Abstract
Changes in DNA methylation frequently accompany cancer development. One prominent change is an apparently genome-wide decrease in methylcytosine that is often ascribed to DNA hypomethylation at retroelements comprising nearly half the genome. DNA hypomethylation may allow reactivation of retroelements, enabling retrotransposition, and causing gene expression disturbances favoring tumor development. However, neither the extent of hypomethylation nor of retroelement reactivation are precisely known. We therefore assessed DNA methylation and expression of three major classes of retroelements (LINE-1, HERV-K, and AluY) in human urinary bladder cancer tissues and cell lines by pyrosequencing and quantitative reverse transcription–polymerase chain reaction, respectively. We found substantial global LINE-1 DNA hypomethylation in bladder cancer going along with a shift toward full-length LINE-1 expression. Thus, pronounced differences in LINE-1 expression were observed, which may be promoted, among others, by LINE-1 hypomethylation. Significant DNA hypomethylation was found at the HERV-K_22q11.23 proviral long terminal repeat (LTR) in bladder cancer tissues but without reactivation of its expression. DNA methylation of HERVK17, essentially absent from normal urothelial cells, was elevated in cell lines from invasive bladder cancers. Accordingly, the faint expression of HERVK17 in normal urothelial cells disappeared in such cancer cell lines. Of 16 additional HERV-Ks, expression of 7 could be detected in the bladder, albeit generally at low levels. Unlike in prostate cancers, none of these showed significant expression changes in bladder cancer. In contrast, expression of the AluYb8 but not of the AluYa5 family was significantly increased in bladder cancer tissues. Collectively, our findings demonstrate a remarkable specificity of changes in expression and DNA methylation of retroelements in bladder cancer with a significantly different pattern from that in prostate cancer.
Keywords: urothelial carcinoma, DNA hypomethylation, retroelements, HERV-K, LINE-1, Alu
Introduction
Human cancers are marked by several genetic and epigenetic changes. In particular, it is well established that aberrant DNA methylation contributes to cancer development. At specific loci, DNA hypermethylation in combination with the gain of a repressive chromatin conformation leads to gene silencing (1). Notably, whereas hypermethylation often targets genes already weakly transcribed in normal tissues, some epigenetically repressed genes exert tumor suppressive functions and their silencing by hypermethylation contributes to tumor development (2). In contrast to hypermethylation affecting single genes or selected genomic regions, a global decrease of methylcytosine occurs in cancers (3, 4). In benign tissues the bulk of the genome is densely DNA methylated. This methylation is believed to be one mechanism of restraining the activity of retroelements that comprise nearly half of the human genome (5). As several retroelements have retained their ability to retrotranspose, hypomethylation was proposed to enable their reactivation and thereby to contribute to the destabilization of the genome in cancer cells (4, 6). Global hypomethylation has been observed in many cancers, albeit occurring at different stages and to varying extents depending on the tumor entity (3).
For instance, many prostate cancers exhibit only a minor decrease in LINE-1 promoter methylation (7) with hypomethylation aggravating in higher stage carcinomas (8). Conversely, urothelial carcinoma of the urinary bladder was reported to show a higher decrease of LINE-1 methylation which was prevalent across all tumor stages (9–11). Likewise, hypomethylation of HERV-K retroelements was observed in bladder cancer cell lines and tissues correlating well with the decline of LINE-1 methylation (9). However, activation of HERV-K elements was not found in that study. In prostate cancers one specific HERV-K element, referred to as HERV-K_22q11.23 (H22q) was strikingly upregulated in cancerous tissues and this overexpression correlated well with hypomethylation of its long terminal repeat (LTR) (7). Remarkably, mRNA expression of the H22q provirus has been reported in several tumor entities and antibodies against H22q Gag protein have been detected in sera from bladder, liver, lung, ovarian, and prostate cancer patients (12). Interestingly, the androgen-responsive LTRs of H22q and HERVK17 are involved in translocation events with ETV1, a member of the ETS family of transcription factors (13, 14). These events represent rare variants of the common translocations in prostate cancer that lead to oncogenic activation of ETS transcription factors.
While the above findings have highlighted the potential role of HERV-K proviruses in prostate carcinogenesis, reports concerning HERV-K expression in bladder cancer remain sporadic. By using massively parallel signature sequencing (MPSS) Stauffer et al. (15) detected HERV-K expression in normal bladder tissue resulting from a subgroup of 17 HERV-K elements. Of particular interest might be the expression of the melanoma-associated antigen HERV-K-MEL in a subset of bladder cancers (16). We have now conducted a broader and detailed analysis of retroelement DNA methylation and expression changes in urothelial carcinomas using mainly established quantitative pyrosequencing and quantitative reverse transcription PCR (qRT-PCR) methods previously applied to prostate cancer. This allows a direct comparison of methylation and expression changes between these genitourinary cancer entities.
Materials and Methods
Tissue samples and cell lines
Patients and tumor characteristics are compiled in Table 1. Patient consent was obtained and the study approved by the Ethics Committee of the Medical Faculty of the Heinrich Heine University. All urothelial cancer cell lines (253J, 5637, 639-V, 647-V, BFTC-905, HT-1376, J82, MGH-U4, RT4, RT-112, SCaBER, SD, SW1710, UMUC3, UMUC6, VMCUB1, T24) and cancer-associated fibroblasts were cultured in DMEM GlutaMax (Gibco, Darmstadt, Germany), supplemented with 10% fetal calf serum as described previously using standard methods (17). The cell lines were obtained from the DSMZ (Braunschweig, Germany), except UMUC3, kindly provided by Dr. Grossman, Houston. The telomerase-immortalized TERT-NHUC cell line was kindly provided by Prof. M. A. Knowles (Leeds, UK) and cultured as described previously (18). The well-differentiated urothelial carcinoma cell line BC61 established in our lab was cultured as described (19). Primary urothelial cells cultures (UP) were established from ureters after nephrectomy and were routinely maintained in keratinocyte serum-free medium (KSFM, Gibco, Darmstadt, Germany) supplemented with 12.5 μg/ml bovine pituitary extract and 0.25 ng/ml epidermal growth factor as described previously (20).
Table 1.
Clinical characterization of tissue sample sets.
| DNA set (n = 23) | RNA set (n = 24) | ||
|---|---|---|---|
| Age | Median | 65 | 66 |
| 95% CI | 61–71 | 63–69 | |
| Range | 41–84 | 45–84 | |
| Gender, n (%) | Female | 7 (30.4) | 5 (20.8) |
| Male | 16 (69.6) | 19 (79.2) | |
| Pathological T stage, n (%) | pTa | 3 (13.0) | 1 (4.2) |
| pT1 | 1 (4.3) | 0 (0) | |
| T2 | 4 (17.4) | 7 (29.2) | |
| T3 | 10 (43.5) | 11 (45.8) | |
| T4 | 5 (21.7) | 5 (20.8) | |
| Nodal status, n (%) | Negative | 11 (47.8) | 13 (54.2) |
| Positive | 7 (30.4) | 8 (33.3) | |
| Unknown | 5 (21.7) | 3 (12.5) | |
| Tumor grading, n (%) | G1 | 1 (4.3) | 0 (0) |
| G2 | 5 (21.7) | 7 (29.2) | |
| G3 | 17 (73.9) | 17 (70.8) |
Nucleic acids extraction and quantitative reverse transcription–polymerase chain reaction
High molecular weight DNA and total RNA were extracted from powdered tissues using standard protocols. Notably, RNA extraction involved acid phenol extraction followed by column purification to minimize DNA contamination. Further DNA contamination was removed by synthesis of complementary DNA including a DNA removal step by DNase using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. In order to estimate the remaining levels of genomic DNA after cDNA preparation, amplification values for three different retroelement specific qPCR assays (HERVK17, LINE-1_5′ and LINE-1_3′) were assessed by quantitative PCR using cDNA preparations from three different bladder cancer cell lines (5637, BC61 and RT4) with or without reverse transcriptase (RT) treatment after DNA removal. As shown in Figure 1B (inset), amplification levels of background genomic DNA were at most around 1% of the total expression of high-copy retroelements (LINE-1_5′ and LINE-1_3′). With an assay for single-copy retroelement (HERVK17) amplification from genomic DNA was essential absent (cf. Figure 1B).
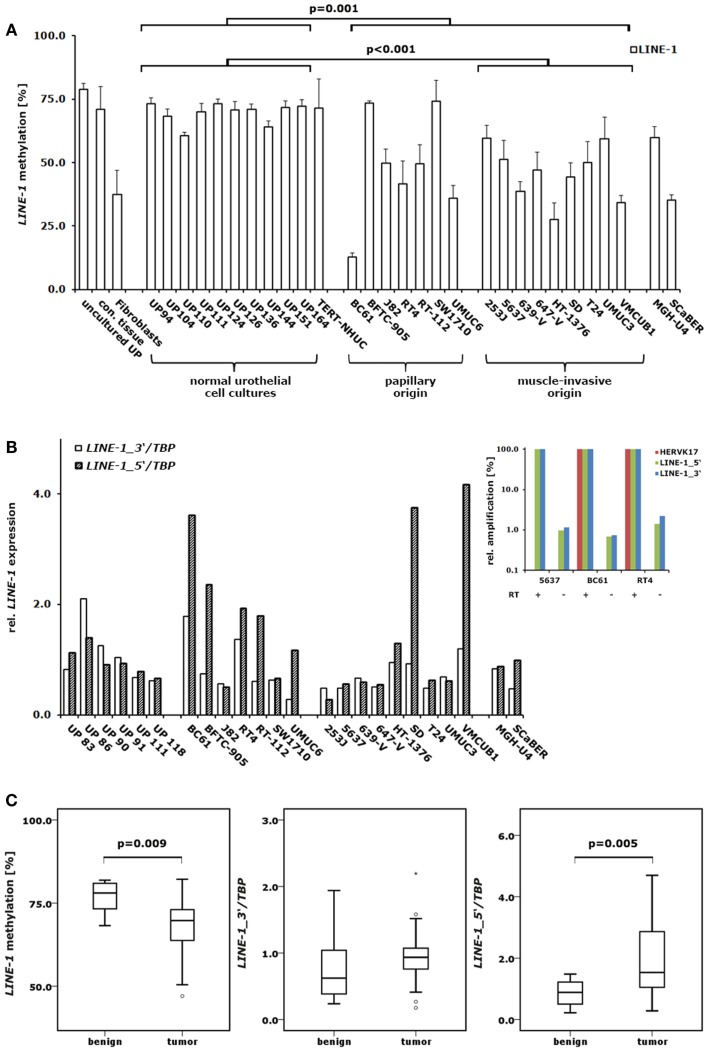
Figure 1.
DNA methylation and expression changes of LINE-1 elements in bladder cancer. (A) DNA methylation in the CpG islets of LINE-1 was quantified by pyrosequencing in a set of 10 normal urothelial cell cultures and 18 bladder cancer cell lines. For comparison, LINE-1 DNA methylation was assessed in immortalized urothelial cells (TERT-NHUC) and in uncultured epithelial cells (uncultured UP) and connective tissue from one ureter. (B) LINE-1 RNA levels from the 5′- and 3′-regions were measured by qRT-PCR in a set of 6 normal urothelial cell cultures and 18 bladder cancer cell lines. Inset: amplification of different retroelements (HERVK17, LINE-1_5′ and LINE-1_3′) was measured in three bladder cancer cell lines using cDNA preparations with (+) or without (−) reverse transcriptase (RT) to assess the impact of genomic DNA contamination. Results were adjusted for each assay and cell line to reverse transcriptase positive preparations set as 100% (C) LINE-1 DNA methylation and expression of the 5′- and 3′-regions were analyzed in a set of 12 benign and 23 cancerous bladder tissues or 11 benign and 24 tumorous bladder tissues, respectively. Methylation is plotted as mean methylation value from four CpGs in percent (A,C). RNA levels were each normalized to TBP and standardized to either the median RNA level of normal urothelial cell cultures (B) or the median RNA level of benign bladder tissues (C) set as 1. p Values calculated by the Mann–Whitney U-test were given above the brackets for significant changes (p < 0.05). Missing p values demonstrate changes without reaching the level of significance.
Quantitative reverse transcription (qRT)–PCR was performed as described previously (7) on a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using QuantiTect SYBR Green PCR Kit (Qiagen). Initial qualitative PCR with specific primers listed in Table 2 was performed as following: initial denaturation step at 95°C for 15 min, followed by 35 amplification cycles consisting of denaturation at 94°C for 15 s, annealing at 56°C for 15 s, and extension at 72°C for 30 s. Samples were analyzed on 1.5% agarose gels. Assays with detectable transcripts in this qualitative PCR were subjected to quantitative PCR analysis. All qRT-PCR data were adjusted to TATA-box-binding protein (TBP) mRNA measured by a specific TBP assay (Table 2). For all other transcripts, specifically designed primers (Table 2) were employed using the following PCR conditions: initial denaturation step at 95°C for 15 min, followed by 40 amplification cycles consisting of denaturation at 95°C for 15 s, annealing for 20 s, and extension at 72°C for 30 s. All measurements were performed in at least duplicates; assay variance was <10%. Relative expression was calculated by a modified ΔΔCt method published by Pfaffl (21).
Table 2.
Oligonucleotides.
| Gene/region | Sequence | Bases (NCBI/hg19) | Primer | Sequence 5′–3′ |
|---|---|---|---|---|
| HERV-K_3p25.3 | Chromosome 3 | 9,895,774–9,895,791 | H3p25_for | GCATCTGTCTCCTGCTTG |
| HERV-K_3p25.3 | Chromosome 3 | 9,895,722–9,895,740 | H3p25_rev | ATCTCAGTAGATGGAATCG |
| HERV-K_3q12.3 | Chromosome 3 | 101,411,166–101,411,184 | H3q12_for | GTGCTGAGGAGGATTAGTG |
| HERV-K_3q12.3 | Chromosome 3 | 101,411,341–101,411,358 | H3q12_rev | AGTATTGCTGCCGGCTTG |
| HERV-K_3q21.2 | Chromosome 3 | 125,609,734–125,609,755 | H3q21_for | ATTAGTAAAAGAGGAAAGAATG |
| HERV-K_3q21.2 | Chromosome 3 | 125,609,821-125,609,842 | H3q21_rev | CATACAATCAGGTTTTATACTG |
| HERV-K_3q27.2 | Chromosome 3 | 185,288,247–185,288,266 | H3q27_for | CATGGTTTCCAGAACAAGAA |
| HERV-K_3q27.2 | Chromosome 3 | 185,288,095–185,288,116 | H3q27_rev | GAAACTGAAACGCTATCTTCTG |
| HERV-K_10p14 | Chromosome 10 | 6,875,319–6,875,337 | H10p14_for | CTCAACTACCCAGGGATAC |
| HERV-K_10p14 | Chromosome 10 | 6,875,183–6,875,200 | H10p14_rev | TTACGGGTGTCGAGCTGC |
| HERV-K_11q22.1 | Chromosome 11 | 101,572,720–101,572,739 | H11q22_for | TCCTATTTGCTTAGGGACAG |
| HERV-K_11q22.1 | Chromosome 11 | 101,572,768–101,572,787 | H11q22_rev | GTACTTCTACCAACCAGTTT |
| HERV-K_12q14.1 | Chromosome 12 | 58,730,155–58,730,176 | H12q14_for | TGTCTCGGTATAAAACCTGACT |
| HERV-K_12q14.1 | Chromosome 12 | 58,729,973–58,729,993 | H12q14_rev | GTCAGCAGACAAACATGTGAA |
| HERV-K_12q24.11 | Chromosome 12 | 111,008,880–111,008,897 | H12q24_for | GACGAGAGATCCCGAGGA |
| HERV-K_12q24.11 | Chromosome 12 | 111,008,929–111,008,947 | H12q24_rev | CCCTAGCTTCTTCCGAGTG |
| HERV-K_10 | Chromosome 5 | 156,085,657–156,085,678 | H10_for | GAAAAGCAAGAGAGATCAAATT |
| HERV-K_10 | Chromosome 5 | 156,085,583–156,085,603 | H10_rev | GCAGAAGAATTTTTCTTAGCA |
| HERV-K_18.2 | Chromosome 1 | 160,661,401–160,661,422 | H18_for | ATCCTCCATATGCTGAACGTTG |
| HERV-K_18.2 | Chromosome 1 | 160,661,489–160,661,510 | H18_rev | TGTTTCTCGTAAGGTGCAATGA |
| HERV-K_102 | AF164610.1 (NCBI) | 819–836 | H102_for | TGGCGGGATCCTCCACAT |
| HERV-K_102 | AF164610.1 (NCBI) | 910–928 | H102_rev | CGTAAGGTGGGATGAGAGA |
| HERV-K_104 | AF164612.1 (NCBI) | 239–295 | H104_for | AGTCATCACCACTCCCTCATC |
| HERV-K_104 | AF164612.1 (NCBI) | 333–355 | H104_rev | GCCATATTTCAGACTATGAAACC |
| HERV-K_108 | AF164614.1 (NCBI) | 778–795 | H108_for | CACCCACAGATGATCAGT |
| HERV-K_108 | AF164614.1 (NCBI) | 908–925 | H108_rev | AAGGTGGGACGAGAGATT |
| HERV-K_113 | AY037928.1 (NCBI) | 565–582 | H113_for | TAGGGAAAAACCGCCTCA |
| HERV-K_113 | AY037928.1 (NCBI) | 694–711 | H113_rev | CGTGAACAAAGGTCTTGG |
| TBP | CR456776.1 (NCBI) | 11–29 | TBP_for | ACAACAGCCTGCCACCTTA |
| TBP | CR456776.1 (NCBI) | 111–130 | TBP_rev | GAATAGGCTGTGGGGTCAGT |
Primer assays for expression analysis of AluYa5, AluYb8, H22q, HERVK17, HERV-K_11q23.3, HERV-K_22q11.21, LINE-1_5′, LINE-1_3′ and pyrosequencing assays for H22q, HERVK17, LINE-1 were published previously (7).
Bisulfite treatment and DNA methylation analyses
Bisulfite conversion was performed using the EZ DNA Methylation-Gold Kit (Zymo Research, Hiss Diagnostics, Freiburg, Germany) according to the manufacturer’s instructions. Bisulfite-treated DNA samples were used for PCR with the indicated primers (Table 2) using HotStartTaq (Qiagen) under the following conditions: initial denaturation step at 95°C for 15 min, followed by 45 amplification cycles consisting of denaturation at 95°C for 15 s, annealing for 20 s, and extension at 72°C for 45 s. Pyrosequencing was carried out on a PyroMark Q24 instrument (Qiagen) according to the manufacturer’s instruction. Corresponding sequencing primers are listed in Table 2. Methylation values were calculated as the mean value of 4 (LINE-1) or 6 (H22q; HERVK17) CpG sites, respectively.
Statistics
p Values were calculated by the Mann–Whitney U-test using SPSS Statistics 20 (IBM) software. Correlation coefficients and their respective level of significance were calculated by non-parametric Spearman’s rank correlation coefficient (Spearman’s ρ) using SPSS Statistics 20 (IBM) software.
Results
LINE-1 DNA methylation and expression in urothelial cell cultures and cell lines
LINE-1 promoter DNA methylation was assessed in a set of 10 primary urothelial cell cultures and 18 bladder cancer cell lines by pyrosequencing of bisulfite-treated DNA using an assay described previously (7). Briefly, the assay interrogates four CpG sites within the internal promoter of approximately 400 full-length LINE-1 elements distributed across the genome. The bladder cancer cell lines are all derived from urothelial carcinomas with the exception of SCaBER (squamous cell carcinoma of the urinary bladder) and MGH-U4 (urothelial dysplasia). Additionally, bisulfite-treated DNA from bladder cancer-associated fibroblasts, immortalized urothelial cells (TERT-NHUC), and connective tissue originating from a freshly dissected ureter were used for comparison.
While primary normal urothelial cell cultures exhibited the high and homogenous LINE-1 promoter methylation expected of normal cells, bladder cancer cell lines showed heterogeneous changes, but with overall significantly decreased methylation levels (Mann–Whitney U-test; p = 0.001) (Figure 1A). By grouping bladder carcinoma cell lines into those either originating from papillary (n = 7) or muscle-invasive (n = 9) cancers we found those with muscle-invasive cancer origin displaying more significant LINE-1 hypomethylation compared to cultured normal urothelial cells (Mann–Whitney U-test; p < 0.001) (Figure 1A). Notably, the well-differentiated papillary cancer cell line BC61 showed the most prominent hypomethylation displaying only 14% mean LINE-1 promoter DNA methylation. Conversely, the bladder papillary cell lines BFTC-905 and SW1710 retained high methylation at LINE-1 promoters comparable with the levels in normal urothelial cells (Figure 1A). Immortalized urothelial cells (TERT-NHUC), uncultured epithelial cells and cells from connective ureter tissue exhibited the same LINE-1 methylation levels found in urothelial cell cultures, whereas cancer-associated fibroblasts had comparably low methylation.
Expression analysis of LINE-1 elements was performed on a set of 6 primary urothelial cell cultures and 18 bladder cancer cell lines from different origins (7 papillary; 9 muscle-invasive, 2 others) using two assays described previously (7) that detect either unspliced, full-length LINE-1 transcripts (LINE-1_5′; ∼400 elements), or spliced and unspliced LINE-1 transcripts (LINE-1_3′; ∼2000 elements). The LINE-1_3′-assay revealed decreased median transcript levels in bladder cancer cells compared to cultured normal urothelial cells but the changes were overall not significant (Figure 1B). In contrast, several cell lines showed increased expression by the LINE-1_5′-assay. Accordingly, we detected a prominent shift toward unspliced, full-length LINE-1 transcripts in several bladder cancer cell lines. The bladder cancer cell lines BC61, BFTC-905, RT-112, UMUC6, SD, and VMCUB1 exhibited 2.0- to 4.2-fold higher normalized LINE-1_5′ transcript levels compared to the respective LINE-1_3′ mRNA levels. However, this shift was not found across all cell lines and was therefore not overall significant.
Correlation analyses of the LINE-1 expression detected a robust and significant positive correlation between the two assessed LINE-1 transcript variants in bladder cancer cell lines (Spearman’s ρ = 0.628; p = 0.005). In bladder cancer cell lines, LINE-1 transcription correlated inversely with LINE-1 DNA methylation without reaching the level of significance. Of note, inverse correlation of LINE-1 DNA methylation with expression measured by the 5′-assay (Spearman’s ρ = 0.443; p = 0.066) was substantially better than that with the 3′-assay (Spearman’s ρ = 0.255; p = 0.307).
LINE-1 DNA methylation and expression in benign and bladder cancer tissues
To analyze LINE-1 promoter DNA methylation and LINE-1 transcript expression in benign and cancerous bladder tissues we performed methylation and expression analyses using our established pyrosequencing and quantitative RT-PCR assays on a set of 12 benign and 23 tumor probes and 11 benign and 24 cancer samples, respectively. Unfortunately, the DNA and RNA samples came from different studies with only limited overlap. LINE-1 promoter DNA methylation was highly significantly decreased in bladder cancer specimen (Mann–Whitney U-test; p = 0.009) compared to normal tissues with striking differences in their percent median values (Δmedian = 8.5%) (Figure 1C). Like the decrease in DNA methylation, LINE-1 expression changes were also similar in bladder tumor tissues to those found in cultured cells. The median levels of transcripts assessed by the LINE-1_3′-assay tended to be slightly higher in bladder cancer specimen, but the changes were not significant (Mann–Whitney U-test; p = 0.123) (Figure 1C). In contrast, analyses of full-length LINE-1 transcripts using the LINE-1_5′-assay revealed a significant increase of full-length transcript levels in bladder tumor tissues (Mann–Whitney U-test; p = 0.005) (Figure 1C). Taken together, these changes resulted in a shift toward full-length LINE-1 expression. Due to the limited overlap of DNA and RNA samples the analysis of the correlation between DNA methylation and expression was not possible.
AluYa5 and AluYb8 expression in benign and bladder cancer samples
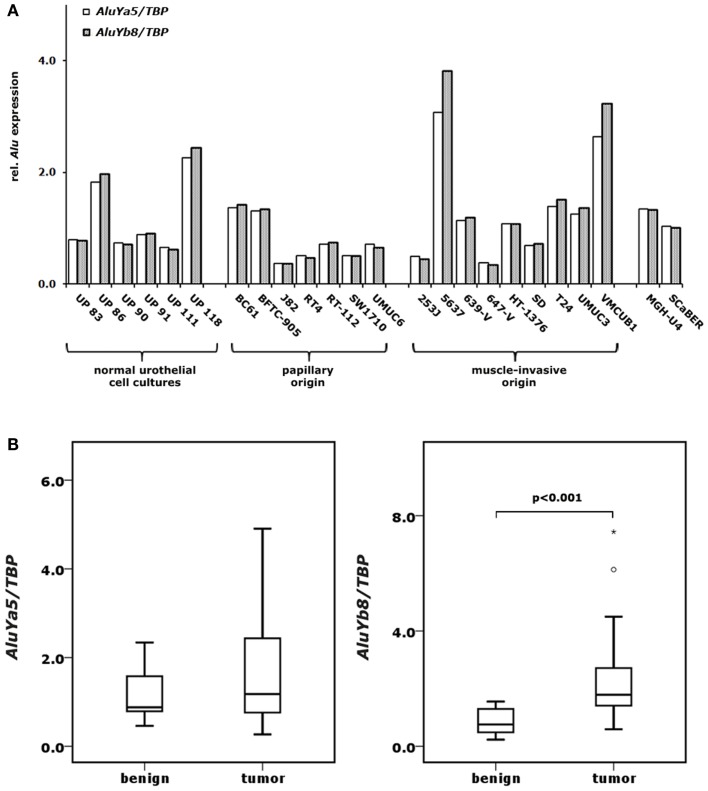
Additionally, we investigated the expression of the two most commonly active retroelements of the AluY family (AluYa5 and AluYb8) in our set of primary urothelial cell cultures and bladder cancer cell lines. We found robust expression of both elements in the primary urothelial cell cultures (Figure 2A). The expression of both elements tended to be diminished in cancer cell lines of papillary origin and was slightly increased in cell lines from muscle-invasive carcinomas without the difference reaching the level of significance (Figure 2A). Of note, the expression of both elements correlated strikingly throughout all samples (Spearman’s ρ = 0.990; p < 0.001). By applying the same assays to our set of benign and bladder cancer tissues we found no significant changes in the expression of the AluYa5 retroelements. Instead, AluYb8 transcript levels were highly significantly increased in bladder cancer specimens (Mann–Whitney U-test; p < 0.001) (Figure 2B). Other than in the cell lines, RNA levels of AluYb8 showed only a weaker and non-significant correlation to AluYa5 expression in the cancer tissues (Spearman’s ρ = 0.354; p = 0.090).
Figure 2.
Expression changes of AluYa5 and AluYb8 in bladder cancer. AluYa5 and AluYb8 RNA levels were measured by qRT-PCR in 6 normal urothelial cell cultures and 18 bladder cancer cell lines (A) as well as in 11 benign and 24 bladder cancer samples (B). RNA levels were each normalized to TBP and standardized to either the median RNA level of normal urothelial cell cultures (A) or the median RNA level of benign bladder tissues (B) set as 1. p Values calculated by the Mann–Whitney U-test were given above the brackets for significant changes (p < 0.05). Missing p values demonstrate changes without reaching the level of significance.
DNA methylation of HERV-K LTRs in benign and bladder cancer probes
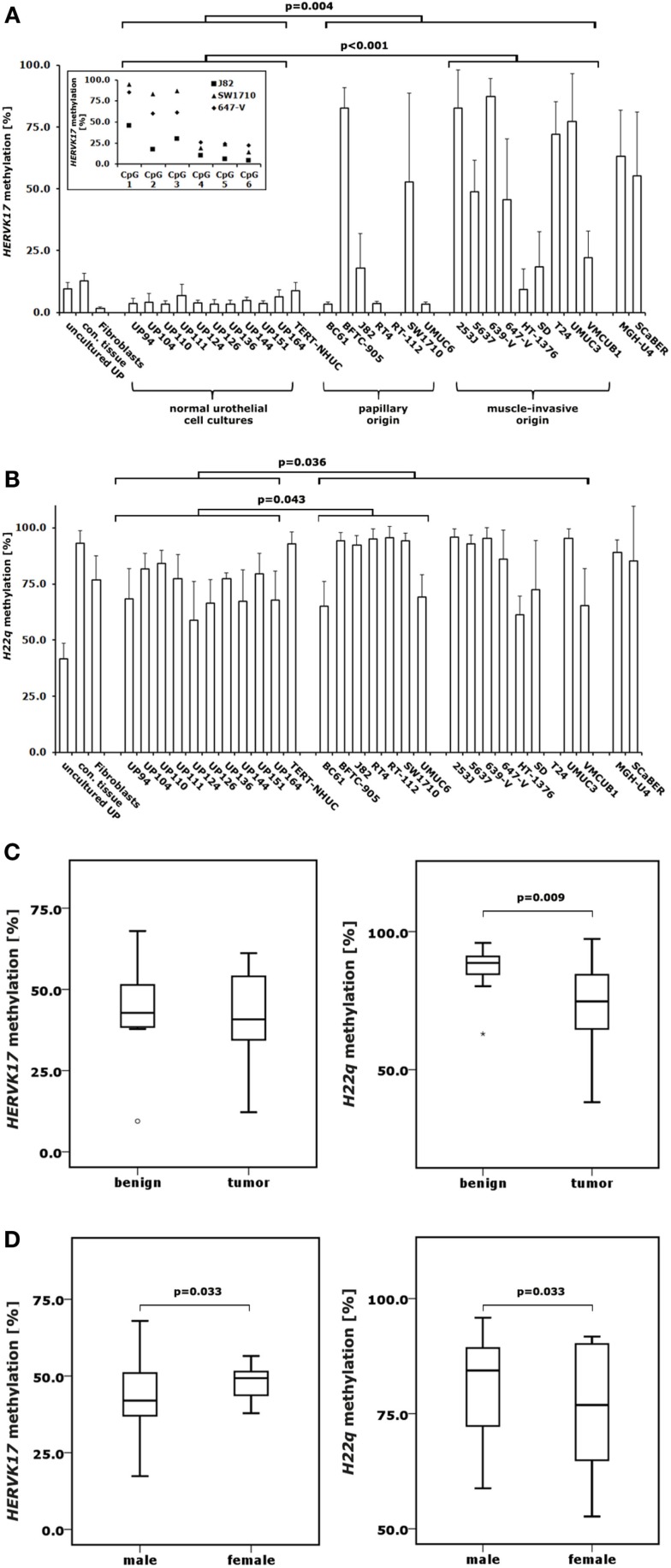
In order to investigate DNA methylation at HERV-K LTRs in urothelial samples, we used two previously established pyrosequencing assays to analyze HERVK17 and H22q methylation in bisulfite-converted DNA samples from the normal urothelial cell cultures, bladder cancer cell lines, benign and bladder cancer tissues also investigated for LINE-1 methylation. Intriguingly, we found the HERVK17 LTR to be essentially demethylated in normal urothelial cell cultures, but becoming hypermethylated in bladder cancer cells (Figure 3A). Noteworthy, DNA methylation levels in the HERVK17 LTR remained low in most bladder cell lines of papillary origin with no significant changes compared to cultured urothelial cells (Mann–Whitney U-test; p = 0.635). Significantly elevated HERVK17 DNA methylation values were instead regularly found in cancer cells derived from muscle-invasive bladder carcinomas (Mann–Whitney U-test; p < 0.001) (Figure 3A). Interestingly, HERVK17 LTR methylation was considerably higher in normal bladder tissues (median: 44.9%) compared to normal urothelial cell cultures (median: 3.7%), remaining on the same level in bladder cancer tissues (Figures 3A,C).
Figure 3.
DNA methylation changes in proviral HERVK17 and H22q LTRs in bladder cancer. DNA methylation in the LTRs of HERVK17 (A) and H22q (B) were analyzed by pyrosequencing in 10 normal urothelial cell cultures and 18 bladder cancer cell lines. Additionally, HERVK17 and H22q DNA methylation was assessed in immortalized urothelial cells (TERT-NHUC) and in uncultured epithelial cells (uncultured UP) and connective tissue from one ureter. (C) DNA methylation of HERVK17 and H22q LTRs were each analyzed in a set of 12 benign and 23 cancerous bladder tissues. (D) DNA methylation of HERVK17 and H22q LTRs from tumor samples were each plotted against patients’ gender. Methylation is plotted as mean methylation value from six CpGs each in percent. The high standard deviation in some samples results from differences in the methylation within the HERVK17 sequence, where the first three CpGs are generally higher methylated as exemplified for data from J82, SW1710, and 647-V bladder cancer cell lines in the insert (A). p Values calculated by the Mann–Whitney U-test are given above the brackets for significant changes (p < 0.05). Missing p values demonstrate changes without reaching the level of significance.
DNA methylation of the H22q proviral LTR was high in benign bladder tissues (88.6%) and declined significantly in bladder cancer specimens (72.4%, Mann–Whitney U-test; p = 0.009) (Figure 3C). Overall, LTR DNA methylation of both HERV-K proviruses correlated well and highly significantly (Spearman’s ρ = 0.669; p < 0.001) in bladder cancer tissues. Although overall comparable DNA methylation changes were found for H22q and LINE-1 no correlation was detectable. Unexpectedly, the H22q provirus was not hypomethylated, but significantly hypermethylated in bladder cancer cell lines compared to cultured normal urothelial cells (Mann–Whitney U-test; p = 0.036) (Figure 3B). Additionally, hypermethylation of the H22q LTR was more prominent in papillary cancer cell lines.
DNA methylation of H22q LTR was slightly but significantly lower in bladder cancer tissues originating from female patients (Mann–Whitney U-test; p = 0.033) (Figure 3D). Conversely, LTR methylation of the HERVK17 provirus was significantly higher in female cancers (Mann–Whitney U-test; p = 0.033). In contrast, LINE-1 promoter methylation showed no significant gender-specific differences in cancers.
Expression analyses of different HERV-K proviruses
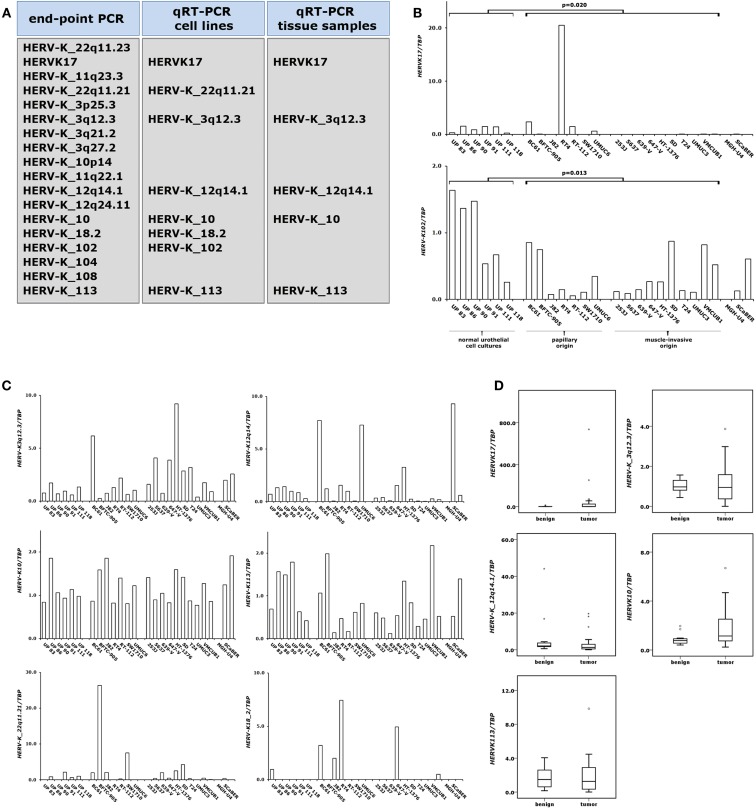
To assess HERV-K expression in benign and cancerous urothelial samples we conducted qRT-PCR analyses on our set of normal urothelial cell cultures, bladder cancer cell lines, benign and bladder tumor tissues. Initially, we performed expression analyses of the four HERV-K retroelements which had previously been investigated in prostate samples by our group (7). Then, we established qRT-PCR assays for 14 additional HERV-K elements which had been described as possibly expressed in bladder tissue by using massively parallel signature sequencing (MPSS) (15). The strategy for analysis of the expression of these HERV-K elements is illustrated in Figure 4A. First, we performed standard end-point PCR on our set of cultured normal urothelial and bladder cancer cells. Transcriptionally active HERV-K elements were subjected to quantitative RT-PCR using the same sample set. Those HERV-K elements exhibiting detectable RNA levels in normal cultured and urothelial cancer cells were analyzed for their expression in benign and cancerous bladder tissues. We then conducted qRT-PCR analyses on the eight HERV-K elements detectable in our set of normal urothelial cell cultures and bladder cancer cell line.
Figure 4.
Expression changes of proviral HERV-K elements in bladder cancer. Expression of different HERV-K elements was assessed by end-point PCR and qRT-PCR as indicated in (A). HERV-K RNA levels were measured by qRT-PCR in 6 normal urothelial cell cultures and 18 bladder cancer cell lines (B,C) and in a set of 11 benign and 24 cancerous bladder tissues (D). p Values calculated by the Mann–Whitney U-test were given above the brackets for significant changes (p < 0.05). Missing p values demonstrate changes without reaching the level of significance.
In general, expression of these HERV-K elements was rather low in these samples bordering on the limit of reliable quantification (Figures 4B,C). Two of the analyzed HERV-K elements (HERVK17 and HERV-K102) showed significant expression changes between normal urothelial cell cultures and bladder cancer cell lines. HERV-K102 was significantly downregulated (Mann–Whitney U-test; p = 0.013) in bladder cancer cells independent of the tumor type of origin, but expression was on the limit of detection (Figure 4B). In general, expression of the HERVK17 provirus was downregulated as well. In cultured normal urothelial cells its transcript level was low and these low expression levels were preserved in most papillary carcinoma cell lines (Figure 4B). Exceptionally, the RT4 cell line showed a high increase of HERVK17 expression. Notably, all papillary cell lines positive for HERVK17 expression (BC61, RT4, and UMUC6) (Figure 4B) showed low methylation levels at the HERVK17 LTR comparable to those found in cultured normal urothelial cells (Figure 3A). In cell lines originating from muscle-invasive bladder carcinoma HERVK17 expression was essentially absent (Figure 4B) fitting well with the hypermethylation found at the HERVK17 locus in the respective cell lines (Figure 3A). Expression of the other HERV-K elements was low and no significant expression changes were observed in bladder cancer cell lines (Figure 4C).
In benign bladder samples expression of the HERVK17 provirus was low or absent with one exception showing significant expression (Figure 4D). Likewise, most of the bladder cancers exhibited low or absent expression of the HERVK17 locus, whereas a few samples showed strikingly increased expression (up to 700-fold). Across all samples, the expression of the HERVK17 provirus was not significantly changed (Mann–Whitney U-test; p = 0.687). Expression levels of the other HERV-K retroelements (HERV-K10, HERV-K_3q12.3, HERV-K_12q14.1, HERV-K113) assessed in our bladder tissue set were rather low and no significant expression increases were found in cancerous tissues (Figure 4D).
Discussion
In the present study we describe the impact of global methylation changes in bladder cancers tissues and cell lines on the most important classes of active retroelements in the human genome. With respect to LINE-1 sequences, which make up 17% of the genome, the quantitative methylation data obtained in this study confirm previous finding of widespread hypomethylation in bladder cancers (9–11). Results of the DNA methylation analyses in bladder cancer cell lines revealed a tendency toward exacerbation in high-grade and high-stage tumors, but also changes in cell lines from papillary tumors. Evidently, LINE-1 hypomethylation is an early and very frequent change in bladder cancer. Quantitative changes of LINE-1 methylation were comparable to those in colorectal cancers where LINE-1 hypomethylation also occurs early, but are more pronounced compared to those in prostate cancer where LINE-1 hypomethylation is a later event during carcinogenesis (7, 8, 22–24).
Nevertheless, the hypomethylation of the LINE-1 promoter found in bladder cancer cell lines did not result in overall increased LINE-1 expression, but went along with a shift toward full-length LINE-1 expression as previously observed in prostate cancers (7). A previous report has described the spliced form of LINE-1 transcripts as the predominant isoform in normal prostate tissue (25), which also seems to be the case in the urinary bladder. In bladder cancer tissues the more prominent hypomethylation compared to prostate cancer went along with a more general increase in LINE-1 expression but only the expression of the full-length LINE-1 isoform increased significantly. Notably, in bladder cancer cell lines the correlation of LINE-1 promoter methylation with the expression of the full-length transcript assessed by the 5′-assay was better than the correlation with the 3′-assay. Taken together, the evidence suggests that the hypomethylation of the LINE-1 promoter in cancers may contribute to a shift toward full-length LINE-1 expression. The most obvious explanation for that observation is that most LINE-1 transcripts in normal tissues originate from truncated elements that represent parts of longer transcripts from host gene promoters, whereas hypomethylation in cancer may allow a degree of transcription initiation from LINE-1 5′-internal promoters. In addition, LINE-1 promoter DNA hypomethylation may enable expression from the LINE-1 antisense promoter (ASP) resulting in cancer-specific chimeric transcripts (26). Noteworthy, DNA hypomethylation of a specific LINE-1 element within the MET oncogene was recently shown to enable expression of a chimeric L1-MET transcript starting from the ASP in bladder cancers (10). As the LINE-1 ASP is located in the L1Hs 5′-UTR between nucleotides 400–600 (27), transcripts starting from the ASP are detectable by our LINE-1_5′-assay. Hence, elevated expression detected by the LINE-1_5′ assay may partially reflect LINE-1 ASP activation.
Whereas TP53 mutations are relatively infrequent in prostate carcinoma, in invasive bladder cancers TP53 missense mutations are found in around 50% of the cases. Additional alterations of “upstream” or “downstream” factors within the p53 network contributing to p53 inhibition are common (28). Accordingly, the more prominent impairment of LINE-1 expression in bladder cancers compared to prostate cancers may be partially explained by p53 mediated regulation of LINE-1 expression (29, 30).
Notably, only full-length LINE-1 transcripts contain information for both LINE-1 open reading frames (ORF1 and ORF2) required for retrotransposition of LINE-1 (31, 32) and other non-autonomous retroelements like SINEs and processed pseudogenes. Up to now roughly 100 disease-causing retrotransposition events, usually occurring during early embryonic or germ line development, are known, predominantly elicited by retrotransposed AluY elements (33). Additionally, within the last few years several reports described examples of LINE-1 retrotransposition in different cancers (34–37). Analyses for bladder cancers are still missing, but will presumably be enabled soon by the data from an ongoing whole-genome sequencing project.
Surprisingly, in contrast to the hypomethylation observed at the LINE-1 promoter in bladder cancer cell lines as compared to normal urothelial cells the LTRs of HERVK17 and H22q showed overall significantly increased methylation levels. Nevertheless, expression of the H22q provirus remained undetectable in any tumor. This is in good accordance to the study by Stauffer et al. (15). Here, H22q expression was not detectable by MPSS in bladder, but only in prostate samples. Predominant prostatic expression of this provirus was confirmed by others (7, 12, 13). In contrast, antibodies reacting with H22q Gag protein were found in the sera from bladder cancer patients (12). As in the same study H22q mRNA was not found in bladder carcinoma specimen, the positive antibody reaction may be due to cross-reactivity of a serum antibody to a different protein resembling the H22q Gag. HERVK17 showed expression only in bladder cancer cell lines of papillary origin whereas expression of the provirus was nearly absent in muscle-invasive cell lines. Noteworthy, expression was only detectable in cell lines with low HERVK17 methylation suggesting that DNA methylation may constitute one factor limiting its expression.
Many studies published in the last decade emphasize the strongly tissue- and cancer-specific expression pattern of HERV-K elements (7, 12, 38–41). The mechanisms underlying this pattern are still poorly understood, although tissue-specific transcription factors and epigenetic regulation are clearly implicated. In our present study expression of eight specific HERV-Ks was detectable in urothelial cells by end-point PCR, whereas that of nine others was not. Quantification of these HERV-K transcript levels revealed generally low expression in normal bladder which is in good concordance to previously published results assessed by MPSS (15). Among the faintly expressed elements was the HERV-K113 provirus. Its expression in nearly all bladder samples does not fit with previous observations that HERV-K113 occurs in a small part of the human population. HERV-K113 was likely acquired in Africa during or after the migration by Homo sapiens north and eastward and showed the highest frequencies in individuals from central Africa (∼20–30%) (42, 43). A large study assessing more than 2800 individuals in the UK found HERV-K113 allele frequency of approximately 7% (44). Most likely, the weak HERV-K113 expression in our data was at least partially caused by cross-reactivity of the used assay with another very closely related HERV-K element. Except for HERV-K102 and HERVK17 (as discussed above) significant cancer-specific expression changes of these elements were detectable neither in bladder cancer cell lines nor tissues. Transcripts of the proviruses HERV-K_11q23.3 and HERV-K_22q11.21 are strongly expressed in testicular cancers (38) but not in prostate tissues (7). Of these, only HERV-K_22q11.21 showed detectable expression in bladder tissues underlining again the strongly tissue-specific expression of distinct HERV-K elements.
In contrast to the methylation changes in bladder cancer cell lines HERVK17 LTR methylation was decreased in bladder tumor tissues with a good correlation to H22q methylation changes. Puzzlingly, HERVK17 LTR exhibited significant higher methylation in normal bladder tissues compared to cultured urothelial cells. In order to exclude that the LTR becomes demethylated during culture, we analyzed freshly prepared, uncultured urothelial cells, which showed only slightly higher methylation than the cultured cells. In addition, residual connective tissue from a ureter after removal of the epithelial layer also exhibited lower HERK17 DNA methylation than benign bladder tissues. Instead, the HERVK17 mean methylation value in benign bladder tissue is rather comparable to that found in benign prostate tissues [mean = 35.1 vs. 44.9%; (7)]. The difference toward cultured cells could therefore result from an admixture of other cell types, such as infiltrating immune cells that are prominent in cancer-carrying bladders or may reflect one of the few distinctive differences between ureter and bladder urothelium tissue (45).
DNA methylation of both HERV-K LTRs but not of the LINE-1 promoter showed significant correlation to patient gender. One explanation for these differences may be the well-known influence of androgens on the development of bladder cancer (46). The correlation could also result from a higher fraction of smokers in the male population. Smoking is a main risk factor for urothelial cancers accounting partly for its higher incidence in males (47). As smoking induces several epigenetic changes in urothelial cells (48) it may also affect HERV methylation and contribute to aberrant HERV expression. Additionally, HERVs were reported to become induced by smoking in urothelial cell lines and tissues (49) which may be causative for HERVK17 expression in a few cancer tissues. As the smoking status was not consistently assessed in our patient cohort we cannot confirm these assumptions.
To unravel the puzzle of the regulation of specific HERV elements high-throughput transcript analyses of HERV expression are highly desirable. Likewise, detailed studies are needed to investigate the tissues-specific regulators of HERV expression as published by us and others (7, 50–53). In this respect, the present study provides a framework for studies on urothelial tissue.
Expression of AluYa5 and AluYb8 SINEs was not significantly altered in bladder cancer cell lines. In contrast, in bladder cancer tissues AluYb8 but not AluYa5 expression was highly significantly elevated. It is generally assumed that Alu induction is related to several different kinds of cellular stresses (54). Human Alu and rodent B2 SINE are activated in response to heat shock (55, 56) and are consecutively involved in heat shock-related stress response (57). Alu expression was elevated during hypoxic stress in human glioblastoma cells, whereas tRNA genes and B2 elements were inhibited in response to hypoxia in rat cardiomyocytes, even though tRNA genes and SINEs have very similar promoters (58, 59). As standardized cell culture conditions are unlikely to induce heat shock or hypoxic stresses, it is plausible to assume that only basal level of Alu transcription were observed in cultured cells. In bladder cancer tissues, a likely inducer of AluYb8 expression is hypoxic stress as hypoxia is a well-known feature in this solid tumor (60). In contrast, AluYa5 expression was only slightly altered and may not respond to this kind of cellular stress. The factors regulating SINE expression in stressed cells and the reasons why these factors do not affect the transcription of other small RNAs with comparable promoters are largely unknown (54). Moreover, our data hint at an element-specific regulation of Alu expression in response to cellular stresses. Alu elements are characterized by their huge number with limited diversity (61), which complicates methylation analyses and calls for genome-wide high-throughput approaches. Recently, such global sequencing approaches for Alu methylation analyses have revealed tissue-specific methylation of certain Alu elements (62) and a decline of Alu DNA methylation in several cancers which was most pronounced for members of the AluY family (63–66). In benign tissues the methylation level of specific Alu elements and the degree of their methylation heterogeneity is dependent on their genomic location and their adjacent sequence motifs and heterogeneity increases in cancer tissues (67). Interestingly, recent whole-genome sequencing studies suggest that besides LINE-1, retrotranspositions in human cancers substantially involve Alu elements (35). In that respect, our study invites the speculation that LINE-1 activation in bladder cancers may also facilitate AluYb8 retrotransposition in particular.
In summary, our study provides the first survey of expression and methylation of the most active retroelement classes in bladder cancer. Even though changes in LINE-1 methylation and expression are more pronounced in this cancer type than those observed in our previous study on prostate cancer (7), the results further support our conclusion that apparently global DNA hypomethylation affects retroelements to very different extents and does not lead to wholesale reexpression. Rather, changes in methylation and expression may have to be investigated at many individual elements to identify those that contribute to genomic instability and deregulation of gene expression in each cancer type.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Rainer Engers for histopathological assessment of tissue samples and to Christiane Hader for experimental support. The project was funded by a grant from the Deutsche Forschungsgemeinschaft (DFG; Schu604/16-3) and the Reinhard-Nagel-Foundation.
References
- 1.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell (2013) 153:38–55 10.1016/j.cell.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell (2007) 128:683–92 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol (2005) 83:296–321 10.1139/o05-036 [DOI] [PubMed] [Google Scholar]
- 4.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta (2007) 1775:138–62 [DOI] [PubMed] [Google Scholar]
- 5.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell (2008) 135:23–35 10.1016/j.cell.2008.09.022 [DOI] [PubMed] [Google Scholar]
- 6.Schulz WA. L1 retrotransposons in human cancers. J Biomed Biotechnol (2006) 2006:83672. 10.1155/JBB/2006/83672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goering W, Ribarska T, Schulz WA. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis (2011) 32:1484–92 10.1093/carcin/bgr181 [DOI] [PubMed] [Google Scholar]
- 8.Florl AR, Steinhoff C, Muller M, Seifert HH, Hader C, Engers R, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer (2004) 91:985–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer (1999) 80:1312–21 10.1038/sj.bjc.6690524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet (2010) 6:e1000917. 10.1371/journal.pgen.1000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Bemmel D, Lenz P, Liao LM, Baris D, Sternberg LR, Warner A, et al. Correlation of LINE-1 methylation levels in patient-matched buffy coat, serum, buccal cell, and bladder tumor tissue DNA samples. Cancer Epidemiol Biomarkers Prev (2012) 21:1143–8 10.1158/1055-9965.EPI-11-1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida T, Obata Y, Ohara N, Matsushita H, Sato S, Uenaka A, et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun (2008) 8:15. [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature (2007) 448:595–9 10.1038/nature06024 [DOI] [PubMed] [Google Scholar]
- 14.Hermans KG, Van Der Korput HA, Van Marion R, Van De Wijngaart DJ, Ziel-Van Der Made A, Dits NF, et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res (2008) 68:7541–9 10.1158/0008-5472.CAN-07-5930 [DOI] [PubMed] [Google Scholar]
- 15.Stauffer Y, Theiler G, Sperisen P, Lebedev Y, Jongeneel CV. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun (2004) 4:2. [PubMed] [Google Scholar]
- 16.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res (2002) 62:5510–6 [PubMed] [Google Scholar]
- 17.Koch A, Hatina J, Rieder H, Seifert HH, Huckenbeck W, Jankowiak F, et al. Discovery of TP53 splice variants in two novel papillary urothelial cancer cell lines. Cell Oncol (Dordr) (2012) 35:243–57 10.1007/s13402-012-0082-8 [DOI] [PubMed] [Google Scholar]
- 18.Chapman EJ, Williams SV, Platt FM, Hurst CD, Chambers P, Roberts P, et al. Integrated genomic and transcriptional analysis of the in vitro evolution of telomerase-immortalized urothelial cells (TERT-NHUC). Genes Chromosomes Cancer (2009) 48:694–710 10.1002/gcc.20672 [DOI] [PubMed] [Google Scholar]
- 19.Seifert HH, Meyer A, Cronauer MV, Hatina J, Muller M, Rieder H, et al. A new and reliable culture system for superficial low-grade urothelial carcinoma of the bladder. World J Urol (2007) 25:297–302 10.1007/s00345-007-0166-7 [DOI] [PubMed] [Google Scholar]
- 20.Swiatkowski S, Seifert HH, Steinhoff C, Prior A, Thievessen I, Schliess F, et al. Activities of MAP-kinase pathways in normal uroepithelial cells and urothelial carcinoma cell lines. Exp Cell Res (2003) 282:48–57 10.1006/excr.2002.5647 [DOI] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res (2001) 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst (2008) 100:1734–8 10.1093/jnci/djn359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res (2008) 68:8954–67 10.1158/0008-5472.CAN-07-6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer (2010) 9:125. 10.1186/1476-4598-9-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res (2010) 38:3909–22 10.1093/nar/gkq132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics (2009) 94:397–406 10.1016/j.ygeno.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 27.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol (2001) 21:1973–85 10.1128/MCB.21.6.1973-1985.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz WA. Understanding urothelial carcinoma through cancer pathways. Int J Cancer (2006) 119:1513–8 10.1002/ijc.21852 [DOI] [PubMed] [Google Scholar]
- 29.Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotransposition-competent human LINE-1 induces apoptosis in cancer cells with intact p53. J Biomed Biotechnol (2004) 2004:185–94 10.1155/S1110724304403131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, et al. p53 responsive elements in human retrotransposons. Oncogene (2009) 28:3857–65 10.1038/onc.2009.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran JV, Holmes SE, Naas TP, Deberardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell (1996) 87:917–27 10.1016/S0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- 32.Evans JD, Peddigari S, Chaurasiya KR, Williams MC, Martin SL. Paired mutations abolish and restore the balanced annealing and melting activities of ORF1p that are required for LINE-1 retrotransposition. Nucleic Acids Res (2011) 39:5611–21 10.1093/nar/gkr171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev (2012) 22:191–203 10.1016/j.gde.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell (2010) 141:1253–61 10.1016/j.cell.2010.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, III, et al. Landscape of somatic retrotransposition in human cancers. Science (2012) 337:967–71 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solyom S, Ewing AD, Hancks DC, Takeshima Y, Awano H, Matsuo M, et al. Pathogenic orphan transduction created by a nonreference LINE-1 retrotransposon. Hum Mutat (2012) 33:369–71 10.1002/humu.21663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell (2013) 153:101–11 10.1016/j.cell.2013.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics (2008) 9:354. 10.1186/1471-2164-9-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res (2008) 68:5869–77 10.1158/0008-5472.CAN-07-6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nellaker C, Li F, Uhrzander F, Tyrcha J, Karlsson H. Expression profiling of repetitive elements by melting temperature analysis: variation in HERV-W gag expression across human individuals and tissues. BMC Genomics (2009) 10:532. 10.1186/1471-2164-10-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst (2012) 104:189–210 10.1093/jnci/djr540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol (2001) 11:1531–5 10.1016/S0960-9822(01)00455-9 [DOI] [PubMed] [Google Scholar]
- 43.Moyes DL, Martin A, Sawcer S, Temperton N, Worthington J, Griffiths DJ, et al. The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics (2005) 86:337–41 10.1016/j.ygeno.2005.06.004 [DOI] [PubMed] [Google Scholar]
- 44.Moyes DL, Goris A, Ban M, Compston A, Griffiths DJ, Sawcer S, et al. HERV-K113 is not associated with multiple sclerosis in a large family-based study. AIDS Res Hum Retroviruses (2008) 24:363–5 10.1089/aid.2007.0196 [DOI] [PubMed] [Google Scholar]
- 45.Catto JW, Yates DR, Rehman I, Azzouzi AR, Patterson J, Sibony M, et al. Behavior of urothelial carcinoma with respect to anatomical location. J Urol (2007) 177:1715–20 10.1016/j.juro.2007.01.030 [DOI] [PubMed] [Google Scholar]
- 46.Waliszewski P, Waliszewska MK, Hemstreet GP, III, Hurst RE. Expression of sex steroid receptor genes and comodulation with retinoid signaling in normal human uroepithelial cells and bladder cancer cell lines. Urol Oncol (1997) 3:141–7 10.1016/S1078-1439(98)00011-8 [DOI] [PubMed] [Google Scholar]
- 47.Kiriluk KJ, Prasad SM, Patel AR, Steinberg GD, Smith ND. Bladder cancer risk from occupational and environmental exposures. Urol Oncol (2012) 30:199–211 10.1016/j.urolonc.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 48.Brait M, Munari E, Lebron C, Noordhuis MG, Begum S, Michailidi C, et al. Genome-wide methylation profiling and the PI3K-AKT pathway analysis associated with smoking in urothelial cell carcinoma. Cell Cycle (2013) 12:1058–70 10.4161/cc.24050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabriel U, Steidler A, Trojan L, Michel MS, Seifarth W, Fabarius A. Smoking increases transcription of human endogenous retroviruses in a newly established in vitro cell model and in normal urothelium. AIDS Res Hum Retroviruses (2010) 26:883–8 10.1089/aid.2010.0014 [DOI] [PubMed] [Google Scholar]
- 50.Christensen T. HERVs in neuropathogenesis. J Neuroimmune Pharmacol (2010) 5:326–35 10.1007/s11481-010-9214-y [DOI] [PubMed] [Google Scholar]
- 51.Fuchs NV, Kraft M, Tondera C, Hanschmann KM, Lower J, Lower R. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J Virol (2011) 85:3436–48 10.1128/JVI.02539-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh I, Mirova A, Kurata S, Murakami Y, Horikawa K, Nakakuki N, et al. Activation of the long terminal repeat of human endogenous retrovirus K by melanoma-specific transcription factor MITF-M. Neoplasia (2011) 13:1081–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ, Jr, et al. Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol (2012) 86:7790–805 10.1128/JVI.07215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikitina TV, Tischenko LI, Schulz WA. Recent insights into regulation of transcription by RNA polymerase III and the cellular functions of its transcripts. Biol Chem (2011) 392:395–404 10.1515/BC.2011.049 [DOI] [PubMed] [Google Scholar]
- 55.Fornace AJ, Jr, Mitchell JB. Induction of B2 RNA polymerase III transcription by heat shock: enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res (1986) 14:5793–811 10.1093/nar/14.14.5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene (2001) 276:135–41 10.1016/S0378-1119(01)00637-0 [DOI] [PubMed] [Google Scholar]
- 57.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell (2008) 29:499–509 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 58.Ernens I, Goodfellow SJ, Innes F, Kenneth NS, Derblay LE, White RJ, et al. Hypoxic stress suppresses RNA polymerase III recruitment and tRNA gene transcription in cardiomyocytes. Nucleic Acids Res (2006) 34:286–94 10.1093/nar/gkj402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pal A, Srivastava T, Sharma MK, Mehndiratta M, Das P, Sinha S, et al. Aberrant methylation and associated transcriptional mobilization of Alu elements contributes to genomic instability in hypoxia. J Cell Mol Med (2010) 14:2646–54 10.1111/j.1582-4934.2009.00792.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ord JJ, Streeter EH, Roberts IS, Cranston D, Harris AL. Comparison of hypoxia transcriptome in vitro with in vivo gene expression in human bladder cancer. Br J Cancer (2005) 93:346–54 10.1038/sj.bjc.6602666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet (2002) 3:370–9 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- 62.Xie H, Wang M, Bonaldo Mde F, Smith C, Rajaram V, Goldman S, et al. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res (2009) 37:4331–40 10.1093/nar/gkp393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer (2009) 124:81–7 10.1002/ijc.23849 [DOI] [PubMed] [Google Scholar]
- 64.Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene (2009) 448:151–67 10.1016/j.gene.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho YH, Yazici H, Wu HC, Terry MB, Gonzalez K, Qu M, et al. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res (2010) 30:2489–96 [PMC free article] [PubMed] [Google Scholar]
- 66.Xiang S, Liu Z, Zhang B, Zhou J, Zhu BD, Ji J, et al. Methylation status of individual CpG sites within Alu elements in the human genome and Alu hypomethylation in gastric carcinomas. BMC Cancer (2010) 10:44. 10.1186/1471-2407-10-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie H, Wang M, De Andrade A, Bonaldo Mde F, Galat V, Arndt K, et al. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res (2011) 39:4099–108 10.1093/nar/gkr017 [DOI] [PMC free article] [PubMed] [Google Scholar]