Abstract
The RON receptor tyrosine kinase, a member of the MET proto-oncogene family, is a pathogenic factor implicated in tumor malignancy. Specifically, aberrations in RON signaling result in increased cancer cell growth, survival, invasion, angiogenesis, and drug resistance. Biochemical events such as ligand binding, receptor overexpression, generation of structure-defected variants, and point mutations in the kinase domain contribute to RON signaling activation. Recently, functional crosstalk between RON and signaling proteins such as MET and EFGR has emerged as an additional mechanism for RON activation, which is critical for tumorigenic development. The RON signaling crosstalk acts either as a regulatory feedback loop that strengthens or enhances tumorigenic phenotype of cancer cells or serves as a signaling compensatory pathway providing a growth/survival advantage for cancer cells to escape targeted therapy. Moreover, viral oncoproteins derived from Friend leukemia or Epstein-Barr viruses interact with RON to drive viral oncogenesis. In cancer cells, RON signaling is integrated into cellular signaling network essential for cancer cell growth and survival. These activities provide the molecular basis of targeting RON for cancer treatment. In this review, we will discuss recent data that uncover the mechanisms of RON activation in cancer cells, review evidence of RON signaling crosstalk relevant to cancer malignancy, and emphasize the significance of the RON signaling addiction by cancer cells for tumor therapy. Understanding aberrant RON signaling will not only provide insight into the mechanisms of tumor pathogenesis, but also lead to the development of novel strategies for molecularly targeted cancer treatment.
Keywords: Receptor tyrosine kinase (RON), signaling pathway, activation mechanism, signaling crosstalk, oncogene addiction, tumorigenesis
INTRODUCTION
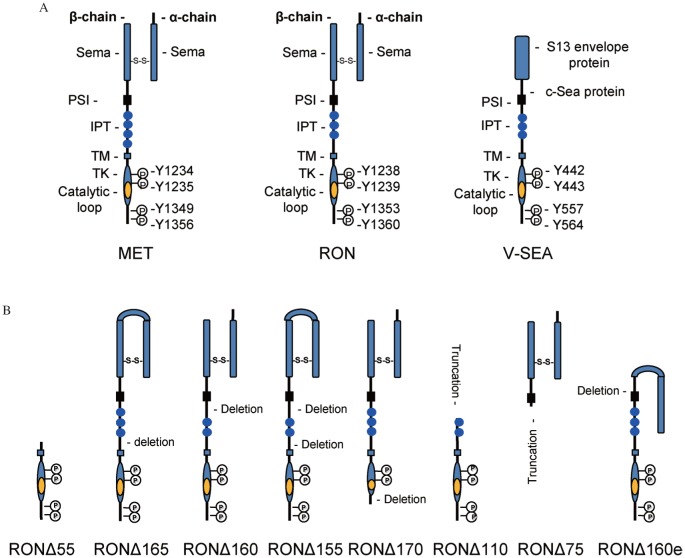
Discoveries of recepteur d'origine nantais (RON) occurred in 1993[1]. Molecular cloning of the human RON cDNA revealed that RON is a receptor protein tyrosine kinase (RTK) belonging to the C-MET proto-oncogene family (Fig. 1A)[2],[3]. Shortly thereafter in 1994, the cDNA coding the mouse homology of RON was cloned and named as stem-cell derived tyrosine kinase receptor[4]. Human RON gene resides in the chromosome 3p21 region[1] and is highly conserved in different species including human, mouse, feline, chicken, zebrafish, and xenopus[1],[4]–[11]. Interestingly, in avian erythroblastosis retrovirus S13 that causes chicken sarcoma, erythroblastosis, and anemia, a viral oncoprotein namely V-SEA was identified (Fig. 1A)[12],[13]. V-SEA is a hybrid protein containing the chicken SEA kinase domain fused with viral envelope sequences[9],[12],[13]. The chicken SEA is a homolog of human RON[10]. These findings indicate that RON is evolutionally preserved in different species. In addition, various RON variants have been identified in cancer cells (Fig. 1B). In 1994, macrophage-stimulating protein (MSP, also known as hepatocyte growth factor (HGF)-like protein) was identified as the ligand of RON[14]–[16]. This finding establishes the MSP-RON signaling axis.
Fig. 1. Schematic representation of RON and RON variant.
A: General features of MET, RON, and V-SEA. MET is the classical example of this family. Mature RON consists of a 35 kDa α-chain and a 145 kDa β-chain linked by a disulfide bond. The α-chain resides extracellularly and contains a portion of Semaphorin (Sema). The β-chain comprises a large extracellular domain, a short transmembrane (TM) segment, and a cytoplasmic portion harboring a tyrosine kinase (TK) domain and a C-terminal tail. The Sema domain harbors a ligand-binding pocket for the MSP β-chain. Regulatory tyrosine residues Tyr1238 and Tyr1239 in the TK domain and Tyr1353 and Tyr1360 in the C-terminal tail are marked. V-SEA is an oncogenic protein fused by the avian S13 retroviral envelope protein with the chicken SEA sequences. PSI, Plexins-Semaphorins-Integrins; IPT, immunoglobulin-plexin-transcription. B: Different RON variants. RONΔ55 is derived from alternative initiation at Met913. RONΔ165 is formed by deletion of exon 11 coding 49 amino acids. RONΔ160 has a deletion of exons 5 and 6 coding 109 amino acids. RONΔ155 has a combined deletion of exons 5, 6 and 11. RONΔ170 is derived from deletion of exon 19 in the kinase domain. RONΔ110 is formed by N-terminal truncation at Arg631. RONΔ85 is a free variant with C-terminal truncation at Asp634 caused by insertion. RONΔ160e is derived by deletion of exon 2.
RON signaling in tumorigenesis and therapy has gained steady attention over the last 20 years. Aberrant RON activation, featured by overexpression[17]–[24], isoform generation[25]–[35], and persistent activation of downstream signaling pathways[17]–[35], has been found in various types of cancers[17]–[35]. Moreover, functional crosstalk between RON and signaling proteins contributes to tumorigenic progression and malignancy[36]–[43]. The finding that RON signaling is abnormal in cancer cells provides a rationale for development of RON-targeted cancer therapy. Currently, small molecule inhibitors and therapeutic antibodies are under clinical trials (www.clinicaltrials.gov). Here, we discuss our current knowledge about mechanisms of RON activation, discuss the emerging roles of RON signaling crosstalk in cancer malignancy, and summarize the significance of RON signaling addiction by cancer cells for potential cancer therapy.
MECHANISMS OF RON ACTIVATION
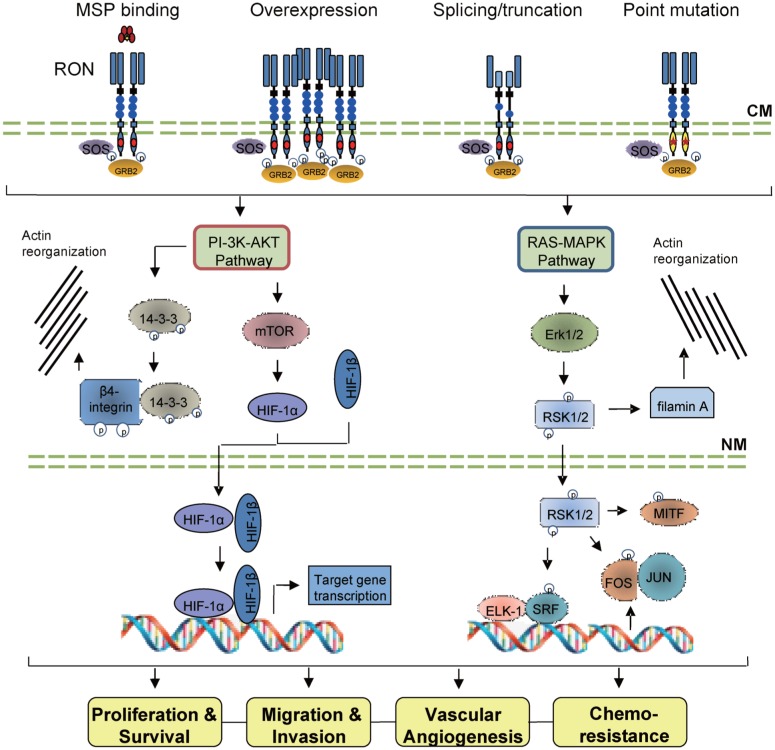
Dimerization of RON in the cell surface is the first step required for RON activation[1],[15],[16]. Four biochemical events are known to activate RON (Fig. 2): specific ligand binding[44],[45], receptor overexpression[17]–[23], generation of oncogenic variants[25],[27], and point mutations in the kinase domain[46],[47]. The feature of RON activation is autophosphorylation at Tyr1238 and Tyr1239 at the A-loop (Phe1227-Pro1250) in the kinase domain[1],[48]–[50]. Phosphorylation of these regulatory residues then activates the tyrosine kinase leading to further phosphorylation of Tyr1353 and Tyr1360 in the C-terminal docking site (Fig. 1 and 2)[48]–[50]. The docking site interacts with downstream signaling proteins triggering classical RAS-MAPK and PI-3K-AKT pathways[28],[34],[51]–[57] (Fig. 2). These pathways are responsible for increased proliferation/ survival[58], epithelial to mesenchymal transition (EMT)[20],[59],[60], motile-invasive activity[51],[59],[61], and chemoresistance[62],[63].
Fig. 2. RON activation mechanisms and classical signaling pathways.
Activation of RON is mediated by MSP binding, overexpression, splicing/truncation, and point mutations. Upon activation, the C-terminal docking site recruits cytoplasmic molecules son of sevenless (SOS) and growth factor receptor-bound protein (GRB2) to initiate two classical signaling pathways, Ras-MAPK and PI-3K-AKT. The RAS-MAPK pathway regulates RON-mediated cell growth, survival, and invasiveness. Activated Erk1/2 also stimulates p90 ribosomal S6 kinase (RSK)-2 to regulate gene transcription and cytoskeleton reorganization to cause EMT. The PI-3K-AKT pathway regulates RON-mediated cell shape change, migration and matrix invasion. It also stimulates mTOR signaling to promote HIF-1α activation for gene transcription. AKT also stimulates 14-3-3 phosphorylation, which regulates α6β4 integrin for cell motility. CM, cell membrane; ELK-1, ETS domain-containing protein-1; Erk, extracellular signal-regulated kinase; MITF, microphthalmia-associated transcription factor; mTOR, mammalian target of rapamycin; NM nuclear membrane, SRF, specific response factors.
Ligand-induced activation
Ligand-induced activation: The binding of MSP to RON is the classical mode to induce RON dimerization leading to signaling activation (Fig. 2)[44],[45]. MSP is the only known physiological ligand that specifically activates RON[15],[16]. As a protein belonging to the HGF family[64]–[66], MSP is a product of hepatocyte, which circulates in blood as a biologically inactive single-chain precursor[64],[66]. Proteolytic conversion results in biologically active/mature MSP[67]–[70], which gains the receptor binding capability[44],[45].
The MSP molecule possesses two-receptor binding sites[44],[45]. The high affinity-binding site is in the MSP β-chain, which binds to an interface in the RON extracellular Sema domain[44],[45],[71]. The MSP α-chain harbors a low affinity-binding site[45]. The location of the corresponding interface in the RON extracellular domain is unknown. Binding by both MSP α- and β-chains is required to activate RON[44],[45]. Crystal structure analysis reveals that a central cleft harboring three residues in the putative catalytic site in the MSP β-chain is essential for the β-chain binding to the RON Sema domain[71]. The binding follows an enzyme-substrate mode conserved in HGF-related growth factors and proteases of the blood clotting pathway[72],[73].
Structural analysis under protein crystal packing reveals that individual molecules of the MSP β-chain do not interact with each other to form a receptor binding moiety[72]. Instead, the central cleft in the single MSP β-chain directly binds to the RON Sema homodimer[72]. This suggests that dimerization of the MSP β-chain is not required for RON activation. In contrast, RON Sema molecules form a homodimer, which creates a ligand-binding interface by two Sema domains[71]. Thus, the interface created by the RON Sema dimer appears to be the high affinity binding pocket for the MSP β-chain.
In light of these discoveries, we propose a model of one MSP molecule interacting with two RON receptors for dimerization. This model depicts that as a monomeric form, MSP uses its high affinity-binding site in the β-chain to bind to the interface in the Sema domain formed the RON homodimer. The binding causes receptor conformational changes and exposes a currently unknown binding pocket in the RON extracellular domain for the low affinity-bind site in the MSP α-chain. The sequential binding of the MSP β- and α-chains initiates triggers autophosphoryla- tion of regulatory tyrosine residues in the RON kinase domain followed by activation of the tyrosine kinase and creation of the C-terminal multifunctional docking site[48]–[50].
Isoform-mediated activation
Generation of constitutively active RON variants is another mechanism activating RON (Fig. 2). Currently, at least eight RON variants have been identified (Fig. 1B), which include RONΔ170[74], RONΔ165[25], RONΔ165.e11p[77], RONΔ160[27], RONE5/6in[30], RONΔ155[27], RON110[76], RONΔ85[29],[78],[79], and RONΔ55[1],[33],[34]. Alternative mRNA splicing is primarily responsible for generation of RON variants[25],[27],[29],[30],[32],[35],[74], although protein truncation and alternative transcription also play a role[34],[75],[76]. RON variants are either constitutively active, oncogenic, or biologically inactive due to defects in various regions[25]–[35]. RON variants also display different cellular localizations either on the cell surface or in the intracellular compartments[25],[27].
The biochemical events that control RON variant activation are largely unknown. Conformational changes due to deletion of amino acids or a particular domain in the RON protein appear to cause spontaneous tyrosine phosphorylation[25],[30],[35]. In the case of RONΔ160, a splicing variant with an in-frame deletion of 109 amino acids coded by exons 5 and 6 for the first IPT motif in the RON extracellular domains[1],[27], deletion results in unbalanced cysteine residues in the extracellular sequences, which leads to spontaneous dimerization of the RON protein[27],[35]. Moreover, deletion converts wild-type RON into an oncogenic variant that transforms cell in vitro and causes tumor growth in vivo[27]. Thus, generation of constitutively active RON variants is a mechanism of RON activation, which has pathological implications in cell transformation and subsequent tumor progression.
Overexpression-induced activation
Overexpression of RON exists in various types of cancers and has prognostic values for patient survival[17]–[24],[80]–[86]. Overexpression is characterized by abnormal accumulation of RON and RON variant proteins at high levels and their constitutive phosphorylation in cancer cells (Fig. 2). The cause of overexpression is complex and still under investigation. Increased RON protein stability and resistance to endocytosis, intracellular proteolysis, and degradation are possible mechanisms leading to RON overexpression[30]. Impairment in the intracellular proteasome degradation pathway in cancer cells is another mechanism resulting in RON accumulation[87]. Moreover, genetic aberrations in the RON gene can lead to RON overexpression[22]. In gastroesophageal adenocarcinoma, the RON gene is highly amplified (22), suggesting that increased gene copy number could be a mechanism of RON overexpression. Finally, cellular conditions surrounding cancer cells such as hypoxia affects RON expression and accumulation[88]. The RON gene transcription is dramatically increased through hypoxia-inducible factor (HIF)-1α in acute hypoxic cancer cells[88]. Thus, overexpression of RON is manifested at various cellular and molecular levels in cancer cells.
Overexpression-induced RON activation appears to be mediated by homodimer of two RON molecules under the condensed conditions[71]. Analysis of RON-RON interaction under crystal packing confirms that the RON Sema domains form homodimer[71]. In cancer cells, abnormal accumulation of RON in the cell surface or in the cytoplasm creates an environment with high density of RON. Such increased density is sufficient to cause formation of the RON homodimer.
Point mutation-mediated activation
Experimental mutation of certain critical residues such as Asp1232 and Met1254 in the RON kinase domain results in RON activation (Fig. 2)[46],[47]. This constitutes the fourth types of RON activation. Asp1232 and Met1254 are two critical residues highly conserved in the kinase domain of RTKs[89],[90]. The same mutations in KIT and RET cause two human malignancies, mastocytosis and multiple endocrine neoplasia type 2B, respectively[91],[92]. In cell lines, Asp1232Val or Met1254Thr substitution in the RON kinase domain is sufficient to convert RON into an oncogenic agent[46],[47]. Moreover, substitution overcomes the requirement for the multifunctional docking site in induction of tumor formation[46],[47].Substitution of Met1254 with Thr in the RON kinase domain causes a conformational rearrangement, which stabilizes a specific open region in the a-loop in the kinase domain[93]. The rearrangement also facilitates the regulatory residue Tyr1238 moving into a position usually reserved for the substrate tyrosine. The localization in the substrate-like position allows the intramolecular or cis phosphorylation of Tyr1238, which eventually activates RON[93]. This mode of intramolecular/cis autophosphorylation provides an insight into the molecular mechanism of RON activation.
CLASSICAL RON SIGNALING PATHWAYS
RON signaling is conventionally transduced by the RAS-MAPK cascade and the PI-3K-AKT pathway (Fig. 2)[16],[18],[94],[51],[54],[95]. This pattern is similar to that activated by MET[96]. Interaction of RON with adaptor proteins including Grb2 and β-arrestin-1 is the first step bridging RON activation with downstream signaling cascades[95],[97],[98]. Various cytoplasmic effector molecules such as PLC-γ[48], PI-3 kinase[53], Src[98], 14-3-3[57], c-Cbl[87],[99], Hsc70[99], protein phosphatase 1[100], plectin[95], and integrin-β4[57],[98] interact with RON through the C-terminal docking site. The differential and selective interactions under different conditions may determine the specificity of RON-mediated signaling in a cell context-dependent manner.
Among tumorigenic activities mediated by RON signaling, the coordinated activation of the RAS-MAPK and PI-3K-AKT pathways is critical for EMT with increased cellular motility[20],[34],[50],[51],[59],[95]. In the MDCK cell model, RON-mediated EMT, featured by spindle morphologies, diminished expression of E-cadherin, and increased appearance of vimentin, is mediated by the RAS-MAPK pathway[59],[101]. Ribosomal protein S6 kinase (RSK)-2, a downstream signaling protein of the MAPK pathway[102],[103], is the principal molecule linking RON signaling to EMT (Fig. 2)[101]. Genetic studies confirm that RSK-2 functions as a molecular switch to confer promotile/invasive phenotypes in epithelial cells[102],[103]. The invasive growth is further regulated by RON-mediated PI-3K-AKT signaling, which increases in vitro epithelial cell adhesion, migration, matrix invasion, and in vivo tumor cell invasion, and distant metastasis[50],[51].
CROSSTALK BETWEEN RON AND SIGNALING PROTEINS
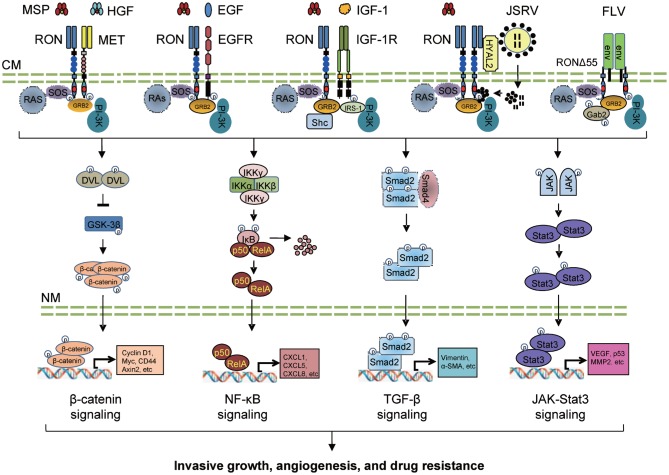
At the cell surface, RON is engaged in active crosstalk with other RTKs such as EGFR, MET, and IGF-1R (Fig. 3)[36]–[43]. RON also crosstalks with viral oncoproteins derived from Friend leukemia virus (FLV)[104]–[106], Jaagsiekte sheep retrovirus (JSRV)[107],[108], and Epstein-Barr virus (EBV) (Fig. 3)[109]. Such crosstalk has emerged as a mechanism for regulating tumorigenic phenotype and chemoresistance[36]–[43],[104]–[112].
Fig. 3. Functional crosstalk between RON and signaling protein.
The crosstalk of RON with MET, EGFR, and IGF-1R occurs in various cancer cells and cause increased tumorigenic activity. RON also crosstalks with viral envelope oncoproteins derived from JSRV and FLV to cell transformation and proliferation. At least four signaling pathways are activated upon crosstalking. The β-catenin pathway is stimulated through RON-mediated PI-3K-AKT pathway that activates protein dishevel (DVL) and inactivates glycogen synthase kinase (GSK)-3β leading to cytoplasmic β-catenin accumulation and nuclear translocation. The crosstalk between RON and the NF-κB pathway causes cancer cell growth, angiogenesis, and survival. NF-κB also directly binds the RON promoter, increases RON transcription, and enhances RON-mediated cancer cell migration. In epithelial cells, RON crosstalks with TGF-β signaling to induce EMT for cancer cell invasiveness. Moreover, RONΔ55 binds the FLV envelope protein and interacts with the JAK-Stat3 pathway to induce erythropoietin-independent proliferation of erythroid cells. CM, cell membrane; CXCL, Chemokine (C-X-C motif) ligand; Gab, GRB2-associated-binding protein; IKK, IκB Kinase; IRS-1, insulin receptor substrate-1; JAK, Janus kinase; MMP, matrix metallopeptidase; NM nuclear membrane. SMA, smooth muscle actin; Smad, mothers against decapentaplegic homolog; Stat, signal transducer and activator of transcription; and VEGF, vascular endothelial growth factor.
The crosstalk between RON and MET is evident by the presences of RON-MET heterodimer on the cell surface[36],[37]. RON also directly associates with EGFR, irrespective of ligand stimulation[38],[42]. HGF-induced MET activation results in transphosphorylation of RON at Tyr1238 and Tyr1239 residues. Similarly, MSP stimulation causes MET transphosphorylation at Tyr1234 and Tyr1235[36],[37]. Such transphosphorylation up-regulates the kinase activity of RON and MET, respectively. Similarly, transphosphorylation also occurs between RON and EGFR or PDGFR[38],[39],[42],[43].
As a signaling regulatory feedback loop, the crosstalk between RON and MET enhances or attenuates MET and RON-mediated tumorigenic activity. In cancer cells, kinase-inactive RON impairs MET-mediated cellular-transforming activity[36],[37]. Moreover, RON kinase transphosphorylation is able to sustain MET oncogene addiction with increased tumorigenic activities[37]. The similar effect also is observed between RON and EGFR[38,428,43]. Considering the fact that various RON variants are expressed in various types of cancer cells, the crosstalk between RON/RON variants and other types of RTKs should have a significant regulatory effect on tumorigenic signaling and their associated biological activities.
Crosstalk between RON and signaling proteins also serves as a signaling-compensatory mechanism (Fig. 3). In sarcoma cells with acquired resistance to IGF-1R targeted therapeutics antibodies, RON expression/activation has emerged as a survival mechanism[40]. In these sarcoma cells, RON is unusually expressed at high levels. Inhibition of RON expression impairs activation of ribosomal protein S6, a critical IGF-1R signaling component for acquired resistance. Furthermore, knockdown of RON expression by specific siRNA restores sensitivities of drug-resistant cells in response to IGF-1R kinase inhibitor BMS-536924. Thus, the crosstalk between RON and IGF-1R represents an escaping strategy for tumor cells in response to IGF-1R targeted cancer therapy.
RON signaling crosstalk also is manifested for viral oncogenesis (Fig. 3)[104]–[110]. In B cell transformation induced by the latent membrane protein (LMP)-1 of EBV, the crosstalk between LMP-1-induced NF-κB and RON expression promotes the growth of transformed lymphoblastoid cells[108]. In JSRV envelope protein-induced sheep lung adenocarcinoma, which is morphologically similar to human bronchioloalveolar carcinoma[111], RON is found to be directly associated with the JSRV envelope protein[107],[108]. The interaction appears to be RON specific because EGFR or CD4 does not form complex with JSRV envelope protein[1073,108]. In addition, association of RON with hyaluronidase (HYAL)-2, a cell surface protein serving as the entry receptor for JSRV[111],[112], also is reported[107]. In FLV-infected cells, RONΔ55 covalently interacts with the FLV viral protein to activate downstream signaling pathways[110]. These findings strongly suggest that RON signaling crosstalk is vital for virus-mediated cell transformation and tumorigenic activity.
RON SIGNALING ADDICTION BY CANCER CELLS
Involvement of RON signaling in cancer pathogenesis raises a critical question: are cancer cells fully addicted to RON signaling for growth/survival or is RON only been utilized for tumorigenic activities? The answer to this question is important to establish RON signaling in cancer biology and to provide a rationale for RON-targeted cancer therapy.
The accepted notion from various in vitro studies is that certain cancer cells are addicted to RON signaling for growth and survival[20],[51],[54],[113]. First, knockdown of RON expression by specific siRNA causes phenotypic changes in cancer cells with decreased cell proliferation, significant cell cycle arrest, reduced cell motility, and increased apoptosis[20],[51],[63],[54],[113]. One report even finds it impossible to establish a RON-deficient pancreatic cancer BxPC-3 cell line after stable expression of RON specific shRNA[63]. However, in most cases, RON-specific siRNA-mediated activity only exerts the partial inhibitory effect or affects a small fraction of cancer cells[20],[51],[54],[63],[113],[114]. Studies from in vivo tumor xenograft models also confirms that tumor growth induced by colon HT-29 and pancreatic FG cells with stable RON-specific siRNA was only partially reduced based on measuring tumor volumes[63],[114]. Second, small molecule inhibitors such as PHA665752, compound-I, and BMS-777607 targeting RON/MET are able to block RON-mediated activities leading to increased growth inhibition and cell apoptosis[114]–[119]. Third, specific RON targeting antibodies is able to inhibit or reduce tumor growth caused by cancer cells that overexpress RON[120],[121]. Again, only partial growth inhibition or reduction of tumor volume is observed from these animal work[120],[121]. Thus, RON signaling appears to be integrated at certain levels into the cellular signaling network for cell growth, survival, and motility.
Cancer cells addicted to RON signaling display interesting patterns of gene expression relevant to advanced tumorigenic phenotypes[20],,[51],[54],[55],[62],[63],[122],[113]. Global gene expression patterns indicate that RON signaling mediates a unique transcriptional program with increased expression of genes for growth, survival, and malignancy[63]. Consistent with these observations, stress-induced RON nuclear localization directly binds and regulates various gene transcription known to participate in tress-response network including p53, c-JUK, and PI-3K-AKT[43]. Activation of these stress-related signaling pathways facilitates tumor cell growth and survival under hostile environments[43]. Moreover, cancer cells addicted to RON signaling often show strong crosstalk with other signaling pathways to strengthen their malignant progression[123-125]. One example is RON signaling in crosstalk with the β-catenin pathway in colon and breast cancer cells[54,123-125]. Thus, RON-mediated gene transcription in addicted cancer cells could be a unique molecule marker determining tumorigenic and drug-resistant phenotypes.
PERSPECTIVES
Studies accumulated from the last decade have allowed us to assess the pathogenic roles of RON signaling in epithelial carcinogenesis. Although lacking evidence as a cancer-causative agent, aberrant RON expression/activation is a pathogenic factor associated with tumorigenic behavior and chemoresistance. At present, our studies of RON pathogenesis in cancer have advanced into translational and clinical phases. The knowledge of RON signaling activation, crosstalk, and addiction by cancer cells provides the mechanistic insight for validating RON as a prognostic biomarker and drug target. With continued advanced in this field, the value of aberrant RON expression/activation will be established by successful application of targeted RON therapy for cancer treatment.
Footnotes
This work was supported in part by National Institutes of Health grant R01 CA91980 (MHW) and a grant from the Amarillo Area Foundation (MHW). RWZ was supported by NIH grants R01 CA112029 and CA121211. Supports were also provided by subproject #2011ZZ01 (MHW) from State Key Laboratory for Diagnosis & Treatment of Infectious Diseases in First Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, Zhejiang, P. R. China. The assistance of Ms. Susan Denney (Texas Tech University Health Sciences Center School of Pharmacy, Amarillo, TX) in editing the manuscript is greatly appreciated. In addition, the authors declare no competing financial interests.
References
- 1.Ronsin C, Muscatelli F, Mattei MG, Breathnach R. A novel putative receptor protein tyrosine kinase of the met family. Oncogene. 1993;8:1195–202. [PubMed] [Google Scholar]
- 2.Gherardi E, Sharpe M, Lane K, Sirulnik A, Stoker M. Hepatocyte growth factor/scatter factor (HGF/SF), the c-met receptor and the behavior of epithelial cells. Symp Soc Exp Biol. 1993;47:163–81. [PubMed] [Google Scholar]
- 3.Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci USA. 1987;84:6379–83. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwama A, Okano K, Sudo T, Matsuda Y, Suda T. Molecular cloning of a novel receptor tyrosine kinase gene, STK, derived from enriched hematopoietic stem cells. Blood. 1994;83:3160–9. [PubMed] [Google Scholar]
- 5.De Maria R, Maggiora P, Biolatti B, Prat M, Comoglio PM, Castagnaro M, et al. Feline STK gene expression in mammary carcinomas. Oncogene. 2002;21:1785–90. doi: 10.1038/sj.onc.1205221. [DOI] [PubMed] [Google Scholar]
- 6.Bassett DI. Identification and developmental expression of a macrophage stimulating 1/ hepatocyte growth factor-like 1 orthologue in the zebrafish. Dev Genes Evol. 2003;213:360–2. doi: 10.1007/s00427-003-0339-3. [DOI] [PubMed] [Google Scholar]
- 7.Huitema LF, Renn J, Logister I, Gray JK, Waltz SE, Flik G, et al. Macrophage-stimulating protein and calcium homeostasis in zebrafish. FASEB J. 2012;26:4092–101. doi: 10.1096/fj.11-202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Théry C, Sharpe MJ, Batley SJ, Stern CD, Gherardi E. Expression of HGF/SF, HGF1/MSP, and c-met suggests new functions during early chick development. Dev Genet. 1995;17:90–101. doi: 10.1002/dvg.1020170110. [DOI] [PubMed] [Google Scholar]
- 9.Huff JL, Jelinek MA, Jamieson TA, Parsons JT. Expression and maturation of the cellular sea receptor, a member of the hepatocyte growth factor (HGF) receptor family of protein tyrosine kinases. Oncogene. 1996;12:299–307. [PubMed] [Google Scholar]
- 10.Wahl RC, Hsu RY, Huff JL, Jelinek MA, Chen K, Courchesne P, et al. Chicken macrophage stimulating protein is a ligand of the receptor protein-tyrosine kinase Sea. J Biol Chem. 1999;274:26361–8. doi: 10.1074/jbc.274.37.26361. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura T, Aoki S, Takahashi T, Matsumoto K, Kiyohara T, Nakamura T. Cloning and expression of Xenopus HGF-like protein (HLP) and Ron/HLP receptor implicate their involvement in early neural development. Biochem Biophys Res Commun. 1996;224:564–73. doi: 10.1006/bbrc.1996.1065. [DOI] [PubMed] [Google Scholar]
- 12.Hayman MJ, Kitchener G, Vogt PK, Beug H. The putative transforming protein of S13 avian erythroblastosis virus is a transmembrane glycoprotein with an associated protein kinase activity. Proc Natl Acad Sci USA. 1985;82:8237–41. doi: 10.1073/pnas.82.23.8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DR, Vogt PK, Hayman MJ. The v-sea oncogene of avian erythroblastosis retrovirus S13: another member of the protein-tyrosine kinase gene family. Proc Natl Acad Sci USA. 1989;86:5291–5. doi: 10.1073/pnas.86.14.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang MH, Iwama A, Skeel A, Suda T, Leonard EJ. The murine stk gene product, a transmembrane protein tyrosine kinase, is a receptor for macrophage-stimulating protein. Proc Natl Acad Sci USA. 1995;92:3933–7. doi: 10.1073/pnas.92.9.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang MH, Ronsin C, Gesnel MC, Coupey L, Skeel A, Leonard EJ, et al. Identification of the ron gene product as the receptor for the human macrophage stimulating protein. Science. 1994;266:117–9. doi: 10.1126/science.7939629. [DOI] [PubMed] [Google Scholar]
- 16.Gaudino G, Follenzi A, Naldini L, Collesi C, Santoro M, Gallo KA, et al. RON is a heterodimeric tyrosine kinase receptor activated by the HGF homologue MSP. EMBO J. 1994;13:3524–32. doi: 10.1002/j.1460-2075.1994.tb06659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang MH, Lee W, Luo YL, Weis MT, Yao HP. Altered expression of the RON receptor tyrosine kinase in various epithelial cancers and its contribution to tumorigenic phenotypes in thyroid cancer cells. J Pathol. 2007;213:402–11. doi: 10.1002/path.2245. [DOI] [PubMed] [Google Scholar]
- 18.Thomas RM, Toney K, Fenoglio-Preiser C, Revelo-Penafiel MP, Hingorani SR, Tuveson DA, et al. The RON receptor tyrosine kinase mediates oncogenic phenotypes in pancreatic cancer cells and is increasingly expressed during pancreatic cancer progression. Cancer Res. 2007;67:6075–82. doi: 10.1158/0008-5472.CAN-06-4128. [DOI] [PubMed] [Google Scholar]
- 19.Kanteti R, Krishnaswamy S, Catenacci D, Tan YH, EL-Hashani E, Cervantes G, et al. Differential expression of RON in small and non-small cell lung cancers. Genes Chromosomes Cancer. 2012;51:841–51. doi: 10.1002/gcc.21968. [DOI] [PubMed] [Google Scholar]
- 20.Camp ER, Yang A, Gray MJ, Fan F, Hamilton SR, Evans DB, et al. Tyrosine kinase receptor RON in human pancreatic cancer: expression, function, and validation as a target. Cancer. 2007;109:1030–9. doi: 10.1002/cncr.22490. [DOI] [PubMed] [Google Scholar]
- 21.Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, De Bortoli M, et al. Overexpression of the RON gene in human breast carcinoma. Oncogene. 1998;16:2927–33. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 22.Catenacci DV, Cervantes G, Yala S, Nelson EA, El-Hashani E, Kanteti R, et al. RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer Biol Ther. 2011;12:9–46. doi: 10.4161/cbt.12.1.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren X, Daa T, Yada N, Kashima K, Fujitomi Y, Yokoyama S. Expression and mutational status of RON in neoplastic lesions of the breast: analysis of MSP/RON signaling in ductal carcinoma in situ and invasive ductal carcinoma. APMIS. 2012;120:358–67. doi: 10.1111/j.1600-0463.2011.02841.x. [DOI] [PubMed] [Google Scholar]
- 24.Chou YC, Chen CL, Yeh TH, Lin SJ, Chen MR, Doong SL, et al. Involvement of recepteur d'origine nantais receptor tyrosine kinase in Epstein-Barr virus-associated nasopharyngeal carcinoma and its metastasis. Am J Pathol. 2012;181:1773–81. doi: 10.1016/j.ajpath.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Collesi C, Santoro M. M., Gaudino G., Comoglio P. M. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol. 1996;16:5518–26. doi: 10.1128/mcb.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santoro M. M., Collesi C., Grisendi S., Gaudino G., Comoglio P. M. Constitutive activation of the RON gene promotes invasive growth but not transformation. Mol. Cell Biol. 1996;16:7072–83. doi: 10.1128/mcb.16.12.7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y. Q., He C, Chen Y. Q., Wang D., Wang M. H. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: generation of different splicing RON variants and their oncogenic potential. Oncogene. 2003;22:186–97. doi: 10.1038/sj.onc.1206075. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Zhao L, Derose YS, Lin YC, Bieniasz M, Eyob H, et al. Short-Form Ron Promotes Spontaneous Breast Cancer Metastasis through Interaction with Phosphoinositide 3-Kinase. Genes Cancer. 2011;2:753–62. doi: 10.1177/1947601911421924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckerich C, Schulte A, Martens T, Zapf S, Westphal M, Lamszus K. RON receptor tyrosine kinase in human gliomas: expression, function, and identification of a novel soluble splice variant. J Neurochem. 2009;109:969–80. doi: 10.1111/j.1471-4159.2009.06027.x. [DOI] [PubMed] [Google Scholar]
- 30.Ma Q, Zhang K, Guin S, Zhou YQ, Wang MH. Deletion or insertion in the first immunoglobulin-plexin-transcription (IPT) domain differentially regulates expression and tumorigenic activities of RON receptor Tyrosine Kinase. Mol Cancer. 2010;9:307–136. doi: 10.1186/1476-4598-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X, Hao L, Ni S, Liu Q, Xu J, Correll PH. Altered exon usage in the juxtamembrane domain of mouse and human RON regulates receptor activity and signaling specificity. J Biol Chem. 2005;280:40241–51. doi: 10.1074/jbc.M506806200. [DOI] [PubMed] [Google Scholar]
- 32.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–90. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Fialin C, Larrue C, Vergez F, Sarry JE, Bertoli S, Mansat-De Mas V, et al. The short form of RON is expressed in acute myeloid leukemia and sensitizes leukemic cells to cMET inhibitors. Leukemia. 2013;27:325–35. doi: 10.1038/leu.2012.240. [DOI] [PubMed] [Google Scholar]
- 34.Bardella C, Costa B, Maggiora P, Patane' S, Olivero M, Ranzani GN, et al. Truncated RON tyrosine kinase drives tumor cell progression and abrogates cell-cell adhesion through E-cadherin transcriptional repression. Cancer Res. 2004;64:5154–61. doi: 10.1158/0008-5472.CAN-04-0600. [DOI] [PubMed] [Google Scholar]
- 35.Wang M. H., Kurtz A. L., Chen Y. Q. Identification of a novel splicing product of the RON receptor tyrosine kinase in human colorectal carcinoma cells. Carcinogenesis. 2000;21:1507–12. [PubMed] [Google Scholar]
- 36.Follenzi A, Bakovic S, Gual P, Stella MC, Longati P, Comoglio PM. Cross-talk between the proto-oncogenes Met and Ron. Oncogene. 2000;19:3041–9. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- 37.Benvenuti S, Lazzari L, Arnesano A, Li Chiavi G, Gentile A, Comoglio PM. Ron kinase transphosphorylation sustains MET oncogene addiction. Cancer Res. 2011;71:1945–55. doi: 10.1158/0008-5472.CAN-10-2100. [DOI] [PubMed] [Google Scholar]
- 38.Peace BE, Hill KJ, Degen SJ, Waltz SE. Cross-talk between the receptor tyrosine kinases Ron and epidermal growth factor receptor. Exp Cell Res. 2003;289:317–25. doi: 10.1016/s0014-4827(03)00280-5. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Furukawa Y, Kikuchi J, Ito C, Miyata Y, Muto S, et al. Transactivation of RON receptor tyrosine kinase by interaction with PDGF receptor beta during steady-state growth of human mesangial cells. Kidney Int. 2009;75:1173–83. doi: 10.1038/ki.2009.44. [DOI] [PubMed] [Google Scholar]
- 40.Potratz JC, Saunders DN, Wai DH, Ng TL, McKinney SE, Carboni JM, et al. Sorensen PH. Synthetic lethality screens reveal RPS6 and MST1R as modifiers of insulin-like growth factor-1 receptor inhibitor activity in childhood sarcomas. Cancer Res. 2010;70:8770–81. doi: 10.1158/0008-5472.CAN-10-1093. [DOI] [PubMed] [Google Scholar]
- 41.Jaquish DV, Yu PT, Shields DJ, French RP, Maruyama KP, Niessen S, et al. IGF1-R signals through the RON receptor to mediate pancreatic cancer cell migration. Carcinogenesis. 2011;32:1151–6. doi: 10.1093/carcin/bgr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas CY, Theodorescu D. Collaboration of RON and epidermal growth factor receptor in human bladder carcinogenesis. J Urol. 2006;176:1909–10. doi: 10.1016/j.juro.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 43.Liu HS, Hsu PY, Lai MD, Chang HY, Ho CL, Cheng HL, et al. An unusual function of RON receptor tyrosine kinase as a transcriptional regulator in cooperation with EGFR in human cancer cells. Carcinogenesis. 2010;31:1456–64. doi: 10.1093/carcin/bgq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang MH, Julian FM, Breathnach R, Godowski PJ, Takehara T, Yoshikawa W, et al. Macrophage stimulating protein (MSP) binds to its receptor via the MSP beta chain. J Biol Chem. 1997;272:16999–7004. doi: 10.1074/jbc.272.27.16999. [DOI] [PubMed] [Google Scholar]
- 45.Danilkovitch A, Miller M, Leonard EJ. Interaction of macrophage-stimulating protein with its receptor. Residues critical for beta chain binding and evidence for independent alpha chain binding. J Biol Chem. 1999;274:29937–43. doi: 10.1074/jbc.274.42.29937. [DOI] [PubMed] [Google Scholar]
- 46.Santoro MM, Penengo L, Minetto M, Orecchia S, Cilli M, Gaudino G. Point mutations in the tyrosine kinase domain release the oncogenic and metastatic potential of the Ron receptor. Oncogene. 1998;17:741–9. doi: 10.1038/sj.onc.1201994. [DOI] [PubMed] [Google Scholar]
- 47.Williams TA, Longati P, Pugliese L, Gual P, Bardelli A, Michieli P. MET(PRC) mutations in the Ron receptor result in upregulation of tyrosine kinase activity and acquisition of oncogenic potential. J Cell Physiol. 1999;181:507–14. doi: 10.1002/(SICI)1097-4652(199912)181:3<507::AID-JCP15>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 48.Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77:261–71. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 49.Iwama A, Yamaguchi N, Suda T. STK/RON receptor tyrosine kinase mediates both apoptotic and growth signals via the multifunctional docking site conserved among the HGF receptor family. EMBO J. 1996;15:5866–75. [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao ZQ, Chen YQ, Wang MH. Requirement of both tyrosine residues 1330 and 1337 in the C-terminal tail of the RON receptor tyrosine kinase for epithelial cell scattering and migration. Biochem Biophys Res Commun. 2000;267:669–75. doi: 10.1006/bbrc.1999.2011. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Rajput A, Kan JL, Rose R, Liu XQ, Kuropatwinski K, et al. Knockdown of Ron kinase inhibits mutant phosphatidylinositol 3-kinase and reduces metastasis in human colon carcinoma. J Biol Chem. 2009;284:10912–22. doi: 10.1074/jbc.M809551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhuri A, Xie MH, Yang B, Mahapatra K, Liu J, Marsters S, et al. Distinct involvement of the Gab1 and Grb2 adaptor proteins in signal transduction by the related receptor tyrosine kinases RON and MET. J Biol Chem. 2011;286:32762–74. doi: 10.1074/jbc.M111.239384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang MH, Montero-Julian FA, Dauny I, Leonard EJ. Requirement of phosphatidylinositol-3 kinase for epithelial cell migration activated by human macrophage stimulating protein. Oncogene. 1996;13:2167–75. [PubMed] [Google Scholar]
- 54.Danilkovitch-Miagkova A, Miagkov A, Skeel A, Nakaigawa N, Zbar B, Leonard EJ. Oncogenic mutants of RON and MET receptor tyrosine kinases cause activation of the beta-catenin pathway. Mol Cell Biol. 2001;21:5857–68. doi: 10.1128/MCB.21.17.5857-5868.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JS, Park JH, Khoi PN, Joo YE, Jung YD. MSP-induced RON activation upregulates uPAR expression and cell invasiveness via MAPK, AP-1 and NF-κB signals in gastric cancer cells. Carcinogenesis. 2011;32:175–81. doi: 10.1093/carcin/bgq241. [DOI] [PubMed] [Google Scholar]
- 56.Danilkovitch A, Donley S, Skeel A, Leonard EJ. Two independent signaling pathways mediate the antiapoptotic action of macrophage-stimulating protein on epithelial cells. Mol. Cell Biol. 2000;20:2218–27. doi: 10.1128/mcb.20.6.2218-2227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santoro MM, Gaudino G, Marchisio PC. The MSP receptor regulates alpha6beta4 and alpha3beta1 integrins via 14-3-3 proteins in keratinocyte migration. Dev Cell. 2003;5:257–71. doi: 10.1016/s1534-5807(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 58.Wang MH, Dlugosz AA, Sun Y, Suda T, Skeel A, Leonard EJ. Macrophage-stimulating protein induces proliferation and migration of murine keratinocytes. Exp Cell Res. 1996;226:39–46. doi: 10.1006/excr.1996.0200. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Shen Q, Chen YQ, Wang MH. Collaborative activities of macrophage-stimulating protein and transforming growth factor-beta1 in induction of epithelial to mesenchymal transition: roles of the RON receptor tyrosine kinase. Oncogene. 2004;23:1668–80. doi: 10.1038/sj.onc.1207282. [DOI] [PubMed] [Google Scholar]
- 60.Côté M, Miller AD, Liu SL. Human RON receptor tyrosine kinase induces complete epithelial-to-mesenchymal transition but causes cellular senescence. Biochem Biophys Res Commun. 2007;360:219–25. doi: 10.1016/j.bbrc.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu PT, Babicky M, Jaquish D, French R, Marayuma K, Mose E, et al. The RON-receptor regulates pancreatic cancer cell migration through phosphorylation-dependent breakdown of the hemidesmosome. Int J Cancer. 2012;131:1744–54. doi: 10.1002/ijc.27447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McClaine RJ, Marshall AM, Wagh PK, Waltz SE. Ron receptor tyrosine kinase activation confers resistance to tamoxifen in breast cancer cell lines. Neoplasia. 2010;12:650–8. doi: 10.1593/neo.10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Logan-Collins J, Thomas RM, Yu P, Jaquish D, Mose E, French R, et al. Silencing of RON receptor signaling promotes apoptosis and gemcitabine sensitivity in pancreatic cancers. Cancer Res. 2010;70:1130–40. doi: 10.1158/0008-5472.CAN-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skeel A, Yoshimura T, Showalter SD, Tanaka S, Ap-pella E, Leonard EJ. Macrophage stimulating protein: purification, partial amino acid sequence, and cellular activity. J Exp Med. 1991;173:1227–34. doi: 10.1084/jem.173.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han S, Stuart LA, Degen SJ. Characterization of the DNF15S2 locus on human chromosome 3: identification of a gene coding for four kringle domains with homology to hepatocyte growth factor. Biochemistry. 1991;30:9768–80. doi: 10.1021/bi00104a029. [DOI] [PubMed] [Google Scholar]
- 66.Yoshimura T, Yuhki N, Wang MH, Skeel A, Leonard EJ. Cloning, sequencing, and expression of human macrophage stimulating protein (MSP, MST1) confirms MSP as a member of the family of kringle proteins and locates the MSP gene on chromosome 3. J Biol Chem. 1993;268:15461–8. [PubMed] [Google Scholar]
- 67.Wang MH, Yoshimura T, Skeel A, Leonard EJ. Proteolytic conversion of single chain precursor macrophage-stimulating protein to a biologically active heterodimer by contact enzymes of the coagulation cascade. J Biol Chem. 1994;269:3436–40. [PubMed] [Google Scholar]
- 68.Wang MH, Gonias SL, Skeel A, Wolf BB, Yoshimura T, Leonard EJ. Proteolytic activation of single-chain precursor macrophage-stimulating protein by nerve growth factor-gamma and epidermal growth factor-binding protein, members of the kallikrein family. J Biol Chem. 1994;269:13806–10. [PubMed] [Google Scholar]
- 69.Kawaguchi M, Orikawa H, Baba T, Fukushima T, Kataoka H. Hepatocyte growth factor activator is a serum activator of single-chain precursor macrophage-stimulating protein. FEBS J. 2009;276:3481–90. doi: 10.1111/j.1742-4658.2009.07070.x. [DOI] [PubMed] [Google Scholar]
- 70.Ganesan R, Kolumam GA, Lin SJ, Xie MH, Santell L, Wu TD, et al. Proteolytic activation of pro-macrophage-stimulating protein by hepsin. Mol Cancer Res. 2011;9:1175–86. doi: 10.1158/1541-7786.MCR-11-0004. [DOI] [PubMed] [Google Scholar]
- 71.Chao KL, Tsai IW, Chen C, Herzberg O. Crystal structure of the Sema-PSI extracellular domain of human RON receptor tyrosine kinase. PLoS One. 2012;7:e41912–8. doi: 10.1371/journal.pone.0041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carafoli F, Chirgadze DY, Blundell TL, Gherardi E. Crystal structure of the beta-chain of human hepatocyte growth factor-like/macrophage stimulating protein. FEBS J. 2005;272:5799–807. doi: 10.1111/j.1742-4658.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- 73.Holmes O, Pillozzi S, Deakin JA, Carafoli F, Kemp L, Butler PJ, et al. Insights into the structure/function of hepatocyte growth factor/scatter factor from studies with individual domains. J Mol Biol. 2007;367:395–408. doi: 10.1016/j.jmb.2006.12.061. [DOI] [PubMed] [Google Scholar]
- 74.Wang MH, Lao WF, Wang D, Luo YL, Yao HP. Blocking tumorigenic activities of colorectal cancer cells by a splicing RON receptor variant defective in the tyrosine kinase domain. Cancer Biol Ther. 2007;6:1121–9. doi: 10.4161/cbt.6.7.4337. [DOI] [PubMed] [Google Scholar]
- 75.Angeloni D, Danilkovitch-Miagkova A, Ivanova T, Braga E, Zabarovsky E, Lerman MI. Hypermethylation of Ron proximal promoter associates with lack of full-length Ron and transcription of oncogenic short-Ron from an internal promoter. Oncogene. 2007;26:4499–512. doi: 10.1038/sj.onc.1210238. [DOI] [PubMed] [Google Scholar]
- 76.Lu Y, Yao HP, Wang MH. Multiple variants of the RON receptor tyrosine kinase: biochemical properties, tumorigenic activities, and potential drug targets. Cancer Lett. 2007;257:157–64. doi: 10.1016/j.canlet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 77.Zhang K, Zhou YQ, Yao HP, Wang MH. Alterations in a defined extracellular region of the RON receptor tyrosine kinase promote RON-mediated motile and invasive phenotypes in epithelial cells. Int J Oncol. 2010;36:255–64. [PubMed] [Google Scholar]
- 78.Ma Q, Zhang K, Yao HP, Zhou YQ, Padhye S, Wang MH. Inhibition of MSP-RON signaling pathway in cancer cells by a novel soluble form of RON comprising the entire sema sequence. Int J Oncol. 2010;36:1551–61. doi: 10.3892/ijo_00000642. [DOI] [PubMed] [Google Scholar]
- 79.Angeloni D, Danilkovitch-Miagkova A., Miagkov A., Leonard E. J., Lerman M. I. The soluble sema domain of the RON receptor inhibits macrophage-stimulating protein-induced receptor activation. J Biol Chem. 2004;279:3726–32. doi: 10.1074/jbc.M309342200. [DOI] [PubMed] [Google Scholar]
- 80.Lee WY, Chen HH, Chow NH, Su WC, Lin PW, Guo HR. Prognostic significance of co-expression of RON and MET receptors in node-negative breast cancer patients. Clin Cancer Res. 2005;11:2222–8. doi: 10.1158/1078-0432.CCR-04-1761. [DOI] [PubMed] [Google Scholar]
- 81.Cheng HL, Liu HS, Lin YJ, Chen HH, Hsu PY, Chang TY, et al. Co-expression of RON and MET is a prognostic indicator for patients with transitional-cell carcinoma of the bladder. Br J Cancer. 2005;92:1906–14. doi: 10.1038/sj.bjc.6602593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Compérat E, Roupret M, Chartier-Kastler E, Bitker MO, Richard F, Camparo P, et al. Prognostic value of MET, RON and histoprognostic factors for urothelial carcinoma in the upper urinary tract. J Urol. 2008;179:868–72. doi: 10.1016/j.juro.2007.10.079. [DOI] [PubMed] [Google Scholar]
- 83.Lee CT, Chow NH, Su PF, Lin SC, Lin PC, Lee JC. The prognostic significance of RON and MET receptor coexpression in patients with colorectal cancer. Dis Colon Rectum. 2008;51:1268–74. doi: 10.1007/s10350-008-9297-1. [DOI] [PubMed] [Google Scholar]
- 84.Ferrandina G, Martinelli E, Petrillo M, Prisco MG, Zucconi A, Santaguida S, et al. Prognostic role of the recepteur d'origine nantais (RON) expression in ovarian cancer patients. Gynecol Oncol. 2008;111:237–43. doi: 10.1016/j.ygyno.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 85.Zhou D, Pan G, Zheng C, Zheng J, Yian L, Teng X. Expression of the RON receptor tyrosine kinase and its association with gastric carcinoma versus normal gastric tissues. BMC Cancer. 2008;8:353–9. doi: 10.1186/1471-2407-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tactacan CM, Chang DK, Cowley MJ, Humphrey ES, Wu J, Gill AJ, et al. RON is not a prognostic marker for resectable pancreatic cancer. BMC Cancer. 2012;12:395–403. doi: 10.1186/1471-2407-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Penengo L, Rubin C, Yarden Y, Gaudino G. c-Cbl is a critical modulator of the Ron tyrosine kinase receptor. Oncogene. 2003;22:3669–79. doi: 10.1038/sj.onc.1206585. [DOI] [PubMed] [Google Scholar]
- 88.Thangasamy A, Rogge J, Ammanamanchi S. Recepteur d'origine nantais tyrosine kinase is a direct target of hypoxia-inducible factor-1alpha-mediated invasion of breast carcinoma cells. J Biol Chem. 2009;284:14001–10. doi: 10.1074/jbc.M809320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, et al. Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest. 1993;92:1736–44. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 91.Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, et al. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet. 1996;12:312–4. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- 92.Carlson KM, Dou S, Chi D, Scavarda N, Toshima K, Jackson CE, et al. Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc Natl Acad Sci USA. 1994;91:1579–83. doi: 10.1073/pnas.91.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Steinbacher S, Augustin M, Schreiner P, Epstein D, Mulvihill MJ, et al. The crystal structure of a constitutively active mutant RON kinase suggests an intramolecular autophosphorylation hypothesis. Biochemistry. 2010;49:7972–4. doi: 10.1021/bi100409w. [DOI] [PubMed] [Google Scholar]
- 94.Gaudino G, Avantaggiato V, Follenzi A, Acampora D, Simeone A, Comoglio PM. The proto-oncogene RON is involved in development of epithelial, bone and neuro-endocrine tissues. Oncogene. 1995;11:2627–37. [PubMed] [Google Scholar]
- 95.Li BQ, Wang MH, Kung HF, Ronsin C, Breathnach R, Leonard EJ, et al. Macrophage-stimulating protein activates Ras by both activation and translocation of SOS nucleotide exchange factor. Biochem Biophys Res Commun. 1995;216:110–8. doi: 10.1006/bbrc.1995.2598. [DOI] [PubMed] [Google Scholar]
- 96.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–25. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 97.Zhou YQ, Luo YL, Zhou ZW, Yao HP, Wang MH. β-Arrestin-1-dependent activation of extracellular signal-regulated kinases in RON receptor-mediated tumorigenic activities in colon cancer cells. J. Cancer Mol. 2010;5:55–62. [Google Scholar]
- 98.Danilkovitch-Miagkova A, Angeloni D, Skeel A, Donley S, Lerman M, Leonard EJ. Integrin-mediated RON growth factor receptor phosphorylation requires tyrosine kinase activity of both the receptor and c-Src. J Biol Chem. 2000;275:14783–6. doi: 10.1074/jbc.C000028200. [DOI] [PubMed] [Google Scholar]
- 99.Germano S, Barberis D, Santoro MM, Penengo L, Citri A, Yarden Y, Gaudino G. Geldanamycins trigger a novel Ron degradative pathway, hampering oncogenic signaling. J Biol Chem. 2006;281:21710–9. doi: 10.1074/jbc.M602014200. [DOI] [PubMed] [Google Scholar]
- 100.Santoro M. M., Gaudino G., Villa-Moruzzi E. Protein phosphatase 1 binds to phospho-Ser-1394 of the macrophage-stimulating protein receptor. Biochem J. 2003;376:587–94. doi: 10.1042/BJ20030391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma Q, Guin S, Padhye SS, Zhou YQ, Zhang RW, Wang MH. Ribosomal protein S6 kinase (RSK)-2 as a central effector molecule in RON receptor tyrosine kinase mediated epithelial to mesenchymal transition induced by macrophage-stimulating protein. Mol Cancer. 2011;10:66–76. doi: 10.1186/1476-4598-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, et al. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol Cell. 2009;35:511–22. doi: 10.1016/j.molcel.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smolen GA, Zhang J, Zubrowski MJ, Edelman EJ, Luo B, Yu M, et al. A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes Dev. 2010;24:2654–65. doi: 10.1101/gad.1989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Persons DA, Paulson RF, Loyd MR, Herley MT, Bodner SM, Bernstein A, et al. Fv2 encodes a truncated form of the Stk receptor tyrosine kinase. Nat Genet. 1999;23:159–65. doi: 10.1038/13787. [DOI] [PubMed] [Google Scholar]
- 105.Nishigaki K, Thompson D, Hanson C, Yugawa T, Ruscetti S. The envelope glycoprotein of friend spleen focus-forming virus covalently interacts with and constitutively activates a truncated form of the receptor tyrosine kinase Stk. J Virol. 2001;75:7893–903. doi: 10.1128/JVI.75.17.7893-7903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Umehara D, Watanabe S, Ochi H, Anai Y, Ahmed N, Kannagi M, et al. Role of phosphatidylinositol 3-kinase in friend spleen focus-forming virus-induced erythroid disease. J Virol. 2010;84:7675–82. doi: 10.1128/JVI.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Danilkovitch-Miagkova A, Duh FM, Kuzmin I, Angeloni D, Liu SL, Miller AD, et al. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by jaagsiekte sheep retrovirus. Proc Natl Acad Sci USA. 2003;100:4580–5. doi: 10.1073/pnas.0837136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Varela M, Chow YH, Sturkie C, Murcia P, Palmarini M. Association of RON tyrosine kinase with the Jaagsiekte sheep retrovirus envelope glycoprotein. Virology. 2006;350:347–57. doi: 10.1016/j.virol.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 109.Chou YC, Lin SJ, Lu J, Yeh TH, Chen CL, Weng PL, et al. Requirement for LMP1-induced RON receptor tyrosine kinase in Epstein-Barr virus-mediated B-cell proliferation. Blood. 2011;118:1340–9. doi: 10.1182/blood-2011-02-335448. [DOI] [PubMed] [Google Scholar]
- 110.He S, Ni S, Hegde S, Wang X, Sharda DR, August A, et al. Activation of the N-terminally truncated form of the Stk receptor tyrosine kinase Sf-Stk by Friend virus-encoded gp55 is mediated by cysteine residues in the ecotropic domain of gp55 and the extracellular domain of Sf-Stk. J Virol. 2010;84:2223–35. doi: 10.1128/JVI.02090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Palmarini M, Fan H. Retrovirus-induced ovine pulmonary adenocarcinoma, an animal model for lung cancer. J Natl Cancer Inst. 2001;93:1603–14. doi: 10.1093/jnci/93.21.1603. [DOI] [PubMed] [Google Scholar]
- 112.Liu SL, Miller AD. Oncogenic transformation by the jaagsiekte sheep retrovirus envelope protein. Oncogene. 2007;26:789–801. doi: 10.1038/sj.onc.1209850. [DOI] [PubMed] [Google Scholar]
- 113.Xu XM, Wang D, Shen Q, Chen YQ, Wang MH. RNA-mediated gene silencing of the RON receptor tyrosine kinase alters oncogenic phenotypes of human colorectal carcinoma cells. Oncogene. 2004;23:8464–74. doi: 10.1038/sj.onc.1207907. [DOI] [PubMed] [Google Scholar]
- 114.Zou HY, Li Q, Lee JH, Arango ME, Burgess K, Qiu M, et al. Sensitivity of selected human tumor models to PF-04217903, a novel selective c-Met kinase inhibitor. Mol Cancer Ther. 2012;11:1036–47. doi: 10.1158/1535-7163.MCT-11-0839. [DOI] [PubMed] [Google Scholar]
- 115.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–55. [PubMed] [Google Scholar]
- 116.Yao HP, Zhuang CM, Zhou YQ, Zeng JY, Zhang WR, Wang MH. Oncogenic Variant RON160 Expression in Breast Cancer and its Potential as a Therapeutic Target by Small Molecule Tyrosine Kinase Inhibitor. Curr Cancer Drug Target. 2013 doi: 10.2174/15680096113139990038. (in revision) [DOI] [PubMed] [Google Scholar]
- 117.Zhang Y, Kaplan-Lefko PJ, Rex K, Yang Y, Moriguchi J, Osgood T, et al. Identification of a novel recepteur d'origine nantais/c-met small-molecule kinase inhibitor with antitumor activity in vivo. Cancer Res. 2008;68:6680–7. doi: 10.1158/0008-5472.CAN-07-6782. [DOI] [PubMed] [Google Scholar]
- 118.Schroeder GM, An Y, Cai ZW, Chen XT, Clark C, Cornelius LA, et al. Discovery of N-(4-(2-amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a selective and orally efficacious inhibitor of the Met kinase superfamily. J Med Chem. 2009;52:1251–4. doi: 10.1021/jm801586s. [DOI] [PubMed] [Google Scholar]
- 119.Sharma S, Zeng JY, Zhuang CM, Zhou YQ, Yao HP, Hu X, Zhang RW, Wang MH. Small-Molecule Inhibitor BMS-777607 Induces Breast Cancer Cell Polyploidy with Increased Resistance to Cytotoxic Chemotherapy Agents. Mol Cancer Ther. 2013 Mar 6; doi: 10.1158/1535-7163.MCT-12-1079. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 120.O'Toole JM, Rabenau KE, Burns K, Lu D, Mangalampalli V, Balderes P, et al. Therapeutic implications of a human neutralizing antibody to the macrophage-stimulating protein receptor tyrosine kinase (RON), a c-MET family member. Cancer Res. 2006;66:9162–70. doi: 10.1158/0008-5472.CAN-06-0283. [DOI] [PubMed] [Google Scholar]
- 121.Yao HP, Zhou YQ, Ma Q, Guin S, Padhye SS, Zhang RW, et al. The monoclonal antibody Zt/f2 targeting RON receptor tyrosine kinase as potential therapeutics against tumor growth-mediated by colon cancer cells. Mol Cancer. 2011;10:82–93. doi: 10.1186/1476-4598-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Welm AL, Sneddon JB, Taylor C, Nuyten DS, van de Vijver MJ, Hasegawa BH, et al. The macrophage-stimulating protein pathway promotes metastasis in a mouse model for breast cancer and predicts poor prognosis in humans. Proc Natl Acad Sci USA. 2007;104:7570–5. doi: 10.1073/pnas.0702095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu XM, Zhou YQ, Wang MH. Mechanisms of cytoplasmic {beta}-catenin accumulation and its involvement in tumorigenic activities mediated by oncogenic splicing variant of the receptor originated from Nantes tyrosine kinase. J Biol Chem. 2005;280:25087–94. doi: 10.1074/jbc.M414699200. [DOI] [PubMed] [Google Scholar]
- 124.Wagh PK, Gray JK, Zinser GM, Vasiliauskas J, James L, Monga SP, et al. β-Catenin is required for Ron receptor-induced mammary tumorigenesis. Oncogene. 2011;30:3694–704. doi: 10.1038/onc.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]