Abstract
Recent seminal studies have rapidly advanced the understanding of intestinal epithelial stem cell (IESC) biology in murine models. However, the lack of techniques suitable for isolation and subsequent downstream analysis of IESCs from human tissue has hindered the application of these findings toward the development of novel diagnostics and therapies with direct clinical relevance. This study demonstrates that the cluster of differentiation genes CD24 and CD44 are differentially expressed across LGR5 positive “active” stem cells as well as HOPX positive “facultative” stem cells. Fluorescence-activated cell sorting enables differential enrichment of LGR5 cells (CD24−/CD44+) and HOPX (CD24+/CD44+) cells for gene expression analysis and culture. These findings provide the fundamental methodology and basic cell surface signature necessary for isolating and studying intestinal stem cell populations in human physiology and disease.
Introduction
Lgr5 was the first validated IESC biomarker shown to be expressed in actively cycling mouse crypt base columnar cells (CBCs) 1. Subsequent studies demonstrated a secondary, “reserve” population of mouse IESCs marked by Bmi1, Hopx, mTert, and Lrig 2–5. Emerging evidence indicates overlapping expression of Lgr5 with these “reserve” IESC biomarkers; however, Lgr5-negative cell populations have also been shown to dedifferentiate in response to damage, suggesting the existence of one or more functionally competent ‘facultative’ IESC populations 6–8. Despite these advances in IESC biomarker discovery, FACS isolation and functional characterization of putative ‘“active” and “reserve/facultative” IESC populations from human intestinal tissue has been limited by the lack of validated human IESC biomarkers and in vitro assays to functionally test stemness at the single cell level.
Investigators in other stem cell fields have utilized FACS-based approaches, which rely on multiple cell surface antigens, to isolate target stem cell populations of varying purity. Notably, biomarkers comprised of cluster-of-differentiation (CD) genes have long been used to identify hematopoietic stem cells and their progenitors 9. We recently adopted a similar strategy to demonstrate that low levels of CD24 facilitate FACS of Sox9Low/Lgr5+ murine IESCs capable of forming enteroids in vitro 10. Similarly, CD44 is expressed in the stem cell zone of the murine small intestine and can be used to enrich for Lgr5+ CBCs (Magness et al, unpublished). In this study we explored whether CD24 and CD44 could be used to FACS-isolate human IESCs.
Methods
Patients/Tissue collection and preparation
De-identified tissue from female patients ranging between 33–53 yrs of age with body mass indices of 39–60 kg/m2 was used in this study. Tissue was obtained from laparoscopic roux-en-y gastric bypass surgery and represents jejunal segments of approximately 4 cm in length. Following resection, tissue was placed in a specimen cup on ice until a mucosectomy was performed, aided by injecting ice-cold saline between the mucosa and submucosa prior to careful dissection. Single cell dissociation was carried out on a small portion of the total mucosa (1 cm × 1 cm) for gene expression studies and a larger tissue area (4cm × 4cm) was dissociated for culture experiments. For an informative comparison, the mass of mucosa used for this preparation represents approximately 300- and 1200-times the mucosal mass of an average biopsy from endoscopy or colonoscopy at UNC (13 mg/biopsy; unpublished, Drs. Tope Keku/Robert Sandler), respectively. Following dissection, mucosa was placed in 3 mM EDTA in 1x PBS for 45 min at 4°C on a rocker to remove villi. The villus fraction was discarded (Supplemental Figure 1A) and the remaining mucosa was then transferred into 5 mL of PBS and lightly shaken by hand (approximately 1 shake/sec for 2 min) to remove the remaining epithelium (Supplemental Figure 1B). An equal volume of 2% Sorbitol made in 1x PBS (Sigma, St. Louis, MO) was added. To further deplete the solution of contaminating villi, the epithelial solution was passed through a 70μm filter. This procedure results in a ‘crypt-enriched’ epithelial preparation (Supplemental Figure 1C). The crypts were pelleted at 150x g for 10 min at 4°C. Crypts were then digested to single cells by resuspending the pellet in 5 mL of HBSS containing 0.3 U/mL of dispase (Worthington Biochemical, Lakewood, NJ) followed by incubation at 37°C for 10 min. The crypt solution was then manually shaken for 30 sec (3–4 shakes/sec). The solution was then checked for extent of dissociation to single cells. If cell clumps remained, shaking cycle was repeated every 5 minutes, then checked for extent of dissociation. Shaking cycles were stopped at the earliest time point at which 80–90% of crypts were dissociated to completion or up to 30 min maximum. An average of 1 × 107 cells was obtained from a 1 cm × 1 cm mucosal segment. Single cells were filtered using a 40μm filter to remove undissociated clumps. For FACS, 1 × 107 cells were placed in 500μL of IESC staining media (see Flow cytometry section, below) for antibody staining. Human tissue used in this study was deemed exempt from full Institutional Review Board review (approval #09-2159).
Immunostaining
A 3 cm square piece of jejunum from each case was fixed with freshly made 4% paraformaldehyde (PFA) for a 24–48h at 4°C. The tissues were then prepared for cryosectioning by immersion in 30% sucrose for at least 24h at 4°C. Tissues were embedded in Tissue-Tek optimal cutting temperature (OCT) medium (Sakura, Torrance, CA) and frozen on dry ice. 8–10μm sections were cut on a cryostat and placed on positively charged microscope slides. Prior to immunostaining, tissue sections were rinsed twice in PBS to remove OCT. Non-specific binding was blocked by applying Dako Protein Block (Dako, Carpinteria, CA, X0909) to tissue sections for 30 min at room temperature. Primary antibodies were applied in Dako Antibody Diluent (Dako, S0809) and incubated for 2h at room temperature. Dilutions were as follows: CD326/EpCAM (1:250, clone 9C4, BioLegend, San Diego, CA), CD44 (1:250, clone IM7, BioLegend), CD24 (1:100, clone ML5, Biolegend), Lysozyme (1:500, Diagnostic Biosystems, Pleasanton, CA), Mucin2 (1:100, Santa Cruz Biotechnology, Santa Cruz, CA), Sucrase Isomaltase A-17 (1:100, Santa Cruz), ChromograninA (1:500, Immunostar, Hudson, WI). Anti-Rabbit-Cy3 (1:500 Sigma, St. Louis, MO, C2306) and anti-Rat-Cy3 (1:500 Jackson Immunoresearch, Carlsbad, CA, 112-165-003) secondary detection antibodies were diluted in Dako Antibody Diluent and applied to tissue for 30 min at room temperature. Nuclei were stained for 10 minutes with bisbenzamide (1:1,000, Sigma) diluted in PBS. Background staining was negligible as determined by nonspecific IgG staining. Images were collected by capturing ~1 μm optical sections using a Zeiss LSM 710 confocal microscope.
Flow Cytometry/FACS
Cells were stained for 90 min on ice in IESC Staining Media [Advanced DMEM/F12 (Gibco), N2 (Gibco), B27 (Gibco), Glutamax (Gibco), Penicillin/Streptomycin (Gibco), 10μM Y27632 (Selleck Chemicals, Houston, TX), 500mM N-acetyl-cysteine (Sigma), and 10% FBS (Gemini Biosciences)] with the following antibodies or isotype controls: AlexaFluor 647 (Alexa647) conjugated anti-CD24 [1:100] (clone ML5, Biolegend #311109, San Diego, CA), Pycoerythrin-Cy7 (PE-Cy7) conjugated anti-CD45 [1.6:100] (Biolegend #304015), Fluorescein isothiocyanate (FITC) conjugated anti-EpCAM [4:100](clone 9C4, Biolegend #324203), and Brilliant Violet 421 (BV421) conjugated anti-CD44 [0.6:100] (clone IM7, Biolegend #103039). For initial analysis by flow cytometry, 1 ×106 cells were stained, fixed in 2% PFA for 10 min at room temperature, then rinsed (with PBS) and resupended in 500 μL of PBS for analysis using a Beckman-Coulter (Dako) CyAn ADP (Supplemental Figure 2). For sorting experiments, cells were rinsed and resuspended in IESC Sort/Culture Media [Advanced DMEM/F12 (Gibco), N2 (Gibco), B27 (Gibco), Glutamax (Gibco), Penicillin/Streptomycin (Gibco), 10mM HEPES (Gibco), 10μM Y27632 (Selleck Chemicals), and 500mM N-acetyl-cysteine (Sigma)]. FACS was conducted using an iCyt Reflection (Visionary BioScience) for RNA collection or FACSAria (BD Biosciences, San Jose, CA) for cell culture experiments. Dead cells and debris were first excluded based on size via bivariate plot of forward scatter (FSC) vs. side scatter (SSC) (Supplemental Figure 3A). Doublets/multimers were excluded using a bivariate plot of FSC peak vs. FSC length (Supplemental Figure 3B). Epithelial cells were FACS-enriched by sorting EpCAM (CD326) positive, CD45 negative cells (Supplemental Figure 3C). The remaining cell events were analyzed for CD24 and CD44 expression on a bivariate plot (Supplemental Figure 3D). Five cell populations: CD45−EpCAM+(whole), CD45−EpCAM+CD24−CD44− (negative), CD45−EpCAM+CD24+CD44− (CD24+CD44−), CD45−EpCAM+CD24−CD44+(CD24−CD44+), CD45−EpCAM+CD24+CD44+(CD24+CD44+) were collected directly into 500 μl of RNA lysis buffer (Ambion RNAqueous Micro, Grand Island, NY) for gene expression analysis. For cell culture experiments, cells were collected into 500μL of IESC Staining Media.
Primary isolation of human small intestinal myofibroblasts
Following mucosectomy for epithelial prep, remnant submucosa (~4cm × 4cm) was diced with a sterile razor blade. To eliminate blood cells and remnant epithelium, tissue chunks were washed in 20mL DMEM (Gibco), shaken for 2 minutes, and then allowed to settle before supernatant was removed and discarded. This process was repeated 8 times. The submucosal tissue was then resuspended in 5mL DMEM containing Penicillin/Streptomycin, 0.3U/mL dispase, and 300U/mL collagenase I (Sigma) and rotated for 25 min at room temperature. 10mL of DMEM +10%FBS was added to quench the reaction and the tissue suspension was pipetted vigorously ~50 times to further mechanically dissociate myofibroblasts. The tissue suspension was centrifuged at 300g for 5min and the resulting supernatant and tissue remnants were plated separately in DMEM +10%FBS. Media was changed every 24hrs. Cultures initiated from the supernatant of the prep produced myofibroblasts, which were passaged 3 times before use in IESC culture experiments.
Intestinal epithelial stem cell culture
Cells were pelleted and resuspended in ES-qualified Matrigel (BD Biosciences) containing IESC culture growth factors (see Supplemental Table 2). For feeder-free cultures, 10μL Matrigel droplets were plated in 96-well plates and overlaid with 100μL IESC Sort/Culture Media, with or without 2.5μM CHIR99021 (Selleck Chemical) following 15 minute of polymerization at 37°C. For feeder co-culture, 25μL Matrigel droplets were allowed to polymerize in 12-well transwell inserts (BD Biosciences) before being placed in wells containing fibroblast feeder cells and 500μL IESC Sort/Culture Media. An additional 500μL of the same media was placed in the transwell to prevent drying of the Matrigel droplet. CHIR99021 was not used in co-culture experiments. To facilitate differentiation of enteroids, Wnt3a, SB202190, and nicotinamide were withdrawn at 14 days of culture, as previously described for whole crypt cultures11.
cDNA preparation/real-time PCR analysis
mRNA from sorted cell populations was purified by using RNAqueous Micro Kit (Ambion) according to the manufacturer’s protocols. cDNA was generated using iTaq Reverse Transcription Supermix (Bio-Rad, Hercules, CA). Real-time PCR was conducted for each sample in triplicate on 1/20,000 of the total amount of cDNA generated. Taqman probes [18S, HS99999901; CD24 Hs00273561_s1; CD44, Hs01075861_m1; LGR5 Hs00173664_m1; OLFM4 HS00197437_m1; HOPX Hs04188695_m1; DEFA6 Hs00427001_m1; LYZ Hs00426232; MUC2 Hs00159374_m1; CHGA Hs00900373_m1] for each gene were obtained from Applied Biosystems (Pleasanton, CA) and used in reactions according to the manufacturer’s protocol.
Statistical Analysis
qRT-PCR data was normalized for the expression of 18S. ΔΔCt values were then calculated using the CD24−CD44− (negative) cell population as the comparator. Statistical analysis compared gene expression across all cell populations by gene for each patient via one-way ANOVA followed by Bonferroni post-test for multiple comparisons between the population of interest and all other populations. Statistical analysis was performed in Graph Pad Prism (ver 4.0, LaJolla, CA).
Results and discussion
CD24 and CD44 expression was assessed on human jejunum derived from patients who had undergone roux-en-y gastric-bypass surgery. Immunostaining demonstrates that CD44 is expressed on cells from the base of the crypt to the crypt-villus junction (Figure 1A, C, & E). By contrast, the villus epithelium does not show appreciable CD44 staining (Figure 1B & D). CD24 demonstrates similar expression to CD44 in the epithelium with the notable exception that staining is primarily distributed along the apical membrane (Figure 1F & H, arrows). A minority population of crypt-based cells expresses high levels of cytoplasmic CD24 (Figure 1G & I, arrows). While CD24/44 expression is highly restricted to the stem cell zone in the epithelium, there is broad expression in non-epithelial cells in the lamina propria, sub-mucosa, and muscle (Figure 1A–I). EpCAM (CD326) expression is unique to all crypt and villus epithelial cells, and was deemed useful for positive FACS selection to separate epithelial from non-epithelial CD24/44-expressing cells (Figure 1J–L).
Figure 1. CD24 and CD44 are expressed in the stem cell zone of the human intestinal epithelium.
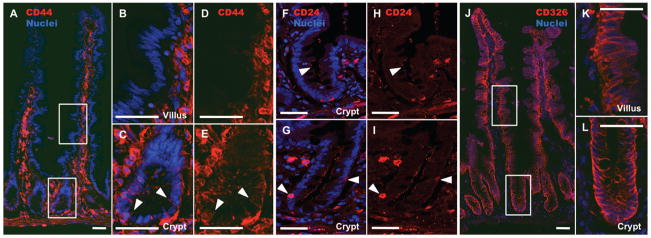
Epithelial expression of CD44 is restricted to the basolateral membranes of crypt cells (A, C, & E), with no expression in villus epithelium (B & D). CD24 is also restricted to the crypt, but is expressed on the apical membrane of epithelial cells (F & H, arrow). A subset of crypt-based epithelial cells expresses high levels of cytoplasmic CD24 (G & I, arrows). CD326 (EpCAM) is expressed throughout both the crypt and villus, but remains restricted to epithelial cells (J–L). Scale bars represent 50μm.
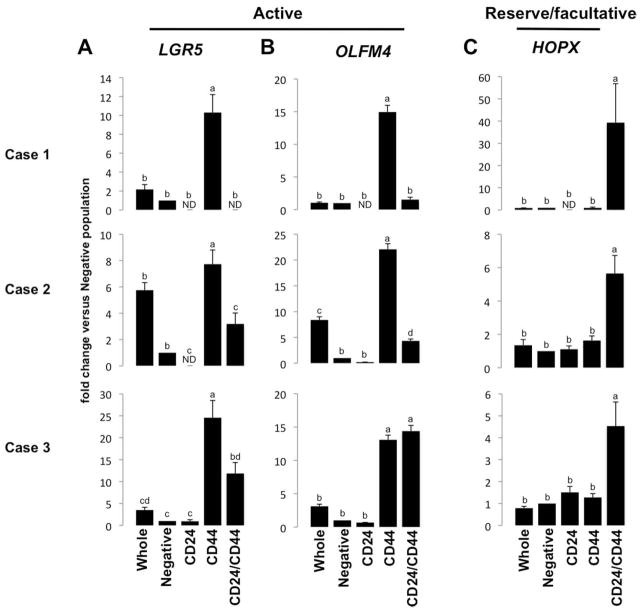
Next, we analyzed CD24, CD44, CD45, and CD326 on dissociated single epithelial cells (Supplemental Figure 2). Positive selection for epithelial cells (EpCAM+) and negative selection for lymphocytes (CD45−) was included in the FACS parameters for robust epithelial enrichment (Supplemental Figure 3C). Flow cytometry data demonstrate four distinct epithelial cell populations based on CD24/44 expression status (Supplemental Figure 3D). Each of the four populations (CD24−/44−; CD24+/44−; CD24−/44+; CD24+/44+) was collected for gene expression analysis. Semi-quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) validated the FACS by showing enriched gene expression for CD24/44 in the appropriate populations (Supplemental Figure 4A & B). The data demonstrate that CD24−/CD44+ populations are most enriched for the “active” cycling IESC markers, LGR5 and OLFM4 (Figure 2A & B); and the CD24+/CD44+ population is most enriched for the “reserve/facultative” IESC marker HOPX (Figure 2C). Interestingly, all CD24+ and CD44+ populations demonstrate significant de-enrichment for Paneth cell markers DefensinA6 (DEFA6) (Supplemental Figure 5A) and Lysozyme (LYZ) (Supplemental Figure 5B), and goblet cell marker, Mucin2 (MUC2) (Supplemental Figure 5C), all of which associate with CD24 expression in mice 7, 12, 13. However, the CD24+/CD44+ population is highly enriched for the enteroendocrine cell marker Chromogranin A (CHGA) which is consistent with observations made in mice (Supplemental Figure 5D) 10.
Figure 2. CD24−/CD44+ and CD24+/CD44+ intestinal epithelial cells are enriched for active and reserve/facultative markers, respectively.
CD24−/CD44+ cells are significantly enriched for active IESC markers (A) LGR5 and (B) OLFM4, while CD24+/CD44+ cells exhibit enrichment for (C) HOPX, which is associated with reserve/facultative IESCs. Letters a–d above each bar indicate data points that are statistically different from each other (p < 0.05).
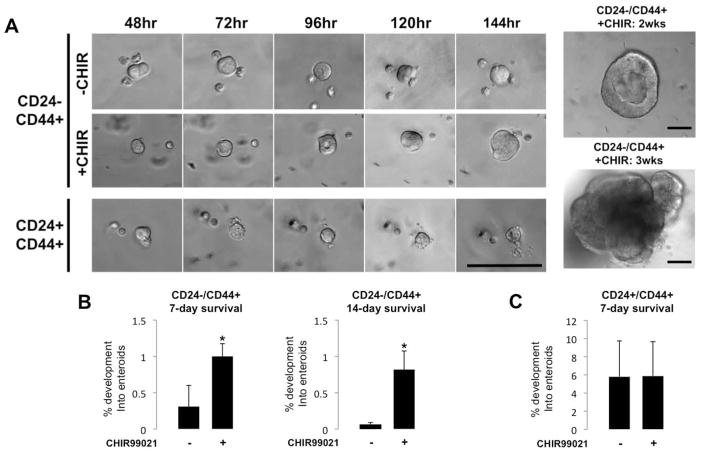
To test if the CD24−/CD44+ and CD24+/CD44+ populations had functional properties of stemness, we subjected isolated single cells to culture conditions similar to those that have been successfully used to grow small intestine crypts and single colonic stem cells from human tissue 11, 14. Both IESC populations formed appreciable cystic enterosphere structures by 48hrs (Figure 3A), while the other populations (CD24−/CD44− and CD24+/CD44−) failed to do so. Enterospheres derived from CD24−/CD44+ cells continued to develop over the first week of culture, while CD24+/CD44+ derived enterospheres exhibited a limited increase in size (Figure 3A). Nevertheless, both populations persisted in culture at 7 days (Figure 3B & C). In an attempt to increase culture efficiency, we added GSK-inhibitor CHIR99021, which promotes self-renewal of embryonic stem cells was recently used to enhance in vitro growth and survival of human colon cancer stem cells and intestinal crypts (Wang, et al unpublished) 15, 16. Interestingly, initial GSK-inhibition significantly improved 7-day enterosphere survival in the CD24−/CD44+ population, but had no appreciable effect on the CD24+/CD44+ population (Figure 3B & C). Initial GSK-inhibition greatly increased 14-day survival of the CD24−/CD44+ population, but was insufficient for the maintenance of CD24+/CD44+ derived enteroids, which did not survive to 14 days, regardless of GSK-inhibition (Figure 3C). Previous studies have shown that Lgr5-negative populations can de-differentiate and function as “facultative” stem cells when presented with the proper extrinsic cues 7, 8. Additionally, co-culture of primary human intestinal crypts with myofibroblasts enhances culture efficiency 17. In an attempt to “activate” the CD24+/CD44+ population, we co-cultured the cells with myofibroblasts isolated from human jejunal submucosa. Surprisingly, when grown in these conditions, both CD24−/CD44+ and CD24+/CD44+ cells produced long-lived enteroids, at rates of 0.07% and 0.76%, respectively (Supplemental Figure 6).
Figure 3. CD24−/CD44+ and CD24+/CD44+ populations generate enteroids in vitro.
By 48hrs, CD24−/CD44+ and CD24+/CD44+ cells form small enteroid structures, which increase in size over the first week of culture (A). GSK inhibition through a single dose of CHIR99021, given when cells are plated, significantly increases 7- and 14-day survival of enteroids derived from CD24−/CD44+ cells (B), but has no noticeable effect on the CD24+/CD44+ population (C). While CD24−/CD44+ cells form long-lived enteroids, the CD24+/CD44+ population does not demonstrate survival at or after 14 days in culture in feeder-free conditions (A). Student’s t test was used to determine significance. Asterisks indicate a P-value of < 0.01. Scale bars represent 100μm.
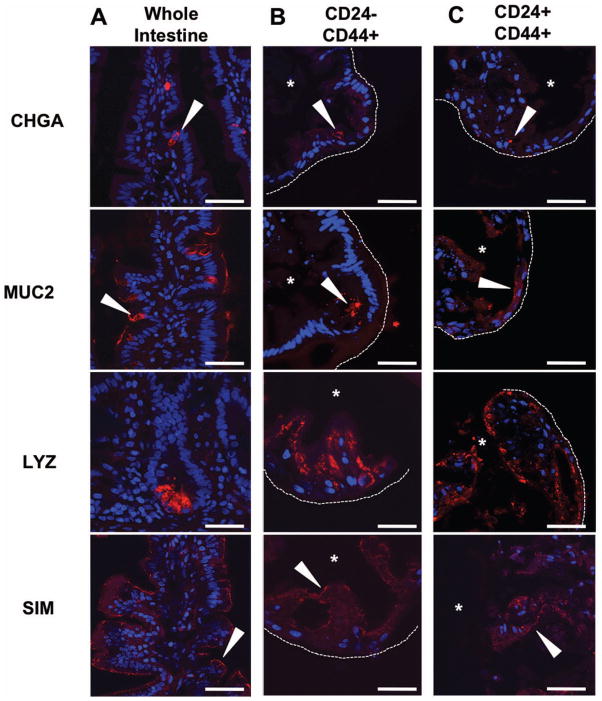
To assess multipotency, we retrieved and processed enteroids for immunohistochemistry at 21 days. CD24−/CD44+ derived enteroids were epithelial in nature (Supplemental Figure 7) and produced enteroendocrine (CHGA), goblet (MUC2), and Paneth cells (LYZ), as well as absorptive enterocytes (SIM) (Figure 4B). CD24+/CD44+ cells grown in co-culture with primary human myofibroblasts exhibited the same characteristics of multipotency as CD24−/CD44+ cells (Figure 4C and Supplemental Figure 7). Enteroids derived from both populations exhibited increased numbers of lysozyme positive cells, consistent with results in mice demonstrating that persistent WNT signaling, specifically through CHIR99021 treatment, increases Paneth cell production in enteroid culture 18. This biased secretory lineage allocation likely explains the rare occurrence of MUC2 positive cells in the enteroids. Importantly, co-culture with myofibroblasts did not induce growth in CD24−/CD44− and CD24+/CD44− populations, demonstrating that functional stemness under all tested growth conditions remains restricted to CD24−/CD44+ and CD24+/CD44+ populations.
Figure 4. Enteroids derived from CD24−/CD44+ and CD24+/CD44+ populations are multipotent.
Enteroendocrine, goblet, and Paneth cells, as well as absorptive enterocytes, are detectable in whole human jejunum by expression of CHGA, MUC2, LYZ, and SIM, respectively (A). Similar expression patterns for each marker are observed by immunofluorescence in enteroids derived from the LGR5-associated CD24−/CD44+ population (B) and the HOPX-associated CD24+/CD44+ population (C). Arrows indicate positive staining, asterisks mark enteroid lumens, and dotted lines denote outer edge of enteroid structures. Scale bars represent 50μm.
The translation of scientific findings made in genetically homogeneous mouse models presents investigators with a daunting challenge when attempting to understand the vast heterogeneity underlying physiology in human populations. This report provides new methodology for epithelial dissociation, isolation, and FACS enrichment of IESCs from human small intestine through the use of a combinatorial CD-marker “signature”. The ability of CD24 and CD44 to enrich for IESCs is highly consistent with observations in mice; however, in stark contrast to findings in mouse models, human Paneth cells do not appear to express high levels of CD24, highlighting the need to validate mouse genetic biomarkers on human samples 7, 10, 12, 13. Furthermore, our results demonstrate variable levels of gene enrichment between some patients. This observation could be attributed to a wide range of factors, including phenotypic heterogeneity between samples. In the present study, tissue procurement protocols required full de-identification of samples; thus, a patient’s dietary habits, state of illness, or medication history could not be correlated to gene expression variations. This highlights the need for comprehensive studies with controlled enrollment criteria, utilizing the foundation for isolation of LGR5 and HOPX-enriched populations, presented here
In addition to FACS-enrichment of ‘active’ LGR5+ IESCs, the CD24/CD44 approach facilitates differential isolation of HOPX+ cells, which emerging evidence suggests may represent facultative IESCs 4. While several of the markers examined here exhibit a degree of variability between samples, it is clear that the population of cells most enriched for LGR5 is distinct from the population most enriched for HOPX, unlike recent observations of Hopx expression in murine Lgr5+ cells 6. Additionally, CD24−/CD44+ and CD24+/CD44+ populations exhibit differential behavior in vitro, both in response to GSK-inhibition and in basal culture conditions required for growth, further supporting the existence of phenotypically distinct IESC populations. Though overlap of LGR5 expression with putative “reserve” IESC markers in mice remains controversial, our data suggests that more distinct populations may exist in the human small intestinal epithelium 4–6, 19. Further work is needed to characterize the CD24+/CD44+ population and determine if it is analogous to proposed “facultative” IESCs observed in mouse models of intestinal damage and regeneration. Multiple reports have demonstrated the ability of Lgr5-negative facultative IESCs to drive regeneration in murine intestine following irradiation, suggesting that an analogous population may be of particular interest in human pathophysiology, especially in patients undergoing radiation treatment for tumors 7, 8. Understanding the genetics and patient-to-patient heterogeneity of phenotypically distinct human IESC populations may provide valuable insight toward the development of novel clinical diagnostics and therapeutic interventions.
Supplementary Material
Supplemental Figure 1: Epithelial preparations enrich for stem cell containing crypts. Initial shaking steps remove large villi (A). After villus fractions are discarded, additional shaking yields both crypts and some remaining villi (B). Filtering results in single isolated crypts, with some debris and single cells from submucosal tissues (C). Scale bars represent 100μm.
Supplemental Figure 2: CD45, CD326, CD44, and CD24 antibodies label positive populations via flow cytometry. Isotype controls and antibody staining by flow cytometry for CD45, EpCAM (CD326), CD44, and CD24 indicate positive staining of populations with negligible non-specific labeling.
Supplemental Figure 3: CD24 and CD44 differentially label human jejunal cells. Gating strategy for FACS isolation of CD24/CD44+ epithelial populations: Removal of debris (A) Singlet gating (B) Selection of epithelial cells by CD45 exclusion and CD326 inclusion (C) Sample of gating strategy based on CD24 (y-axis) and CD44 (x-axis) (D). Composition of total jejunal epithelium by CD24/CD44 expression: table describes % of all gated cells in CD24/CD44 histogram by cell population ± SEM (n=3).
Supplemental Figure 4: CD24/CD44 sorting strategy enriches for CD24− and CD44−positive populations. qPCR for CD24 (A) and CD44 (B) demonstrates population-specific enrichment for each marker, validating FACS parameters. Letters a–c above each bar indicate data points that are statistically different from each other (p < 0.05).
Supplemental Figure 5: CD24/CD44 populations are de-enriched for Paneth and goblet cell markers. Gene expression analysis demonstrates that CD24 and CD44 populations are significantly de-enriched for Paneth cell markers DEFA6 (A) and LYZ (B) and demonstrate enriched expression of these markers in CD24/CD44 negative populations over whole epithelium. De-enrichment is also observed for goblet cell marker MUC2 (C) in two of three cases, with no statistical difference between MUC2 in CD24−/CD44− and CD24+/CD44− populations in Case 1, which may be attributable to patient-patient heterogeneity. High levels of enteroendocrine marker CHGA are observed specifically in the CD24+/CD44+ population in two of three cases, with significant enrichment in CD24+/CD44− and CD24+/CD44+ populations in Case 3 (D), consistent with reports in mice. Letters a–g above each bar indicate data points that are statistically different from each other (p < 0.05).
Supplemental Figure 6: Co-culture with myofibroblasts induces long-lived enteroid formation in CD24−/CD44+ and CD24+/CD44+ populations. Both LGR5-associated CD24−/CD44+ and HOPX-associated CD24+/CD44+ cells form enteroids when co-culture on transwells with myofibroblasts isolated from human jejunal remnants. Enteroids are shown at 14 days post-plating.
Supplemental Figure 7: Human enteroids are composed of epithelial cells. Immunofluorescence for EPCAM (CD326) demonstrates that enteroids derived from CD24−/CD44+ and CD24+/CD44+ populations are epithelial in nature. Scale bars represent 50μm.
Culture conditions for human intestinal epithelial stem cell populations.
Acknowledgments
Funding: This work was funded by the NC TraCS Institute Pilot Grant Program, 2KR271103 (Gracz/Fuller) and 2KR381202 (Gracz), and the Center for Gastrointestinal Biology and Disease, 5P30DK034987-27 (Magness). The NC TraCS Institute is supported by grants UL1RR025747, KL2RR025746, and TLRR025745 from the NIH National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health. The National Institute of Diabetes and Digestive and Kidney Diseases/National Institute of Allergy and Infectious Disease Intestinal Stem Cell Consortium (U01DK085541-SJH; U01DK85507-LL/FW; U01DK85535-MGM/JCYD/MS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
The authors thank the UNC flow cytometry facility (P30CA06086) and the neuroscience confocal facility, which is co-funded by NINDS and the National Institute of Child Health and Human Development (NICHD) (P30NS045892). Drs. Timothy Farrell and Wayne Overby, and Myra Jones, Karen Colton, and Lisa Prestia, for coordinating the collection of de-identified remnant jejunal tissue from gastric bypass operations. The Duke Human Vaccine Institute flow cytometry core, and Dr. John Whitesides and Ian Cumming, for technical assistance. Drs. Tope Keku and Robert Sandler for unpublished data regarding mucosal biopsy weight. Drs. P. Kay Lund, Susan Henning (SJH), and Christopher Dekaney for useful discussions and critical review of the manuscript.
Footnotes
Author contribution:
ADG and STM conceived and designed the study and wrote the manuscript, MKF developed and optimized protocols for epithelial isolation and FACS. FW and LL provided GSK-inhibitor conditions. MS, JCYD, and MGM provided critical review of the manuscript. ADG and STM carried out culture and gene expression experiments.
References
- 1.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery RK, Carlone DL, Richmond CA, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powell AE, Wang Y, Li Y, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda N, Jain R, LeBoeuf MR, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan KS, Chia LA, Li X, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz J, Stange DE, Schepers AG, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Es JH, Sato T, van de Wetering M, et al. Dll1(+) secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012 doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Landeghem L, Santoro MA, Krebs AE, et al. Activation of two distinct Sox9-EGFP expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012 doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 10.Gracz AD, Ramalingam S, Magness ST. Sox9-Expression Marks a Subset of CD24-expressing Small Intestine Epithelial Stem Cells that Form Organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010 doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Stange DE, Ferrante M, et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Furstenberg RJ, Gulati AS, Baxi A, et al. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2010 doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung P, Sato T, Merlos-Suarez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 15.Kemper K, Prasetyanti PR, De Lau W, et al. Monoclonal antibodies against lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378–2386. doi: 10.1002/stem.1233. [DOI] [PubMed] [Google Scholar]
- 16.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahar N, Lei NY, Wang J, et al. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One. 2011;6:e26898. doi: 10.1371/journal.pone.0026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farin HF, Van Es JH, Clevers H. Redundant Sources of Wnt Regulate Intestinal Stem Cells and Promote Formation of Paneth Cells. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Tian H, Biehs B, Warming S, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Epithelial preparations enrich for stem cell containing crypts. Initial shaking steps remove large villi (A). After villus fractions are discarded, additional shaking yields both crypts and some remaining villi (B). Filtering results in single isolated crypts, with some debris and single cells from submucosal tissues (C). Scale bars represent 100μm.
Supplemental Figure 2: CD45, CD326, CD44, and CD24 antibodies label positive populations via flow cytometry. Isotype controls and antibody staining by flow cytometry for CD45, EpCAM (CD326), CD44, and CD24 indicate positive staining of populations with negligible non-specific labeling.
Supplemental Figure 3: CD24 and CD44 differentially label human jejunal cells. Gating strategy for FACS isolation of CD24/CD44+ epithelial populations: Removal of debris (A) Singlet gating (B) Selection of epithelial cells by CD45 exclusion and CD326 inclusion (C) Sample of gating strategy based on CD24 (y-axis) and CD44 (x-axis) (D). Composition of total jejunal epithelium by CD24/CD44 expression: table describes % of all gated cells in CD24/CD44 histogram by cell population ± SEM (n=3).
Supplemental Figure 4: CD24/CD44 sorting strategy enriches for CD24− and CD44−positive populations. qPCR for CD24 (A) and CD44 (B) demonstrates population-specific enrichment for each marker, validating FACS parameters. Letters a–c above each bar indicate data points that are statistically different from each other (p < 0.05).
Supplemental Figure 5: CD24/CD44 populations are de-enriched for Paneth and goblet cell markers. Gene expression analysis demonstrates that CD24 and CD44 populations are significantly de-enriched for Paneth cell markers DEFA6 (A) and LYZ (B) and demonstrate enriched expression of these markers in CD24/CD44 negative populations over whole epithelium. De-enrichment is also observed for goblet cell marker MUC2 (C) in two of three cases, with no statistical difference between MUC2 in CD24−/CD44− and CD24+/CD44− populations in Case 1, which may be attributable to patient-patient heterogeneity. High levels of enteroendocrine marker CHGA are observed specifically in the CD24+/CD44+ population in two of three cases, with significant enrichment in CD24+/CD44− and CD24+/CD44+ populations in Case 3 (D), consistent with reports in mice. Letters a–g above each bar indicate data points that are statistically different from each other (p < 0.05).
Supplemental Figure 6: Co-culture with myofibroblasts induces long-lived enteroid formation in CD24−/CD44+ and CD24+/CD44+ populations. Both LGR5-associated CD24−/CD44+ and HOPX-associated CD24+/CD44+ cells form enteroids when co-culture on transwells with myofibroblasts isolated from human jejunal remnants. Enteroids are shown at 14 days post-plating.
Supplemental Figure 7: Human enteroids are composed of epithelial cells. Immunofluorescence for EPCAM (CD326) demonstrates that enteroids derived from CD24−/CD44+ and CD24+/CD44+ populations are epithelial in nature. Scale bars represent 50μm.
Culture conditions for human intestinal epithelial stem cell populations.