Abstract
The rat has been widely used as a disease model in a laboratory setting, resulting in an abundance of genetic and phenotype data from a wide variety of studies. These data can be found at the Rat Genome Database (RGD, http://rgd.mcw.edu/), which provides a platform for researchers interested in linking genomic variations to phenotypes. Quantitative trait loci (QTLs) form one of the earliest and core datasets, allowing researchers to identify loci harboring genes associated with disease. These QTLs are not only important for those using the rat to identify genes and regions associated with disease, but also for cross-organism analyses of syntenic regions on the mouse and the human genomes to identify potential regions for study in these organisms. Currently, RGD has data on >1,900 rat QTLs that include details about the methods and animals used to determine the respective QTL along with the genomic positions and markers that define the region. RGD also curates human QTLs (>1,900) and houses >4,000 mouse QTLs (imported from Mouse Genome Informatics). Multiple ontologies are used to standardize traits, phenotypes, diseases, and experimental methods to facilitate queries, analyses, and cross-organism comparisons. QTLs are visualized in tools such as GBrowse and GViewer, with additional tools for analysis of gene sets within QTL regions. The QTL data at RGD provide valuable information for the study of mapped phenotypes and identification of candidate genes for disease associations.
Keywords: Rat Genome Database, mapping, QTL
the laboratory rat (Rattus norvegicus) has been extensively used in scientific research for over 150 yr. The rat has been used to study the genetics of hypertension, autoimmune disease, diabetes, obesity, cancer, and many other conditions, because of its usability as a model for human diseases (1, 7). The mapping of simple sequence length polymorphisms (SSLPs) to the genome in the 1990s and their association with different quantitative phenotypes opened a new path for the discovery of quantitative trait loci (QTLs). To determine a QTL, two strains, for example, one disease-susceptible and the other disease-resistant, are crossed, and the phenotypes in the offspring are observed and analyzed (3). Most of these complex phenotypes are associated with multiple genes or genomic elements across the genome. One may think of QTLs as signposts that point toward genomic areas that may contain genes or other elements of interest. QTLs identify relatively narrow regions of the genome that are likely to affect the trait under study, making the search for genetic influences much easier. According to Flint et al. (5) only 1% of QTLs in rodents have been narrowed down to the level of specific genes. The process of gene identification from a QTL utilizes a somewhat cumbersome and time-consuming method involving haplotype, gene expression, and DNA sequence data. However, new technologies and approaches are providing additional methods for gene discovery (4). Generation of congenic strains (6) in which a specific region can be segregated and analyzed has been used as a means to study subregions of broad QTLs. Similarly, generation of mutants in which a candidate QTL gene is targeted has assisted tremendously in narrowing down regions of interest. Now, with the advancement in genome sequencing and the identification of single nucleotide variants, changes in phenotypes based on the difference in a single base pair can be captured.
QTL Traits
A wide variety of traits associated with multiple physiological systems are represented by the QTLs at Rat Genome Database (RGD). The concentration of QTLs in a few significant areas reflects the value of the rat as a model for diseases related to those traits and the experiments conducted by rat researchers to date. Currently RGD has 1,913 rat QTLs. Of these, 492 are associated with cardiovascular traits (Table 1), including those for blood pressure, heart rate, and cardiac mass. There are 213 QTLs for renal morphology and function traits, 116 for tumor- and neoplasm-related traits, 139 for glucose levels, and 28 for insulin levels. Other trait areas include body weight (Table 2), bone structure and strength, joint and spinal cord inflammation, as well as those for alcohol consumption and response, anxiety, activity, memory, and other neurological traits.
Table 1.
Major categories of disease QTLs at RGD: some of the QTLs are associated with more than one disease
| Major QTL Categories Based on Diseases | QTLs, n |
|---|---|
| Metabolic disease/obesity | 794 |
| Diabetes | 790 |
| Cardiovascular disease | 547 |
| Immune and inflammatory disease | 239 |
| Neurological disease | 136 |
| Cancer | 60 |
| Respiratory disease | 3 |
QTL, quantitative trait locus; RGD, Rat Genome Database.
Table 2.
Major categories of abnormal phenotype QTLs at RGD: some of the QTLs are associated with more than one phenotype
| Major QTL Categories Based on Phenotypes | QTLs, n |
|---|---|
| Cardiovascular | 492 |
| Neurological | 240 |
| Glucose homeostasis | 164 |
| Immune and inflammatory | 158 |
| Body weight | 123 |
| Lipid homeostasis | 109 |
QTL Nomenclature, Curation, and Registration
RGD collects and curates all known rat QTLs from direct submissions by rat researchers as well as those published in the current literature. RGD curators have precise searches that are set to retrieve recent published papers weekly from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/pubmed). These searches are based on the different ways in which QTLs are characterized in the literature, on different QTL names, and on prominent rat researchers specializing in rat QTLs. All of the retrieved papers are curated by RGD, so that the database has the latest and most up-to-date record of rat QTLs. During the process of curating a QTL, the name and symbol are assigned, based on the Guidelines for Nomenclature of Genes, Genetic Markers, Alleles, and Mutations in Mouse and Rat instituted by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/gene.shtml). A condensed version of these guidelines is available at RGD (http://rgd.mcw.edu/nomen/quick_guide_qtl_nomenclature.html). The name and symbol are based on the trait to which the QTL is linked using the Vertebrate Trait Ontology (VT) (http://rgd.mcw.edu/rgdweb/ontology/view.html?acc_id=VT:0000001#s). If a researcher does not assign an appropriate symbol and name to the QTL that complies with the nomenclature rules, then RGD assigns official nomenclature and stores the name and symbol used by the author as an alias (old QTL symbol or name). These aliases are indexed in the search table so that the correct QTL report is retrieved even when an alias is used as the keyword (8).
During QTL curation, statistical details such as the logarithm of odds (LOD) score and P value that determine the QTL, chromosome location, and markers that flank the boundaries of the QTL are extracted from the literature or submitted by the researcher. The method used to measure the phenotype, the number of animals used, and the breeding techniques are curated. Any drug, chemical, dietary regime or other condition used to induce or aid progression of the phenotype is also indicated. The references used to extract the data are linked to the QTL record as curated references. QTLs in RGD are annotated with the Mammalian Phenotype Ontology (13) and the RDO disease ontology (based on the MEDIC disease ontology) (2). To define the experimental parameters used to identify the QTL, annotations are also made using the Rat Strain Ontology, the VT, the Clinical Measurement Ontology (CMO), the Measurement Method Ontology (MMO), and the Experimental Condition Ontology (XCO) (11, 12).
RGD encourages rat researchers to submit and register their QTL data prior to publication. This ensures that the researchers name their QTLs correctly and that the data are made available at RGD in a timely manner. During this process of registering QTLs, a QTL symbol with a sequential number, name, and RGD ID are assigned. The researchers are provided with the name and ID and are encouraged to mention the correct nomenclature and RGD ID in their publication. QTLs can be easily submitted for registration at http://rgd.mcw.edu/tools/qtls/qtlRegistrationIndex.cgi. QTLs with similar phenotypes are assigned the same root symbol plus a unique sequential number, so that similar QTLs are grouped together. There are instances where identical QTL symbols and names have been assigned to different QTLs by different researchers, so the registration process helps to eliminate these types of errors. The elapsed time between submitting data and receiving an official name/symbol is typically <24 h. Data registered at RGD can be kept private until they are published or released at the researcher's discretion. Researchers can contact RGD directly by phone or email and also by using the “contact us” link on the RGD website.
Searching for a QTL at RGD
Every QTL in RGD has its own report page, which contains specifications and relevant information for that specific QTL. There are four standard strategies to search for a QTL report in RGD.
General keyword search.
Entering a keyword in the text box at the top right of any RGD page searches the RGD ID, symbol, name, trait, alias, annotations, and notes fields for QTLs, as well as similar fields for other data objects such as genes and strains (Fig. 1A). If the author(s)' name(s) or the PubMed ID of the relevant publication is known, that can also be searched.
Fig. 1.

Quantitative trait locus (QTL) search page. A: the keyword search box is found in the upper right corner of most pages on the Rat Genome Database (RGD) website. B: the keyword search box on the QTL search page specifically searches QTL-associated data fields. The search can also be limited by chromosomal location, genomic assembly, and species.
QTL-specific search.
A link to the QTL-specific search page is found on the RGD Data page, which is accessed by clicking the “Data” tab at the top of most RGD web pages. Entering a term in the QTL keyword search queries the QTL table in the RGD database. Chromosomal position, genomic assembly, and species can also be used to limit the QTL search (Fig. 1B).
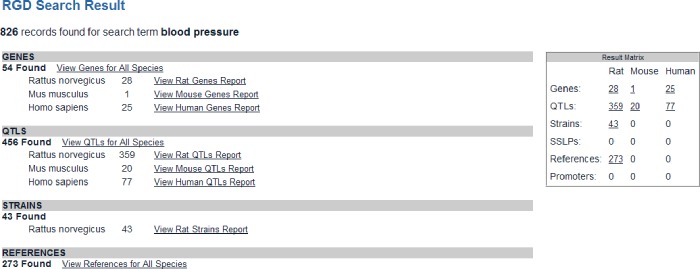
The search result page for general and specific searches displays all the objects, such as genes, QTLs, strains, and references, that have the associated searched term (Fig. 2). A click on the link “View Rat QTLs Report” goes to the list of returned QTLs, with links to their individual report pages.
Fig. 2.
General keyword search result page listing all RGD objects that have the term “blood pressure” in them.
Classic QTL search.
The QTL classic search is accessible from the QTL search page (Fig. 3). In addition to the search constraints on the QTL search page, the Classic Search allows the user to filter by trait, strain, LOD score, P values, start and stop positions on the chromosome, and references.
Fig. 3.
Classic QTL search entry page. The QTL search on this page can be limited by a variety of parameters including chromosome, logarithm of odds (LOD) score, P value, trait, and others.
All of the above mentioned searches lead to the result page (Fig. 2), which has links leading to the QTL report pages, such as the Bp155 report page (see Fig. 5).
Fig. 5.
A: typical RGD QTL report page showing the general information section along with a strain report linked from the QTL page and a PhenoMiner details table linked from the strain page. B: annotation section of the QTL report page displaying candidate gene along with a gene page linked from the QTL page and an allele page linked to the gene page. In addition, the “Experimental Data Annotations” and “RGD Disease Portals” subsections have been expanded to show details. C: region section; D: additional Information section.
Ontology search.
Standardized terms can be used to search for QTLs using the ontology/vocabulary term browser (10) (Fig. 4A). A keyword search (for example, “arterial blood pressure”) leads to a list of ontologies/vocabularies that have terms associated with the search word or phrase. Clicking on one of the listed ontologies/vocabularies (for example CMO) returns all the associated terms for that ontology/vocabulary (Fig. 4B). Clicking on a specific term (for example, “mean arterial blood pressure”) returns an image of the entire genome showing all the objects annotated to this term (Fig. 4C). The QTLs and other object symbols in the accompanying list are linked to their respective report pages.
Fig. 4.
Ontology general search page. A: any of the ontologies/vocabularies used to annotate QTLs can be searched on this page. B: the search for “arterial blood pressure” returns a list of ontologies/vocabularies that have terms containing the searched word or phrase. In this case the Clinical Measurement Ontology (CMO) has been selected, and 5 terms have been returned in the right frame (C). When the term “mean arterial blood pressure” is selected, the Ontology Report page is returned with all objects annotated to that term displayed in the Genome Viewer (GViewer) (chromosome display) and listed below in a table. All QTL symbols in the list are hyperlinked to the respective QTL report pages.
Data in a QTL Report Page
A typical QTL report page consists of the following major sections:
General.
The basic details of blood pressure QTL 155 (Fig. 5) are displayed in the top section (Fig. 5A), including the symbol, name, trait, subtrait, aliases, method used to determine this QTL, the LOD score, P value, and variance.
The map position is determined by the SSLP markers reported in the original journal article. The chromosome and map positions for different assemblies are presented. The cross type and strain(s) used to characterize the QTL are indicated with links to the strain report pages of the parental strains (SHR/N and WKY/N for the Bp155 example in Fig. 5A). The strain pages contain links to the quantitative phenotype values curated from this paper and can be accessed through the section on the strain report page “Phenotype Values via PhenoMiner” (8, 9). To visualize the QTL in the genome browser (GBrowse) (Fig. 6), links can be found at the bottom and upper right portions of the General section, which provide a view of additional data within the QTL region. GBrowse can be used to compare the position of the QTL to candidate genes, markers, and other QTLs. Also, on the upper right side of the report page, a link to the QTL registration page and RGD object information for the QTL can be found.
Fig. 6.
GBrowser at RGD showing the QTLs that lie in the same region as the rat gene Nppa, with a balloon pop-up that gives detailed information related to QTL Bp155 when the cursor hovers over the Bp155 track.
Annotation.
The next section of the report contains information on candidate genes, phenotype, and disease associations, as well as information on the experiment. The candidate genes, “Nppa” and “Nppb” for Bp155 reported in the original QTL publication, are mentioned here (Fig. 5B) under “Candidate Gene Status.” A click on a gene symbol takes the user to the gene report page, which has details about this gene, along with the allele and splice variants reported. These curated alleles in turn have their own report pages, listing the mutant strains from which they were derived. “Disease” and “Phenotype Annotations” are also available on the QTL report page. The “Experimental Data Annotations” section contains annotations with terms from the VT, the CMO, the MMO, and the XCO (8, 9). These annotations are based on how the QTL trait was determined by the authors in the publication used to curate the QTL. The “References” subsection lists the citation of the original publication in which the QTL was determined, as well as citations of any subsequent papers that refer to the same QTL. Each citation is a hyperlink to the reference report page, which has an abstract of the publication and a list of all annotations that came from that paper. The “RGD Disease Portals” subsection lists disease portals (for example, the cardiovascular, diabetes, and the obesity/metabolic syndrome portal) in which the Bp155 QTL appears, with links to the portals (Fig. 5B). The rat and human QTLs are presented as lists in the appropriate disease portal(s) (Fig. 7), based on the diseases that are annotated to them. Specifying a disease category and disease at the top of the disease portal page allows for a filtered search for specific QTLs.
Fig. 7.
GViewer at RGD showing the annotated genes, QTLs, and congenic strains that are associated with the cardiovascular portal.
Information on “Related QTLs” that interact with or map to the same region as the QTL of interest is also provided. For example, the Bp155 QTL maps to the same region to which Cm55, Lanf1, and Strs2 QTLs map, as mentioned by Ye and West (15).
Region and additional information.
The “Region” section (Fig. 5C) provides details of the chromosomal region to which the QTL maps. The subsections include “Genes in Region,” “Markers in Region,” “Position Markers,” and “QTLs in Region”. These data are based on the map positions for the various objects in RGD. The “Genes in Region” subsection lists all the genes that overlap the upstream and downstream coordinates of the QTL. These genes are not included as candidate genes since the original publication did not nominate them, but they do map within the QTL region. This list has options for downloading, printing, or linking to GViewer. The “Markers in Region” subsection lists all the SSLPs that are located within the upstream and downstream coordinates of the QTL. The list has the same reporting and viewing options as the previous subsection. The “Position Markers” subsection displays the peak and flanking markers as reported in the original publication from which the QTL was curated. The markers are represented as the appropriate SSLP or gene symbol with positions listed by specific assemblies and cytogenetic map. The “QTLs in Region” subsection lists all QTLs that overlap any portion of the region of the QTL that is the subject of the report page. The reporting and viewing options of the list are the same as for the “Genes in Region” and “Markers in Region.”
The “External Database Links” (Fig. 5D) subsection of the “Additional Information” section contains a link to the corresponding QTL report page in the NCBI gene database. The “RGD Curation Notes” subsection contains additional free text information about the QTL that is not found elsewhere on the QTL report page.
Visualization of QTL Data With RGD Tools
Software tools are helpful in the process of discovering genes that are involved in specific diseases. QTLs are narrow chromosomal regions that can encompass many genes, so they act as a road map of the genome. The Genome Viewer (GViewer) at RGD is a tool that provides a genome-wide view of all the genes, QTLs, and congenic strains that have been annotated with searched phenotype or disease ontology/vocabulary terms. The Bp155 QTL (Chr5:157,584,492..167,844,579) can be viewed in GViewer along with all the genes, QTLs, and congenic strains (Figs. 4C and 7) annotated with the same ontology term. From the genome-wide view one can zoom in to a smaller region to get additional information. These QTL boundaries can be transferred into GBrowse (Fig. 6) for a more magnified view. Syntenic regions from the rat GBrowse can also be viewed in the human and/or mouse GBrowse, which helps in comparing QTLs among the three species. Disease-related tracks and the syntenic blocks in GBrowse add depth to the analysis of a region (12). In Fig. 6, Bp155 is visualized along with all the other QTLs that are mapped to this region, with the candidate gene Nppa and with the congenic strain that harbors this genomic region as an introgressed fragment.
Summary
The identification of QTLs is a valuable first step in understanding the associations among genes, markers, and phenotypes of complex diseases. RGD provides a valuable resource for QTL data that allows comparative analyses across the rat genome and, through synteny, across mouse and human genomes. RGD is a unique resource for official nomenclature of rat and human QTLs. The QTL report pages provide important details curated for published QTLs, and links to tools for visualization and analysis in a comparative context. RGD allows researchers to focus on what has already been established in regard to QTLs and where they might direct future research.
RGD help pages (http://rgd.mcw.edu/wg/help3) can be accessed throughout the site as a descriptive guide. All aspects of the RGD website, including QTLs, are explained in the help pages, which aid in understanding and exploring the RGD data and tools. Users can subscribe to the Rat Community Forum (http://mailman.mcw.edu/mailman/listinfo/rat-forum), a web-based moderated forum that allows the research community to post questions, answers, and comments related to the rat. This interactive forum is widely used to increase community involvement, which is greatly needed to complement and add value to rat research. Researchers are urged to submit their QTL and other data to RGD prior to publication. Users can also submit their concerns and comments to RGD using the “Contact Us” form (http://rgd.mcw.edu/contact/index.shtml), a platform used for interactions between the rat research community and the RGD team. Being a community resource, RGD has QTL and all other data freely available for download through its ftp site (ftp://rgd.mcw.edu/pub/data_release/).
GRANTS
RGD and PhenoMiner are funded by the National Heart, Lung, and Blood Institute (RGD, HL-64541; PhenoMiner, HL-094271).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: R.N., M.R.D., M.S., and H.J.J. conception and design of research; R.N., S.J.F.L., G.T.H., J.R.S., S.-J.W., T.F.L., and V.P. interpreted results of experiments; R.N. and S.J.F.L. prepared figures; R.N. drafted manuscript; R.N. edited and revised manuscript; R.N., S.J.F.L., G.T.H., J.R.S., S.-J.W., T.F.L., V.P., J.d.P., M.T., W.L., P.J., D.H.M., E.A.W., M.R.D., M.S., and H.J.J. approved final version of manuscript.
REFERENCES
- 1.Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, Gauguier D, Geurts AM, Gould M, Harris PC, Holmdahl R, Hubner N, Izsvak Z, Jacob HJ, Kuramoto T, Kwitek AE, Marrone A, Mashimo T, Moreno C, Mullins J, Mullins L, Olsson T, Pravenec M, Riley L, Saar K, Serikawa T, Shull JD, Szpirer C, Twigger SN, Voigt B, Worley K. Progress and prospects in rat genetics: a community view. Nat Genet 40: 516–522, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Davis AP, Wiegers TC, Rosenstein MC, Mattingly CJ. MEDIC: a practical disease vocabulary used at the Comparative Toxicogenomics Database. Database (Oxford) 2012: bar065, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng Y, Rapp JP. Cosegregation of blood pressure with angiotensin converting enzyme and atrial natriuretic peptide receptor genes using Dahl salt-sensitive rats. Nat Genet 1: 267–272, 1992 [DOI] [PubMed] [Google Scholar]
- 4.Drinkwater NR, Gould MN. The long path from QTL to gene. PLoS Genet 8: e1002975, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet 6: 271–286, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Garrett MR, Rapp JP. Two closely linked interactive blood pressure QTL on rat chromosome 5 defined using congenic Dahl rats. Physiol Genomics 8: 81–86, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Jacob HJ. Functional genomics and rat models. Genome Res 9: 1013–1016, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Laulederkind SJ, Hayman GT, Wang SJ, Lowry TF, Nigam R, Petri V, Smith JR, Dwinell MR, Jacob HJ, Shimoyama M. Exploring genetic, genomic, and phenotypic data at the rat genome database. Curr Protoc Bioinformatics 1: Unit1.14, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laulederkind SJ, Liu W, Smith JR, Hayman GT, Wang SJ, Nigam R, Petri V, Lowry TF, De Pons J, Dwinell MR, Shimoyama M. PhenoMiner: quantitative phenotype curation at the rat genome database. Database (Oxford) 2013: bat015, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laulederkind SJ, Tutaj M, Shimoyama M, Hayman GT, Lowry TF, Nigam R, Petri V, Smith JR, Wang SJ, De Pons J, Dwinell MR, Jacob HJ. Ontology searching and browsing at the Rat Genome Database. Database (Oxford) 2012: bas016, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimoyama M, Nigam R, McIntosh LS, Nagarajan R, Rice T, Rao DC, Dwinell MR. Three ontologies to define phenotype measurement data. Front Genet 3: 87, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimoyama M, Smith JR, Hayman T, Laulederkind S, Lowry T, Nigam R, Petri V, Wang SJ, Dwinell M, Jacob H, Team RGD. RGD: a comparative genomics platform. Hum Genomics 5: 124–129, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 6: R7, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spence JP, Liang T, Liu L, Johnson PL, Foroud T, Carr LG, Shekhar A. From QTL to candidate gene: a genetic approach to alcoholism research. Curr Drug Abuse Rev 2: 127–134, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye P, West MJ. Cosegregation analysis of natriuretic peptide genes and blood pressure in the spontaneously hypertensive rat. Clin Exp Pharmacol Physiol 30: 930–936, 2003 [DOI] [PubMed] [Google Scholar]