Abstract
Insulin self-tolerance is, to a large extent, assured by the expression of small quantities of insulin by medullary thymic epithelial cells (mTECs). Regulation of thymic insulin expression differs from that in pancreas and its therapeutic manipulation could play an important role in the prevention of type 1 diabetes (T1D). Knowledge of the transcriptional regulators involved in the mTEC nuclear environment is essential for the development of such therapeutics. The yeast one-hybrid (Y1H) approach was used in order to identify such mTEC-specific nuclear proteins. We used a target composed of the human insulin gene promoter joined to the upstream class III VNTR allele, which is associated with both protection from T1D and higher thymic insulin expression, and a cDNA library from our insulin-producing mouse mTEC line. The Y1H screening allowed the identification of eleven proteins. An in vitro assay was used to confirm and quantify protein-DNA binding to the human insulin gene promoter alone or joined to a class I or class III VNTR allele, and identified the transcription factors ZBTB7A, JUN and EWSR1 as strong interacting partners. All three proteins could induce insulin expression in transfected HEK-293 cells, but ZBTB7A provided the most robust results especially in the presence of AIRE, with an additional 11-fold increase of the insulin mRNA levels from a co-transfected reporter driven by the class III VNTR allele. Thus, ZBTB7A is identified as a strong candidate for regulation of thymic insulin expression.
Keywords: insulin expression, thymus, transcription factors, type 1 diabetes, yeast one-hybrid
1. Introduction
Type 1 diabetes (T1D) is a complex trait with a strong genetic component (Polychronakos and Li, 2011). So far, more than 40 T1D susceptibility loci have been identified (Ounissi-Benkalha and Polychronakos, 2008). After the major histocompatibility complex (MHC) region (Aly et al., 2006; Anjos and Polychronakos, 2004), the second strongest effect comes from a 4.1-kb region on chromosome 11 and maps to a variable number of tandem repeats (VNTR) located 596 bp upstream of the insulin gene (Ounissi-Benkalha and Polychronakos, 2008; Bennett et al. 1995). It consists of tandem repeats of 14–15 bp bearing the consensus sequence unit ACAGGGGTGTGGGG (Bell et al., 1982; Vafiadis et al., 2001). The class I allele is the shortest and predisposes to T1D (26 to 63 repeats) whereas the long class III allele (140 to 210 repeats) provides a co-dominant protective effect, reducing the risk of T1D of 2- to 4-fold compared to class I/I homozygotes (Anjos and Polychronakos, 2004).
As previously demonstrated in our laboratory, the impact of the VNTR on the disease is related to the expression of insulin in the thymus (Vafiadis et al., 1997), where levels of insulin mRNA were 2 to 3 times higher from chromosomes carrying class III alleles compared to class I, showing that VNTR alleles only marginally influence insulin mRNA levels in the pancreas. It was therefore hypothesized that insulin expression in the thymus could be implicated in the establishment of immune central tolerance. These results were validated by several other studies and insulin was confirmed to be one of the major autoantigens involved in the disease (Durinovic-Belló et al., 2010; Fan et al., 2009; Kent et al., 2005; Nakayama et al. 2005; Taubert et al., 2007). This observation inspired studies that have demonstrated a more general mechanism through which specific autoreactive T cells undergo negative selection due to thymic expression of tissue-restricted antigens (TRAs) (Derbinski et al., 2001). Human and animal-model studies have shown association between thymic insulin expression and autoreactivity toward insulin as lower levels of insulin in the thymus have been correlated to the presence of autoreactive insulin-specific T lymphocytes (Durinovic-Belló et al., 2010; Chentoufi and Polychronakos, 2002; Durinovic-Belló et al., 2005) and accelerated diabetes in the non-obese diabetic (NOD) mouse (Thébault-Baumont et al., 2003).
The selection for self-tolerant T cells takes place in the thymus and requires AIRE, the autoimmune regulator, responsible of the expression of thousands of TRAs in a specialized subset of medullary thymic epithelial cells (mTECs) (Derbinski et al., 2001; Palumbo et al., 2006). By a mechanism that is still poorly understood, AIRE induces the expression of TRAs within the mTECs, which present the antigens for the purpose of negative selection (Anderson et al., 2005; Irla et al., 2008). The AIRE-dependance of thymic insulin expression has been directly demonstrated in the mouse (Anderson et al., 2002) and indirectly in the human (Ahonen et al., 1990).
Although insulin expression in the thymus seems to depend on both AIRE and the VNTR (Taubert et al., 2007; Cai et al., 2011), the transcriptional regulation of thymic gene expression is still poorly understood. In order to address this question, we probed an insulin-producing mTEC line that was previously generated in our laboratory (Palumbo et al., 2006). We used the yeast one-hybrid (Y1H) approach to identify factors that had the capacity to regulate insulin expression by binding to a genomic sequence composed of the human insulin gene promoter joined to the class III VNTR allele. Candidates thus identified were functionally evaluated.
2. Materials and methods
2.1 Yeast one-hybrid screening
The Y1H screening was performed using the Matchmaker™ One-Hybrid Library Construction & Screening Kit from Clontech, as recommended by the manufacturer. The yeast strain Y187, which is unable to synthetize histidine, was used for the assay. The vector containing the DNA target, or bait, was assembled by transferring the bait to vector pHIS2.1 (supplied with the kit). The bait was composed of the class III VNTR allele (E1 clone (Vafiadis et al., 2001)) joined to the minimal promoter of the human insulin gene, which reached nucleotide -188 (with regard to the ATG) at its 3′ end. The cDNA library used for the screening was generated using the Advantage 2 PCR Kit and the vector pGADT7-Rec2, both from Clontech, using RNA from our insulin-producing mTEC line (Palumbo et al., 2006). For the one-hybrid library screening, both the bait and the cDNA library were cotransformed in competent yeast cells and the resulting cells were plated on the appropriate selective medium (synthetically defined (SD) medium containing leucine, tryptophan and histidine). Positive colonies were restreaked on the selective medium to remove contaminating plasmids and, by yeast colony PCR, we amplified the genes responsible for the Y1H interactions. For these amplifications, we used the Expand High FidelityPLUS PCR System from Roche in combination with the primers Matchmaker ADLD from Clontech. PCR products were sequenced for identification of the interacting proteins.
2.2 Protein-DNA binding assay
The Protein-DNA Binding Assay Kit from Clontech was used in this step. Clones to be tested were first transfected in HEK-293 cells and two days later, a whole cell extract was prepared as recommended by the manufacturer. Of this extract, 75 μg of protein (measures by the BCA™ Protein Assay Kit from PIERCE) was mixed with either blocking buffer (for the background reference) or one of the three biotinylated DNA targets. Protein-DNA mixes were then added to wells coated with streptavidin and subsequently washed and revealed using the appropriate substrates. ProLabel activity was quantified using the VICTOR plate reader from PerkinElmer. Luminescence was measured every 15 minutes and results were determined by taking the mean signal:background ratio at 45 and 60 minutes. Every assay, starting from the transfection step, was carried out in triplicates.
2.3 Cloning of the interacting genes and controls
Overall, 19 genes were cloned in frame with the ProLabel tag from the vector pProLabel-C (Clontech). Those included the 11 mouse genes from the Y1H screening, 5 human orthologues of these genes (Ddx1, Zfp90, Jun, Zbtb7a and Ewsr1), and AIRE and Pdx-1 as positive controls, both mouse and human. The Expand High FidelityPLUS PCR System from Roche or the Platinum® Pfx DNA Polymerase from Invitrogen was used for PCR amplifications according to the target. All 19 genes were cloned, transformed and sequenced using standard methods.
2.4 Transient transfections
HEK-293 cells were used for transfections related to the protein-DNA binding assay and to the assay for the modulation of insulin expression as well. The CalPhos™ Mammalian Transfection Kit form Clontech was used to this end. Between 2 and 7 μg of the pProLabel constructs were used for each reaction (depending on the ProLabel tag expression (data not shown)) whereas different amounts of plasmids were used to study the modulation of insulin expression (depending on the type of vector).
2.5 Generation of the DNA targets
Three different DNA targets were generated to carry out the protein-DNA binding assay: the human insulin gene promoter alone, with the first 50 bases of exon 1 (oligo ins389; 389 bp), or joined to the class I (oligo I; 1863 bp) or class III VNTR allele (oligo III; 3393 bp). For the protein-DNA binding assay, the three DNA targets were labeled with biotin using the Biotin 3′ End DNA Labeling Kit from PIERCE, as recommended by the manufacturer. Following each reaction, labeling efficiency was assessed and quantified by slot blot and chemiluminescence using the kit controls.
2.6 Modulation of insulin expression
Modulation of insulin expression by human JUN, EWSR1 and ZBTB7A was assessed in HEK-293 cells, with or without AIRE in triplicate transfections. Constructs containing the promoter regions were different for this assay as they were bearing the complete insulin gene including the introns. Vectors pENTRY-noVNTR, -classI (814 allele) or -classIII (E1 allele) were thus used as reporters for this assay and insulin mRNA levels were measured by quantitative PCR, using the TaqMan® Gene Expression Assay from Applied Biosystems.
3. Results
3.1 Identification of the putative regulators
The Y1H system allows the identification of novel genes encoding proteins that bind to a specific DNA target, in this case, factors regulating insulin expression in the thymus by binding to the human insulin gene promoter or the class III VNTR allele. Following the Y1H screening, 152 clones stemming from our mouse mTEC cDNA library were obtained. Of these, 11 were selected for further analyses because they fulfilled two important criteria: i) they were previously shown to be nuclear proteins, and ii) they were previously shown to display functions related to the immune system, transcription or T1D. These clones are part of different categories of proteins and as expected, most of them are transcription factors (Table 1).
Table I.
List of the eleven proteins selected following the Y1H screening.
| Proteins | Category | Putative Functions |
|---|---|---|
| RBM28 | RNA binding | Associated to endocrinopathy syndrome (Nousbeck et al., 2008) |
| DDX1 | Helicase | Interacts with RelA, a subunit of NF-kappaB, and enhances its transactivation (Ishaq et al., 2009) |
| TAX1BP1 | Immune signalling | Negative regulator of TNF-α and involved in NF-kappaB signalling (Iha et al., 2008) |
| PSMB1 | Adaptor protein | Located in the IDDM8 region, associated with susceptibility to rheumatoid arthritis (Hinks et al., 2006) |
| UBB | Adaptor protein | Binds to UBASH3A (T1D risk factor) (Concannon et al., 2008) |
| EWSR1 | Transcription factor | Can interact with many transcription factors (Law et al., 2006) and binds G-quadruplex structures (Takahama et al., 2011) |
| VEZF1 | Transcription factor | Vascular zinc finger transcription factor, transcription activator in some systems (Gowher et al., 2008) |
| ZBTB7A | Transcription factor | Zinc finger transcription factor, can act as a transcription activator or repressor (Jeon et al., 2008) |
| ZFP90 | Transcription factor | Zinc finger transcription factor with a powerful transcriptional repression activity (Thiel et al., 2001) |
| JUN | Transcription factor | Involved in thymocyte development (Riera-Sans and Behrens, 2007) |
| BLZF1 | Transcription factor | Basic leucine zipper transcription factor that can interact with AP-1 (Tong et al., 1999) |
3.2 Validation of the interactions
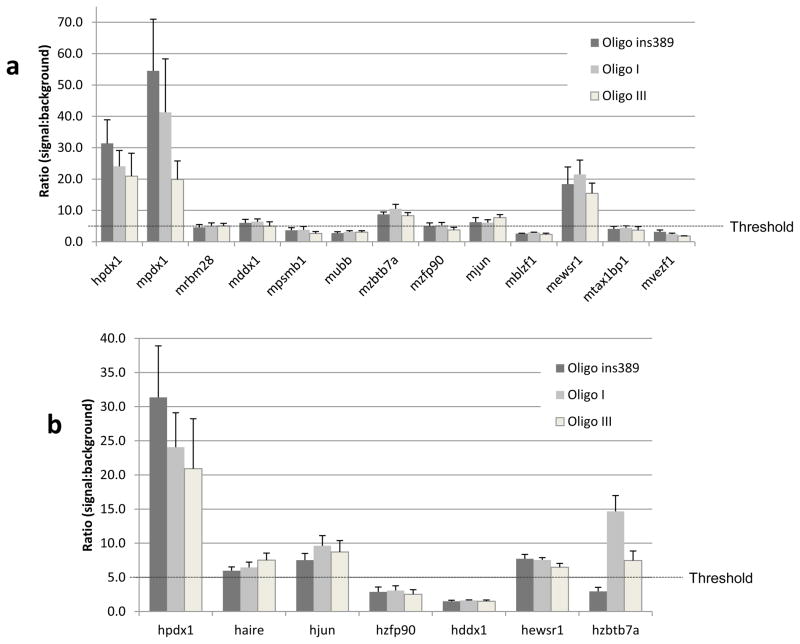
In order to narrow down the number of library proteins, an in vitro assay was used to validate and quantify protein-DNA interactions (Clontech). Human and mouse versions of the transcription factor Pdx-1 were used as controls since they are known to specifically bind the insulin gene promoter. Thus, the 11 previously identified library proteins were submitted to this analysis using three different DNA targets: the human insulin gene promoter alone (oligo ins389), or joined to the class I (oligo I) or class III VNTR allele (oligo III). Results from this analysis are displayed in Figure 1. A signal was considered positive when a signal:background ratio of 5 was reached (Clontech) and five clones fulfilled this criterion: Ddx1, Zbtb7a, Zfp90, Jun and Ewsr1 (Figure 1a). It is noteworthy that both positive controls generated a very strong signal demonstrating that mouse proteins have the ability to recognize and bind to a specific human DNA sequence.
Figure 1.
Results of the protein-DNA binding assay. (a) Experiment involving the 11 mouse library proteins from the Y1H experiment. Mouse and human Pdx-1 were used as controls. For each protein of interest, four reactions were made: one without a DNA target, in order to establish the background level, and three reactions involving the different DNA targets (oligo ins389, oligo I and oligo III). Results are presented as a ratio of the signal generated by a protein-DNA interaction divided by the background signal. A signal was considered positive when a signal:background ratio of 5 was reached (Clontech). (b) Experiment involving the human orthologues of the five best library proteins from the previous assay. Human Pdx-1 was used as control. Legend is displayed in the upper right corner of each graph and every experiment was carried out in triplicates. Error bars represent standard deviation. The letter m or h before the name of the gene respectively means mouse or human.
The five mouse proteins that generated the best signal had their human counterpart cloned in frame with the ProLabel tag in vector pProLabel-C and the same assay was repeated with the same three DNA targets (Figure 1b). Human AIRE was tested as well in this experiment in order to study interactions with this important immune factor. Three transcription factors reached the threshold ratio of 5 and came out of this analysis: JUN, EWSR1 and ZBTB7A. It is noteworthy that AIRE provided a signal above the determined threshold but the significance of this binding remains elusive for the moment. Although AIRE is involved in the expression of several TRAs such as insulin (Derbinski et al., 2001), this result does not necessarily imply a direct binding of AIRE to our DNA targets.
3.3 ZBTB7A synergistically modulates insulin expression
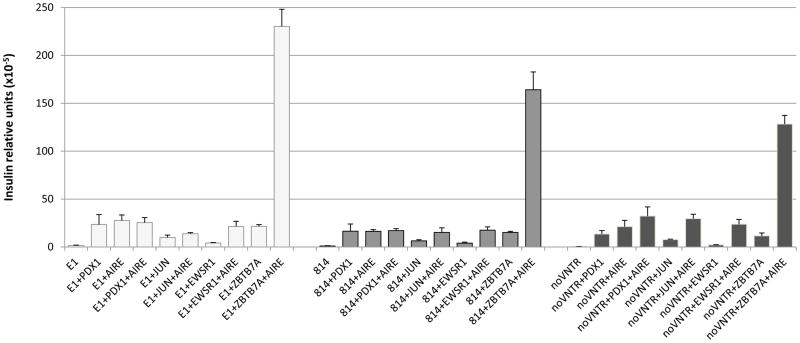
Human JUN, EWSR1 and ZBTB7A were then further analyzed in order to determine their capacity to modulate insulin expression in a human cellular system. To this end, HEK-293 cells were used because they were previously shown to act as a very effective model in similar studies (Ferguson et al., 2009). The three different reporter vectors were co-transfected with one of the three clones, with or without AIRE, and insulin mRNA levels were determined by quantitative PCR (Figure 2). All three clones had the capacity to induce insulin expression from the reporter and this expression was amplified in the presence of AIRE. However, ZBTB7A showed the strongest induction of expression especially in the presence of AIRE, which increased insulin expression by at least 10-fold regardless of the reporter (statistically significant, determined by two-way ANOVA; p<0.01), suggesting that ZBTB7A could play a crucial role in insulin expression in the thymus. In addition, insulin levels reached with ZBTB7A were higher for the class III VNTR allele compared to the class I allele, suggesting a direct role in the genetic effect.
Figure 2.
Insulin expression induced by transfection of the candidate factors in HEK-293 cells. Results are displayed in insulin relative units (x10−5) as mRNA levels were normalized with both GAPDH and AIRE. Cells were transfected with one of the three reporters (E1, 814 or noVNTR), in combination with human Jun, Ewsr1 or Zbtb7a, with or without AIRE. As controls, reporters were transfected alone, with human Pdx-1, with human AIRE or with both Pdx-1 and AIRE. Experiments were carried out in triplicates. Error bars represent standard deviation. Statistics determined by a two-way ANOVA. Reporters: E1 (class III), white; 814 (class I), grey and noVNTR, dark grey.
4. Discussion
Because of the divergent patterns of insulin expression in the pancreas and in the thymus (Vafiadis et al., 1997; Levi and Polychronakos, 2009), we started with the assumption that the transcriptional regulators involved in thymic insulin expression differ between the two organs. The three transcription factors that came out of the Y1H screening and were shown capable of modulating insulin expression within a non-thymus cellular system with or without AIRE, may be involved in determining these differences. ZBTB7A is of particular interest as it drastically increases insulin expression in that system especially in the presence of AIRE, with a 12-fold increase of the insulin mRNA levels in conjunction with the class III VNTR allele. Although the actual Y1H screening assessed a mouse cDNA library against a human genomic DNA sequence, it was likely that positive interactions would be identified since we knew transcription from the human insulin promoter could be activated in the mouse (Ounissi-Benkalha and Polychronakos, unpublished). Moreover, it was also known that transcription factors are usually very well conserved among species and so are their binding domains (Lowry and Atchley, 2000). Even though the insulin VNTR is primate-specific, it is unlikely that new transcription factors have evolved between mouse and human to interact with them.
Although some characterization remains to be done, our results strongly suggest that this factor could play a prominent role in the prevention of the autoimmunity observed in T1D. The strong synergy between ZBTB7A and AIRE, as well as the enhanced expression with the class III vs. class I construct suggest a specific role in thymic insulin expression. This is in agreement with a recent study by Cai et al. in which it was demonstrated that VNTRs and AIRE could synergistically modulate insulin expression in mTECs (Cai et al., 2011) and supported by the preferential expression pattern of ZBTB7A in the thymus (data not shown), suggesting that this transcription factor is one of the key elements involved in protection from T1D.
Four years ago, Ferguson et al. isolated an engineered zinc finger protein (ZFP) from a library based on its ability to activate transcription of the endogenous insulin gene (Ferguson et al., 2009). ZBTB7A is also a ZFP, consistent with such functions in the thymus. Even though the engineered ZFP identified in this study was shown to act through the VNTRs and that we identified a related Zbtb7a binding domain within the insulin gene minimal promoter, we cannot exclude that ZBTB7A interacts with the VNTRs given the results obtained following the in vitro binding assay (Figure 1).
Transcription factors displaying the ability to induce thymic expression of a specific TRA have already been identified and this is the case of IRF8 (Giraud et al., 2007). This protein is responsible for the expression of CHRNA1, a gene encoding the alpha-subunit of the muscle acetylcholine receptor, the main autoimmune target in myasthenia gravis. More recently, MafA was identified as a transcription factor that has the ability to modulate insulin expression in the thymus (Noso et al., 2010). Although MafA is also involved in pancreatic insulin expression, the authors stated that MafA was sufficient to induce thymic expression even though it is known to be a weak transactivator (Nishizawa et al., 2003). The promoter activity of MafA, which was proportional to the Ins2 levels in their mTEC line, was inversely proportional to the length of the VNTRs, which contradicts the trend usually observed (Cai et al., 2011). Moreover, expression of TRAs, including Ins2, in mTECs has been shown to depend on transcriptional regulators and transcriptional start sites different from those used in peripheral cells (Villaseñor et al., 2008).
5. Conclusions
Although Zbtb7a is usually known to be involved in cellular differentiation or oncogenic transformation, we showed here that it plays a role in thymic expression of insulin and maybe other TRAs. Given this feature of the protein, it is highly likely that Zbtb7a influences susceptibility to T1D and that it could be used in various therapeutic approaches for prevention of the disease.
Acknowledgments
The study was supported by grants to Constantin Polychronakos from the Juvenile Diabetes Foundation and the National Institutes of Health (USA). Julien St-Jean was a recipient of a postdoctoral research fellowship from the Fonds de la Recherche en Santé du Québec (FRSQ) and Constantin Polychronakos is a recipient of the Sessenwein Award from the Department of Paediatrics, McGill University.
Abbreviations
- T1D
type 1 diabetes
- MHC
major histocompatibility complex
- VNTR
variable number of tandem repeats
- TRA
tissue-restricted antigen
- AIRE
autoimmune regulator
- mTEC
medullary thymic epithelial cell
- Y1H
yeast one-hybrid
- SNP
single nucleotide polymorphism
- ZFP
zinc finger protein
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest in relation to this publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahonen P, Myllärniemi S, Sipilä I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. New England Journal of Medicine. 1990;322:1829–1836. doi: 10.1056/NEJM199006283222601. [DOI] [PubMed] [Google Scholar]
- 2.Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, Babu SR, Yu L, Miao D, Erlich HA, Fain PR, Barriga KJ, Norris JM, Rewers MJ, Eisenbarth GS. Extreme genetic risk for type 1A diabetes. Proceedings of the National Academy of Sciences. 2006;103:14074–14079. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an Immunological Self Shadow Within the Thymus by the Aire Protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 5.Anjos S, Polychronakos C. Mechanisms of genetic susceptibility to type I diabetes: beyond HLA. Molecular Genetics and Metabolism. 2004;81:187–195. doi: 10.1016/j.ymgme.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Bell GI, Selby MJ, Rutter WJ. The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature. 1982;295:31–35. doi: 10.1038/295031a0. [DOI] [PubMed] [Google Scholar]
- 7.Bennett ST, Lucassen AM, Gough SCL, Powell EE, Undlien DE, Pritchard LE, Merriman ME, Kawaguchi Y, Dronsfield MJ, Pociot F, Nerup J, Bouzekri N, Cambon-Thomsen A, Rønningen KS, Barnett AH, Bain SC, Todd JA. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nature Genetics. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 8.Cai CQ, Zhang T, Breslin MB, Giraud M, Lan MS. Both Polymorphic Variable Number of Tandem Repeats and Autoimmune Regulator Modulate Differential Expression of Insulin in Human Thymic Epithelial Cells. Diabetes. 2011;60:336–344. doi: 10.2337/db10-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chentoufi AA, Polychronakos C. Insulin Expression Levels in the Thymus Modulate Insulin-Specific Autoreactive T-Cell Tolerance: The Mechanism by Which the IDDM2 Locus May Predispose to Diabetes. Diabetes. 2002;51:1383–1390. doi: 10.2337/diabetes.51.5.1383. [DOI] [PubMed] [Google Scholar]
- 10.Concannon P, Onengut-Gumuscu S, Todd JA, Smyth DJ, Pociot F, Bergholdt R, Akolkar B, Erlich HA, Hilner JE, Julier C, Morahan G, Nerup J, Nierras CR, Chen WM, Rich SS Type 1 Diabetes Genetics Consortium. A Human Type 1 Diabetes Susceptibility Locus Maps to Chromosome 21q22.3. Diabetes. 2008;57:2858–2861. doi: 10.2337/db08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nature Immunology. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 12.Durinovic-Belló I, Jelinek E, Schlosser M, Eiermann T, Boehm BO, Karges W, Marchand L, Polychronakos C. Class III alleles at the insulin VNTR polymorphism are associated with regulatory T-cell responses to proinsulin epitopes in HLA-DR4, DQ8 individuals. Diabetes. 2005;54(Suppl 2):S18–24. doi: 10.2337/diabetes.54.suppl_2.s18. [DOI] [PubMed] [Google Scholar]
- 13.Durinovic-Belló I, Wu RP, Gersuk VH, Sanda S, Shilling HG, Nepom GT. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes and Immunity. 2010;11:188–193. doi: 10.1038/gene.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Y, Rudert WA, Grupillo M, He J, Sisino G, Trucco M. Thymus-specific deletion of insulin induces autoimmune diabetes. EMBO Journal. 2009;28:2812–2824. doi: 10.1038/emboj.2009.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson LA, Docherty H, MacKenzie A, Docherty K. An engineered zinc finger protein reveals a role for the insulin VNTR in the regulation of the insulin and adjacent IGF2 genes. FEBS Letters. 2009;583:3181–3186. doi: 10.1016/j.febslet.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 16.Giraud M, Taubert R, Vandiedonck C, Ke X, Lévi-Strauss M, Pagani F, Baralle FE, Eymard B, Tranchant C, Gaidos P, Vincent A, Willcox N, Beeson D, Kyewski B, Garchon HJ. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–937. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 17.Gowher H, Stuhlmann H, Felsenfeld G. Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes and Development. 2008;22:2075–2084. doi: 10.1101/gad.1658408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinks A, Barton A, John S, Shephard N, Worthington J. Fine mapping of genes within the IDDM8 region in rheumatoid arthritis. Arthritis Research and Therapy. 2006;8:R145. doi: 10.1186/ar2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, Starost MF, Yedavalli V, Heyninck K, Dikic I, Beyaert R, Jeang KT. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-kappaB activation. EMBO Journal. 2008;27:629–641. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irla M, Hugues S, Gill J, Nitta T, Hikosaka Y, Williams IR, Hubert FX, Scott HS, Takahama Y, Holländer GA, Reith W. Autoantigen-Specific Interactions with CD4+ Thymocytes Control Mature Medullary Thymic Epithelial Cell Cellularity. Immunity. 2008;29:451–463. doi: 10.1016/j.immuni.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Ishaq M, Ma L, Wu X, Mu Y, Pan J, Hu J, Hu T, Fu Q, Guo D. The DEAD-box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB-mediated transcription. Journal of Cellular Biochemistry. 2009;106:296–305. doi: 10.1002/jcb.22004. [DOI] [PubMed] [Google Scholar]
- 22.Jeon B-N, Yoo J-Y, Choi W-I, Lee C-E, Yoon H-G, Hur M-W. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) Represses Transcription of the Tumor Suppressor Rb Gene via Binding Competition with Sp1 and Recruitment of Co-repressors. Journal of Biological Chemistry. 2008;283:33199–33210. doi: 10.1074/jbc.M802935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, Hering BJ, Hafler DA. Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature. 2005;435:224–228. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- 24.Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Briefings in Functional Genomics and Proteomics. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- 25.Levi D, Polychronakos C. Regulation of insulin gene expression by cytokines and cell-cell interactions in mouse medullary thymic epithelial cells. Diabetologia. 2009;52:2151–2158. doi: 10.1007/s00125-009-1448-y. [DOI] [PubMed] [Google Scholar]
- 26.Lowry JA, Atchley WR. Molecular Evolution of the GATA Family of Transcription Factors: Conservation within the DNA-Binding Domain. Journal of Molecular Evolution. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, Yu L, Wegmann DR, Hutton JC, Elliott JF, Eisenbarth GS. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishizawa M, Kataoka K, Vogt PK. MafA has strong cell transforming ability but is a weak transactivator. Oncogene. 2003;22:7882–7890. doi: 10.1038/sj.onc.1206526. [DOI] [PubMed] [Google Scholar]
- 29.Noso S, Kataoka K, Kawabata Y, Babaya N, Hiromine Y, Yamaji K, Fujisawa T, Aramata S, Kudo T, Takahashi S, Ikegami H. Insulin Transactivator MafA Regulates Intrathymic Expression of Insulin and Affects Susceptibility to Type 1 Diabetes. Diabetes. 2010;59:2579–2587. doi: 10.2337/db10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nousbeck J, Spiegel R, Ishida-Yamamoto A, Indelman M, Shani-Adir A, Adir N, Lipkin E, Bercovici S, Geiger D, van Steensel MA, Steijlen PM, Bergman R, Bindereif A, Choder M, Shaley S, Sprecher E. Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. American Journal of Human Genetics. 2008;82:1114–1121. doi: 10.1016/j.ajhg.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ounissi-Benkalha H, Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends in Molecular Medecine. 2008;14:268–275. doi: 10.1016/j.molmed.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Palumbo MO, Levi D, Chentoufi AA, Polychronakos C. Isolation and Characterization of Proinsulin-Producing Medullary Thymic Epithelial Cell Clones. Diabetes. 2006;55:2595–2601. doi: 10.2337/db05-1651. [DOI] [PubMed] [Google Scholar]
- 33.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nature Reviews Genetics. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 34.Riera-Sans L, Behrens A. Regulation of alphabeta/gammadelta T cell development by the activator protein 1 transcription factor c-Jun. Journal of Immunology. 2007;178:5690–5700. doi: 10.4049/jimmunol.178.9.5690. [DOI] [PubMed] [Google Scholar]
- 35.Takahama K, Kino K, Arai S, Kurokawa R, Oyoshi T. Identification of Ewing’s sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS Journal. 2011;278:988–998. doi: 10.1111/j.1742-4658.2011.08020.x. [DOI] [PubMed] [Google Scholar]
- 36.Taubert R, Schwendemann J, Kyewski B. Highly variable expression of tissue-restricted self-antigens in human thymus: Implications for self-tolerance and autoimmunity. European Journal of Immunology. 2007;37:838–848. doi: 10.1002/eji.200636962. [DOI] [PubMed] [Google Scholar]
- 37.Thébault-Baumont K, Dubois-Laforgue D, Krief P, Briand JP, Halbout P, Vallon-Geoffroy K, Morin J, Laloux V, Lehuen A, Carel JC, Jami J, Muller S, Boitard C. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. Journal of Clinical Investigation. 2003;111:851–857. doi: 10.1172/JCI16584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel G, Lietz M, Bach K, Guethlein L, Cibelli G. Biological activity of mammalian transcriptional repressors. Biological Chemistry. 2001;382:891–902. doi: 10.1515/BC.2001.111. [DOI] [PubMed] [Google Scholar]
- 39.Tong JH, Duprez E, Lanotte M. JEM-1, a novel nuclear cofactor: localisation and functional interaction with AP-1. Leukemia. 1999;13:1982–1992. doi: 10.1038/sj.leu.2401560. [DOI] [PubMed] [Google Scholar]
- 40.Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nature Genetics. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 41.Vafiadis P, Ounissi-Benkalha H, Palumbo M, Grabs R, Rousseau M, Goodyer CG, Polychronakos C. Class III Alleles of the Variable Number of Tandem Repeat Insulin Polymorphism Associated with Silencing of Thymic Insulin Predispose to Type 1 Diabetes. Journal of Clinical Endocrinology and Metabolism. 2001;86:3705–3710. doi: 10.1210/jcem.86.8.7733. [DOI] [PubMed] [Google Scholar]
- 42.Villaseñor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: Probabilistic, monoallelic, misinitiated. Proceedings of the National Academy of Sciences. 2008;105:15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]