Abstract
Steroid hormone receptors (SHRs) are hormone-activated transcription factors involved in numerous cellular functions and in health and disease. Their activities depend on the cellular level of the receptor, the presence of coregulator proteins and the cell signaling pathways that are active in the cell. SHRs and their coregulators are phosphorylated on multiple sites by a wide variety of kinases. Each site may contribute to multiple functions and the net effect of an individual phosphorylation depends on the activating kinase. Here we discuss functions of known SHR phosphorylation sites, kinase regulation, evidence of translational relevance, and cross-talk between SHRs and cell signaling pathways. Understanding how cell signaling pathways regulate SHRs might yield novel therapeutic targets for multiple human diseases.
Keywords: steroid receptor, phosphorylation, coactivator, cell signaling
Steroid hormone receptors: the basics
The nuclear receptor (NR) superfamily is a large family of transcription factors that includes NRs that bind steroid hormones (SHRs), other ligand-activated transcription factors such as the thyroid hormone receptor (TR) and vitamin D receptor (VDR), and orphan receptors for which endogenous ligands have yet to be identified such as RAR-related orphan receptors (RORs) and chicken ovalbumin upstream transcription factor (COUP-TF). The focus of this review is the SHR subfamily. Members of the SHR subfamily include the estrogen receptor (ER), glucocorticoid receptor (GR), progesterone receptor (PR), androgen receptor (AR) and mineralocorticoid receptor (MR) [2]. ER exists in two isoforms (ERα and ERβ) derived from distinct genes that mediate the action of estrogens in the mammary gland and the reproductive tract, as well as the central nervous, skeletal, and cardiovascular systems [1]. Cortisol and other glucocorticoids activate GR to regulate metabolism as well as the immune, central nervous, digestive, renal, and reproductive systems [2]. There are two main isoforms of PR (PR-A and PR-B) that are derived from the same gene and mediate the actions of progestins in reproduction, mammary gland development, and sexual behavior [2]. Androgens, specifically testosterone and dihydrotestosterone, activate AR to regulate reproduction in both males and females [3]. Aldosterone binds to MR to regulate serum levels of sodium and other electrolytes and to regulate cardiovascular function [2].
Multiple Mechanisms of SHR action
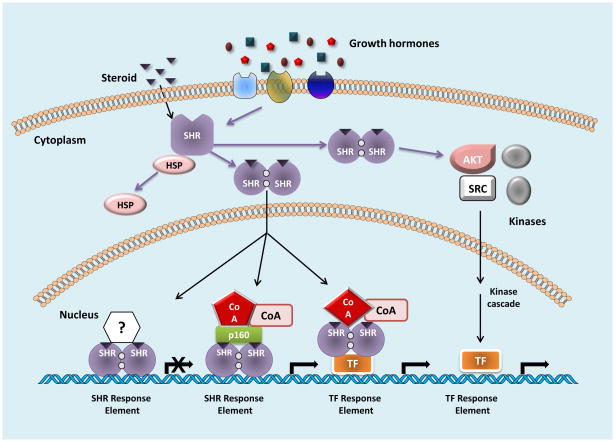
While NRs function as either homo and/or hetero-dimers with other NRs, SHRs function predominantly as homodimers. SHRs can be activated in either a ligand- dependent or ligand-independent manner to regulate transcription (Figure 1). In the former, classical genomic model of SHR action, ligands diffuse across the cell membrane and bind the receptor inducing a conformational change that promotes dissociation of the receptor from chaperone complexes, translocation to the nucleus, and binding of receptor homodimers to hormone response elements (HREs, see figure 1). This binding is followed by dynamic and sequential recruitment of coactivator complexes that modify chromatin to facilitate transcription [4]. All transcription factors depend upon coactivator complexes to regulate transcription; most, if not all, coactivators regulate many classes of transcription factors. A variety of SHR coactivators have been identified; these include the p160 steroid receptor coactivators (SRCs), chromatin remodelers such as the ATP-dependent helicase SMARCA4 (BRG1) and the androgen receptor-interacting protein (ARIP4), the histone modifiers K(lysine) acetyltransferase 2B (PCAF) among others, and the coactivator-associated arginine methyltransferase 1 (CARM1) and protein arginine N-methyltransferase 1 (PRMT1). SHR coactivators also include factors that catalyze post-translational modifications (PTMs) of the receptor itself, other coregulators, and components of the transcriptional machinery, for example protein kinases such as cyclin-dependent kinases (Cdks) and protein kinase B (Akt), regulators of desumoylation such as sentrin-specific protease 1(SENP-1), PRMT1, and regulators of ubiquitination such as E6-AP E3 ubiquitin- protein ligase [4]. SHRs also stimulate transcription through protein-protein interactions with other DNA-bound transcription factors such as activator protein 1 (AP-1), specificity protein 1 (Sp-1) and nuclear factor-kappa B (NFkB), resulting in gene transcription in the absence of direct binding of SHR to the DNA [5] (Figure 1). SHRs are also known to cause transcriptional repression; GR-mediated repression mechanisms are the best characterized and include binding to negative GREs (nGRE) resulting in displacement of other transcription factors (e.g. in the prolactin promoter) [6]. GR also represses transcription through protein-protein interactions with other transcription factors such as AP-1 and NFkB. In the case of the collagenase 3 promoter, GR binds to AP-1 and recruits TIF2/GRIP1/SRC-2 in a conformation that exposes a repressor surface rather than its better known activation function [7]. Finally, SHRs can rapidly activate kinases and resulting downstream cell signaling pathways, for example epidermal growth factor receptor (EGFR), c-Src kinase (Src), and Akt, that mediate biological responses independent of SHR nuclear localization. This form of activation, originally termed non-genomic, is often termed “rapid” and is more accurately described as extranuclear because the common characteristic of this form of signaling is extranuclear (cytoplasmic or membrane-bound) receptor mediated activation of a kinase independent of RNA or protein synthesis. The final downstream event may be altered transcription, but the SHR does not interact with the target gene [8].
Figure 1. Mechanisms of steroid hormone receptor (SHR) action.
In the absence of ligand, SHRs are bound to heat shock protein (HSP) complexes. Upon binding of ligand and/or activation of cell signaling pathways, SHRs dissociate from these complexes, form homodimers, translocate to the nucleus, and bind to SHR response elements. Following this binding, coactivator complexes (CoA) are recruited, facilitating transcription of target genes. Alternatively, SHRs can bind to other transcription factors (TF) sitting on TF response elements, resulting in the recruitment of CoAs and induction of target gene expression. SHRs can also inhibit gene transcription. Finally, SHRs also interact with and activate kinases, such as protein kinase B/Akt (AKT) and c-Src (SRC) resulting in phosphorylation and activation of other TFs bound to TF response elements and altered target gene transcription.
Phosphorylation of SHRs
Identification of phosphorylation sites
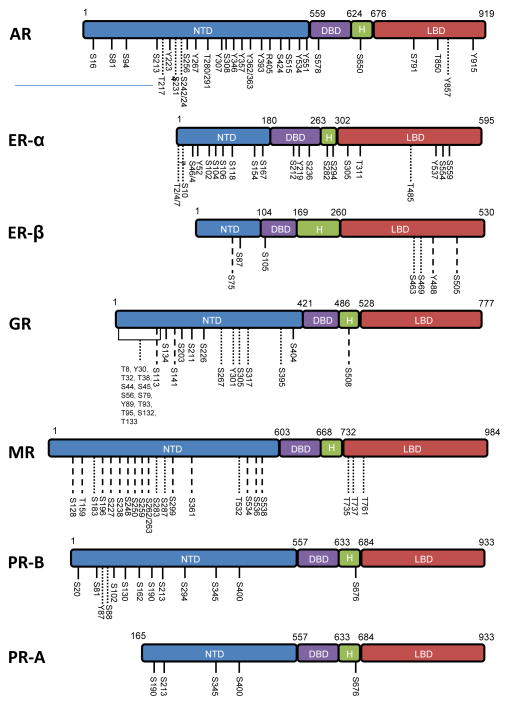
All SHRs contain multiple phosphorylation sites. Many sites have been reported (see Figure 2) and there may be additional sites. Sites initially identified in SHRs were characterized as being phosphorylated under basal conditions or in response to hormone treatment, and investigators relied on the characterization of radiolabeled proteins and site-directed mutagenesis for site identification. Subsequent studies showed that additional sites are phosphorylated in response to activated signaling pathways. Sites also have been identified using mass spectrometry to characterize peptides derived from purified proteins. Others have been identified in general phosphoproteomic screens, suggested by homology to sites in SHRs from other species, or identified as candidate sites from in vitro phosphorylation studies; it is less certain that these sites are phosphorylated in vivo under typical physiological conditions. Sites identified using the various methods and respective references are summarized in Figure 2. Although most sites are not positionally conserved among SHRs, there is one such conserved phosphorylation site in the hinge region of chicken PR (serine (Ser) 530), human PR-B and PR-A (Ser676), human AR (Ser650), and human ERα (Ser294) (Figure 2). Despite this apparent conservation, a common function for this site has not been identified. In chicken PR, the site sensitizes PR to lower concentrations of hormone without changing affinity for progesterone [9], and in human AR phosphorylation of the site promotes nuclear export [10].
Figure 2. Structure and phosphorylation sites of SHRs.
The numbers indicate the amino acids that mark the domain boundaries in the individual receptors between the NTD, DBD=, hinge region (H), and LBD. Solid lines indicate sites that have been determined using radiolabeling, protein sequencing, or mass spectrometry analysis of sites in purified proteins; dotted lines indicate sites identified by phosphoproteomic approaches, and dashed lines indicate sites identified by in vitro phosphorylation or by homology with sites in rodents. References for identification of the sites, with the exception of the MR sites Ser128, Thr159, and Ser250 [80] can be found at www.phosphosite.org. Note that the proposed Ser/Thr sites reported on www.phosphosite.org that have not been detected by at least one of these direct methods are not included in the figure.
AR, androgen receptor; ER, estrogen receptor; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; PR, progesterone receptor. S, serine; T, threonine.
Kinases and SHR phosphorylation
A variety of strategies have been used to identify kinases that phosphorylate SHR. These include in vitro phosphorylation studies and the use of kinase activators and inhibitors in cell culture studies. These studies suggest that SHR are substrates for the mitogen activated protein kinase (MAPK) family (p42/p44 MAPK, p38 MAPK, and c- Jun N-terminal kinases (JNK)), Cdks, casein kinase 2 (Ck2), glycogen synthase kinase 3 (GSK-3) and a number of other serine/threonine kinases. Although SHRs are phosphorylated on tyrosine residues in response to specific signaling pathways, more is known about the effect of site-specific phosphorylation on serine and threonine residues. It should be noted that multiple kinases can phosphorylate the same residue under various conditions (see Table 1) and this likely facilitates the integration of signals from multiple pathways. In addition, kinases can phosphorylate other proteins, so inhibition or activation of a kinase may have effects on SHR function not directly dependent on SHR phosphorylation.
Table 1.
Candidate kinases and functions for some SHR Ser/Thr phosphorylation sites
| SHR | Phosphorylation site | Kinase(s) | Ligand-dependent activity | Ligand-independent activity * | Activity (other) | Physiological relevance | Reference |
|---|---|---|---|---|---|---|---|
| AR | Ser81 | Cdk9 | Gene-specific effect on ligand- dependent transcription | Promotes prostate cancer cell growth | [11] | ||
| Cdk5 | Increased protein stability | Promotes prostate cancer cell growth | [13] | ||||
| Cdk1/Cdk9 | B | DNA binding; Cytoplasmic localization | ND | [12] | |||
| Ser213 | Akt | Decreased protein stability | ND | [14] | |||
| Pim-1 | C | Possible marker of Pim-1 activity in prostate cancer | [15] | ||||
| Pim-1S | C | Decreased protein stability | Promotes prostate cancer growth (low androgen) | [16] | |||
| Pim-1L | Promotes prostate cancer growth (low androgen); Gene-specific effect on ligand- dependent transcription | [16] | |||||
| Ser515 | Cdk7 | A | Decreased protein stability | ND | [65] | ||
| Ser650 | p38 MAPK | Nuclear export | ND | [10] | |||
| JNK | Nuclear export | ND | [10] | ||||
| Thr850 | Pim-1L | A | Increased protein stability | Prostate cancer cell growth (low androgen) | [16] | ||
| ERα | Ser104/106 | Cyclin A- Cdk2 | A | ND | [66] | ||
| MAPK | A | Tamoxifen resistance | [67] | ||||
| Ser118 | ND (hormone- mediated) | A | ND | [68] | |||
| ND (hormone- mediated) | B | ND | [19] | ||||
| ND | Increased protein stability (Pin1) | ND | [69] | ||||
| MAPK | A | ND | [49] | ||||
| MAPK | A | ND | [67] | ||||
| Cdk7 | A | ND | [70] | ||||
| Ikkα | DNA binding; Coregulator recruitment | ND | [71] | ||||
| Ser167 | p90 RSK | A | ND | [72] | |||
| Akt | B | B | Tamoxifen resistance | [73] | |||
| Ikkε | A, B | DNA binding | Tamoxifen resistance | [74] | |||
| Ser282 | Ck2 | C (basal transcription) | ND | [24] | |||
| Ser294 | ND (hormone- mediated) | A | ND | [24] | |||
| Cdk (hormone-mediated) | B | Increased protein stability | ND | [75] | |||
| p38 MAPK | Decreased protein stability (Skp2) | ND | [26] | ||||
| Cdk2 | A | Mediates receptor interaction with Pin1; Regulates phosphorylation of Ser118 and Ser167 | ND | [25] | |||
| Ser305 | Pak1 | A | Regulation of cell proliferation (mouse mammary gland) | ND | [76] | ||
| PKA | Blocks acetylation of K303 | Enhances estrogen sensitivity of (K303R mutant) | [77] | ||||
| Akt | Aromatase inhibitor resistance (K303R mutant) | [78] | |||||
| Ser559 | Ck2 | C, D (basal transcription) | ND | [24] | |||
| ERβ | Ser105 | ND (hormone- mediated) | Associated with good prognosis in breast cancer patients | [79] | |||
| MAPK | A | Decreased breast cancer cell migration and invasion (phospho- mimetic) | ND | [27] | |||
| GR | Ser134 | p38 MAPK | Interaction with 14-3-3-zeta; Modulates promoter selectivity; Gene-specific effect on ligand- dependent transcription | ND | [35] | ||
| Ser203 | ND (basal and hormone- mediated) | Cytoplasmic localization; DNA binding | ND | [31] | |||
| Ser211 | ND (hormone- mediated) | C, D | Receptor conformational change; Enhanced interaction with MED14; Gene- specific effect on transcription | ND | [29] | ||
| p38 MAPK | A | Regulation of apoptosis | ND | [28] | |||
| p38 MAPK | Receptor conformational change; Coregulator interaction | ND | [30] | ||||
| Ser226 | ND (hormone- mediated) | C, D | ND | [29] | |||
| JNK | C | Enhanced nuclear export (ligand- independent) | ND | [32] | |||
| Ser404 | GSK3β | Enhanced nuclear export; Decreased protein stability; Decreased coregulator interaction; Gene-specific effect on transcription | ND | [34] | |||
| MR | Ser128/Thr159/Ser250 | Cdk5 | C | ND | [80] | ||
| PR | Ser81 | Ck2 | B (S79/81) | B (basal transcription; S79/81); DNA binding | ND | [43] | |
| Ser294 | ND (hormone- mediated) | Decreased protein stability (ligand- dependent) | ND | [39] | |||
| MAPK | A, B | ND | [38] | ||||
| MAPK | Nuclear localization (ligand- independent) | ND | [81] | ||||
| Ser345 | MAPK/Src | Interaction with Sp1 (S344/345); Gene-specific effect on ligand- dependent transcription | [42] | ||||
| Ser400 | Cdk2 | A | Nuclear translocation (ligand- independent) | ND | [44] |
Phosphorylation is associated with A = increased expression of a reporter gene; B = increased endogenous gene expression; C = decreased expression of a reporter gene; D = decreased endogenous gene expression.
Ligand-independent activation is defined as activation by a kinase signaling cascade and does not refer to basal activity.
ND, not determined.
Interpreting the function of SHR phosphorylation sites: approaches and caveats
Phosphorylations play roles in many functions. They may be “activating” or “inactivating”, change stability, localization, or protein/protein interactions and serve as signals for additional PTMs that alter protein function. Both direct (site-directed mutagenesis) and indirect (activation/inhibition of signaling pathways) approaches have been used to assess the role of phosphorylation. One striking feature of phosphorylation is that it can integrate signals from multiple pathways. As indicated in Table 1, SHR sites may be substrates for multiple kinases. If kinase A and B both phosphorylate a specific site, but kinase A also activates a protein that interacts with the phosphorylated SHR, the biological outcome may be different from that observed by activation of kinase B. Diverse and sometimes contradictory functions have been reported for SHR phosphorylation sites (for example, see the role of AR Ser213 below). In many cases, these differences may be due to signaling pathway or cell type specific differences. Alternatively, the amino acid substitution in mutagenesis experiments may cause a change in activity independent of the supposed change in phosphorylation. There is no perfect substitute to mimic a non-phosphorylated or phosphorylated amino acid. Substitution of an alanine (Ala) for a phospho-serine or phenylalanine (Phe) for a phospho-tyrosine is the most common approach to eliminate site-specific phosphorylation. Other choices such as glycine (Gly) for serine or Ala for tyrosine (Tyr) typically induce conformational changes that can alter activity independent of the change due to phosphorylation. Glutamic acid (Glu) or aspartic acid (Asp) substitutions can mimic a phosphorylated serine/threonine (Ser/Thr) if the major role of the phosphorylation is to introduce a negative charge. However, phosphorylations that serve as recognition motifs for binding proteins including those that serve as a signal for additional PTMs often cannot be mimicked by substitution of a negatively charged amino acid. A final caveat in interpreting these studies is that mutation of additional nearby amino acids either for convenience in early site-directed mutagenesis strategies or “in case” there is an alternate phosphorylation site generated by the primary substitution may lead to phenotypes that are not strictly a result of elimination of SHR phosphorylation. Selected examples of roles of individual sites are presented below. For a summary of the proposed roles for additional Ser/Thr sites in SHR see Table 1 and the references therein.
Effect of SHR phosphorylation on SHR function
AR phosphorylation
The reported functions of specific AR phosphorylation are kinase-dependent. Phosphorylation of Ser81 is required for induction of endogenous AR target genes [11,12], but not for artificial promoter activation [12]. Cdk9 phosphorylates AR on Ser81 [11] leading to regulation of promoter selectivity, chromatin binding, and cellular distribution of AR [11,12]. Cdk5-dependent stabilization of AR also is dependent on Ser81 phosphorylation [13]. Phosphorylation of AR also plays a role in nuclear localization. Activation of p38 MAPK or JNK results in Ser650 phosphorylation; mutation of this site is associated with reduced nuclear export of AR [10]. The role of Ser213 phosphorylation has been controversial. Akt can phosphorylate this site, but effects of Akt on AR activity in LNCaP cells appear to be cell passage dependent [14]. More recently, two groups have shown that this site is phosphorylated by serine/threonine protein kinase pim-1 (PIM-1) [15,16]. Effects of Ser213 phosphorylation and PIM-1 expression are target gene dependent and PIM-1 isoform dependent. There are two forms of PIM-1. PIM-1S phosphorylates Ser213 leading to recruitment of the E3 ubiquitin-protein ligase Mdm2 and AR degradation. In contrast, PIM-1L phosphorylates Ser213 and Thr850, inducing recruitment of the E3 ubiquitin-protein ligase RNF6, stabilizing and activating AR [16]. Overexpression of PIM-1S inhibits R1881 (synthetic androgen)-dependent reporter activity, and the Ser213Ala mutant is resistant to this repression [15]. In contrast, over-expression of PIM-1L enhances AR-dependent reporter activity in androgen-depleted medium, and mutation of either Thr850 or Ser213 eliminates the stimulation [16]. Depletion of endogenous PIM-1 in VCaP prostate cancer cells enhances androgen-dependent PSA expression, but overexpression of PIM-1S enhances androgen-dependent interleukin 6 (IL6) expression in LNCaP prostate cancer cells [15]. Nuclear Ser213 phosphorylation is higher in castration-resistant prostate cancer compared to hormone-sensitive tumors. Interestingly, cytoplasmic phospho-Ser213 staining in prostate cancer was correlated with shorter time to progression [15].
ER phosphorylation
Phosphorylation has been implicated in many ERα functions. Substitution of Ala for serines 104,106 and 118 reduce transcriptional activity measured using an ER-responsive reporter [17], and reduces coactivation of activation function-1 (AF-1) by p160 and CREB-binding protein (CBP) coactivators [18]. The Ser118Ala mutant transfected into HeLa cells shows somewhat reduced pS2 activation and much decreased cyclin D1 and c-myc induction relative to wild type ER, as well as a different pattern of recruited coactivator complexes at a single time-point [19]. Whether these changes in coactivators are kinetic or a reflection of differential recruitment remains to be determined. There is evidence that phosphorylation of Ser118 in breast cancer tissues correlates with responsiveness to tamoxifen therapy [20]. In contrast, phosphorylation of a site in the ligand-binding domain (LBD), Ser305, mediated by p21- activated kinase 1 (PAK1) leads to tamoxifen resistance [21,22]. Mutation of Ser305 decreases activation of ERα by ~40% in the absence or presence of hormone [23]. Phosphorylation of the hinge site, Ser294, enhances ER activity measured using a reporter [24]. Cdk2-mediated phosphorylation of Ser294 provides a binding site for peptidyl-prolyl isomerase (Pin1) and a resulting increase in Ser118 and Ser167 phosphorylation [25]. In addition, p38 MAPK-mediated phosphorylation of Ser294 stimulates ubiquitination and turnover of ER [26].
Functional studies of human ERβ phosphorylation are limited. In some contexts, a Ser105Ala mutant shows reduced reporter activity relative to wild type, and a Ser105Glu mutant exhibits enhanced reporter activity, as well as the ability to reduce migration of breast cancer cells [27].
GR phosphorylation
Although a wide variety of candidate sites in human GR have been identified directly, or suggested by phosphoproteomics or homology, the functions of relatively few sites have been examined. Ser211 in the amino-terminal domain (NTD) of GR is the best-characterized phosphorylation site; mutation to Ala decreases transcriptional activity by 25–70% measured using a GR-responsive reporter [28] or endogenous target gene expression [29], reduces its ability to induce GR-dependent apoptosis [28], and alters the conformation of the amino terminus of GR [30]. In contrast, phosphorylation of either Ser203 or Ser226 typically reduces GR activity. Ser203 phosphorylated GR is predominantly cytoplasmic and is not recruited to GR-responsive promoters as assessed by chromatin immunoprecipitation (ChIP) assays [31]. Ser226 is phosphorylated by JNK [32]; mutation to Ala increases GR transcriptional activity [29,33]. Phosphorylation of Ser404 alters target gene dependent gene regulation with both loss and gain of function. Elimination of the phosphorylation site enhances GR-dependent repression of a variety of genes [34]. In addition, phosphorylation at this site reduces GR stability, and may enhance nuclear export [34]. Cell stresses such as starvation or oxidative stress induce p38 MAPK-dependent phosphorylation of Ser134. This phosphorylation mediates GR interaction with 14-3-3 zeta, a protein associated with oxidative stress, binding to select gene promoters and alters the GR-mediated target gene profile [35]. Aberrant GR phosphorylation has been proposed to play a role in disease. For example some glucocorticoid-resistant asthma patients become responsive when p38 MAPK inhibitors are given with reduced Ser226 phosphorylation as one of the results of kinase inhibition [36]. In women, the ratio of nuclear phospho-Ser211/phospho-Ser226 measured in peripheral blood mononucleocytes (PMBCs) is inversely correlated with depression [37].
PR phosphorylation
Functions for several of the sites in PR have been reported. Substitution of Ala for Ser294 in the NTD decreases PR transcriptional activity by 50–90% and in a target gene specific manner [38], increases protein stability [39] and enhances PR sumoylation at K388 [40]. Interestingly, although this amino acid is common to both PR-B and PR-A, only the longer PR-B form is efficiently phosphorylated on this site [41]. Phosphorylation of Ser345 promotes association of PR with Sp1 in target genes that lack canonical progesterone-responsive elements; PR with Ala substitutions at both Ser 344 and Ser345 are not recruited to Sp1 binding sites in the p21 promoter [42]. A S79/81A double mutant shows no defect in activity when measured by using a PR-responsive reporter, but shows gene- specific changes in activation of target genes. For example, the levels of baculoviral IAP repeat-containing protein 3 (BIRC3) are increased when PR is expressed in PR negative cells, and expression is increased further with R5020, a synthetic progestin and a PR agonist. In contrast, both basal and hormone- induced BIRC3 levels are reduced in the S79/81A line [43]. Basal, but not R5020-dependent soft agar colony formation was reduced in the S79/81A clone relative to the wild type clone. The difference in basal activity both for target genes and colony formation is somewhat surprising since the level of Ser81 phosphorylation is quite low in the absence of hormone [43]. Ser400 is a substrate for Cdk2 and expression of constitutively active Cdk2 enhances PR activity in the absence or presence of hormone. Interestingly, a Ser400Ala mutant fails to show the basal increase in activity, although its response to R5020 in the presence of Cdk2 is not compromised [44].
Cross-talk between cell signaling pathways and SHRs
Although it is well established that cell signaling pathways modify SHR activity as summarized above, there is evidence that under some conditions, activated cell signaling pathways can eliminate the requirement for hormone. Conversely, SHRs can activate kinases independent of classical upstream growth factor receptors. Finally, there is increasing evidence for receptor-mediated recruitment of kinases to chromatin, facilitating local phosphorylation of factors that regulate transcription.
Transcriptional activation of SHRs in steroid depleted medium
Some of the most striking evidence for cross-talk between cell signaling pathways and SHRs is the finding that some SHRs are activated in the absence of their cognate ligands by activators of cell signaling pathways such as pharmacologic activators of cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (PKA), growth factor signaling (e.g. EGF, human epidermal growth factor 2 (HER2), insulin-like growth factor (IGF)), and cytokines (IL-6). Changes in SHR or coactivator phosphorylation have been identified in some cases, while in others the targets have not been identified. O’Malley’s group first reported that 8-Bromo cAMP, a PKA activator, induced chicken PR-mediated transcription of a PR-responsive reporter, in the absence of progesterone [45]. Human PR is not activated under the same treatment conditions, but PKA signaling does cause the partial PR antagonist, RU486, to act as an agonist [46]. In some contexts, constitutively active Cdk2 stimulates basal PR activity [44] and heregulin can activate PR [47]. Human ERα responds to multiple activators of cell signaling pathways including growth factors [48]. The mechanisms have not been fully elucidated. For example, substitution of Ala for Ser118 eliminates EGF-dependent, but not hormone-dependent activation of ER in some cell types [49]. Substitution of a Glu for the Ser restores EGF-dependent activation, showing that a negative charge at Ser118 is required, but not sufficient for EGF-dependent activation. AR also is responsive to multiple signaling pathways. Activation of AR through PKA signaling [50], IL-6 [51], and growth factors have all been reported. Phosphorylation of SRC-1, a known AR coactivator, has been implicated as one of the factors in IL-6-dependent activation of AR [51].
Activation of kinases by SHR
Activation of kinases by SHR-mediated extranuclear signaling is important in many tissues. For example, rapid signaling of ER is important for bone biology, the central nervous system, and the cardiovascular system. In endothelial cells, estradiol stimulates production of nitric oxide through the rapid activation of the small G protein subunit, Gαi, by ERα, leading to activation of Erk and phosphoinositide 3-kinase (PI3K) [52,53]. This was confirmed by studies utilizing estrogen dendrimer conjugates (EDCs), bulky synthetic ligands that are excluded from the nucleus and activate only extranuclear pathways [54], and ERα mutants that cannot interact with Gαi [55]. AR also has been implicated in extranuclear signaling through Gβγ, cAMP, MAPK, and EGFR pathways to affect oocyte maturation, spermatogenesis, bone turnover, and proliferation/survival of prostate cancer cells (reviewed in [56]). Human PR mediates extranuclear signaling in breast cancer cells through direct interactions with Src or through release of EGF ligands that activate EGFR [57,58]. Extranuclear signaling of GR is important for neural progenitor cell function [59], as well as pituitary, hippocampal, cardiovascular, and immune function (reviewed in [60]). Finally, extranuclear signaling effects of aldosterone in vascular and kidney cells may be mediated, in part, by MR (reviewed in [60]). Although activation of kinases is independent of nuclear actions of SHRs, the two forms of signaling often are integrated to mediate SHR function in target tissues.
SHR-dependent recruitment of kinases to chromatin
SHR also facilitates the phosphorylation of substrates by kinases by recruiting the kinases to chromatin. For example, PR recruits cyclin A/Cdk2 to PR-responsive promoters [61] resulting in Cdk2-dependent phosphorylation of poly (ADP-ribose) polymerase 1 (PARP-1) and subsequent displacement of histone H1 [62]. PR also recruits MAPK and mitogen- and stress-activated protein kinase 1 (MSK1) [63] as well as Ck2 [43] to promoters regulated by PR. Recruitment of kinases by SHR is not restricted to PR. ER also recruits MAPK [64] indicating that SHR-dependent recruitment of kinases to chromatin may be a common feature of SHR action.
Concluding remarks and future perspectives
SHR signaling is complex and numerous cell signaling pathways can impact SHR function. Site-specific phosphorylation modulates both ligand-dependent and ligand-independent SHR activity through the regulation of overall transcriptional activity, protein stability, cellular localization, chromatin binding and interaction with coregulators. Phosphorylation of coregulators and other DNA-binding proteins further fine-tunes SHR transcriptional activity. Thus, phosphorylation, along with other PTMs, defines a “code” that ultimately impacts biological response of SHRs under various physiological conditions. As noted above, a few studies have demonstrated the physiological relevance and/or correlation of these modifications to human disease. Studies have begun to elucidate the importance of site-specific phosphorylation but much is still unknown. Identification of phosphorylation sites in the receptors and their interacting proteins is incomplete. New, more sensitive technologies will aid in filling the gaps. Receptor phosphorylation clearly is not an on/off switch for global activity. Rather, it modulates several functions of the receptor; specific phosphorylations likely are critical for sub-sets of genes or for tissue-specific actions. Additional studies using methods that measure global gene regulation and receptor binding to chromatin followed by mechanistic studies of specific target genes are needed. Microarrays have been employed with some success [34], but global RNA-Seq and ChIP-Seq experiments using phosphorylation-specific antibodies have not been done. Phosphorylation- specific antibodies are extremely useful tools, but also have limitations particularly in immunohistochemical analyses. Many of the antibodies distinguish well between phosphorylated and unphosphorylated receptors, but may cross react with other proteins. Techniques such as proximity ligation assays (PLAs), which give a positive signal only when two antibodies (e.g. SHR and phospho-specific) are in close proximity may overcome these limitations for immunohistochemical studies. A second limitation is the inability to distinguish between an occluded epitope and failure to recruit a protein in ChIP assays. Antibodies specific to the dephosphorylated epitope would permit the investigator to distinguish between differential recruitment of the phosphorylated and dephosphorylated protein and epitope occlusion. To address the role of phosphorylation in vivo, approaches such as site-specific elimination of phosphorylation in mice are needed. In some cases, phosphorylation acts as a signal for subsequent modifications that alter function. Although there is compelling evidence that receptors are regulated by other types of PTMs, the identity, function, and regulation of these modifications is incomplete. Thus, there also is a need to identify and characterize other PTMs. This knowledge will not only yield insight into the mechanisms of cross-talk between SHR signaling and cell signaling pathways, but might also help develop novel therapeutic strategies for multiple human diseases.
TEXT BOX 1. Structure of SHRs.
Almost all NRs share a common structure comprised of a highly variable amino-terminal domain (NTD), DNA-binding domain (DBD), a flexible hinge domain, and a ligand-binding domain (LBD) responsible for binding of specific ligands (Figure 2). An additional region important for ligand-dependent receptor transcriptional activity exists within the LBD; it is termed activation function -2 (AF-2). This modular structure facilitates allosteric regulation of receptor activity [82].
In the absence of hormone, SHRs are bound to heat shock proteins and immunophilins in a heterocomplex (Figure 1). Some SHRs are predominantly cytoplasmic (GR and AR), while others are predominantly nuclear (ER and PR), but all can shuttle between the two compartments. Ligand binding induces conformational changes that alter receptor-DNA and receptor-protein interactions, ultimately regulating receptor activity. The LBD also contains sequences that serve as an interface for dimerization, particularly for ER and GR (reviewed in [82]). Ligand-induced conformational changes also expose the nuclear localization signal (NLS) that resides in the hinge region, leading to translocation of the receptor-steroid complex into the nucleus, binding to DNA, and transactivation of target genes. The DBD, which contains two zinc finger motifs, binds to hormone response elements (HREs) on the DNA that partly dictate cell or promoter-selective SHR function. This binding can also influence which coregulatory complexes are recruited by the receptor. Compared to the LBD and DBD, there is little homology among the NTDs of SHRs. The NTD contains an activation function region (AF-1) that is ligand-independent.
Intrinsically disordered SHR regions
Unlike the LBD and the DBD, the hinge and NTD are intrinsically disordered (ID) regions that are capable of undergoing disorder-to-order transitions under specific physiological conditions (reviewed in [82]). This conformational flexibility may create large interaction surfaces that allow specific, but reversible interactions with target molecules [82]. Disorder-to-order transitions may be mediated through post-translational modifications (PTMs) of the receptor, including phosphorylation [30,83]. In fact, ID regions have a higher frequency of known phosphorylation sites and there is a strong correlation between known phosphorylation residues within AR-, ERα-, GR-, and PR-NTD and predicted intrinsic disorder [82]. Pin1, an enzyme that binds to proteins and plays a role as a post phosphorylation control in regulating protein function, recognizes phosphorylated serine/threonine-proline (Ser/Thr-Pro) motifs, and induces a conformational change in target proteins. Pin1 interactions with AR phosphorylated at Ser81 [84] and ERα phosphorylated at Ser118 [85] are important for transcriptional activity of the receptors.
Highlights.
Multiple kinases phosphorylate steroid hormone receptors and their coregulators
Site-specific phosphorylation modulates selective actions of receptors
Activation of cell signaling pathways impacts biological response of receptors
Regulation of receptor function by cell signaling is relevant to human disease
Acknowledgments
This study was supported by CA57539 (NLW) and F32GM103080 (LST).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 2.Lu NZ, et al. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev. 2006;58:782–797. doi: 10.1124/pr.58.4.9. [DOI] [PubMed] [Google Scholar]
- 3.Walters KA, et al. Molecular insights into androgen actions in male and female reproductive function from androgen receptor knockout models. Hum Reprod Update. 2010;16:543–558. doi: 10.1093/humupd/dmq003. [DOI] [PubMed] [Google Scholar]
- 4.Bulynko YA, O’Malley BW. Nuclear receptor coactivators: structural and functional biochemistry. Biochem. 2011;50:313–328. doi: 10.1021/bi101762x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biddie SC, et al. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol Metab. 2010;21:3–9. doi: 10.1016/j.tem.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos GM, et al. Negative regulation by nuclear receptors: a plethora of mechanisms. Trends Endocrinol Metab. 2011;22:87–93. doi: 10.1016/j.tem.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogatsky I, et al. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammes SR, Levin ER. Recent advances in extranuclear steroid receptor actions. Endocrinol. 2011;152:4489–4495. doi: 10.1210/en.2011-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai W, et al. Phosphorylation of Ser530 facilitates hormone-dependent transcriptional activation of the chicken progesterone receptor. Mol Endocrinol. 1994;8:1465–1473. doi: 10.1210/mend.8.11.7877616. [DOI] [PubMed] [Google Scholar]
- 10.Gioeli D, et al. Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol Endocrinol. 2006;20:503–515. doi: 10.1210/me.2005-0351. [DOI] [PubMed] [Google Scholar]
- 11.Gordon V, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol. 2010;24:2267–2280. doi: 10.1210/me.2010-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, et al. Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. J Biol Chem. 2012;287:8571–8583. doi: 10.1074/jbc.M111.325290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu FN, et al. Regulation of androgen receptor and prostate cancer growth by cyclin-dependent kinase 5. J Biol Chem. 2011;286:33141–33149. doi: 10.1074/jbc.M111.252080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin HK, et al. Suppression versus induction of androgen receptor functions by the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer LNCaP cells with different passage numbers. J Biol Chem. 2003;278:50902–50907. doi: 10.1074/jbc.M300676200. [DOI] [PubMed] [Google Scholar]
- 15.Ha S, et al. Phosphorylation of the androgen receptor by PIM1 in hormone refractory prostate cancer. Oncogene. 2012 doi: 10.1038/onc.2012.412. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linn DE, et al. Differential regulation of androgen receptor by PIM-1 kinases via phorphorylation-dependent recruitment of distinct ubiquitin E3 ligases. J Biol Chem. 2012;287:22959–22968. doi: 10.1074/jbc.M111.338350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeGoff P, et al. Phosphorylation of the human estrogen receptor. Identification of hormone-regulated sites and examination of their influence on transcriptional activity. J Biol Chem. 1994;269:4458–4466. [PubMed] [Google Scholar]
- 18.Dutertre M, Smith CL. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-α: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol Endocrinol. 2003;17:1296–1314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- 19.Duplessis TT, et al. Phosphorylation of estrogen receptor α at serine 118 directs recruitment of promoter complexes and gene-specific transcription. Endocrinol. 2011;152:2517–2526. doi: 10.1210/en.2010-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy LC, et al. Phospho-serine-118 estrogen receptor-alpha expression is associated with better disease outcome in women treated with tamoxifen. Clin Cancer Res. 2004;10:5902–5906. doi: 10.1158/1078-0432.CCR-04-0191. [DOI] [PubMed] [Google Scholar]
- 21.Rayala SK, et al. P21-activated kinase 1 regulation of estrogen receptor-alpha activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Res. 2006;66:1694–1701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- 22.Holm C, et al. Phosphorylation of the oestrogen receptor alpha at serine 305 and prediction of tamoxifen resistance in breast cancer. J Pathol. 2009;217:372–379. doi: 10.1002/path.2455. [DOI] [PubMed] [Google Scholar]
- 23.Tharakan R, et al. Phosphorylation of estrogen receptor alpha, serine residue 305 enhances activity. Mol Cell Endocrinol. 2008;295:70–78. doi: 10.1016/j.mce.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Williams CC, et al. Identification of four novel phosphorylation sites in estrogen receptor alpha: impact on receptor-dependent gene expression and phosphorylation by protein kinase CK2. BMC Biochem. 2009;10:36. doi: 10.1186/1471-2091-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucchetti C, et al. The prolyl isomerase Pin1 acts synergistically with CDK2 to regulate the basal activity of estrogen receptor α in breast cancer. PLoS ONE. 2013;8:e55355. doi: 10.1371/journal.pone.0055355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatt S, et al. Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor α turnover and functional activity via the SCFSkp2 proteasomal complex. Mol Cell Biol. 2012;32:1928–1943. doi: 10.1128/MCB.06561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam HM, et al. Phosphorylation of human estrogen receptor-beta at serine 105 inhibits breast cancer cell migration and invasion. Mol Cell Endocrinol. 2012;358:27–35. doi: 10.1016/j.mce.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller AL, et al. p38 mitogen-activated protein kinase (MAPK) is a key mediator in glucocorticoid-induced apoptosis of lymphoid cells: correlation between p38 MAPK activation and site-specific phosphorylation of the human glucocorticoid receptor at serine 211. Mol Endocrinol. 2005;19:1569–1583. doi: 10.1210/me.2004-0528. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, et al. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol Endocrinol. 2008;22:1754–1766. doi: 10.1210/me.2007-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garza AM, et al. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol. 2010;30:220–230. doi: 10.1128/MCB.00552-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blind RD, Garabedian MJ. Differential recruitment of glucocorticoid receptor phospho-isoforms to glucocorticoid-induced genes. J Steroid Biochem Mol Biol. 2008;109:150–157. doi: 10.1016/j.jsbmb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh M, et al. Nuclear export of glucocorticoid receptor is enhanced by c-Jun N-terminal kinase-mediated phosphorylation. Mol Endocrinol. 2002;16:2382–2392. doi: 10.1210/me.2002-0144. [DOI] [PubMed] [Google Scholar]
- 33.Rogatsky I, et al. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc Natl Acad Sci USA. 1998;95:2050–2055. doi: 10.1073/pnas.95.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galliher-Beckley AJ, et al. Glycogen synthase kinase 3beta-mediated serine phosphorylation of the human glucocorticoid receptor redirects gene expression profiles. Mol Cell Biol. 2008;28:7309–7322. doi: 10.1128/MCB.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galliher-Beckley AJ, et al. Ligand-independent phosphorylation of the glucocorticoid receptor integrates cellular stress pathways with nuclear receptor signaling. Mol Cell Biol. 2011;31:4663–4675. doi: 10.1128/MCB.05866-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercado N, et al. Restoration of corticosteroid sensitivity by p38 mitogen activated protein kinase inhibition in peripheral blood mononuclear cells from severe asthma. PLoS ONE. 2012;7:e41582. doi: 10.1371/journal.pone.0041582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simic I, et al. A preliminary evaluation of leukocyte phospho-glucocorticoid receptor as a potential biomarker of depressogenic vulnerability in healthy adults. Psychiatry Res. 2013 doi: 10.1016/j.psychres.2013.02.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Shen T, et al. Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol Cell Biol. 2001;21:6122–6131. doi: 10.1128/MCB.21.18.6122-6131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange CA, et al. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc Natl Acad Sci USA. 2000;97:1032–1037. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel AR, et al. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol Endocrinol. 2007;21:2890–2906. doi: 10.1210/me.2007-0248. [DOI] [PubMed] [Google Scholar]
- 41.Clemm DL, et al. Differential hormone-dependent phosphorylation of progesterone receptor A and B forms revealed by a phosphoserine site specific monoclonal antibody. Mol Endocrinol. 2000;14:52–65. doi: 10.1210/mend.14.1.0413. [DOI] [PubMed] [Google Scholar]
- 42.Faivre EJ, et al. Progesterone receptor rapid signaling mediates serine 345 phosphorylation and tethering to specificity protein 1 transcription factors. Mol Endocrinol. 2008;22:823–837. doi: 10.1210/me.2007-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagan CR, et al. ck2-dependent phosphorylation of progesterone receptors (PR) on Ser81 regulates PR-B isoform-specific target gene expression in breast cancer cells. Mol Cell Biol. 2011;31:2439–2452. doi: 10.1128/MCB.01246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierson-Mullany LK, Lange CA. Phosphorylation of progesterone receptor serine 400 mediates ligand-independent transcriptional activity in response to activation of cyclin-dependent protein kinase 2. Mol Cell Biol. 2004;24:10542–10557. doi: 10.1128/MCB.24.24.10542-10557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denner LA, et al. Regulation of progesterone receptor-mediated transcription by phosphorylation. Science. 1990;250:1740–1743. doi: 10.1126/science.2176746. [DOI] [PubMed] [Google Scholar]
- 46.Beck CA, et al. The progesterone antagonist RU486 acquires agonist activity upon stimulation of cAMP signaling pathways. Proc Natl Acad Sci USA. 1993;90:4441–4445. doi: 10.1073/pnas.90.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labriola L, et al. Heregulin induces transcriptional activation of the progesterone receptor by a mechanism that requires functional ErbB-2 and mitogen-activated protein kinase activation in breast cancer cells. Mol Cell Biol. 2003;23:1095–1111. doi: 10.1128/MCB.23.3.1095-1111.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ignar-Trowbridge DM, et al. Peptide growth factors elicit estrogen receptor-dependent transcriptional activation of an estrogen-responsive element. Mol Endocrinol. 1993;7:992–998. doi: 10.1210/mend.7.8.8232319. [DOI] [PubMed] [Google Scholar]
- 49.Bunone G, et al. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 50.Nazareth LV, Weigel NL. Activation of the human androgen receptor through a protein kinase A signaling pathway. J Biol Chem. 1996;271:19900–19907. doi: 10.1074/jbc.271.33.19900. [DOI] [PubMed] [Google Scholar]
- 51.Ueda T, et al. Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J Biol Chem. 2002;277:38087–38094. doi: 10.1074/jbc.M203313200. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z, et al. Estrogen receptor alpha mediates the nongenomic activation of endothelial nitric oxide synthase by estrogen. J Clin Invest. 1999;103:401–406. doi: 10.1172/JCI5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar P, et al. Direct interactions with G αi and G βγ mediate nongenomic signaling by estrogen receptor α. Mol Endocrinol. 2007;21:1370–1380. doi: 10.1210/me.2006-0360. [DOI] [PubMed] [Google Scholar]
- 54.Chambliss KL, et al. Non-nuclear estrogen receptor alpha signaling promotes cardiovascular protection but not uterine or breast cancer growth in mice. J Clin Invest. 2010;120:2319–2330. doi: 10.1172/JCI38291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Q, et al. Point mutations in the ERα Gai binding domain segregate nonnuclear from nuclear receptor function. Mol Endocrinol. 2013;27:2–11. doi: 10.1210/me.2011-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sen A, et al. Understanding extranuclear (nongenomic) androgen signaling: what a frog oocyte can tell us about human biology. Steroids. 2011;76:822–828. doi: 10.1016/j.steroids.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boonyaratanakornkit V, et al. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol Cell. 2001;8:269–280. doi: 10.1016/s1097-2765(01)00304-5. [DOI] [PubMed] [Google Scholar]
- 58.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol Cell Biol. 2007;27:466–480. doi: 10.1128/MCB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samarasinghe RA, et al. Cooperativity and complementarity: synergies in non-classical adn classical glucocorticoid signaling. Cell Cycle. 2012;11:2819–2827. doi: 10.4161/cc.21018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammes SR, Levin ER. Extranuclear steroid receptors: nature and actions. Endocr Rev. 2007;28:726–741. doi: 10.1210/er.2007-0022. [DOI] [PubMed] [Google Scholar]
- 61.Narayanan R, et al. Cyclin-dependent kinase activity is required for progesterone receptor function: novel role for cyclin A/Cdk2 as a progesterone receptor coactivator. Mol Cell Biol. 2005;25:264–277. doi: 10.1128/MCB.25.1.264-277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright RH, et al. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012;26:1972–1983. doi: 10.1101/gad.193193.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vicent GP, et al. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 64.Madak-Erdogan Z, et al. Genomic collaboration of estrogen receptor α and extracellular signal-regulated kinase 2 in regulating gene and proliferation programs. Mol Cell Biol. 2011;31:226–236. doi: 10.1128/MCB.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chymkowitch P, et al. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO. 2011;30:468–479. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rogatsky I, et al. Potentiation of human estrogen receptor α transcriptional activation through phosphorylation of serines 104 and 106 by the cyclin A-CDK2 complex. J Biol Chem. 1999;274:22296–22302. doi: 10.1074/jbc.274.32.22296. [DOI] [PubMed] [Google Scholar]
- 67.Thomas RS, et al. Phosphorylation at serines 104 and 106 by Erk1/2 MAPK is important for estrogen receptor-alpha activity. J Mol Endocrinol. 2008;40:173–184. doi: 10.1677/JME-07-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ali S, et al. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12:1153–1160. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajbhandari P, et al. Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene. 2013 doi: 10.1038/onc.2013.78. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen D, et al. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell. 2000;6:127–137. [PubMed] [Google Scholar]
- 71.Park KJ, et al. Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol Cel. 2005;18:71–82. doi: 10.1016/j.molcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 72.Joel PB, et al. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of ser-167. Mol Cell Biol. 1998;18:1978–1984. doi: 10.1128/mcb.18.4.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell RA, et al. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- 74.Guo JP, et al. IKKepsilon phosphorylation of estrogen receptor a Ser-167 and contribution to tamoxifen resistance in breast cancer. J Biol Chem. 2010;285:3676–3684. doi: 10.1074/jbc.M109.078212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Held JM, et al. Ligand binding promotes CDK-dependent phosphorylation of ER-alpha on hinge serine 294 but inhibits ligand-independent phosphorylation of serine 305. Mol Cancer Res. 2012;10:1120–1132. doi: 10.1158/1541-7786.MCR-12-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang RA, et al. p21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. EMBO J. 2002;20:5437–5447. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui Y, et al. Phosphorylation of estrogen receptor alpha blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004;64:9199–9208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- 78.Barone I, et al. Phosphorylation of the mutant K303R estrogen receptor alpha at serine 305 affects aromatase inhibitor sensitivity. Oncogene. 2010;29:2404–2414. doi: 10.1038/onc.2009.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamilton-Burke W, et al. Phosphorylation of estrogen receptor beta at serine 105 is associated with good prognosis in breast cancer. Am J Pathol. 2010;177:1079–1086. doi: 10.2353/ajpath.2010.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kino T, et al. Cyclin-dependent kinase 5 modulates the transcriptional activity of the mineralocorticoid receptor and regulates expression of brain-derived neurotrophic factor. Mol Endocrinol. 2010;24:941–952. doi: 10.1210/me.2009-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu M, et al. Mitogen activated protein kinase regulates nuclear association of human progesterone receptors. Mol Endocrinol. 2003;17:628–642. doi: 10.1210/me.2002-0378. [DOI] [PubMed] [Google Scholar]
- 82.Kumar R, McEwan IJ. Allosteric modulators of steroid hormone receptors: structural dynamics and gene regulation. Endocr Rev. 2012;33:271–299. doi: 10.1210/er.2011-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunker AK. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 84.La Montagna R, et al. Androgen receptor serine 81 mediates Pin1 interaction and activity. Cell Cycle. 2012;11:3415–3420. doi: 10.4161/cc.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rajbhandari P, et al. Regulation of estrogen receptor α N-terminus conformation and function by peptidyl prolyl isomerase Pin1. Mol Cell Biol. 2012;32:445–457. doi: 10.1128/MCB.06073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]