Abstract
Objective
The freshwater snail Biomphalaria glabrata is the principal intermediate host for the parasite Schistosoma mansoni within Brazil. We assessed the potential effects of snail population dynamics on parasite transmission dynamics via population genetics.
Methods
We sampled snail populations located within the confines of three schistosome-endemic villages in the state of Minas Gerais, Brazil. Snails were collected from individual microhabitats following seasonal periods of flood and drought over the span of one year. Snail spatio-temporal genetic diversity and population differentiation of 598 snails from 12 sites were assessed at 7 microsatellite loci.
Results
Average genetic diversity was relatively low, ranging from 4.29 to 9.43 alleles per locus and, overall, subpopulations tended to exhibit heterozygote deficits. Genetic diversity was highly spatially partitioned among subpopulations, while virtually no partitioning was observed across temporal sampling. Comparison with previously published parasite genetic diversity data indicated that S. mansoni populations are significantly more variable and less subdivided than those of the B. glabrata intermediate hosts.
Discussion
Within individual Brazilian villages, observed distributions of snail genetic diversity indicate temporal stability and very restricted gene flow. This is contrary to observations of schistosome genetic diversity over the same spatial scale, corroborating the expectation that parasite gene flow at the level of individual villages is likely driven by vertebrate host movement.
Keywords: Biomphalaria glabrata, Schistosoma mansoni, population genetics, microsatellite, gene flow, Brazil
Introduction
Researchers in both parasitology and medicine have long been interested in elucidating the transmission of disease agents among hosts. In their own ways, both groups are striving to understand the historical or evolutionary associations of host and parasite that have shaped current dynamics, as well as the degree to which various biotic and abiotic factors continue to influence coevolutionary landscapes and epidemiological outcomes. With the advent of molecular ecology and the increasing availability of molecular tools, many researchers in both fields have taken to investigating parasite and host genetics as either direct or proxy measurements of parasite transmission dynamics and/or the factors influencing patterns of transmission. One of the most frequently used tools is estimation of genetic variation and the partitioning of that variation using neutral polymorphic loci like microsatellites (variable number tandem repeat markers). Generally, when neutral genetic diversity is high and/or individuals are genetically similar across putative population groups, we conclude that mating is random (in the case of sexually reproducing organisms) and that gene flow is high and not spatially or temporally restricted. For pathogens, such an outcome might produce conclusions of widespread transmission that is not limited to distinct foci.
These measures of genetic variation have been routinely applied to the study of natural populations of the trematode parasites in the genus Schistosoma (Steinauer et al. 2010), which are estimated to cause disease in over 200 million people worldwide (Engels et al. 2002, Chitsulo et al. 2004). In these studies observed levels of parasite genetic diversity and its spatial partitioning often lead to conclusions that the vertebrate definitive hosts, whether human or otherwise, are the primary agents of gene flow within the parasite population. In studies utilizing non-human definitive hosts, this can be supported by direct measurements of both host and adult worm genetic variation (Prugnolle et al. 2005). Moreover, numerous studies of the molluscan intermediate host population genetics (Woolhouse et al. 1992, Langand et al. 1999, Sire et al. 2001, Webster et al. 2001, Charbonnel et al. 2002a&b, Mavárez et al. 2002a&b, Wethington et al. 2007) and demographics and ecology (Utzinger et al. 1997, Utzinger and Tanner 2000, Erko et al. 2006), as a whole, support the assumption that snail populations are subdivided over both temporal and increasingly limited spatial scales. However, given the role of schistosomiasis as a tropical disease with global implications, much of the interest in transmission dynamics is focused on endemic human populations. Clearly studies of human populations have ethical and logistical limitations that restrict investigation of host influence on parasite movement to observations of water contact and usage patterns (e.g. Kloos et al. 1998 2001 2006) and assessments of genetic variation are restricted to collection of parasite offspring from human hosts (Brouwer et al. 2001; Curtis et al. 2002; Thiele et al. 2008; Standley et al. 2010). These studies on parasites of humans have found that parasite gene flow ranges along a continuum that can reasonably be associated with human movements, but cannot eliminate alternative factors, such as hydrogeography and the influence of alternative or intermediate hosts. In this study, we sought to supplement previous investigations of parasite transmission dynamics within human populations by assessing the role that snail host populations may play in the establishment of observed parasite population genetics. More specifically, we aimed to do so at a scale most relevant to the local epidemiology of schistosomes – that of the spatial confines of single villages.
We have investigated both the temporal and spatial variation in snail host genetic diversity within three villages in a region of endemic schistosomiasis in Brazil. Although Brazil’s schistosomiasis control program has enjoyed success, the disease remains an important factor in the public health and economy of the country (Coura and Amaral 2004, Amaral et al. 2006). Estimates of prevalence, though admittedly imprecise, include over 6 million people (Katz & Peixoto 2000). In contrast to previous studies of snail genetic diversity, the work presented here aims to investigate patterns of snail movement at the epidemiologically relevant spatial scale of the individual village, replicated over three distinct villages and over a temporal scale that encompasses a year’s seasonal population fluctuations to better estimate the actual role that snail movements may have in the establishment of observed patterns in parasite population genetics. These results are also presented in the context of patterns of parasite genetic variation previously observed in one of the Brazilian villages.
Methods and Materials
Site Descriptions and Snail Collection
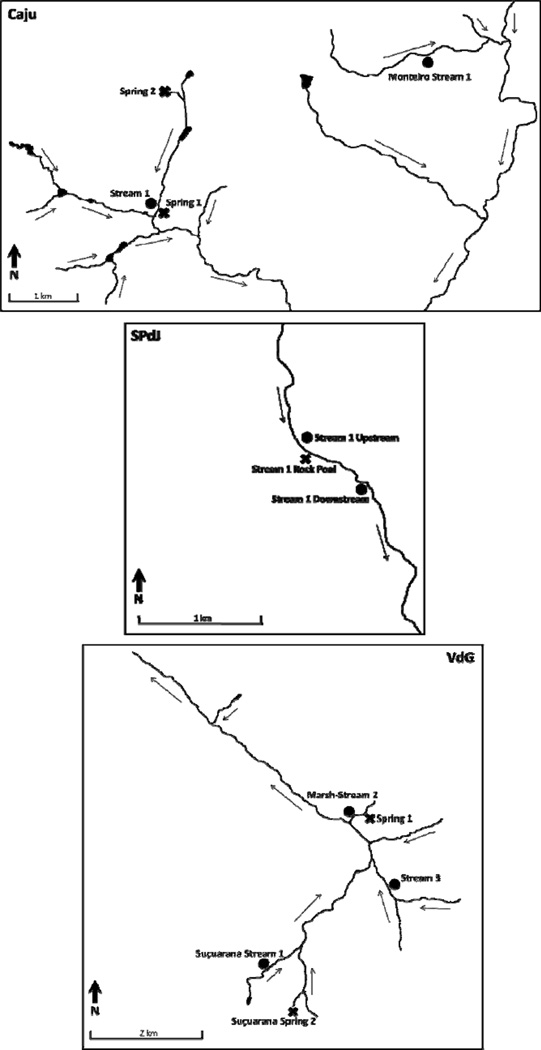
Natural populations of Biomphalaria glabrata were sampled in three villages endemic for schistosomiasis in the Jequitinhonha Valley of northeastern Minas Gerais, Brazil. The villages of Caju (16° 21' S; 41° 18' W), São Pedro do Jequitinhonha (SPdJ, 16° 30' S; 41° 20' W), and Virgem das Graças (VdG, 16° 56' S; 41° 21' W) can all be broadly characterized as semi-arid locales subject to high levels of poverty and urban migration. Lack of water delivery or sanitary infrastructure and reliance on subsistence agriculture ensures widespread human use, and often fecal contamination, of all habitat types suitable for B. glabrata, guaranteeing maintenance of the schistosome life cycle. All three villages have a prevalence of schistosomiasis in excess of60% in human hosts (Kloos et al. 2004; Carlos dos Reis et al. 2010). Geographically, all three villages are characterized by well-defined drainages, however, the degree of confluency and the scale across which snails were sampled varied for each one (Figure 1). Within the sampled region, there is a precipitation pattern of distinct periods of high and low rainfall (i.e., rainy versus dry seasons), with the rainiest period being from late October to March. To investigate the temporal effects of season on snail populations and levels of genetic variation, sites within each village were sampled once at the end of each season in 2006, with the post-rainy season collection in April and post-dry season collection in early November.
Figure 1.
Maps of the villages in which snails were sampled. Individual sites are indicated by the assigned site name and symbols to indicate the habitat designation as “closed” or “open”. Open habitats (circles) are expected to allow relatively equal rates of snail influx and efflux, while closed habitats (X’s) are expected to principally serve as source populations. Permanent/semi-permanent waterways are represented by the lines, with arrows indicating water flow direction.
To assess spatial variation in snail genetic diversity at an epidemiologically relevant level, sampling was confined to the immediate area of each village. Sample sites were selected based on the known existence of B. glabrata populations and expectation of B. glabrata presence based on the suitability of habitat types (Kloos et al. 2001). Most sites sampled were used by both the molluscan intermediate and human definitive hosts. Sampling at all sites in all villages was performed with a standard FIOCRUZ metal scoop and large forceps and was standardized according to sampling area and sampling effort (10 minutes within each of 3–5 transects of approximately 1 m2 per site at depths ≤1m). All snails were transported to the malacology lab at the Centro de Pesquisas René Rachou in Belo Horizonte, Minas Gerais, where they were examined for parasite infection over a period of 3–4 weeks, and then later to Purdue University where they were re-assessed for infection after tissue was collected for genetic analysis.
Microsatellite Analysis
Total genomic DNA was extracted from individual snails’ head-foot tissue via a modified Puregene extraction method (Gentra). Briefly, tissue was digested in 200µL of cell lysis solution (100mM Tris-HCl, 10mM EDTA, 100mM NaCl, 1% SDS, 0.06mg Proteinase K, 1.5mM dithiothreitol), followed by protein precipitation with 30% v/v of 10M potassium acetate. DNA was then separated from the supernatant via standard ethanol precipitation, dried, resuspended in sterile nuclease-free water, and quantified with a NanoDrop 1000 (Thermo Fisher Scientific).
Individual snails were genotyped across 7 microsatellites (Table 1; BgC7, BgC8, BgE4, BgE5 [Mavárez et al. 2000] and Bgµ15, Bgµ16, µBg1 [Jones et al. 1999]) in 2 multiplex reactions using the QIAGEN Type-It PCR Kit (Qiagen). Each 5 µL reaction contained 0.2 µM each of 3–4 fluorescently labeled primers (6-FAM, HEX [Integrated DNA Technologies], or NED [Applied Biosystems]), 1X each of the proprietary master mix and Q solutions, and 10 ng of total genomic DNA. Thermal cycling was performed in an Eppendorf Mastercycler ep thermocycler under the following conditions: 15 min at 95 °C; 35 repetitions of 30 s at 94 °C, 90 s at 57 °C, and 60 s at 72 °C; 30 min final extension at 60 °C; and a final hold at 4 °C. PCR products were genotyped with an ABI 3130 automated sequencer (Applied Biosystems) and scored with GeneMapper v.4.0 software (Applied Biosystems).
Table 1.
Microsatellite primers utilized for assessment of B. glabrata genetic variation.
| Name | Repeat Motif | Acc. # | Size Range | Citation |
|---|---|---|---|---|
| BgC7 | (AG)3G(GA)8GGGAGG(GA)5 | AF216280 | 317–321 | Mavárez et al. 2000 |
| BgC8 | (AG)4N23(AG)8G(GA)3N13(AG)5 | AF216274 | 269–271 | Mavárez et al. 2000 |
| BgE4 | (GATA)13 | AF216272 | 185–246 | Mavárez et al. 2000 |
| BgE5 | (GATA)34 | AF216271 | 273–345 | Mavárez et al. 2000 |
| Bgµ15 | (GA)14(G)11 | AF157700 | 182–162 | Jones et al. 1999 |
| Bgµ16 | (TC)24(TATC)6 | AF157701 | 138–124 | Jones et al. 1999 |
| µBg1 | (TC)20 | AF157703 | 200–160 | Jones et al. 1999 |
Data Analysis
A total of 598 snails were genotyped from across the 3 villages. Prior to assessing the standard estimates of genetic diversity, the data were evaluated with the freely available programs MICRO-CHECKER (van Oosterhout et al. 2004) and GENETIX (Belkhir et al. 2004). MICRO-CHECKER helps validate allelic designations by evaluating the dataset for evidence of typographical errors and the presence of null alleles, allelic dropout, and stutter-related scoring errors. Because related and morphologically similar species of Biomphalaria (e.g. B. tenagophila) can coexist with B. glabrata, genetic data were screened with a 2D factorial correspondence analysis in GENETIX to identify genetic outliers. Individuals that were clearly separated from the village-wide assortment (i.e. not just from their putative subpopulation) were eliminated from the data set to avoid potential inclusion of related species in population genetic analyses.
To assess the genetic diversity of snail populations at each site within each village and for each sampling event, number of alleles per locus and unbiased heterozygosity (Hs; Nei 1987) were calculated in FSTAT version 2.9.3.2 (Goudet 2001). The Weir and Cockerham (1984) estimator f of FIS was calculated for each locus and across all loci to test for deviations from Hardy-Weinberg equilibrium (HWE). The hypothesis that the snail populations are not panmictic (i.e., f ≠ 0) was tested in SPAGeDi version 1.3 (Hardy and Vekemans 2002) via a two-tailed test of 10,000 permutations of alleles among individuals, with significance set at P < 0.05. Linkage disequilibrium between pairs of loci was tested in FSTAT, with the nominal significance value set to 0.05. Significance of multiple comparisons was determined after the sequential Bonferroni correction (Rice 1989).
Genetic differentiation of snails was assessed with several measures of subdivision. The standard estimator θ of FST (Weir & Cockerham 1984) was calculated to assess the variance in allele frequencies among subpopulations. Significance of genetic differentiation was assessed with the G-based test (Goudet et al. 1996) following 50,000 permutations of host genotypes, and 95% confidence intervals were calculated by bootstrapping over loci in FSTAT. Because differing levels of heterozygosity and marker-based variability can limit the ability to compare measures of FST among studies (Hedrick 2005), all measures of FST were standardized using the recoding method of Meirmans (2006) to represent the proportion of Fmax comprised by Fobs (i.e. F′ST). Both traditional and standardized representations are presented here and all quantitative comparisons with previous studies, whether of B. glabrata or other organisms, utilize F′ST.
To evaluate the proportion of genetic variance explained by population and/or seasonal effects, analysis of molecular variance (AMOVA) was performed with ARLEQUIN version 3.11 (Excoffier et al. 2005). Patterns of isolation-by-distance were investigated within each village and collection event using the method of Rousset (1997). To determine if the distribution of genetic variation (measured as θ/(1- θ)) was significantly correlated to the distance between populations, a partial Mantel test with 10,000 permutations was performed in FSTAT. Finally, the Bayesian clustering analysis of STRUCTURE version 2.2 (Pritchard et al. 2000; Falush et al. 2003) was also utilized to test for subdivision of snails into distinct genetic clusters. Clustering of populations was performed within individual villages, both within and across seasons. The maximum number of clusters (K) was set at twice the number of sampling sites so as to include the maximum number of populations reasonably expected if snail microhabitats are representative of demes. Under the assumptions of admixture and allele frequency independence, each K-value was replicated 10 times with burn-in and Markov chain Monte Carlo lengths of 10,000. Estimation of the true value of K followed the procedure ofEvanno et al. (2005).
The influence of village, season, and habitat type on measures of genetic diversity was assessed by 5000 permutations of individual snails between designated seasonal or habitat groupings in FSTAT. Habitat groupings were created by categorizing sampling sites according to estimates of habitat openness (i.e. amount of expected inflow and outflow of water and, by extension, snails). Sites were either designated as open (expected to allow relatively equal influx and efflux of snails) or closed (expected to principally serve only as source populations of snails). Open habitats were primarily streams or marshes located in the midst of a clear drainage path, while closed habitats were generally springs or wells that were structurally and/or elevationally prohibitive to passive snail migration.
Results
Snail Sampling and Infection Prevalence
Snail abundance varied widely between sites, with a minimum density of 0.4 snails/m3 in a wet season VdG stream and a maximum of 414.7 snails/m3 in a dry season Caju spring (Table 3). Likewise, abundance varied between seasons within individual sites, sometimes quite dramatically, though there was no association between snail density and season or openness of the site. Prevalence of schistosome infection in the snails, overall, was 1.8%. Infected snails were found in only 3 of the 24 sampling events and the majority of infections occurred in snails collected during the dry season from Caju’s “Monteiro Stream 1” (7 infected snails versus 1 and 2 snails from Caju’s “Stream 1” and SPdJ’s “Stream 1 Rock Pool”, respectively).
Table 3.
Genetic diversity and number of snails genotyped for each collection event and site. Parenthetical numbers below number of snails genotyped indicate snail density per cubic meter at each collection site. Superscript O indicates habitat classified as “open”, while X indicates habitat classified as “closed”. Values in bold are significantly different from zero (α = 0.05).
| Collection Site | Season | # Snails | NA | HS | f | |
|---|---|---|---|---|---|---|
| Caju | Stream 1 O | Wet | 51 (303.3) |
3.57 | 0.38 | 0.16 |
| Dry | 14 (23.21) |
2.43 | 0.39 | 0.06 | ||
| Spring 1 X | Wet | 6 (4.9) |
1.43 | 0.16 | 0.36 | |
| Dry | 64 (414.7) |
1.86 | 0.17 | −0.17 | ||
| Spring 2 X | Wet | 24 (15.7) |
2.00 | 0.22 | 0.33 | |
| Dry | 12 (10.9) |
1.57 | 0.22 | −0.09 | ||
| Monteiro Stream 1 O | Wet | 3 (113.3) |
1.57 | 0.26 | −0.09 | |
| Dry | 61 (168.3) |
3.86 | 0.39 | 0.09 | ||
| SPdJ | Stream 1 Upstream O | Wet | 11 (18.7) |
5.86 | 0.70 | 0.06 |
| Dry | 18 (74.8) |
7.29 | 0.76 | 0.18 | ||
| Stream 1 Downstream O | Wet | 31 (356.7) |
6.57 | 0.72 | 0.20 | |
| Dry | 22 (10) |
6.86 | 0.73 | 0.20 | ||
| Stream 1 Rock Pool X | Wet | 17 (58) |
2.71 | 0.41 | 0.40 | |
| Dry | 46 (170.7) |
2.00 | 0.23 | 0.06 | ||
| VdG | Marsh-Stream 2 O | Wet | 7 (0.6) |
3.12 | 0.43 | −0.05 |
| Dry | 15 (190.4) |
3.43 | 0.56 | 0.02 | ||
| Stream 3 O | Wet | 6 (3.9) |
2.57 | 0.48 | 0.11 | |
| Dry | 11 (3.6) |
2.43 | 0.14 | 0.10 | ||
| Spring 1 X | Wet | 69 (28.5) |
3.29 | 0.48 | 0.01 | |
| Dry | 28 (17.1) |
3.43 | 0.57 | 0.01 | ||
| Suçuarana Stream 1 O | Wet | 5 (0.4) |
2.29 | 0.40 | 0.15 | |
| Dry | 11 (1.5) |
2.57 | 0.41 | 0.22 | ||
| Suçuarana Spring 2 X | Wet | 52 (34.4) |
1.29 | 0.01 | 0 | |
| Dry | 14 (3.5) |
1.14 | 0.01 | 0 | ||
NA, number of alleles
HS, Nei’s (1987) unbiased heterozygosity
f, Weir and Cockerham’s (1984) estimate of inbreeding
Genetic Diversity
Screening of genetic data with MICRO-CHECKER revealed no indication of null alleles, dropout, or scoring errors. GENETIX 2D factorial correspondence analysis indicated six outlier individuals from five different collection sites (2 sites in the village of Caju and 3 in SPdJ). These individual snails were removed from further genetic analyses. Overall, a range of 1 to 13 alleles was observed per locus, with the average number of alleles per locus per population ranging from 4.29 to 8.29 after the wet season and 5.43 to 9.43 after the dry season (Table 2). Mean overall unbiased heterozygosity (HS) ranged from 0.26 to 0.61 and 0.29 to 0.57 following the wet and dry seasons, respectively. Overall estimates of inbreeding (f) ranged from 0.01 to 0.21 and deviated significantly from 0 for three of the six sampling events, indicating significant heterozygote deficits following the wet season in Caju and both seasons in SPdJ (Table 2).
Table 2.
Measures of global genetic diversity within each village and sampling season. Numbers in parentheses and italics represent ranges and 95% C.I., respectively.
| Caju | SPdJ | VdG | ||||
|---|---|---|---|---|---|---|
| Post-Wet | Post-Dry | Post-Wet | Post-Dry | Post-Wet | Post-Dry | |
| # Pop. | 4 | 3 | 5 | |||
| # Snails | 84 (3–51) |
151 (12–64) |
59 (11–31) |
86 (18–46) |
139 (5–69) |
79 (11–28) |
| NA | 4.29 (1–7) |
5.43 (3–8) |
8.29 (5–11) |
9.43 (2–13) |
5.00 (2–8) |
5.86 (2–10) |
| HS | 0.26 (0.16–0.38) |
0.29 (0.17–0.39) |
0.61 (0.41–0.72) |
0.57 (0.23–0.75) |
0.36 (0.01–0.48) |
0.39 (0.01–0.57) |
| f | 0.21*
0.06–0.38 |
0.02 −0.13–0.25 |
0.19 * 0.12–0.27 |
0.16 * 0.03–0.33 |
0.01 −0.03–0.07 |
0.06 −0.03–0.13 |
| θ | 0.47 * 0.36–0.59 |
0.49 * 0.30–0.66 |
0.18 * 0.12–0.24 |
0.30 * 0.22–0.39 |
0.62 * 0.53–0.70 |
0.41 * 0.34–0.48 |
| F′ST | 0.68 | 0.69 | 0.47 | 0.61 | 0.88 | 0.71 |
Indicates p < 0.0001.
NA, number of alleles
HS, Nei’s (1987) unbiased heterozygosity
f, Weir and Cockerham’s (1984) estimate of inbreeding
θ, Weir and Cockerham’s (1984) estimate of genetic differentiation
F′ST, standardized value of FST (Hedrick, 2005; Meirmans, 2006)
At the level of individual collection sites (12 putative subpopulations, nsnails = 5–69), the mean number of alleles per locus ranged from 1.43 to 3.86 in Caju, 2 to 7.29 in SPdJ, and 1.14 to 3.43 in VdG (Table 3). Two sampling sites, springs located in Caju and VdG were monomorphic for all but 1–3 loci, regardless of season (Table 3). Average unbiased heterozygosity (HS) for each collection site ranged from 0.16 to 0.39 in Caju, 0.23 to 0.73 in SPdJ, and 0.01 to 0.57 in VdG. In Caju only one population deviated significantly from Hardy-Weinberg expectations, while all but two populations in SPdJ deviated significantly, and no populations indicated significant deviation in VdG (Table 3). In all cases, significant deviations from HWE were positive and reflective of inbreeding, and while individual loci may not have differed significantly from zero, the majority were positive and indicative of inbreeding.
In all villages, permutation of individuals between populations grouped by season indicated that there was no significant difference in either mean unbiased heterozygosity or deviation from HWE between seasons (all p > 0.3 after 5000 permutations). However, significant differences in genetic diversity associated with habitat type were found within villages. Within the village of Caju, populations collected from stream sites (open habitat) exhibited significantly higher gene diversity than those collected from springs (closed habitats) (HS open 0.40, HS closed 0.20, p = 0.05), though average FIS did not differ significantly (FIS open 0.16, FIS closed 0.05, p = 0.59). The same habitat-specific differences were found in SPdJ (HS open 0.73, HS closed 0.28, p = 0.02; FIS open 0.18, FIS closed 0.18, p >0.99), but there was no significant difference in genetic diversity between habitat types in VdG (HS open 0.46, HS closed 0.30, p = 0.64; FIS open 0.08, FIS closed 0.01, p = 0.12).
Genetic Differentiation
In all villages and in all seasons, the estimate of genetic differentiation, FST, was significantly greater than zero and indicative of significant genetic subdivision among individual sampling sites (Table 2). The lowest levels of subdivision were observed in SPdJ (θ = 0.18 and 0.30 after the wet and dry seasons, respectively), while θ for both Caju and VdG was 0.41 or greater. Given the observed levels of heterozygosity, the observed θ for all sites was equal to or greater than 47% of the maximum obtainable FST (calculated F′ST: 0.47 – 0.88; Table 2). In all villages, the level of genetic partitioning did not differ significantly between seasons (all p ≥ 0.66 after 5000 permutations). This was corroborated by analysis of molecular variance, which indicated that seasons did not comprise a significant component of the partitioning of genetic variation in any sampled village (all p ≥ 0.93). Rather, in all villages, a significant proportion of the variance was described both by the distribution of genetic variation among snail populations within each village and season and by variation within individuals (Table 4). In no village was there a significant association between genetic level of genetic differentiation and distance between populations.
Table 4.
Analysis of molecular variance (AMOVA) of B. glabrata populations across seasons and microhabitat site (population)
| Variance component | % Variation | P - value | |
|---|---|---|---|
| Caju | Among seasons | −16.98 | 0.93 |
| Among populations within seasons | 59.12 | <0.0001 | |
| Among individuals within populations | 0.27 | 0.48 | |
| Within individuals | 57.59 | <0.0001 | |
| SPdJ | Among seasons | −8.89 | 1 |
| Among populations within seasons | 26.48 | <0.0001 | |
| Among individuals within populations | 5.85 | 0.005 | |
| Within individuals | 76.56 | <0.0001 | |
| VdG | Among seasons | −17.67 | 0.93 |
| Among populations within seasons | 65.38 | <0.0001 | |
| Among individuals within populations | −0.44 | 0.61 | |
| Within individuals | 52.73 | <0.0001 | |
Additional support for high levels of genetic subdivision among snail populations at the spatial scale of a single village was provided by Bayesian cluster analysis. In all villages, under the most likely value of K (Caju = 4, SPdJ = 2, VdG = 3), a majority of individuals from each site (≥ 94%) were assigned to one cluster with >70% probability. In Caju each sampling site formed its own cluster, regardless of distance between sampling sites (Figure 1). In SPdJ snails from the two sites along the same stream clustered together, while the closed habitat formed an independent cluster. VdG sites “Spring 1” and “Marsh-Stream 2” shared the same cluster, as did the disparate sites “Stream 3” and “Suçuarana Stream 1”, while “Suçuarana Spring 2” clustered independently. Partitioning did not extend to the temporal component in any site or village, such that the majority of snails collected from the same microhabitat were always assigned to the same cluster, regardless of collection season.
Discussion
Our goal in this research was to assess spatial and temporal genetic variation of Biomphalaria glabrata, the intermediate host of the human parasite Schistosoma mansoni. In particular, we aimed to investigate this variation over a spatial scale most likely to be relevant to the epidemiology of the parasite (individual villages) and compare the results to the pattern of variation previously observed in parasite populations. Prior studies of genetic variation among schistosomes in Brazil indicated low to medium levels of genetic differentiation among human host infrapopulations and that patterns of differentiation could be influenced by environmental factors – namely distribution of waterways (Curtis et al. 2002 and Thiele et al. 2008). Here we observed that levels of snail subdivision remain high across the various hydrogeologies and spatial scales of three villages. For example, despite the single primary drainage in VdG and the fact that several sampled populations are situated along it, VdG B. glabrata populations exhibited similar levels of differentiation to those from the village of Caju, where populations are located on at least 3 different drainages. Likewise, we observed that this differentiation can occur over scales easily traversable by even passive migration of snails (<5 m). Though collections were not performed in the same year, we were able to compare levels of snail differentiation with those of previously published parasite data for the village of VdG (Thiele et al. 2008). We found and demonstrate here (Table 5) that levels of standardized pairwise differentiation among snail populations in VdG are significantly greater than those of human schistosome infrapopulations collected from the same regions of the village (Mann-Whitney U: Z = −5.25, p < 0.0001). Unfortunately, no infected snails were found within VdG, preventing genetic comparison of snail and human infrapopulations.
Table 5.
Comparison of pairwise F’ST calculated for previously collected VdG S. mansoni infrapopulations (human hosts, Thiele et al. 2008) and B. glabrata populations – a) wet season snail sampling and b) dry season snail sampling. Values below the diagonal represent pairwise standardized differentiation of snails and values above the diagonal represent pairwise standardized differentiation of parasite infrapopulations.
| a) Wet Season | |||||
|---|---|---|---|---|---|
| Marsh- Stream 2 |
Stream 3 | Spring 1 | Suçuarana Stream 1 |
Suçuarana Spring 2 |
|
| Marsh-Stream 2 | ------ | 0.13 | 0.06 | 0.12 | 0.13 |
| Stream 3 | 0.65 | ------ | 0.06 | 0.12 | 0.14 |
| Spring 1 | 0.19 | 0.75 | ------ | 0.07 | 0.09 |
| Suçuarana Stream 1 | 0.81 | 0.67 | 0.79 | ------ | 0.17 |
| Suçuarana Spring 2 | 0.97 | 0.96 | 0.91 | 0.98 | ------ |
|
b) Dry Season | |||||
|---|---|---|---|---|---|
| Marsh- Stream 2 |
Stream 3 | Spring 1 | Suçuarana Stream 1 |
Suçuarana Spring 2 |
|
| Marsh-Stream 2 | ------ | 0.13 | 0.06 | 0.12 | 0.13 |
| Stream 3 | 0.50 | ------ | 0.06 | 0.12 | 0.14 |
| Spring 1 | 0.13 | 0.67 | ------ | 0.07 | 0.09 |
| Suçuarana Stream 1 | 0.65 | 0.67 | 0.68 | ------ | 0.17 |
| Suçuarana Spring 2 | 0.85 | 0.88 | 0.86 | 0.90 | ------ |
It is often assumed, and has been shown in other species (Nadler 1995; Blouin et al. 1995 1999; McCoy et al. 2003; Criscione and Blouin 2004) and in sylvatic systems of schistosome transmission (Sire et al. 2001, Prugnolle et al. 2005), that vertebrate hosts are primarily responsible for parasite migration and gene flow. However, assessments of snail population patterns at spatial and temporal levels most relevant to human disease (i.e. that of single villages) are limited. The results shown here are in general agreement with other studies of Biomphalaria population genetics (Langand et al. 1999; Sire et al. 2001; Charbonnel et al. 2002a&b; Mavárez et al. 2002a&b; Wethington et al. 2007), with the added benefit of assessment over an epidemiologically relevant scale and comparability to parasite infrapopulations collected from the same regions of one village. Moreover, we have shown that the observed pattern of snail population differentiation is maintained across seasons and, at least within the span of a single year, there is little genetic evidence for significant demographic flux. This conclusion supports the view that snail intermediate hosts are not primarily responsible for parasite gene flow on epidemiologically relevant levels. This does not rule out the possibility that heavy rains/flooding events may play a role in parasite migration, via passive movement of either infected snails or free-swimming parasite larvae. But it should be noted that the topography of the region (high elevational variation with well-defined drainages) makes it likely that the majority of such movements would follow a pattern of effectively unidirectional migration along highly constrained paths. Moreover, this funneling of runoff could be expected to generate water flows that promote flushing of populations, rather than mixing. Such an effect would not be expected to homogenize snail populations (or their parasite infrapopulations) across the expanse of these villages, explaining in part why snail populations along the same stream can remain highly subdivided and why we detected no seasonal influence.
Overall, in questions of genetic structuring of populations, scale matters. In the three Brazilian villages studied here, our assessments of genetic variation indicate that the population dynamics of the snail host operate on a spatial scale that is much more limited than the parasite and that snail population genetics may be more temporally stable than we expected. This is evolutionarily significant in that it has been shown that asymmetrical patterns of host and parasite migration can promote local adaptation (Gandon and Michalakis 2002). Our findings here, taken with those of previous observations (Curtis et al. 2002; Thiele et al 2008), indicate that B. glabrata snails have significantly lower levels of gene flow than their schistosome parasites. Given this asymmetry, local adaptation is a feasible outcome at the level of individual villages in this host-parasite system. So while snails may not be expected to be responsible for the regular transport of parasites across large spatial scales, it may be reasonable to expect that snails would have a role as genetic filters and that the outcome of a snail-schistosome interaction (e.g. a parasite’s transmission) could be influenced by where in the environment (village) a parasite finds itself deposited. Thus, snail population structure may be implicated in the coevolutionary trajectory between hosts and parasites and impact transmission dynamics. The degree to which snails and their genetic variation influence these outcomes continues to be a valid question and one that warrants continued investigation.
Acknowledgements
We would like to thank Gregory J. Sandland, Alice V. Foster, and Jared Wilkinson for assistance with snail collections and genotyping. We are also indebted to Liana Kanovaloff Jannotti Passos, Delza de Moura Soares Reis, Lidiane Bento Braga, and Sueleny Silva Ferreira Teixeira for their assistance with snail maintenance at CPqRR. Manuscript comments by the Minchella lab group, Richard D. Howard, Morris Levy, and J. Andrew DeWoody were especially valuable. This work was supported by a National Institutes of Health (USA) Research Grant to D.J.M., a Fogarty International Center (USA) Grant to D.J.M and G.O., the National Council for Research and Development (Brazil), and the Research Foundation of the State of Minas Gerais (to G.O.).
References
- Amaral RSd, Tauil PL, Lima DD, Engels D. An analysis of the impact of the Schistosomiasis Control Programme in Brazil. Memorias do Instituto Oswaldo Cruz. 2006;101(Suppl. 1):79–85. doi: 10.1590/s0074-02762006000900012. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Genetix 4.05, Logiciel sous Windows pour la Génétique des Populations. Montpellier, France: Laboratoire Génome, Populations, Interactions: CNRS UMR 5000, Université de Montpellier II; 2004. [Google Scholar]
- Blouin MS, Yowell CA, Courtney CH, Dame JB. Host movement and the genetic-structure of populations of parasitic nematodes. Genetics. 1995;141:1007–1014. doi: 10.1093/genetics/141.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin MS, Liu J, Berry RE. Life cycle variation and the genetic structure of nematode populations. Heredity. 1999;83:253–259. doi: 10.1038/sj.hdy.6885420. [DOI] [PubMed] [Google Scholar]
- Brouwer KC, Ndhlovu P, Munatsi A, Shiff CJ. Genetic diversity of a population of Schistosoma haematobium derived from schoolchildren in East Central Zimbabwe. The Journal of Parasitology. 2001;87(4):762–769. doi: 10.1645/0022-3395(2001)087[0762:GDOAPO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Carlos dos Reis D, Kloos H, King C, Quites HFO, Matoso LF, Coelho KR, Gazzinelli A. Accessibility to and utilisation of schistosomiasis-related health services in a rural area of state of Minas Gerais, Brazil. Memórias do Instituto Oswaldo Cruz. 2010;105(4):587–597. doi: 10.1590/s0074-02762010000400039. [DOI] [PubMed] [Google Scholar]
- Charbonnel N, Angers B, Rasatavonjizay R, Bremond P, Jarne P. Evolutionary aspects of the metapopulation dynamics of Biomphataria pfeifferi, the intermediate host of Schistosoma mansoni. Journal of Evolutionary Biology. 2002a;15(2):248–261. [Google Scholar]
- Charbonnel N, Quesnoit M, Razatavonjizay R, Brémond P, Jarne P. A spatial and temporal approach to microevolutionary forces affecting population biology in the freshwater snail Biomphalaria pfeifferi. The American Naturalist. 2002b;160(6):741–755. doi: 10.1086/343875. [DOI] [PubMed] [Google Scholar]
- Chitsulo L, Loverde P, Engels D. Schistosomiasis. Nature reviews. Microbiology. 2004;2(1):12–13. doi: 10.1038/nrmicro801. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Life cycles shape parasite evolution: comparative population genetics of salmon trematodes. Evolution. 2004;58(1):198–202. doi: 10.1111/j.0014-3820.2004.tb01587.x. [DOI] [PubMed] [Google Scholar]
- Coura JR, Amaral RS. Epidemiological and control aspects of schistosomiasis in Brazilian endemic areas. Memorias do Instituto Oswaldo Cruz. 2004;99(Suppl. 1):13–19. doi: 10.1590/s0074-02762004000900003. [DOI] [PubMed] [Google Scholar]
- Curtis J, Sorensen RE, Minchella DJ. Schistosome genetic diversity: the implications of population structure as detected with microsatellite markers. Parasitology. 2002;125:S51–S59. doi: 10.1017/s0031182002002020. [DOI] [PubMed] [Google Scholar]
- Engels D, Chitsulo L, Montresor A, Savioli L. The global epidemiological situation of schistosomiasis and new approaches to control and research. Acta Tropica. 2002;82(2):139–146. doi: 10.1016/s0001-706x(02)00045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erko B, Balcha F, Kifle D. The ecology of Biomphalaria sudanica in Lake Ziway, Ethiopia. African Journal of Ecology. 2006;44:347–352. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier, Laval LG, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. Journal of Evolutionary Biology. 2002;15:451–462. [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001 Available from http://www2.unil.ch/popgen/softwares/fstat.htm. Updated from Goudet (1995)
- Goudet J, Raymond M, de Meeüs T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144(4):1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Jones CS, Lockyer AE, Rollinson D, Piertney SB, Noble LR. Isolation and characterization of microsatellite loci in the freshwater gastropod, Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Molecular Ecology. 1999;8:2149–2151. doi: 10.1046/j.1365-294x.1999.00802-5.x. [DOI] [PubMed] [Google Scholar]
- Katz N, Peixoto SV. Análise crítica da estimativa do número de portadores de esquistossomose mansoni no Brasil. Revista da Sociedade Brasileira da Medica Trópica. 2000;33:303–308. doi: 10.1590/s0037-86822000000300009. [DOI] [PubMed] [Google Scholar]
- Kloos H, de Souza C, Gazzinelli A, Soares Filho BS, Temba PDC, Bethony J, Page K, et al. The distribution of Biomphalaria spp. in different habitats in relation to physical, biological, water contact and cognitive factors in a rural area in Minas Gerais, Brazil. Memórias do Instituto Oswaldo Cruz. 2001;96(Suppl):57–66. doi: 10.1590/s0074-02762001000900008. [DOI] [PubMed] [Google Scholar]
- Kloos H, Gazzinelli A, Van Zuyle P. Microgeographical patterns of schistosomiasis and water contact behavior; examples from Africa and Brazil. Memórias do Instituto Oswaldo Cruz. 1998;93(Suppl I):37–50. doi: 10.1590/s0074-02761998000700006. [DOI] [PubMed] [Google Scholar]
- Kloos H, Passos LKJ, LoVerde P, Oliveira RC, Gazzinelli A. Distribution and Schistosoma mansoni infection of Biomphalaria glabrata in different habitats in a rural area in the Jequitinhonha Valley, Minas Gerais, Brazil: environmental and epidemiological aspects. Memórias do Instituto Oswaldo Cruz. 2004;99(7):673–681. doi: 10.1590/s0074-02762004000700002. [DOI] [PubMed] [Google Scholar]
- Kloos H, Rodrigues JCAP, Pereira WR, Velásquez-Meléndez G, Loverde P, Oliveira RC, Gazzinelli A. Combined methods for the study of water contact behavior in a rural schistosomiasis-endemic area in Brazil. Acta Tropica. 2006;97(1):31–41. doi: 10.1016/j.actatropica.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Langand J, Théron A, Pointier JP, Delay B, Jourdane J. Population structure of Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni in Guadeloupe island, using RAPD markers. Journal of Molluscan Studies. 1999;65:425–433. [Google Scholar]
- Mavárez J, Amarista M, Pointier J-P, Jarne P. Fine-scale population structure and dispersal in Biomphalaria glabrata, the intermediate snail host of Schistosoma mansoni, in Venezuela. Molecular Ecology. 2002a;11(5):879–889. doi: 10.1046/j.1365-294x.2002.01486.x. [DOI] [PubMed] [Google Scholar]
- Mavárez J, Pointier J-P, David P, Delay B, Jarne P. Genetic differentiation, dispersal and mating system in the schistosome-transmitting freshwater snail Biomphalaria glabrata. Heredity. 2002b;89:258–265. doi: 10.1038/sj.hdy.6800127. [DOI] [PubMed] [Google Scholar]
- McCoy KD, Boulinier T, Tirard C, Michalakis Y. Host-dependent genetic structure of parasite populations: differential dispersal of seabird tick host races. Evolution. 2003;57:288–296. doi: 10.1111/j.0014-3820.2003.tb00263.x. [DOI] [PubMed] [Google Scholar]
- Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- Morgan JAT, Dejong RJ, Adeoye GO, Ansa EDO, Barbosa CS, Brémond P, Cesari IM, et al. Origin and diversification of the human parasite Schistosoma mansoni. Molecular Ecology. 2005;14(12):3889–3902. doi: 10.1111/j.1365-294X.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- Nadler SA. Microevolution and the genetic structure of parasite populations. The Journal of Parasitology. 1995;81(3):395–403. [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Théron A, Pointier J-P, Jabbour-Zahab R, Jarne P, Durand P, de Meeûs T. Dispersal in a parasitic worm and its two hosts: consequence for local adaptation. Evolution. 2005;59(2):296–303. [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire C, Langand J, Barral V, Théron A. Parasite (Schistosoma mansoni) and host (Biomphalaria glabrata) genetic diversity: population structure in a fragmented landscape. Parasitology. 2001;122:545–554. doi: 10.1017/s0031182001007727. [DOI] [PubMed] [Google Scholar]
- Standley CJ, Kabatereine NB, Lange CN, Lwambo NJS, Stothard JR. Molecular epidemiology and phylogeography of Schistosoma mansoni around Lake Victoria. Parasitology. 2010;137:1937–1949. doi: 10.1017/S0031182010000788. [DOI] [PubMed] [Google Scholar]
- Steinauer ML, Blouin MS, Criscione CD. Applying evolutionary genetics to schistosome epidemiology. Infection, Genetics and Evolution. 2010;10:433–443. doi: 10.1016/j.meegid.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele EA, Sorensen RE, Gazzinelli A, Minchella DJ. Genetic diversity and population structuring of Schistosoma mansoni in a Brazilian village. International Journal for Parasitology. 2008;38(3–4):389–399. doi: 10.1016/j.ijpara.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzinger J, Mayombana C, Smith T, Tanner M. Spatial microhabitat selection by Biomphalaria pfeifferi in a small perennial river in Tanzania. Hydrobiologia. 1997;356:53–60. [Google Scholar]
- Utzinger J, Tanner M. Microhabitat preferences of Biomphalaria pfeifferi and Lymnaea natalensis in a natural and a man-made habitat in southeastern Tanzania. Memórias do Instituto Oswaldo Cruz. 2000;95(3):287–294. doi: 10.1590/s0074-02762000000300002. [DOI] [PubMed] [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Resources. 2004;4(3):535–538. [Google Scholar]
- Webster JP, Davies CM, Hoffman JI, Ndamba J, Noble LR, Woolhouse MEJ. Population genetics of the schistosome intermediate host Biomphalaria pfeifferi in the Zimbabwean highveld: implications for co-evolutionary theory. Annals of Tropical Medicine and Parasitology. 2001;95(2):203–214. doi: 10.1080/00034980120041062. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wethington AR, Zavodna M, Smith MK, Oliveira G, Lewis F, Minchella DJ. Population genetic structure of Biomphalaria glabrata in a schistosomiasis-endemic region in Brazil. Journal of Molluscan Studies. 2007;73:45–52. [Google Scholar]
- Woolhouse MEJ. Population biology of the freshwater snail Biomphalaria pfeifferi in the Zimbabwe Highveld. Journal of Applied Ecology. 1992;29(3):687–694. [Google Scholar]