Abstract
The allergen Act d 11, also known as kirola, is a 17 kDa protein expressed in large amounts in ripe green and yellow-fleshed kiwifruit. Ten percent of all kiwifruit-allergic individuals produce IgE specific for the protein. Using X-ray crystallography, we determined the first three-dimensional structures of Act d 11, produced from both recombinant expression in E. coli and from the natural source (kiwifruit). While Act d 11 is immunologically correlated with the birch pollen allergen Bet v 1 and other members of the pathogenesis-related protein family 10 (PR-10), it has low sequence similarity to PR-10 proteins. By sequence Act d 11 appears instead to belong to the major latex/ripening-related (MLP/RRP) family, but analysis of the crystal structures shows that Act d 11 has a fold very similar to that of Bet v 1 and other PR-10 related allergens regardless of the low sequence identity. The structures of both the natural and recombinant protein include an unidentified ligand, which is relatively small (about 250 Da by mass spectroscopy experiments) and most likely contains an aromatic ring. The ligand-binding cavity in Act d 11 is also significantly smaller than those in PR-10 proteins. The binding of the ligand, which we were not able to unambiguously identify, results in conformational changes in the protein that may have physiological and immunological implications. Interestingly, residue corresponding to Glu45 in Bet v 1 (Glu46), which is important for IgE binding to the birch pollen allergen, is conserved in Act d 11, even though it is not in other allergens with significantly higher sequence identity to Bet v 1. We suggest that the so-called Gly-rich loop (or P-loop), which is conserved in all PR-10 allergens, may be responsible for IgE cross-reactivity between Bet v 1 and Act d 11.
Keywords: Act d 11, kirola, allergen, food allergy, kiwifruit, Bet v 1
1. Introduction
Food allergies affect 2–3% of adults and 8% of children and their prevalence is influenced by dietary habits and other factors (Rona et al., 2007; Sicherer and Sampson, 2010; Zuidmeer et al., 2008). Frequently, food allergies are caused by cow’s milk, eggs, peanuts, wheat, soy, tree nuts, fish, shellfish, and fruits. The first reports on allergic reactions to kiwifruit appeared three decades ago (Cumplido-Laso et al., 2012). Clinical reports show that the kiwi allergy causes mostly oral symptoms, but in some cases the reactions are more severe and may include life-threatening anaphylaxis (Fine, 1981; Miyawaki et al., 2012). Currently, there are fourteen kiwifruit allergens that are registered by WHO/IUIS Allergen Nomenclature Sub-committee. Eleven allergens were identified in green kiwifruit (Actinidia deliciosa) and three in yellow-fleshed kiwifruit (Actinidia chinensis).
Kirola, according to the official allergen nomenclature termed Act d 11, is a 17-kDa protein found in high amounts in ripe green and yellow-fleshed kiwifruit (Ciardiello et al., 2009). Ten percent of kiwifruit allergic individuals have IgE that recognizes Act d 11 (D’Avino et al., 2011; Knopf, 1991). This protein belongs to the major latex protein/ripening-related protein family (MLP/RRP), and is the first protein from this family identified as an allergen (D’Avino et al., 2011). Act d 11 is immunologically related to Bet v 1-like allergens that are members of the PR-10 protein family. MLP/RRP and PR-10 families both belong to the Bet v 1 superfamily, but the sequence identity between the members of the two protein groups is rather low (<25%) (Osmark et al., 1998). However, it was shown that despite the low sequence identity, Act d 11 is able to inhibit, at least partially, binding of IgE to Bet v 1, Cor a 1, Dau c 1 and Mal d 1, suggesting that these allergens share some IgE epitopes (D’Avino et al., 2011).
Allergens belonging to the Bet v 1 allergen family are the main cause of pollen-related food allergies (so called pollen-food allergy syndrome) (Cano, 1991). In general, Bet v 1 related allergens are characterized as labile proteins, in contrast to other food allergens, which are more resistant to heating and proteolysis (Bollen et al., 2010). The physiological role of these proteins in plants is not well understood. They may function as ligand carriers, as one of the most distinctive features of the Bet v 1 fold is a large hydrophobic cavity (Cano, 1991). In the case of the archetype Bet v 1 protein, its high degree of structural similarity with the START domain of the human MLN64 protein (Radauer et al., 2008), which binds steroids, led to the hypothesis that Bet v 1 may be involved in steroid binding. The ability of Bet v 1 to bind these kinds of molecules was later confirmed and it was suggested that Bet v 1 may serve as a plant steroid carrier (Markovic-Housley et al., 2003). However, it was also shown that the protein is able to bind many other structurally and chemically divergent compounds, and it is possible that Bet v 1 is involved in many different biological processes (Kofler et al., 2012; Mogensen et al., 2002).
Here we present a thorough structural analysis of Act d 11 that was purified from both its natural source and as a recombinant protein from E. coli. Results of the structural analysis are discussed within the context of other Bet v 1-like proteins including allergens belonging to this group.
2. Materials and methods
2.1. Cloning, expression and purification
A synthetic Act d 11 gene was ordered from GenScript and re-cloned from the pUC57 vector into the pMCSG7 vector, a derivative of pET21a (Novagen), using a ligation-independent cloning protocol. The pMCSG7 vector encodes a hexahistidyl-tag followed by a spacer and a tobacco etch virus (TEV) protease cleavage site at the N-terminus of the expressed protein. The amino acid triplet Ser-Asn-Ala remains at the N-terminus of the protein after cleavage of the tag by the TEV protease. Purified plasmid was transformed into the E. coli strain BL21-CodonPlus(DE3) RIL which harbors an extra plasmid encoding three rare tRNAs (AGG and AGA for Arg and ATA for Ile; Stratagene, Inc.).
The cells were grown in lysogeny broth (LB) medium at 37°C to an optical density (at 600 nm) of approximately 1.2 then induced with IPTG to a final concentration of 1 mM. After induction, the cells were incubated overnight with shaking at 16°C. Cells expressing Act d 11 were harvested, resuspended in binding buffer [500 mM NaCl, 50 mM Tris (pH=7.5), 5% glycerol, and 5 mM imidazole] and lysed by sonication after the addition of ‘complete’ EDTA-free protease inhibitor cocktail (Roche). The lysate was clarified by centrifugation (30 min at 17,000g), and the liquid fraction was applied to a Ni-NTA resin (Qiagen) pre-equilibrated with binding buffer. The resin with bound protein was washed with wash buffer [500 mM NaCl, 50 mM Tris (pH=7.5), 5% glycerol, and 30 mM imidazole] to remove weakly binding contaminants and then eluted from the column with elution buffer [500 mM NaCl, 5% glycerol, 50 mM Tris (pH 7.5), and 250 mM imidazole]. The metal affinity tag was cleaved from the protein by treatment with recombinant His-tagged TEV protease. The cleavage reaction was conducted during the dialysis [500 mM NaCl, 50 mM Tris (pH=7.5), 5% glycerol, 0.5 mM EDTA and 1mM TCEP] to remove the imidazole. The cleaved protein was then separated from the cleaved His-tag and the His-tagged protease by passing the mixture through a second Ni2+-chelate affinity column, followed by gel filtration in crystallization buffer [150 mM NaCl, 10 mM Tris (pH=7.5)]. Pure protein fractions were pooled together and concentrated to about 3.4 mg/mL for use in crystallization.
Natural Act d 11 was purified according to a previously described protocol (D’Avino et al., 2011).
2.2. Crystallization and data collection
Crystallization experiments were performed at 293 K using the hanging drop vapor diffusion method and NeXtal plates (Qiagen). A solution of the natural protein (9 mg/mL; in 10 mM Tris-HCl, 250 mM NaCl, pH 7.5) was mixed with the well solution in a 1:1 ratio. The Index (Hampton Research) screen was used to find initial crystallization conditions. Crystallization experiments were tracked and analyzed with the XTALDB crystallization system (Cymborowski et al., 2010; Zimmerman et al., 2005). Crystallization and cryocooling conditions for the recombinant protein are summarized in Supplementary Table 1. For crystallization, the recombinant protein was used at a concentration of 3.4 mg/mL. Data collection was performed at the 19-ID beamline of the Structural Biology Center (Rosenbaum et al., 2006) at the Advanced Photon Source (APS). Data collection from a HgCl2-soaked crystal was performed at 19-BM beamline. Data were collected at 100K and processed with HKL-2000 (Otwinowski, 1997). Data collection statistics are reported in Table 1.
Table 1.
Data collection and refinement statistics. Ramachandran plot was calculated using MOLPROBITY. Numbers in parenthesis refer to the highest resolution shell. AU=asymmetric unit
| PDB code | - | 4IGV | 4IGW | 4IGX | 4IGY | 4IH0 | 4IH2 | 4IHR |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Wavelength (Å) | 0.9792 | 0.9793 | 0.9786 | 0.9791 | 0.9786 | 0.9793 | 0.9793 | 0.9772 |
| Unit cell (Å,°) | a=b=72.1; c=114.6 | a=b=71.8, c=113.8 | a=b=60.0, c=339.0 | a=38.7, b=43.0, c=94.1 | a=56.1, b=56.1, c=65.0 | a=b=70.8, c=114.0 | a=b=71.6, c =114.5 | a=b=72.1, c=113.6 |
| α=β=90,γ=120 | α=β=90, γ=120 | α=β=90, γ=120 | α=76.7, β=85.7, γ=83.5 | α=66.4, β=85.4, γ=65.2 | α=β=90, γ=120 | α=β=90, γ=120 | α=β=90, γ=120 | |
| Space group | P6322 | P6322 | P6122 | P1 | P1 | P6322 | P6322 | P6322 |
| Solvent content (%) | 50 | 49 | 51 | 43 | 49 | 48 | 49 | 49 |
| Protein chains in AU | 1 | 1 | 2 | 4 | 4 | 1 | 1 | 1 |
| Resolution range (Å) | 50.0–2.4 | 50–1.5 | 50–2.55 | 50–2.35 | 50–2.92 | 50.0–1.75 | 50.0–2.00 | 50.0–1.60 |
| Highest resolution shell (Å) | 2.44–2.40 | 1.53–1.50 | 2.59–2.55 | 2.39–2.35 | 2.97–2.92 | 1.78–1.75 | 2.03–2.00 | 1.63–1.60 |
| Unique reflections | 7377(352) | 28517(1380) | 12244(602) | 237411058 | 14019(688) | 17592(861) | 12396(590) | 23683(1141) |
| Redundancy | 25.0(27.4) | 5.7(5.8) | 6.5(6.4) | 3.4(3.1) | 2.1(2.0) | 10.3(10.8) | 10.8(11.2) | 20.9(17.2) |
| Completeness (%) | 99.7(100.0) | 99.9(100.0) | 94.6(95.3) | 97.1(84.8) | 98.3(94.5) | 98.5(100.0) | 99.5(100.0) | 100.0(100.0) |
| Rmerge (%) | 12.0(36.1) | 5.2(53.8) | 8.5(61.0) | 8.6(34.0) | 10.3(29.8) | 8.5(64.1) | 9.5(62.2) | 6.0(83.4) |
| Average I/σ(I) | 41.4(14.5) | 30.8(2.5) | 21.4(3.4) | 18.3(2.3) | 8.3(2.1) | 44.1(6.8) | 44.6(5.8) | 55.2(3.4) |
| FOMMLPHARE | 0.17 | - | - | - | - | - | - | - |
| FOMDM | 0.77 | - | - | - | - | - | - | - |
| Refinement | ||||||||
| R (%) | 16.9 | 21.8 | 18.4 | 21.0 | 19.0 | 19.7 | 17.7 | |
| Rfree (%) | 20.3 | 25.3 | 24.2 | 25.8 | 21.6 | 24.4 | 21.2 | |
| Mean B value (Å2) | 24.1 | 55.8 | 48.2 | 41.4 | 24.9 | 36.8 | 21.4 | |
| B from Wilson plot (Å2) | 17.4 | 54.2 | 46.0 | 58.0 | 18.8 | 28.0 | 16.1 | |
| RMS deviation bond lengths (Å) | 0.020 | 0.020 | 0.015 | 0.015 | 0.016 | 0.016 | 0.020 | |
| RMS deviation bond angles (°) | 1.7 | 1.7 | 1.4 | 1.4 | 1.7 | 1.7 | 1.8 | |
| Number of amino acid residues | 149 | 295 | 576 | 589 | 145 | 148 | 149 | |
| Number of water molecules | 177 | 61 | 62 | 33 | 143 | 98 | 179 | |
| Ramachandran plot | ||||||||
| Most favored regions (%) | 97.3 | 97.9 | 98.2 | 96.6 | 99.3 | 98.0 | 98.3 | |
| Additional allowed regions (%) | 2.7 | 2.1 | 1.8 | 3.4 | 0.7 | 2.0 | 1.3 | |
During the search of possible Act d 11 ligands compounds used for crystal soaking were added directly to the drop in solid form.
2.3. Structure determination, refinement and validation
Single-wavelength anomalous diffraction (SAD) was used for structure determination. Structure determination was performed with HKL-3000 (Minor et al., 2006) which integrates SHELXC/D/E (Sheldrick, 2008), MLPHARE (Otwinowski, 1991), DM (Cowtan and Main, 1993), RESOLVE (Terwilliger, 2004), ARP/wARP (Perrakis et al., 1999) and selected programs from the CCP4 package (Winn et al., 2011). Single Hg-site was used to generate an initial map. The partial model, which was built with RESOLVE using the 2.4Å data collected from the HgCl2 soaked crystal, was later used as a starting model for ARP/wARP. The high-resolution native (P6322) data set was used for model building with ARP/wARP. The model was later updated with COOT (Emsley and Cowtan, 2004) and refined with REFMAC (Murshudov et al., 2011). TLS was used in the final stages of the refinement and TLS groups were determined using TLSMD server (Painter and Merritt, 2006). MOLPROBITY (Davis et al., 2007) and ADIT (Yang et al., 2004) were used for structure validation. The high-resolution model was used later as a starting model in determination of other Act d 11 crystal forms. Molecular replacement was performed with HKL-3000 and MOLREP (Vagin and Teplyakov, 1997). Refinement and validation for other Act d 11 structures were done using the same methodology as in case of the high-resolution structure. Refinement statistics are summarized in Table 1. Coordinates and structure factors have been deposited to the PDB with the following accession codes: 4IGV, 4IGW, 4IGX, 4IGY, 4IH0, 4IH2 and 4IHR.
2.4. Mass spectrometry
Native mass spectrometry experiments were carried out on a Synapt HDMS (Waters Ltd, Manchester, UK) mass spectrometer (Pringle et al., 2007) operated in positive ion mode. The instrument was mass calibrated using a 33 μM solution of cesium iodide. Act d 11 isolated from natural sources was buffer exchanged twice into 100 mM ammonium acetate pH 6.9, using Biospin-6 columns (Bio-Rad) and diluted to a final concentration of 10 μM. Samples (2–3 μL) were delivered to the mass spectrometer by means of nanoelectrospray ionization using gold-coated capillaries prepared in house. Typical instrument parameters were as follows: capillary voltage 0.9–1.1 kV, cone voltage 40 V, trap energy 6 V, transfer energy 4 V, bias, 2.0 V, backing pressure 5.5 mbar, source temperature 40 °C. Mass spectra were smoothed (mean filter 5/5), and peak-centered (centroid top 90 / 12) in MassLynx v4.1 (Waters). Centroid peaks were used as the input for an in-house script implementing the mass measurement approach previously published by Tito et al. (2000) in order to calculate the mass of species present in the mass spectrum.
2.5. Sequence analysis
Representative sequences of allergens from the PR10-related protein family (Act c 8, Ara h 8, Cor a 1, Mal d 1, Pru p 1, Que a 1, Dau c 1, Pyr c 1, Rub i 1, Act d 8, Bet v 1, Fra a 1, Pru ar 1, Api g 1, Car b 1, Gly m 4, Pru av 1, Vig r 1) and sequences from MLP/RRP protein family (Act d 11, Q7Y082, B2WS86, ML168, Q71HN2, Q9SMF5, O65884, Q9AXU0, O65178, MLP43, Q949M0) were used as queries for position specific iterative BLAST (PSI-BLAST) (Altschul et al., 1997) searches against NCBI’s non-redundant sequences (nr) BLAST database with default options. Results were retrieved with an expectation value (e-value) 10−3 and each sequence from the first round was used for reciprocal PSI-BLAST searches against the nr database and thus distinguished between false positive and true positive hits. After the second round of PSI-BLAST searches non-redundant sequences were obtained and clustered using CLANS (Frickey and Lupas, 2004).
2.6. Docking studies
Docking experiments were performed with Glide (Schrödinger, LLC, New York, NY, Suite 2008) in Extreme Precision (XP) docking mode (Friesner et al., 2006) as described previously. The target was prepared using the Protein Preparation Wizard in Maestro. The Virtual Screening Workflow script was used for the docking.
Briefly, Glide default parameters were used and no binding constraints were defined for docking. Receptor grids were generated and centered on the residues defining the ligand-binding site. LigPrep (Greenwood et al., 2010) and Epik (Shelley et al., 2007) were used for preparing both protein and ligands, which were docked flexibly. Compounds from the library were docked using the HTVS, SP and finally the XP scoring function.
2.7. Other computational calculations
Volumes of binding cavities were calculated with CASTp (Dundas et al., 2006). If a structure contained more than one chain in the asymmetric unit, or was determined by NMR, the first protein chain was taken for binding cavity volume calculations. PISA (Krissinel and Henrick, 2007) was used to calculate information on oligomeric state of Act d 11 in different crystal forms. The SSM algorithm (Krissinel and Henrick, 2004) implemented in COOT was used to superpose macromolecular models. All figures presenting macromolecular structures were prepared with PYMOL (DeLano W., 2002). Protein Variability Server (Garcia-Boronat et al., 2008) was used to map the sequence variability onto the Bet v 1 structure.
3. Results
3.1. Sequence analysis
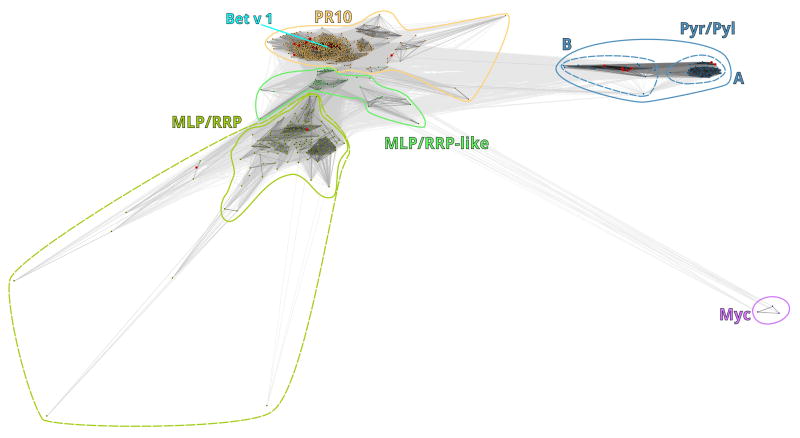
Sequence similarity-based clustering of Act d 11 and Bet v 1 homologs (Fig. 1) showed that PR-10-related proteins created a dense cluster (PR-10) with distinguishable subclusters corresponding to groups of closest homologs of different allergens. MLP/RRP family members including Act d 11 created a separate dispersed cluster (MLP/RRP) with some satellite sequences, with Act d 11 placed in the central part of it. There were a number of uncharacterized Bet v 1-like sequences between the two clusters (MLP/RRP-like) that could not be unambiguously assigned to any of the two groups at the threshold P-value used. However, they were pulled much closer to MLP/RRP cluster when more stringent (1e-20) P-values were applied, and disconnected from PR-10 group when using an even more stringent threshold (1e-30). The third group that formed during clustering seemed to be a little more distantly related to the other two than they are to each other. Two subgroups could be distinguished in that cluster (Pyr/Pyl), one (A) contained abscisic acid receptors Pyr-1, Pyl-8 and Pyl-9, whereas the other (B) contained the abscisic acid receptor Pyl-2. There were also three sequences of cyclase/dehydrase from Mycobacterium species, that were distant to all other groups (Myc), but reciprocal searches of structure and sequence databases confirmed their closest similarity to Bet v 1-like superfamily members.
Figure 1.
Two-dimensional representation of the CLANS clustering results. Proteins are shown as black dots. Proteins with known structures are shown as red dots. Lines indicate sequence similarities detected by BLAST and are colored by a grayscale spectrum according to BLAST’s P-value (black: P-value < 10−255, light grey: P-value < 10−5).
3.2. Structure of Act d 11
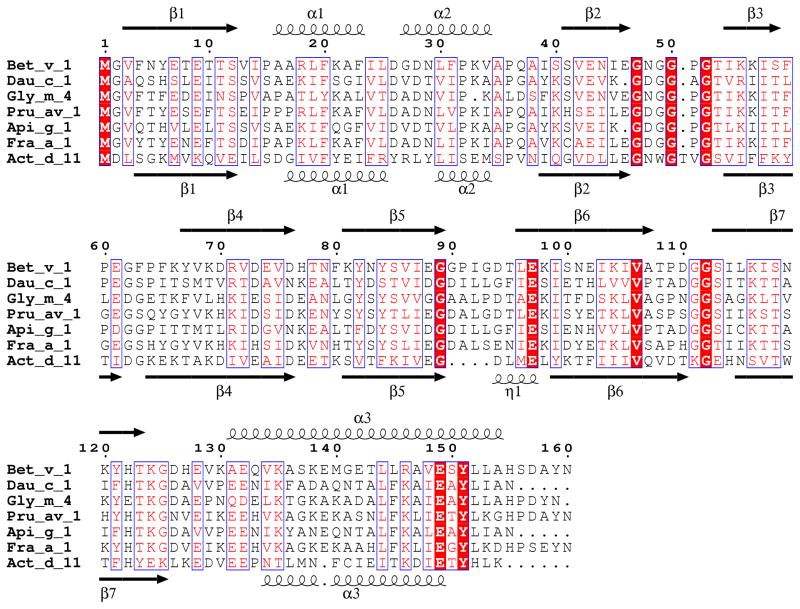
Natural Act d 11 crystallized in four crystal forms (Table 1), in which protein monomers were packed in different ways. The Act d 11 molecule, which is composed of 150 residues, including an acetylated N-terminal methionine (D’Avino et al., 2011), has an overall fold similar to Bet v 1. It is composed of a curved, seven-stranded anti-parallel β-sheet that ‘grasps’ three αhelices (Fig. 2). Two shorter helices (α1 and α2) are located between the β1 and β2 strands, which are flanking the β-sheet. The longest helix (α3) is located at the C-terminus of the protein. In different crystal forms, Act d 11 molecules have very similar conformations. Their Cα atoms superpose with root mean square deviation (rmsd) values of 0.4–0.6 Å. There are no significant differences between structures of the natural (PDB code: 4IGV) and the recombinant protein (PDB code: 4IHR). Their models superpose with a rmsd value of 0.3 Å. The biggest differences are observed in a loop region (residues 107–112) that connects strands β6 and β7. DALI (Uematsu et al., 1991) identified a phytohormone binding protein from Medicago truncatula (PDB code: 3US7; rmsd 1.9 Å), a cytokinin-specific binding protein from Vigna radiata (PDB codes: 3C0V and 2FLH; rmsd values 1.7–1.9 Å) and a pathogenesis-related protein LIPR-10.2B from Lupinus luteus (PDB code: 3E85, rmsd value 1.9 Å) as having the most similar structures to Act d 11. These three proteins are followed by Bet v 1 (PDB codes: 3K78, 1BV1, 1FM4, 1FSK and 1QMR; rmsd values 1.8–2.1 Å) and Api g 1 (PDB code: 2BK0; rmsd value 2.1 Å). Other PR-10 related allergens with determined structures (Dau c 1, Fra a 1, Gly m 4 and Pru av 1) also show high structural and significant sequence similarities to Act d 11 (Fig. 3). However, Dau c 1 (PDB code: 2WQL) and Pru av 1 (PDB code: 1H2O) superpose with Act d 11 with lower rmsd values (~2.2 Å) than Fra g 1 (PDB code: 2LPX) and Gly m 4 (PDB code: 2K7H), for which the rmsd values are 2.5 Å and 2.6 Å respectively. Disregarding differences in length of particular secondary structural elements, the most significant structural difference between Act d 11 and the aforementioned structures are related to the conformation of a motif composed of the strands of β3, β4 and the loop that connects them. In the case of Act d 11, the main chain of this fragment bows toward the longest helix (α3) (Fig. 4). In this respect, Act d 11 is more similar to the abscisic acid receptor (PDB codes: 3KL1 and 3KAZ) than, for example, to other Bet v 1 related allergens.
Figure 2.
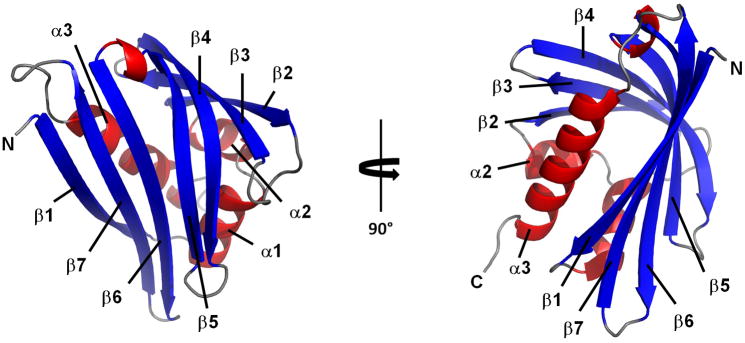
Overall structure of Act d 11. Ribbon representation is shown in two orientations related by 90° rotation. β-Strands are shown in blue, α-helices in red, loop regions are colored in gray.
Figure 3.
Sequence alignment of Bet v 1-related allergens for which structures were determined and are available in the PDB.
Figure 4.
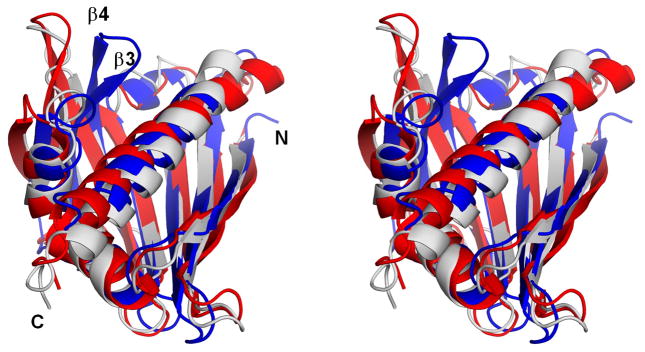
Stereoview of the superposition of Act d 11 (in blue; PDB code: 4IGV), Gly m 4 (in red; PDB code: 2K7H) and Pru av 1 (in gray; PDB code: 1H2O).
A BLAST search with the sequence of Act d 11 against the Protein Data Bank identifies an Arabibidopsis thaliana protein At1g70830 (PDB code: 2I9Y), which belongs to the MLP protein family, as the most similar in terms of sequence to Act d 11 (42% sequence identity and 67% of sequence similarity). The structure of natural Act d 11 and structure of At1g70830 superposes with rmsd of 2.6Å over 120 Cα atoms. However, the two structures differ significantly in regions corresponding to helix α2 and strand β2 of Act d 11, with this region being disordered and lacking any secondary structure in the case of At1g70830.
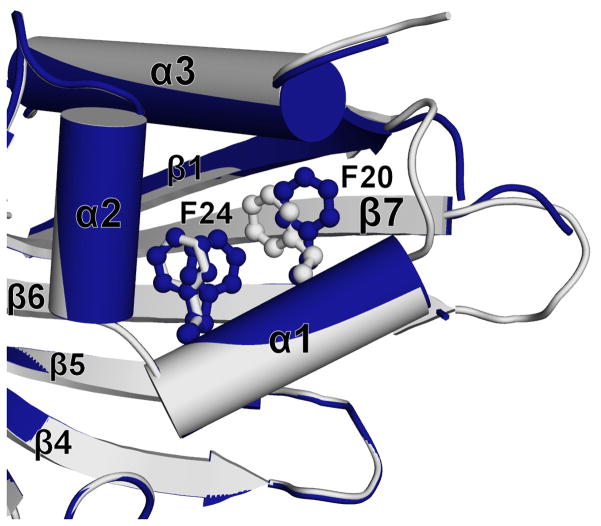
The central part of the Act d 11 molecule contains a relatively large cavity with a volume of approximately 430 Å3. However, it is relatively small in comparison with cavities observed for Bet v 1 and Pru av 1, which are approximately four times larger. The walls of the cavity are composed of both β-sheet and α-helical parts of the protein (Fig. 5). The pocket comprises 20 residues where Asp71 is the only charged residue and Cys137 contains a free thiol. Crystal structures of Act d 11 revealed the presence of an unidentified molecule. Unfortunately, even in the highest resolution structure (1.5 Å) (PDB code: 4IGV), the shape of the electron density does not allow for an unambiguous identification of the ligand (Fig. 6). Attempts to replace the endogenous ligand by soaking Act d 11 crystals with small molecular compounds were unsuccessful. During this analysis the following small molecular compounds were tested: 2-aminopurine, 3-butyric acid, gibberelin, hydrocortisone, 3-iodo-L-Tyr, indole, L-Phe, L-Trp, progesterone, serotonin and zeatin. None of the tested compounds could be identified in the obtained electron density maps. In the case of the experiment with 2-aminopurine, the shape of the electron density changed somewhat (PDB code: 4IH2). It is possible that after the soaking experiment the electron density corresponds to the 2-aminopurine, but there is no unambiguous way to place this molecule into the electron density map. However, the shape of the electron density suggests, that the 2-aminopurine is oriented parallel to side chain of Phe58 with aromatic rings being approximately 4.2Å apart. Soaking Act d 11 crystals with serotonin also resulted in a change of the electron density in the binding site, and in addition it resulted in several conformational changes (Fig. 7). Subsequent to soaking, Phe24 was initially observed in a double conformation. Increased mobility of the Phe24 side chain is correlated with change of the Phe20 side chain conformation. Most likely the conformational change of Phe20 resulted in the destabilization of the loop formed by residues 14–17, and thus this loop became disordered. In this structure the neighboring loop formed by residues 107–111 was also disordered.
Figure 5.
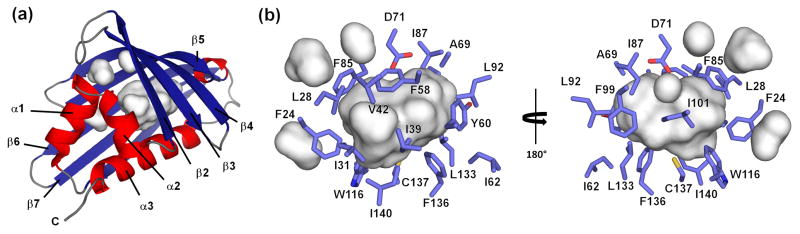
Cavities in Act d 11. (a) Localization of cavities in respect to the secondary structural elements. (b) Residues forming the biggest cavity. The residues are represented by sticks with only their side chains shown.
Figure 6.
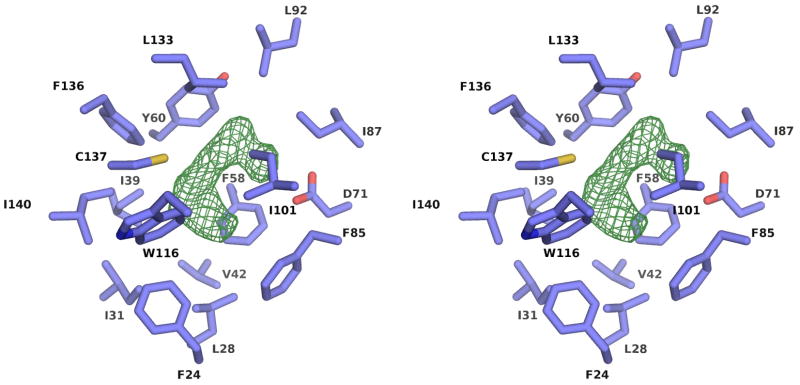
Stereoview of the residues forming the central cavity with the difference map (in green) indicating the presence of an unknown ligand. The difference map is contoured at the 3σ level.
Figure 7.
Conformational changes of Act d 11 after interactions with serotonin (dark blue). Structure of Act d 11 with unknown ligand is shown in grey.
Assuming that the endogenous ligand, present in the structures of Act d 11 isolated from the natural source, is characteristic for plants we decided to use recombinant version of Act d 11 to pursue ligand binding. However, structure determination of the recombinant protein also revealed presence of an unidentified ligand. Moreover, the shape of the electron density corresponding to the ligand (Fig. 6) is very similar for both natural and recombinant protein. Attempts of protein unfolding and re-folding in order to get an apo-form of Act d 11 were unsuccessful (data not shown).
3.3. Ligand identification
To try and identify the nature of the ligand bound to the protein we analysed Act d 11 isolated from natural sources by means of native mass spectrometry. Using native mass spectrometry, non-covalent complexes between proteins and other molecules can be successfully maintained and transferred to the mass spectrometer for mass analysis (Heck, 2008). A mass spectrum of Act d 11 after the protein had been buffer exchanged into 100 mM ammonium acetate is shown in figure 3S and reveals that the protein exists as a monomer. A narrow charge state distribution indicates that the protein is maintained in a near-native state during our analysis, as proteins that have been unfolded in solution exhibit a broader range of charge states (Scarff et al., 2008). The major charge state series in the spectrum corresponds to the N-acetylated version of Act d 11. The mass for this series is 17446.043 ± 0.028 Da, which is in excellent agreement with the mass calculated from the protein sequence (17446.09 Da). A less abundant charge state series was observed which had a mass of 256.09±0.06 Da higher than that of the protein. To disrupt the protein ligand complex and, therefore, obtain a more accurate measurement for the ligand, the sample was buffer exchanged into a denaturing buffer (50% MeOH, 1% HCOOH final concentration). We were not able to obtain a mass spectrum from this sample due to protein precipitation.
3.4. Docking results
To gain insights into the identity of the natural ligand bound to Act d 11, we performed virtual docking using the XP Scoring Function of Glide (Schrödinger, LLC, New York, NY, Suite 2008). Ligands from a library of natural products (InterBioScreen Ltd., Chernogolovka, Russia, 2008 ) were used. The top one hundred molecules were chosen from the docking experiment. The binding scores ranged from −12.5 (for the best) to −10.7 (for the worst). Among the top one hundred compounds, most contained aromatic rings. Over 20% of these compounds contain an indole or similar ring structure. In the first 24 compounds (Supplementary Table 2, Supplementary Figures 1S and 2S), there are four close homologs of tryptamine. For example, 2-(5,7-dichloro-1H-indol-3-yl)ethanamine and 3-(5-chloro-1H-indol-3-yl)propan-1-amine are listed as the 3rd and 6th on the list (Supplementary Table 1 and Figures 1S and 2S). The tryptamine derivatives are docked in a similar manner to that observed for 2-aminopurine in the crystal structure (PDB code: 4IH2). These compounds docked with the aromatic system being parallel to Phe58 with distance 4.3–3.6 Å between ring planes. For all tryptamine derivatives the amine group forms a hydrogen bond with Asp 71.
4. Discussion
Our structural analysis revealed that, despite the low sequence identity, Act d 11 has a very similar fold to Bet v 1 and other related allergens (e.g. Api g 1 (Schirmer et al., 2005), Dau c 1 (Markovic-Housley et al., 2009), Fra a 1 (Seutter von Loetzen et al., 2012), Gly m 4 (Berkner et al., 2009) and Pru av 1 (Neudecker et al., 2001)) for which structures were determined and reported to the PDB. Both proteins contain ligand-binding cavities and are composed of a seven-stranded anti-parallel β-sheet and three α-helices. However, the volume of the Bet v 1 ligand-binding site is significantly bigger than in Act d 11. The ligand-binding cavity is very hydrophobic and contains only one hydrophilic residue (Asp71). In addition, the cavity contains a cysteine residue (Cys137) that is on the opposite side of the cavity in relation to Asp71. It is very likely that Act d 11 has completely different ligand specificity in comparison to Bet v 1 and other PR-10 proteins.
Unfortunately, we were not able to identify the ligand bound to either natural or recombinant Act d 11. This suggests that the ligand is either present in both plants and bacteria, or that the ligand binding cavity is able to accommodate different small molecules. Using native mass spectrometry, we observe ligand-bound species, indicating that this ligand binds in solution and is not an artifact introduced during crystalisation. The intensity of the ligand-bound species is much lower compared to the unbound species, at the pH and ionic strength solution conditions tested here. The mass of the ligand, calculated from our experiments, is not accurate enough to allow for unambiguous identification from a database search. In addition, none of the ligands identified from our virtual screen have a mass that matches that of the experimentally determined mass. This could be because the ligand present in solution is not in the ligand database that we used. Another possibility is that Act d 11, like Bet v 1 accommodates two ligands in its binding pocket. To answer this we isolated the Act d 11 ligand-bound species in the gas phase, and attempted to dissociate the ligand from the protein by means of collision induced dissociation. However, due to the low signal of this species, these experiments were not successful.
A structural analysis of the crystals soaked in different compounds showed that Act d 11 might change its conformation after ligand binding. We speculate that the differences in the protein conformation prior to and after ligand binding may play a role in protein-protein recognition/interactions.
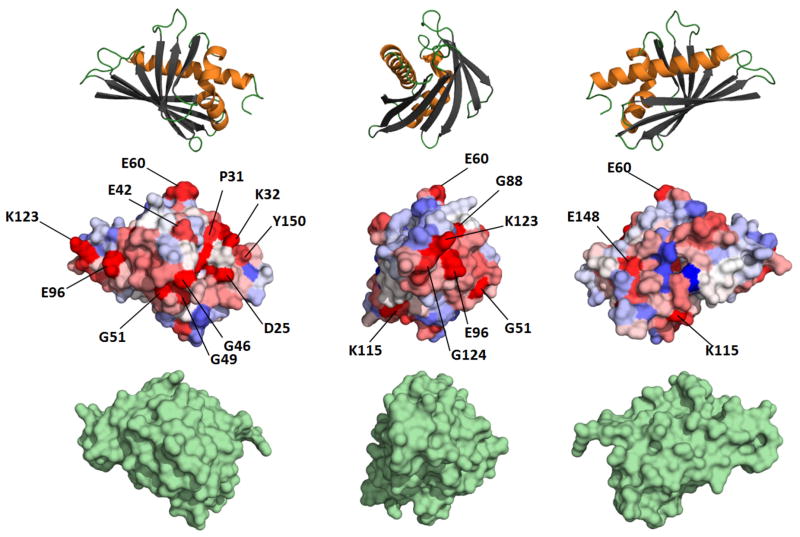
On the basis of sequence alignments, Osmark and Brisson pointed out that several residues were conserved in both PR-10 and MLP proteins (Fig. 8). The conserved residues include aromatic residues in position 21 (Tyr; numbering according to Act d 11), Gly47, Gly53, Gly90 and aromatic residues in position 120 (Tyr). While the previously mentioned residues are conserved in Act d 11, two other residues Gly88 and Ser122 (Bet v 1 numbering) were found to be not conserved in Act d 11 after the structural alignment had been conducted.
Figure 8.
Sequence conservation of Bet v 1-like allergens. Upper part of the panel shows Bet v 1 molecules in ribbon representation at the same orientation as the middle part. The middle part of the panel shows conservation of surface residues mapped on Bet v 1 (red – the most conserved residues, blue - the least conserved residues). The most conserved residues are labeled. Lower part of the panel shows Act d 11 in surface representation. The orientation of Act d 11 is the same as Bet v 1.
The loop connecting strands β2 and β3 (so called Gly-rich loop) is characteristic of PR-10-related proteins. This loop shows extraordinary rigidity (Fernandes et al., 2009), moreover the residues forming this loop have a sequence similar to the Walker A motif (P-loop motif) composed of the following residues: Gx(4)GK[S, T] (Prosite syntax). In many cases, the Walker A motif is involved in interactions with the phosphate group of nucleotides. The Walker A motif should be placed between a β-strand and an α-helix. However, the role of the Gly-rich loop in the PR-10-related proteins is not known. Analysis of the 30 allergenic proteins of the PR-10 family included in the AllFam database (www.meduniwien.ac.at/allergens/allfam/) revealed that this region is also highly conserved and can be described by the following pattern [SV][DEQ]x(0,1)[ILV][EGK]G[ND][GW]Gx(1,2)G[ST][ILV]. Only Act d 11 has a tryptophan residue instead of glycine. Most likely, the tryptophan residue present in the loop of Act d 11 stabilizes this region and improves its rigidity, as it is anchored between several hydrophobic residues.
It was shown that the dominating IgE-binding epitope in Bet v 1 includes Glu45 (marked in the pattern above in bold and italics), and a mutation of this residue resulted in a 50% reduction in the binding of human polyclonal IgE (Spangfort et al., 2003). The region of the Gly-rich loop was also shown to be responsible for the IgE cross-reactivity to Bet v 1 and its homologs (Mittag et al., 2006; Neudecker et al., 2001). In addition, analysis of two isoforms, Api g 1.01 and Api g 1.02, of the celery allergen Api g 1 which differ in their capacity to bind IgE showed that the Gly-rich loop plays an important role in the IgE binding of celery allergic subjects (Wangorsch et al., 2007). The Gly-rich loop in Act d 11 is very similar to that observed for Bet v 1, including Glu46 (Glu45 in Bet v 1).
In comparison with other PR-10 related allergens, the significantly smaller volume of the ligand-binding cavity in Act d 11 is very striking. It was shown that Bet v 1 was able to accommodate very divergent compounds in different regions of the ligand-binding cavity, while proteins from mung bean (Pasternak et al., 2006) or lupin (Fernandes et al., 2009) were able to simultaneously bind several molecules of the same type. The difference between the sizes of the binding cavities of Act d 11 and other PR-10 like allergens can be explained when assuming that the protein from the kiwifruit binds only one ligand molecule. In addition, one could speculate that Act d 11 might also bind different ligands, which is in agreement with the results of our crystal soaking experiments. It also cannot be excluded that in solution Act d 11 has a somewhat different conformation and the volume of the binding cavity is bigger. Unfortunately the presence of an unknown ligand or ligands in both the recombinant protein and the natural protein isolated from its source makes the analysis of the crystal structures difficult and does not allow an unambiguous interpretation. It is also possible that the Act d 11 ligand is somewhat disordered due to the mainly hydrophobic cavity in which only Asp71 may “anchor” the ligand through hydrogen bonds.
Changes in the protein’s conformation after soaking the crystals with serotonin not only indicate that the protein interacted with the ligand, but also suggest that the conformational changes induced by a physiological ligand might be important for the protein’s function. It is possible that the increased mobility of the Phe24 side chain in conjunction with the change of the Phe20 side chain conformation results in the destabilization of the loop formed by residues 14–17. The difference in protein conformations between apo and liganded forms may be important for interactions with other proteins, as observed in the case of the PYR-PYL-RCAR family of the abscisic acid receptors (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009).
It is also possible that the type of the ligand bound may play a role in the interaction of the PR-10 related proteins with the human immune system. For example, it was suggested that Bet v 1 should be considered a “dressed allergen”, which through isoform-dependent ligand complexes may interact in different ways with our immune system (Kofler et al., 2012). This idea could also be used to explain the existence of low and high IgE binding forms of Bet v 1. This example clearly shows that the human immune system encounters allergen molecules that may not only differ in sequence, presence or absence of post translational modifications, but that they may also carry dissimilar ligands.
5. Conclusions
Structural analysis revealed that, in spite of the low sequence identity, Act d 11 has a fold very similar to that of Bet v 1 and other PR-10 related allergens. The ligand-binding cavity in Act d 11 is significantly smaller than in similar PR-10 allergens. Both the natural and recombinant protein carry an unidentified ligand, which is relatively small and most likely contains an aromatic ring. The binding of this ligand induces conformational changes in the protein. Glu46 (Glu45 in Bet v 1), which was shown to be important for IgE binding in Bet v 1, is conserved in Act d 11, while it is not conserved in other allergens with significantly higher sequence identity to the birch pollen allergen. Most likely, the so called Gly-rich loop (P-loop), which includes Glu46, may be responsible for the IgE cross-reactivity between Bet v 1 and Act d 11.
Supplementary Material
We determined structure of kiwifruit allergen Act d 11.
Act d 11 and Bet v1 have a very similar fold.
Act d 11 has smaller ligand-binding cavity than PR-10 allergens.
Gly-rich loop may cause the IgE cross-reactivity between Bet v 1 and Act d 11.
Acknowledgments
The authors would like to thank Matt Demas, Lesa Offermann, and Rob Solberg for valuable discussions. The work described in the paper was supported by GM53163 grant and internal funds from University of South Carolina.
The structural results shown in this report are derived from work performed at Argonne National Laboratory, at the Structural Biology Center of the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Abbreviations
- MLP
Major latex protein
- PDB
Protein Data Bank
- PR-10
Pathogenesis-related proteins family 10
- RMSD
Root Mean Square Deviation
- RRP
Ripening Related Protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner H, Neudecker P, Mittag D, Ballmer-Weber BK, Schweimer K, Vieths S, Rosch P. Biosci Rep. 2009;29:183–192. doi: 10.1042/BSR20080117. [DOI] [PubMed] [Google Scholar]
- Bollen MA, Wichers HJ, Helsper JPFG, Savelkoul HFJ, van Boekel MAJS. Food Chemistry. 2010;119:241–248. [Google Scholar]
- Cano MP. J Agr Food Chem. 1991;39:1786–1791. [Google Scholar]
- Ciardiello MA, Giangrieco I, Tuppo L, Tamburrini M, Buccheri M, Palazzo P, Bernardi ML, Ferrara R, Mari A. J Agric Food Chem. 2009;57:1565–1571. doi: 10.1021/jf802966n. [DOI] [PubMed] [Google Scholar]
- Cowtan KD, Main P. Acta Crystallogr D Biol Crystallogr. 1993;49:148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- Cumplido-Laso G, Medina-Puche L, Moyano E, Hoffmann T, Sinz Q, Ring L, Studart-Wittkowski C, Caballero JL, Schwab W, Munoz-Blanco J, Blanco-Portales R. J Exp Bot. 2012;63:4275–4290. doi: 10.1093/jxb/ers120. [DOI] [PubMed] [Google Scholar]
- Cymborowski M, Klimecka M, Chruszcz M, Zimmerman MD, Shumilin IA, Borek D, Lazarski K, Joachimiak A, Otwinowski Z, Anderson W, Minor W. J Struct Funct Genomics. 2010;11:211–221. doi: 10.1007/s10969-010-9092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino R, Bernardi ML, Wallner M, Palazzo P, Camardella L, Tuppo L, Alessandri C, Breiteneder H, Ferreira F, Ciardiello MA, Mari A. Allergy. 2011 doi: 10.1111/j.1398-9995.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WS. The PyMOL Molecular Graphics System 2002 [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. Nucleic Acids Res. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fernandes H, Bujacz A, Bujacz G, Jelen F, Jasinski M, Kachlicki P, Otlewski J, Sikorski MM, Jaskolski M. Febs J. 2009;276:1596–1609. doi: 10.1111/j.1742-4658.2009.06892.x. [DOI] [PubMed] [Google Scholar]
- Fine AJ. J Allergy Clin Immunol. 1981;68:235–237. doi: 10.1016/0091-6749(81)90189-5. [DOI] [PubMed] [Google Scholar]
- Frickey T, Lupas A. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. Nucleic Acids Res. 2008;36:W35–41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JR, Calkins D, Sullivan AP, Shelley JC. J Comput Aided Mol Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- Heck A. Nature Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- Knopf M. Psychol Res-Psych Fo. 1991;53:203–211. [Google Scholar]
- Kofler S, Asam C, Eckhard U, Wallner M, Ferreira F, Brandstetter H. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z, Basle A, Padavattan S, Maderegger B, Schirmer T, Hoffmann-Sommergruber K. Acta Crystallogr D Biol Crystallogr. 2009;65:1206–1212. doi: 10.1107/S0907444909034854. [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H. J Mol Biol. 2003;325:123–133. doi: 10.1016/s0022-2836(02)01197-x. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, Wang Y, Li J, Peterson FC, Jensen DR, Yong EL, Volkman BF, Cutler SR, Zhu JK, Xu HE. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Mittag D, Batori V, Neudecker P, Wiche R, Friis EP, Ballmer-Weber BK, Vieths S, Roggen EL. Mol Immunol. 2006;43:268–278. doi: 10.1016/j.molimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Fukuoka S, Kadomura Y, Hamaoka H, Mito T, Ohuchi H, Schwab W, Noji S. Plant Biotechnol-Nar. 2012;29:271–277. [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, Yoshida T, Kodaira KS, Yamaguchi-Shinozaki K, Tanokura M. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Mogensen JE, Wimmer R, Larsen JN, Spangfort MD, Otzen DE. J Biol Chem. 2002;277:23684–23692. doi: 10.1074/jbc.M202065200. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker P, Schweimer K, Nerkamp J, Scheurer S, Vieths S, Sticht H, Rosch P. J Biol Chem. 2001;276:22756–22763. doi: 10.1074/jbc.M101657200. [DOI] [PubMed] [Google Scholar]
- Osmark P, Boyle B, Brisson N. Plant Mol Biol. 1998;38:1243–1246. doi: 10.1023/a:1006060224012. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. In: Editor. Isomorphous replacement and anomalous scattering. Wolf W, Evans PR, Leslie AGW, editors. Warrington, UK: Daresbury Laboratory; 1991. [Google Scholar]
- Otwinowski Z, Minor W. Methods in enzymology: Macromolecular crystallography, part A. Vol. 276. Academic Press; New York: 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- Pasternak O, Bujacz GD, Fujimoto Y, Hashimoto Y, Jelen F, Otlewski J, Sikorski MM, Jaskolski M. Plant Cell. 2006;18:2622–2634. doi: 10.1105/tpc.105.037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int J Mass Spectrometry. 2007;261(1):1–12. [Google Scholar]
- Radauer C, Lackner P, Breiteneder H. BMC Evol Biol. 2008;8:286. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rona RJ, Keil T, Summers C, Gislason D, Zuidmeer L, Sodergren E, Sigurdardottir ST, Lindner T, Goldhahn K, Dahlstrom J, McBride D, Madsen C. J Allergy Clin Immunol. 2007;120:638–646. doi: 10.1016/j.jaci.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. J Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. Rapid Comm Mass Spectrometry. 2008;22:3297–3304. doi: 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- Schirmer T, Hoffimann-Sommergrube K, Susani M, Breiteneder H, Markovic-Housley Z. J Mol Biol. 2005;351:1101–1109. doi: 10.1016/j.jmb.2005.06.054. [DOI] [PubMed] [Google Scholar]
- Seutter von Loetzen C, Schweimer K, Schwab W, Rosch P, Hartl-Spiegelhauer O. Biosci Rep. 2012;32:567–575. doi: 10.1042/BSR20120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- Sicherer SH, Sampson HA. J Allergy Clin Immunol. 2010;125:S116–125. doi: 10.1016/j.jaci.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Spangfort MD, Mirza O, Ipsen H, Van Neerven RJ, Gajhede M, Larsen JN. J Immunol. 2003;171:3084–3090. doi: 10.4049/jimmunol.171.6.3084. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- Tito MA, Tars K, Valegard K, Hajdu J, Robinson CV. J Am Chem Soc. 2000;122:3550–3551. [Google Scholar]
- Uematsu C, Murase M, Ichikawa H, Imamura J. Plant Cell Rep. 1991;10:286–290. doi: 10.1007/BF00193143. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Wangorsch A, Ballmer-Weber BK, Rosch P, Holzhauser T, Vieths S. Mol Immunol. 2007;44:2518–2527. doi: 10.1016/j.molimm.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Acta Crystallographica Section D. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- Zimmerman MD, Chruszcz M, Koclega KD, Otwinowski Z, Minor W. Acta Crystallogr Sect A. 2005;61:c178–c179. [Google Scholar]
- Zuidmeer L, Goldhahn K, Rona RJ, Gislason D, Madsen C, Summers C, Sodergren E, Dahlstrom J, Lindner T, Sigurdardottir ST, McBride D, Keil T. J Allergy Clin Immunol. 2008;121:1210–1218. e1214. doi: 10.1016/j.jaci.2008.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.