordinated to the same. In the H285/L240 pocket, however, the interactions were primarily simple van der Waals interactions; the peripheral residues in the latter pocket tend to be of the non-polar variety, with V346, H285, L240, and F222 encompassing the sides of the pocket, while I291 and F292 bracket the upper portion.
Examination of the poses with substrate bound following an MD simulation revealed consistent changes in the receptor geometry in the active site. The hydrogen-bonding interaction between H115 and H134 was frequently disrupted, with the leaving group from the OP occupying the space. This was particularly common for the V-series agents, with the larger thioalkylamino fragment, which is due to a preference for the charged ammonium group of these agents to hydrogen-bond with D183, thereby occupying the portion of the binding site which would otherwise be occupied by the H115-H134 dyad (Figure 10).
Figure 10.
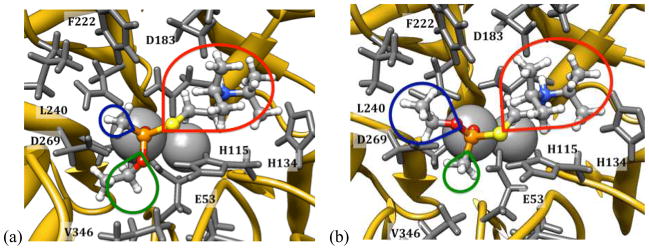
Minimum energy poses for (a) (R)- and (b) (S)-VX bound in the active site of the G2E6 protein model. The colored lobes indicate the relative sizes and orientations of the leaving group (red) and O-alkyl and methyl substituents.
The hydrogen bond between H115 and E53, however, was generally preserved even in cases where the H115-H134 dyad was broken. Indeed, the calcium coordination environment was also quite stable, even with the coordination of an OP on top of the calcium ion (see Supporting Information for details).
MM-PBSA and MM-GBSA calculations were performed to estimate the free energy of binding of the substrates into the protein active site following MD. From these calculations, a comparative evaluation of the energetics for several binding poses was possible. For the V-series agents, poses were evaluated for the two primary binding modes identified in docking; that is, with the leaving groups positioned anti to D269 and, alternatively, oriented toward the L240/F222 pocket. In the former binding mode, the V-agents typically orient the phosphoryl oxygen on the calcium ion, with the thioalkylamino group oriented toward D183, with which the protonated amino group hydrogen bonds either directly or via a bridging water. The O-alkyl fragment is oriented either toward V346 and H285, or toward F222 and L240, depending upon the configuration at phosphorus. Specifically, V-agents with an (R)-configuration place the O-alkyl group toward V346 and H285; (S)-configuration agents orient the O-alkyl group toward L240 and F222 (Figure 10). The size of the pockets in the active site of the G2E6 model of huPON1 are such that the PR configuration molecules are significantly more sterically congested in the active site cavity, and in some instances, stable bound poses were not obtained which maintained the hydrogen bond with D183. In contrast, for the agents with a PS configuration, the O-alkyl fragment is oriented into the larger L240/F222 region.
The stereoselectivity of huPON1 toward hydrolysis of V-series nerve agents is still somewhat unclear; if it follows similar trends as for the G-series agents, huPON1 would likely be more selective for the less toxic P+ (PR) enantiomers, although this remains unverified to-date. In light of our modeling data, we propose that the orientation and bulk of the O-alkyl substituents of the V-agents may influence the binding and/or turnover in a stereoselective fashion, and that modification of the spatial constraints in proximity to L69 and V346 might modulate the turnover of the PR enantiomers. The lowest energy binding modes predicted to orient the O-alkyl fragments in proximity to D269/E53, which suggests that interactions with and/or occlusion of the approaching nucleophile, be it one of the carboxylates or a coordinated water molecule, by the O-alkyl fragments of the V-agents could be impeding turnover and reducing kcat. The reader should note, however, that these predictions are based only on binding information. This is consistent with both the direct hydrolysis of substrate by D269, as well as the general-base hydrolysis mechanisms for huPON1.
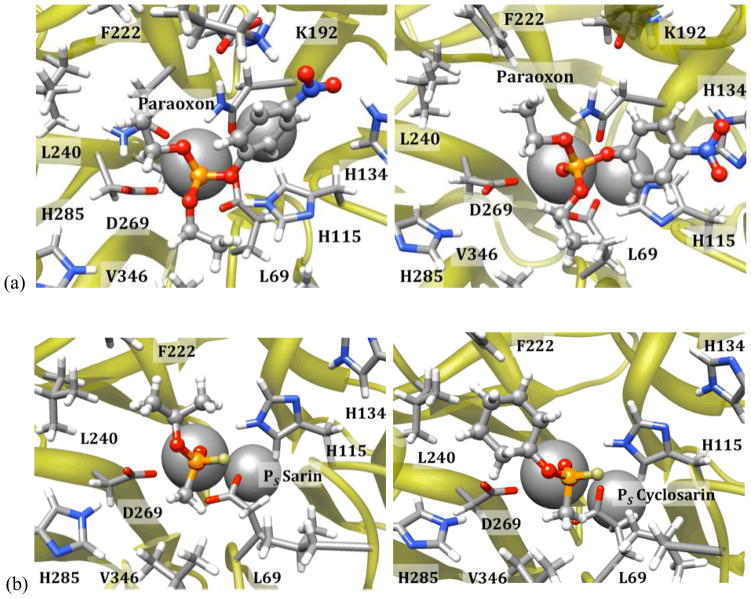
Interestingly, work by Furlong et al.xxxvii on huPON1 ‘status’ suggests that the polymorphic residue 192, which in G2E6 is modified to lysine from arginine or glutamine in the WT human protein, significantly alters the activity of PON1 for hydrolysis of OP pesticides. In the context of the study, it was found that 192R was more protective than 192Q for exposure to paraoxon. In light of the predicted binding modes for the V-series agents orienting the protonated amino fragment in proximity to D183 and K192, we propose a molecular-level rationalization for this modulation: competition for the hydrogen bond to D183, which is reduced in the 192Q polymorph. In contrast, the lowest energy binding modes for paraoxon placed the p-nitrophenoxide group in proximity to K192 itself, hydrogen bonding directly with the protonated amino group, and with an alternative pose with similar energy orienting the leaving group parallel to H134 in a π-stacking mode (Figure 11). An alternative theory regarding the K192 mutation, suggesting that it mitigates charge repulsion with D183 via hydrogen bonding, is also consistent with our modeling results.xl
Figure 11.
Stable, bound poses derived from MD simulations for (a) paraoxon, and (b) the PS isomers of sarin and cyclosarin in the G2E6 model.
These results suggest the role of residue 192 in the active site for substrates with large, polar/charged groups may be related to orientation of the substrate prior to hydrolysis. However, for the G-series nerve agents, which lack a large polar leaving group, there are no clear interactions with this portion of the active (Figure 11). Instead, the primary constraints on binding for these agents are the orientation of the large non-polar peripheral groups, which generally interact with non-polar residues in the active site, such as L69, V346, F222, and L240, some of which have been identified as modulating the stereoselectivity of the protein.xli For G-series nerve agents, these residues influence the fit of substrate into the active site and likely contribute to the stereoselectivity of the protein. Based on our binding studies, it appears that orientations which allow the bulky substituents to orient in proximity to L240 and F222 are favorable, so it is the resulting orientation of the leaving group which influences turnover. For the PS agents, orientation of the leaving group anti to the D269 pocket is possible in these poses, whereas the PR agents place the leaving group in the opposite direction in such a pose. While stable, bound orientations were identified for PS G-series agents that placed the O-alkyl substituents in proximity to L69 and V346 (and thus, with the leaving groups anti to the D269 region), these poses tended to be higher in energy than the more favorable mode by between 3 to 5 kcal/mol, or slightly higher than the margin of error (see Supporting Information). Given that huPON1 does turn over both enantiomers of cyclosarin, as well as all four diastereomers of soman, the observation of ‘productive’ binding modes with the leaving group anti to the D269 region for the PR isomers is not inconsistent with experiment.xlii
It should be noted that one interaction which has been proposed between receptor and substrate in huPON1 is the ‘capping’ of the active site by residue Y71, found on the flexible surface loop that was unresolved in the protein crystal structure.xl This loop, while highly flexible in our simulations, was not universally observed to participate in such an interaction in bound poses. Some poses of bound agent, particularly for the V-agents, did show capping of the bound substrate by Y71 in a manner similar to that reported (see Supporting Information). It remains possible that the Y71 interaction is vital for OP binding, or alternatively, that Y71 is involved in substrate mobility into the active site; however, such mobility was not observed over the time scales sampled in this study, and additional simulations are necessary to determine if this is the case.
An alternative method for examining the binding of substrates to huPON1 involves the use of umbrella samplingxliii to estimate the potential energy surface for removal of substrate from coordination to the calcium ion, or from the active site overall.xliv In this methodology, we utilized a conformational sampling protocol in conjunction with parallel, constrained MD trajectories, to estimate a probability distribution for a given geometric parameter in the active site, from which the free energy can be extracted. In the context of our study, the calcium-phosphoryl oxygen bond coordinate was chosen for sampling, starting from the bound geometries obtained from MD refinement of the V-series agents, as described above. The output from several such runs are shown in Figure 12, for both (R)- and (S)- configuration VX. The minimum energy bond distance in both cases was estimated to be 2.7 Å, which is somewhat longer than the calcium-phosphate distance of 2.2 Å observed in the x-ray crystal structure of G2E6,iv but which is consistent with the stable, bound orientations calculated above. A plateau observed in the potential surface for the S- configuration agents corresponds to a movement of the phosphoryl oxygen away from calcium toward the calcium-coordinating residue N168, which is able to form a hydrogen bond and to stabilize the partially dissociated OPs. This may represent an intermediate stage in the substrate association/dissociation potential energy surface, and more simulations are in progress to fully explore this possibility.
Figure 12.
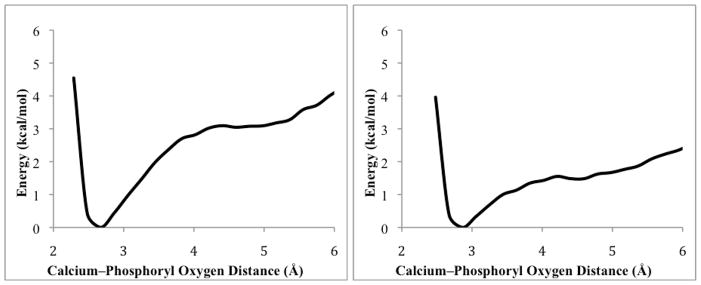
Potential energy surfaces for loss of coordination to calcium calculated for (PR)–VR (left), and (PS)–VX (right), calculated using umbrella sampling methods from bound orientations.
Interestingly, a similar analysis on poses which showed a lack of stability in calcium coordination found no minima at ~2.7 Å corresponding to the bound state, suggesting that this methodology is also useful in validating the stability of bound substrate to the metal ion. When applied to the V-series nerve agents studied herein, the correlation between poses that were stably bound to calcium over the course of MD and those with minima in the dissociation potentials for bound poses was excellent (see Supporting Information).
Discussion
Human paraoxonase 1 (huPON1) is a protein that promiscuously catalyzes the hydrolysis of a variety of electrophilic substrates. It is tolerant of a variety of alkyl and aromatic esters and coumarin substrates, as well as organophosphorus compounds. In this study, we have attempted to model the protein using computational chemistry techniques to both gain insight into the structure and function of the enzyme as well as provide guidance for the development of novel protein mutants via rational design.
We have produced a structural model of the G2E6 variant of huPON1, including a loop above the putative active site, which was unresolved in the published x-ray crystal structure, as part of a cooperative program aimed at enhancing the catalytic function of the protein for use as a bioscavenger. In the active site itself, the largest change from the published structure is in the hydrogen bonding of H115, which migrates considerably closer to E53, and maintains a tight hydrogen bond; this interaction alters the distance between these residues from ~4 Å in the crystal structure to 2.5 Å in the G2E6 computational model. Other than this divergence, the differences between the G2E6 computational model and the published x-ray structureiv are in the surface loops, and these changes do not significantly alter the active site geometry; furthermore, these surface loops are likely highly flexible in the absence of HDL association.
From the molecular docking studies, the observed interactions between residues 183, 192, and polar groups with the leaving groups for the bound OP substrate suggest a potential role of these residues in substrate binding and orientation in the active site, which is consistent with the available experimental data. Additional work is necessary, both in developing new protein mutants from which additional kinetic information can be obtained, and in terms of additional sampling of the conformational motion of these residues, before the exact nature of the interactions can be conclusively determined. This is particularly true given the lack of a reference orientation of substrate in any experimentally obtained structure of PON1 to date.
The lowest energy binding modes observed for V-series agents and paraoxon orient the leaving group anti to the carboxylate residues which coordinate the catalytic calcium, and this observation supports several of the proposed reaction mechanisms, above. In this pose, direct hydrolysis of the OP by one of the two carboxylates is possible, with average distances on the order of 3.5 – 4.5 Å from the coordinated OP to D269 and/or E53. However, for E53 to act as a nucleophile, attack would be necessarily precluded by loss of a hydrogen bond with H115, which is only rarely observed in the MD trajectories of either the free or ligand-bound proteins. The interaction between H285 and D269 is significantly more labile, which suggests that the latter residue might be more available for attack; however, the nucleophilicity of an aspartate coordinated to a calcium ion is likely significantly diminished. A third possibility is the attack by an activated water molecule, possibly as part of a catalytic dyad or triad in the active site. The observation of coordinated water molecules in proximity to both D269 and E53 in ligand-bound complexes supports the possibility that deprotonation of water by either one of the carboxylates, or possibly the basic nitrogen of H285 might be able to generate hydroxide in a suitable orientation for hydrolysis of substrate. The lack of detectible ‘aging’ of huPON1 also supports this notion, as a general base mechanism would not produce a covalent protein-ligand conjugate. However, molecular dynamics simulations alone are insufficient to probe this possibility. To examine reaction pathways explicitly, and to obtain energetic parameters to compare and contrast them, hybrid quantum mechanical/molecular mechanical (QM/MM) simulations are in progress; the primary complication in this approach is the large number of degrees of freedom in the active site, as well as the numerous potential geometries for water clusters. The results, however, will reduce some of the ambiguity present in the literature with regard to the mechanism of huPON1 hydrolysis of OPs.
The binding modes identified also correlate well with published results on mutations that influence the selectivity of huPON1. Modification of peripheral active site residues such as L69, H115, F222, and V346 have all been identified in the course of directed evolution by Tawfik et al.xli to modify the selectivity of the protein, and these residues are generally in proximity to the peripheral groups of ligands bound to the huPON1 model. While the exact molecular-level role of these mutations on substrate binding and turnover remains speculative at this stage, the development of improved models for the protein opens the door for development of in silico mutants with the potential to both calibrate and refine the models and potentially drive the development of protein mutants with altered selectivity and activity.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support of this research by the National Institutes of Health (U54-NS058183). Generous computational resources have been provided by the Ohio Supercomputer Center. We also acknowledge fruitful and insightful discussions with Professor Thomas Magliery (OSU). The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the United States Army or the Department of Defense.
Footnotes
Additional details regarding model preparation, molecular docking results, molecular dynamics simulations are included in the supporting information.
References
- i.(a) Furlong CE. In: Paraoxonases: An historical perspective, Proteins and Cell Regulation. Mackness B, Mackness M, Aviram M, Paragh G, editors. Vol. 6. Springer; NY: 2008. [Google Scholar]; (b) Khersonsky O, Tawfik DS. ChemBioChem. 2006;7:49–53. doi: 10.1002/cbic.200500334. [DOI] [PubMed] [Google Scholar]; (c) Yeung DT, Lenz DE, Cerasoli DM. In: The Paraoxonases: Their Role in Disease, Development and Xenobiotic Metabolism. Proteins and Cell Regulation. Mackness B, Mackness M, Aviram M, Paragh G, editors. Vol. 6. Springer; NY: 2008. pp. 151–170. [Google Scholar]; (d) Lenz DE, Yeung DT, Smith RJ, Sweeney RE, Lumley LA, Cerasoli DE. Toxicol. 2007;233:31–39. doi: 10.1016/j.tox.2006.11.066. [DOI] [PubMed] [Google Scholar]
- ii.Lenz DE, Brimfield AA, Cook LA. The Development of Immunoassays for Detection of Chemical Warfare Agents. In: Aga D, Thurman EM, editors. Development and Applications of Immunoassays for Environmental Analysis. ACS Books; Washington, DC: 1997. pp. 77–86. [Google Scholar]
- iii.Yeung DT, Josse D, Nicholson JD, Khanal A, McAndrew CW, Bahnson BJ, Lenz DE, Cerasoli DM. Biochim Biophys Acta. 2004;1702:67–77. doi: 10.1016/j.bbapap.2004.08.002. [DOI] [PubMed] [Google Scholar]
- iv.Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RBG, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Nature Struct Mol Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- v.Blum M, Timperley CM, Williams GR, Thiermann GRH, Worek HF. Biochemistry. 2008;47:5216–5224. doi: 10.1021/bi702222x. [DOI] [PubMed] [Google Scholar]
- vi.Blum M, Koglin A, Rüterjans R, Schoenborn B, Langan P, Chen JCH. Acta Crystallogr F. 2007;63:42–45. doi: 10.1107/S1744309106052924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vii.Worek F, Eyer P, Szinicz L. Arch Toxicol. 1998;72:996–998. doi: 10.1007/s002040050546. [DOI] [PubMed] [Google Scholar]
- viii.Saxena A, Doctor BP, Maxwell DM, Lenz DE, Radic Z, Taylor P. Biochem Biophys Rsh Communications. 1993;197:343–349. doi: 10.1006/bbrc.1993.2481. [DOI] [PubMed] [Google Scholar]
- ix.Fleming CD, Edwards CC, Kirby SD, Maxwell DM, Potter PM, Cerasoli DM, Redinbo MR. Biochemistry. 2007;46:5063–5071. doi: 10.1021/bi700246n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- x.(a) Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–197. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]; (b) Houk KN, Cheong PH. Nature. 2008;455:309–313. doi: 10.1038/nature07368. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xi.Sorenson RC, Bisgaier CL, Aviram M, Hsu C, Billecke S, La Du BN. Arterioscler Thromb Vasc Biol. 1999;19:2214–2225. doi: 10.1161/01.atv.19.9.2214. [DOI] [PubMed] [Google Scholar]
- xii.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. Nucleic Acids Res. 2004;32:665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xiii.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- xiv.Wang J, Wolf RM, Caldwell JW, Kollamn PA, Case DA. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- xv.Aqvist J. J Phys Chem. 1990;94:8021–8024. [Google Scholar]
- xvi.Blum M, Löhr F, Richardt A, Rüterjans H, Chen GC. J Am Chem Soc. 2006;128:12750–12757. doi: 10.1021/ja061887n. [DOI] [PubMed] [Google Scholar]
- xvii.(a) Mahoney MW, Jorgensen WL. J Chem Phys. 2000;112:8910–8922. [Google Scholar]; (b) Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- xviii.(a) Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco: 2006. [Google Scholar]; (b) Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Seetin MG, Sagui C, Babin V, Schafmeister WS, Ross C, Kollman PA. AMBER 10. University of California; San Francisco: 2008. [Google Scholar]
- xix.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- xx.Harvey SC, Tan RK, Cheatham TE., III J Comput Chem. 1998;19:726–740. [Google Scholar]
- xxi.Chachra R, Rizzo RC. J Chem Theory Comput. 2008;4:1526–1540. doi: 10.1021/ct800068v. [DOI] [PubMed] [Google Scholar]
- xxii.Miyamoto S, Kollman PA. J Comput Chem. 1992;13:952–962. [Google Scholar]
- xxiii.Toukmaji A, Sagui C, Board J, Darden T. J Chem Phys. 2000;113:10913–10927. [Google Scholar]
- xxiv.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- xxv.Breneman CM, Wiberg KB. J Comput Chem. 1990;11:361–373. [Google Scholar]
- xxvi.(a) Becke AD. J Chem Phys. 1993;98:5648–5656. [Google Scholar]; (b) Lee C, Yang W, Parr RG. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- xxvii.Labanowski J. Density Functional Methods in Chemistry. Springer-Verlag; Heidelberg: 1991. [Google Scholar]; (b) Parr RG, Weitao Y. Density-Functional Theory in Atoms and Molecules. Oxford University Press; New York: 1989. [Google Scholar]
- xxviii.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Jr, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima Y, Honda O, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas MC, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision B.04. Gaussian, Inc; Pittsburgh PA: 2003. [Google Scholar]
- xxix.(a) Stevens PJ, Devlin FJ, Chablowski CF, Frisch MF. J Phys Chem. 1994;98:11623–11627. [Google Scholar]; (b) Becke AD. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]; (c) Becke AD. J Chem Phys. 1992;96:2155–2160. [Google Scholar]; (d) Becke AD. J Chem Phys. 1992;97:9173–9177. [Google Scholar]
- xxx.(a) Proft FD, Martin JML, Geerlings P. Chem Phys Lett. 1996;250:393–401. [Google Scholar]; (b) Martin F, Zipse H. J Comput Chem. 2004;26:97–105. doi: 10.1002/jcc.20157. [DOI] [PubMed] [Google Scholar]
- xxxi.Luo R, David L, Gilson MK. J Comput Chem. 2002;23:1244–1253. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- xxxii.Weiser J, Shenkin PS, Still WC. J Comput Chem. 1999;20:217–230. doi: 10.1002/(SICI)1096-987X(199905)20:7<688::AID-JCC4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- xxxiii.Sanner MF, Olson AJ, Spehner J. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- xxxiv.Rizzo RC, Aynechi T, Case DA, Kuntz ID. J Chem Theory Comput. 2006;2:128–139. doi: 10.1021/ct050097l. [DOI] [PubMed] [Google Scholar]
- xxxv.Kuhn B, Gerber P, Schulz-Gasch T, Stahl M. J Med Chem. 2005;48:4040–4048. doi: 10.1021/jm049081q. [DOI] [PubMed] [Google Scholar]
- xxxvi.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- xxxvii.Furlong CE, Cole TB, Jarvik GP, Pettan-Brewer C, Geiss GK, Richter RJ, Shih DM, Tward AD, Lusis AJ, Costa LG. NeuroToxicology. 2005;26:651–659. doi: 10.1016/j.neuro.2004.08.002. [DOI] [PubMed] [Google Scholar]
- xxxviii.Otto TC, Harsch CK, Yeung DT, Magliery TJ, Cerasoli DM, Lenz DE. Biochemistry. 2009;48:10416–10422. doi: 10.1021/bi901161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxxix.Khersonsky O, Tawfik DS. J Biol Chem. 2006;281:7649–7656. doi: 10.1074/jbc.M512594200. [DOI] [PubMed] [Google Scholar]
- xl.Hu X, Jiang X, Lenz DE, Cerasoli DM, Wallqvist A. Proteins. 2009;75:486–498. doi: 10.1002/prot.22264. [DOI] [PubMed] [Google Scholar]
- xli.Amitai G, Gaidukov L, Adani R, Yishay S, Yacov G, Kushnir M, Teitlboim S, Lindenbaum M, Bel P, Khersonsky O, Tawfik DS, Meshulam H. FEBS J. 2006;273:1906–1919. doi: 10.1111/j.1742-4658.2006.05198.x. [DOI] [PubMed] [Google Scholar]
- xlii.Yeung DT, Smith JR, Sweeny RE, Lenz DE, Cerasoli DM. FEBS J. 2007;274:1183–1191. doi: 10.1111/j.1742-4658.2006.05650.x. [DOI] [PubMed] [Google Scholar]
- xliii.Torrier GM, Valleau JP. Chem Phys Lett. 1974;28:578–581. [Google Scholar]
- xliv.Kottalam J, Case DA. J Am Chem Soc. 1988;110:7690–7697. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.