Abstract
Context:
Septicemia in neonates refers to generalized bacterial infection documented by positive blood culture in the first four weeks of life and is one of the four leading causes of neonatal mortality and morbidity in India.
Aim:
To isolate and identify the bacterial etiologic agents responsible for neonatal sepsis and to determine the susceptibility pattern of isolates in a tertiary care hospital in North Karnataka.
Materials and Methods:
Six hundred eighty-three blood samples were collected and processed from patients in accordance with standard protocols. Antibiotic susceptibility of the isolates was done by disc diffusion method according to National Committee for Clinical Laboratory Standards (NCCLS) recommendations.
Results:
Blood culture reports were positive in 19.2% cases. Among the culture positive cases, there were 65.5% males and 34.5% females. Early-onset sepsis was present in 74.8% and late-onset sepsis was observed in 25.2% of the cases. Best overall sensitivity among Gram-negative isolates was to imipenem (93%), followed by amikacin (52%) and netilmicin (41%). Gram-positive isolates had sensitivity of 91% to linezolid, 68% to tetracycline, 64% to piperacillin/tazobactam erythromycin, and 52% to ciprofloxacin.
Conclusion:
Gram-negative organisms (Klebsiella, Acinetobacter), coagulase-negative staphylococci, and Staphylococcus aureus are the leading cause of neonatal sepsis in this study and most of them are resistant to multiple antibiotics. Therefore the results of this study suggest that, surveillance of antimicrobial resistance in our hospital is necessary.
Keywords: Antimicrobial resistance, antibiotics, neonatal septicemia
INTRODUCTION
Septicemia in neonates refers to generalized bacterial infection documented by positive blood culture in the first four weeks of life[1] and is one of the four leading causes of neonatal mortality and morbidity in India.[2,3,4] Neonatal septicemia continues to be a major problem for neonates in neonatal intensive care units around the world.[5]
Neonatal mortality rate is one of the indicators for measuring the health status of a nation.[6] There could be various reasons for neonatal mortality but septicemia continues to be a major cause of neonatal mortality and morbidity worldwide. Incidence varies from country to country, but it is much higher in developing countries than in developed nations.[6] According to World Health Organization (WHO) estimates, there are about 5 million neonatal deaths a year, with 98% occurring in developing countries.[7]
Neonatal sepsis is broadly divided into two types according to age of onset: Early-onset sepsis (<72 hrs) and late-onset sepsis (≥72 hrs-28 days). Early-onset sepsis is acquired during fetal life, delivery, or at the nursery.[8] Neonatal sepsis is caused by a variety of Gram-positive as well as Gram-negative bacteria, and sometimes yeasts.[5] The spectrum of organisms that causes neonatal sepsis changes over times and varies from region to region. This is due to the changing pattern of antibiotic use and changes in lifestyle.[9]
Periodic evaluation of organisms responsible for neonatal sepsis is essential for the appropriate management of neonates. Therefore, this study was undertaken to determine the profile and antibiotic sensitivity patterns of aerobic isolates from blood cultures of neonates in a tertiary care hospital in Bijapur, India.
MATERIALS AND METHODS
An analysis was conducted on all blood culture reports obtained between January 2008 and December 2010 from newborns admitted to the Department of Pediatrics and the Neonatal Intensive Care Unit (NICU) at Shri B M Patil Medical College, Bijapur. Blood culture was done for all neonates suspected to have septicemia.
Blood culture sample included a single sample collected from a peripheral vein or artery under aseptic conditions. The local site was cleansed with 70% alcohol and povidone iodine (1%), followed by 70% alcohol again. Blood cultures were done in a brain heart infusion biphasic medium. Approximately, 3 ml of blood was inoculated into the brain heart infusion broth and incubated at 37°C. Subcultures were done on sheep blood agar and MacConkey agar at the earliest visual detection of turbidity or blindly on days 1, 4, and 7 if the bottles did not show turbidity. Isolate was identified by their characteristic appearance on their respective media, Gram staining and confirmed by the pattern of biochemical reactions using the standard method.[10] Members of the family enterobacteriaceae were identified by indole production, H2S production, citrate utilization, motility test, urease test, oxidase, carbohydrate utilization tests, and other tests. For Gram-positive bacteria, coagulase, catalase, bacitracin and optochin susceptibility tests and other tests were used. Blood culture broth that showed no microbial growth within seven days was reported as culture negative, only after result of routine subculture on blood, MacConkey, and chocolate agar.[10]
Antimicrobial susceptibility testing was performed for all blood culture isolates by Kirby–Bauer disc diffusion method as recommended in the National Committee for Clinical Laboratory Standards (NCCLS) guidelines.[11]
The drugs for disc diffusion testing were in the following concentrations: Ampicillin (10 μg), cloxacillin (1 μg), lomefloxacin (10 μg), amoxiclav (20/10 μg), cephalexin (30 μg), cefuroxime (30 μg), ciprofloxacin (5 μg), erythromycin (15 μg), gentamicin (10 μg), (30 μg), penicillin (10 units), tetracycline (30 μg), co-trimoxazole (1·25 μg trimethoprim/23·75 μg sulfamethoxazole), amikacin (30 μg), ofloxacin (5 μg), sparfloxacin (5 μg), pefloxacin (5 μg), cefoperazone (75 μg), netilmicin (30 μg), imipenem (10 μg), piperacillin/tazobactam (100/10 μg), azithromycin (15 μg), and linezolid (30 μg). The discs were obtained from Himedia (India) Laboratories.
Data analysis was done using Statistical Package for Social Sciences (SPSS) software version 14.0. The level of significance for tests was set at P < 0.05.
RESULTS
During the study period, a total of 683 newborns with clinical sepsis were admitted. Blood culture reports were positive in 131 cases (19.2%). Among the culture positive cases, there were 86 (65.5%) male and 45 (34.5%) female neonates with the male-to-female ratio of 1.9:1. Early-onset sepsis cases were found to be three times higher than late-onset sepsis. Out of 131 cases, 98 (74.8%) had early-onset sepsis and 33 (25.2%) had late-onset sepsis.
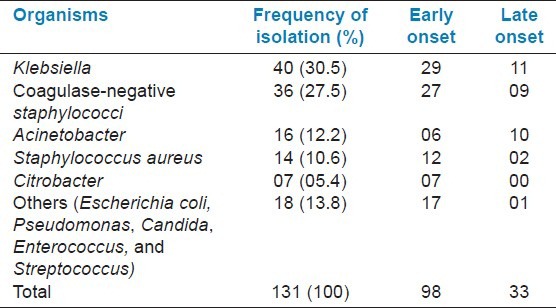
Detailed aetiology of the 131 isolates is provided in Table 1. These included Gram-negative bacilli (73/131, 55.7%) and Gram-positive cocci (58/131, 44.3%). Klebsiella spp. and coagulase-negative staphylococci (CONS) were the most common Gram-negative and Gram-positive organisms.
Table 1.
Distribution of isolated organisms
Tables 2 and 3 show the antibiotic susceptibility pattern in Gram-negative and Gram-positive isolates. Best overall sensitivity among Gram-negative isolates was to imipenem (93%), followed by amikacin (52%) and netilmicin (41%). Gram-positive isolates had sensitivity of 91% to linezolid, 68% to tetracycline, 64% to piperacillin/tazobactam erythromycin, and 52% to ciprofloxacin.
Table 2.
Antibiotic susceptibility of Gram-negative organisms
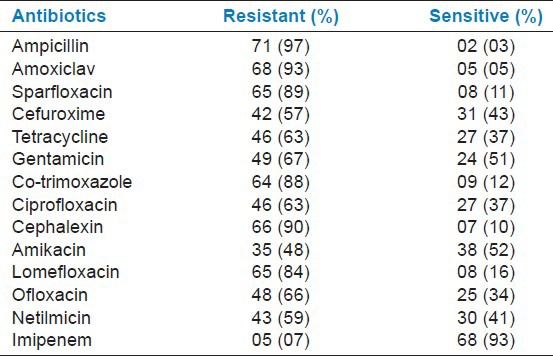
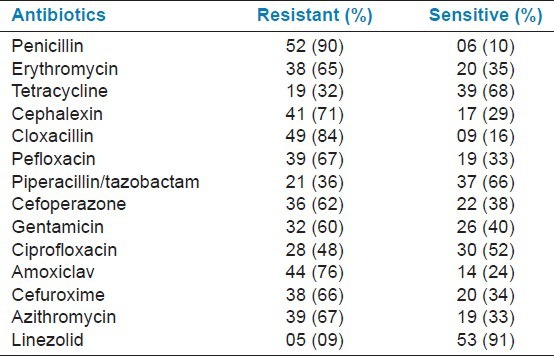
Table 3.
Antibiotic susceptibility of Gram-positive organisms
DISCUSSION
The uncertainty surrounding the clinical approach to treatment of neonatal septicemia can be minimized by periodic epidemiological surveys of aetiological agents and their antibiotic sensitivity patterns leading to recognition of the most frequently encountered pathogens in a particular geographical area. For effectual management of septicemia cases, study of bacteriological profile along with the antimicrobial sensitivity pattern plays a noteworthy role.[12,13,14] Out of the 683 clinically suspected cases of sepsis in our study, 131 were culture positive with a blood culture positivity rate of 19%. The incidence of Gram-negative and Gram-positive organisms was 55.7% and 44.3%, respectively. There were 98 (74.8%) isolates from early onset septicemia cases, while 33 (25.2%) were from late-onset illness.
In this study, a male predominance with male-to-female ratio of 1.9:1 was found in our study, which agrees with previous reports. This might be because of the importance given to the male infants and also because of more number of male infants born compared to female infants born. Culture-positivity for aerobic organisms in neonates vary from 25% to 60%.[15,16,17] In this study, blood culture-positivity rate is 19%. This finding is comparable with other reports.[9] However, a high blood culture-positivity rate in septicemic children (56%) had been reported by Sharma et al.[18] and Jain et al.[19]
A low blood culture isolation rate could be due to administration of antibiotic before blood collection from the primary centers or the possibility of infection with anaerobes. A negative blood culture does not exclude sepsis and about 26% of all neonatal sepsis could be due to anaerobes.[9]
The pathogens most often implicated in neonatal sepsis in developing countries differ from those seen in developed countries. Overall, Gram-negative organisms are more common and are mainly represented by Klebsiella, Escherichia coli, Pseudomonas, and Salmonella. Of the Gram-positive organisms, Staphylococcus aureus, CONS, Streptococcus pneumonia, and S. pyogenes are most commonly isolated.[7]
Gram-negative and Gram-positive septicemia was encountered in 56% and 44% of the culture-positive cases in this study, which is comparable to a study conducted by Agnihotri et al.,[1] which reported that Gram-negative and Gram-positive organisms were responsible for 59% and 41% of the septicemia cases, respectively. Similar observations were made by other workers.[4,6]
The report of the National Neonatal-Perinatal database showed Klebsiella as the predominant (29%) pathogen.[15] Klebsiella spp.(31%) was the predominant Gram-negative species isolated in this study, which agrees with previous reports.[2,9] Of the total 131 cases of neonatal sepsis, 98 (74.8%) were early-onset sepsis in this study, which is comparable to previous studies.[1,6]
Antibiotic resistance is today a global problem. Reports of multi-resistant bacteria causing neonatal sepsis in developing countries are increasing. The wide availability of over-the-counter antibiotics and the inappropriate use of broad-spectrum antibiotics in the community may explain this situation. It is difficult to compare antibiotic resistance between countries because the epidemiology of neonatal sepsis is extremely variable.[7]
Antibiotic susceptibility pattern was studied for all isolates causing neonatal sepsis. The analysis of drug resistance pattern showed that, among Gram-negative isolates, maximum numbers (97%) were resistant to ampicillin and lowest to imipenem (7%). Resistance was observed to be against commonly used antibiotics such as ampicillin, amoxiclav, cephalexin, and co-trimoxazole. Among Gram-positive isolates, high resistance was seen to penicillin (90%), cloxacillin (84%), and amoxiclav (76%). Least resistance was seen to linezolid (9%), followed by tetracycline (32%), and piperacillin/tazobactam (36%). The greater prevalence of resistance to commonly used antibiotics has also been reported by other studies.[2,4] Among aminoglycosides, amikacin was found to have an edge over netilmicin and gentamicin in Gram-negative septicemia, with sensitivity of 52%, 41%, and 33%, respectively. Similar observations have been made by previous group of workers.[20]
In this study, maximum sensitivity (93%) was observed in imipenem and linezolid (91%). Sensitivity to imipenem and linezolid was much higher than that to other antibiotics and the difference was statistically significant (P < 0.05), but these two drugs should not be used indiscriminately and be kept as a reserve drugs, otherwise resistance to these drugs may develop, thereby threatening the treatment.
CONCLUSION
It is evident from this study that Gram-negative organisms (Klebsiella, Acinetobacter), CONS, and S. aureus are the leading cause of neonatal sepsis in this study, and most of them are resistant to multiple antibiotics. Therefore, the authors suggest that surveillance of antimicrobial resistance is necessary. Also, an antibiotic policy should be formulated in the hospital. Depending on the antibiotic sensitivity pattern of the isolates, antibiotics should be used. Furthermore, we advise that health education be provided to the public on the dangers of indiscriminate use of antibiotics, which is currently considered to be a menace in our society and which has been responsible for the ineffectiveness of most commonly used antibiotics such as penicillin and ampicillin, as observed in our study.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Agnihotri N, Kaistha N, Gupta V. Antimicrobial susceptibility of isolates from neonatal septicemia. Jpn J Infect Dis. 2004;57:273–5. [PubMed] [Google Scholar]
- 2.Tsering DC, Chanchal L, Pal R, Kar S. Bacteriological profile of septicemia and the risk factors in neonates and infants in Sikkim. J Global Infect Dis. 2011;3:425. doi: 10.4103/0974-777X.77295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain A, Awasthi AK, Kumar M. Etiological and antimicrobial susceptibility profile of nosocomial blood stream infections in a neonatal intensive care unit. Indian J Med Microbiol. 2007;25:299–300. doi: 10.4103/0255-0857.34783. [DOI] [PubMed] [Google Scholar]
- 4.Kumhar GD, Ramachandran VG, Gupta P. Bacteriological analysis of blood culture isolates from neonates in a tertiary care hospital in India. J Health Popul Nutr. 2002;20:343–7. [PubMed] [Google Scholar]
- 5.Gomaa HHA, Udo EE, Rajaram U. Neonatal septicemia in Al-Jahra hospital, Kuwait: Etiologic agents and antibiotic sensitivity patterns. Med Princ Pract. 2001;10:145–50. [Google Scholar]
- 6.Kaistha N, Mehta M, Singla N, Garg R, Chander J. Neonatal septicemia isolates and resistance patterns in a tertiary care hospital of North India. J Infect Dev Ctries. 2009;4:55–7. doi: 10.3855/jidc.625. [DOI] [PubMed] [Google Scholar]
- 7.Vergnano S, Sharland M, Kazembe P, Mwansambo C, Heath PT. Neonatal sepsis: An international perspective. Arch Dis Child Fetal Neonatal Ed. 2005;90:F220–4. doi: 10.1136/adc.2002.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puopolo KM. Bacterial and fungal infection. In: Cloherty JP, Eichenwald EC, Stark AR, editors. Manual of neonatal care. 6th ed. Philadelphia: Lippincott William and Wilkins; 2008. pp. 274–300. [Google Scholar]
- 9.Shrestha P, Das BK, Bhatta NK, Jha DK, Das B, Setia A, et al. Clinical and bacteriological profiles of blood culture positive sepsis in newborns. J Nepal Paediatr Soc. 2008;27:64–7. [Google Scholar]
- 10.Collee JG, Marr W. Culture of Bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New York: Churchill Livingstone; 1996. pp. 113–29. [Google Scholar]
- 11.Wayne, PA: NCCLS; 2004. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disc susceptibility testing. Fourteenth informational supplement (M100-S14) [Google Scholar]
- 12.Zakariya BP, Bhat V, Harish BN, Arun Babu T, Joseph NM. Neonatal sepsis in a tertiary care hospital in South India: Bacteriological profile and antibiotic sensitivity pattern. Indian J Pediatr. 2011;78:413–7. doi: 10.1007/s12098-010-0314-8. [DOI] [PubMed] [Google Scholar]
- 13.Dutta S, Reddy R, Sheikh S, Kalra J, Ray P, Narang A. Intrapartum antibiotics and risk factors for early onset sepsis. Arch Dis Child Fetal Neonatal Ed. 2010;95:F99–103. doi: 10.1136/adc.2009.163220. [DOI] [PubMed] [Google Scholar]
- 14.Jiang JH, Chui NC, Huang FY, Kao HA, Hsu CH, Hung HY, et al. Neonatal sepsis in the neonatal intensive care unit: Characteristics of early versus late onset. J Microbial Immunol Infect. 2004;37:301–6. [PubMed] [Google Scholar]
- 15.Neonatal morbidity and mortality; report of the National Neonatal-Perinatal Database. Indian Pediatr. 1997;34:1039–42. [PubMed] [Google Scholar]
- 16.Mathur NB. Neonatal sepsis. Indian Pediatr. 1996;33:663–74. [PubMed] [Google Scholar]
- 17.Mathur M, Shah H, Dixit K, Khambadkone S, Chakrapani A, Irani S. Bacteriological profile of neonatal septicemia cases (for the year 1990-91) J Postgrad Med. 1994;40:18–20. [PubMed] [Google Scholar]
- 18.Sharma PP, Halder D, Dutta AK, Dutta R, Bhatnagar S, Bali A, et al. Bacteriological profile of neonatal septicemia. Indian Pediatr. 1987;24:1011–7. [PubMed] [Google Scholar]
- 19.Jain NK, Jain VM, Maheshwari S. Clinical profile of neonatal sepsis. Kathmandu Univ Med J. 2003;1:117–20. [PubMed] [Google Scholar]
- 20.Bhat YR, Lewis LE, Vandana KE. Bacterial isolates of early-onset neonatal sepsis and their antibiotic susceptibility pattern between 1998 and 2004: An audit from a center in India. Ital J Pediatr. 2011;37:32. doi: 10.1186/1824-7288-37-32. [DOI] [PMC free article] [PubMed] [Google Scholar]


