Abstract
Vitiligo is a complex disorder in which autoimmune destruction of melanocytes results in white patches of skin and overlying hair. Over the past several years, extensive genetic studies have outlined a biological framework of vitiligo pathobiology that underscores its relationship to other autoimmune diseases. This biological framework offers insight into both vitiligo pathogenesis and perhaps avenues towards more effective approaches to treatment and even disease prevention.
Keywords: autoimmune destruction, disease prevention, vitiligo, vitiligo genetics, vitiligo pathogenesis
INTRODUCTION
Vitiligo is a complex disorder in which white patches of skin and overlying hair result from autoimmune destruction of melanocytes within the lesions. Vitiligo appears to be of multifactorial causation, involving multiple underlying susceptibility genes and environmental triggers. Over the past several years there has been considerable progress in defining the genetic epidemiology and genetic pathogenesis of vitiligo, and its relationships to other autoimmune diseases. To date, almost all genetic studies have been of generalized or “non-segmental” vitiligo. Recently, advances by these genetic studies have outlined a biological framework that offers real insight into vitiligo pathogenesis and perhaps to more effective approaches to treatment and even disease prevention.
EARLY HISTORY
Human pigmentation diseases present the most visually striking of all disorders, and the phenotypes of oculocutaneous albinism, piebaldism and others have been known for thousands of years.1 Vitiligo itself has had a remarkable history of discovery and re-discovery of key observations overlooked or forgotten over the passage of time. Certainly, patients with various forms of patchy leukoderma were recognized during ancient times; however, vitiligo per se was not described as a specific medical entity until the mid-18th century.2
The first clue to vitiligo pathogenesis came from the description and illustration of a patient with concomitant vitiligo, adrenal insufficiency and pernicious anemia,3 highlighting a link between these three autoimmune diseases. Similar patients with multiple autoimmune diseases were later reported by Schmidt4 and codified by Neufeld and Blizzard.5 Further evidence of immune or inflammatory influences in vitiligo came from the recognition that new lesions often occur at sites of skin injury,6 a phenomenon first described in 1872 by Köbner in the context of psoriasis and which came to bear his name.7 Perhaps the most important observation regarding vitiligo pathogenesis also came from Kaposi, who illustrated histological lack of cells containing pigment granules within vitiligo skin lesions,8 later re-described by Hu et al.9 and yet again by Breathnach et al.10
Probably the landmark early study of vitiligo was that of Lerner,11 who reported on clinical, epidemiological and genetic characteristics of 200 cases, thereby delineating many important clinical and epidemiological aspects of the disease, establishing the clinical terminology and classification scheme that is basically still in use today. This study provided an analytic framework that was adopted by many subsequent epidemiological surveys of vitiligo patients around the world,12–21 providing key information in formulating the genetic underpinnings of the disorder.
GENETIC EPIDEMIOLOGY
The earliest formal considerations of the genetic basis of vitiligo appear to have been by Stuttgen,22 Teindel23 and later by Lerner,11 all of whom noted familial aggregation of cases not clearly consistent with Mendelian single-gene inheritance. Stuttgen,22 in particular, was remarkable in suggesting the simultaneous involvement of both recessive and dominant contributory influences, predating the modern genetic concept of “complex inheritance” by decades. Subsequently, Das et al.,24,25 Bhatia et al.26 and Majumder et al.27 likewise all noted frequent familial clustering of vitiligo cases in a non-Mendelian pattern. Majumder et al.28 and Nath et al.29 suggested a multilocus recessive model, Arcos-Burgos et al.30 postulated multiple forms of vitiligo with different genetic underlying models, whereas most other investigators favored polygenic, multifactorial inheritance,12,26,31–33 which is now generally accepted.
Close relatives of vitiligo patients have elevated risk of vitiligo, as well as other autoimmune diseases, with relative risks for first-degree relatives estimated at approximately 6–18-fold elevated.12,24,27,33–35 Estimates of vitiligo heritability range from 46% in India to 16% in China,33 and Alkhateeb et al.12 found that the concordance for vitiligo in monozygotic twins was 23%, supporting roles for both genetic and non-genetic factors in disease pathogenesis.
PRE-MOLECULAR GENETIC STUDIES
The earliest attempts to identify specific genes that may contribute to vitiligo were genetic association studies of vitiligo using a variety of protein polymorphic markers, including ABO and other blood groups,15,25,36–42 blood group secretor status,15,43 and a number of other serum proteins.25,44,45 Essentially, all of these studies proved negative.
Subsequently, there were a number of case–control genetic association studies of histocompatibility antigen (human leukocyte antigen, HLA) types and vitiligo.46–55 While several of these studies reported weak associations, the results were inconsistent, partly because of inadequate study designs and partly because of likely genetic heterogeneity among the many different populations analyzed. Nevertheless, meta-analysis of data from 11 of these studies56 subsequently identified association of vitiligo with HLA-A2 (odds ratio, 2.07).
MOLECULAR GENETIC ERA
Modern genetic studies of vitiligo have principally entailed five scientific approaches: genetic linkage analysis of families with multiple affected relatives (multiplex families), candidate gene association studies comparing relatively small numbers of vitiligo patients (cases) to unaffected controls, genome-wide association studies (GWAS) comparing far larger numbers of cases to controls, DNA sequencing studies and gene expression studies. Genetics researchers give highest credence to disease gene discoveries derived from genome-wide linkage and GWAS, as these approaches are relatively free of biases and are robust to several important sources of potential error. Candidate gene association studies, by contrast, are largely discounted by modern geneticists as an approach to gene discovery, as the vast majority report false-positives resulting from chance fluctuation due to insufficient statistical power, population stratification and the impossibility of adequate correction for multiple testing given publication bias of positive results.57,58 DNA sequencing studies largely involve candidate genes previously identified by other means, and thus are subject to the caveats appropriate to the identification of these genes in the first place. Gene expression studies are generally considered invalid as a means of disease gene discovery as they cannot distinguish between primary causal differences versus secondary changes that are the result of pathway dysregulation or disease, but can be useful as a means of confirming the biological relevance of previous genetic findings.
Genetic linkage studies
The first vitiligo genetic linkage studies were of candidate genes. A linkage study of the HLA region in a large family with polyglandular autoimmune disease type II (Schmidt syndrome), including with vitiligo, was negative.59 Tripathi et al.60 tested linkage of vitiligo to MITF, again with negative results. The first positive results came from targeted linkage analysis of the major histocompatibility complex (MHC) on chromosome 6p, detecting linkage of vitiligo with microsatellite polymorphic markers in HLA gene regions in families from several different populations.30,61,62
The first detection of novel vitiligo loci by genome-wide study came from linkage analysis of families with systemic lupus erythematosus (SLE) that included at least one case of vitiligo, detecting the SLEV1 locus on chromosome 17p13.63SLEV1 was subsequently confirmed by direct genome-wide study of Caucasian multiplex vitiligo families,64 and the corresponding causal gene was eventually identified as NLRP1,65 encoding a key regulator of the innate immune response. Genome-wide analysis of a unique Caucasian family with auto-somal dominant vitiligo detected AIS1 on chromosome 1p31.3-32.2,66 and the corresponding gene was subsequently identified as FOXD3,67 encoding a developmental regulator of melanoblast differentiation; thus far, additional families with dominant vitiligo and FOXD3 mutations have not been described. Comprehensive genome-wide linkage analyses of other multiplex vitiligo families also detected a number of other vitiligo susceptibility loci, on chromosomes 1, 7, 8, 9, 11, 13, 19 and 21 in Caucasians,64,68 and on chromosomes 4, 6 and 22 in Chinese,61,69 some of which may correspond to genes detected in subsequent GWAS. Ren et al.70 studied XBP1 as a candidate gene within the 22q12 linkage region, though this assignment has not yet been confirmed. Xu et al.71 studied PDGFRA as a candidate gene within the 4q12-q21 linkage region, finding more variation in familial vitiligo cases than controls. However, PDGFRA is not known to play a role in pigment cell biology, and the closely adjacent KIT gene would seem a better candidate for this linkage signal.
Candidate gene association studies
The earliest candidate gene association studies of vitiligo were of loci that previously had been shown to be genetically linked and/or associated with other autoimmune diseases. Though candidate gene association studies are not considered a valid approach to primary discovery of disease genes, this approach is considered acceptable for confirmation of established genes.
The first such study reported genetic association of vitiligo with single nucleotide polymorphisms (SNP) within CTLA4,72 a locus that had been previously associated with several other autoimmune diseases.73 Even in that initial report, CTLA4 was only associated with vitiligo in patients who also had concomitant autoimmune diseases, as confirmed in subsequent studies.74–76 This has been interpreted as perhaps indicating that there are multiple genetically distinct subtypes of vitiligo76 or that CTLA4 is not actually causal for vitiligo, with apparent genetic association being secondary to epidemiological association of vitiligo with other autoimmune diseases for which CTLA4 is causal.74 Subsequently, Canton et al.77 reported association of vitiligo with PTPN22, a gene that also had been previously associated with a number of other autoimmune diseases.78 This association was subsequently confirmed by other investigators,79,80 most importantly by the first GWAS of vitiligo.81 A number of investigators reported association of vitiligo with loci in the MHC.82–85 However, because of complex patterns of long-range linkage disequilibrium across the MHC, it has been difficult to assign genetic association to specific genes within the MHC.
Over the following years, a large number of candidate gene studies of vitiligo were published, many based on little or no compelling biological rationale. Birlea et al.75 published a comprehensive review of 33 claimed vitiligo candidate genes (ACE, AIRE, CAT, CD4, CLEC11A, COMT, CTLA4, C12orf10, DDR1, EDN1, ESR1, FAS, FBXO11, FOXD3, FOXP3, GSTM1, GSTT1, IL1RN, IL10, KITLG, MBL2, NFE2L2, PDGFRA-KIT, PTGS2, STAT4, TAP1-PSMB8, TGFBR2, TNF, TSLP, TXNDC5, UVRAG, VDR, XBP1), finding support for only three (TSLP, XBP1, FOXP3) in analysis of a genome-wide vitiligo GWAS dataset.81 Additional claimed candidate gene associations with vitiligo include AHR86, COX2,87GSTP1,88IL4,89IL19 and IL20RB,90INOS,91PRO2268,92TLR2 and TLR4,93 while candidate genes reported not to be associated include CD28,94ICOS,94IL20, IL24e and IL20RA,90MTHFR,95 and SMOC2.96 Some of these claimed novel candidate gene associations may be valid, though most are likely to represent false-positives.
GWAS
Genome-wide association studies are currently the “gold standard” of genetic studies of complex diseases. The first vitiligo GWAS was an analysis of patients from a population isolate in Romania with a high prevalence of vitiligo and other autoimmunity,97 detecting association with SMOC2 located at distal chromosome 6q27.98 While association with SMOC2 has not been detected in other GWAS, it is located very close to CCR6 in chromosome 6q27, which was detected in GWAS of vitiligo in both Caucasians81 and Chinese,99 and it may be that all of three reports represent the same association signal.
Three large GWAS of vitiligo have been reported thus far; two in Caucasians 81,100,101 and one in Chinese,99,102 while a small gene-centric GWAS of vitiligo has been reported in Indian–Pakistani patients.103 These studies have detected 30 vitiligo susceptibility loci in Caucasians (Table 1): PTPN22, RERE, IFIH1, CTLA4 (only in patients with other autoimmune diseases), FOXP1, CD80, LPP, CLNK, TSLP, HLA-A, MHC class II (c6orf10-BTNL2-DRB1-DQA1), BACH2, CCR6, TG/SLA, IL2RA, CASP7, CD44, TYR, a gene desert at 11q21, IKZF4, SH2B3, GZMB, OCA2, MC1R, TICAM1, UBASH3A, XBP1, C1QTNF6, TOB2, and FOXP3.81,100,101,104 The parallel studies of Chinese have detected nine vitiligo susceptibility loci (Table 1): LPP, MHC class I (HLA-B-HLA-C), RNASET2-FGFR1OP-CCR6, IL2RA, ZMIZ1, IKZF4, IL2RB-C1QTNF6, an intergenic interval at 10q22.1 between SLC29A3 and CDH23, and an intergenic interval at 11q23.3 between DDX6 and CXCR5.99,102 Thus, vitiligo association with at least LPP, the HLA class I gene region, CCR6, IL2RA, IKZF4 and C1QTNF6 appears to be shared between the Caucasian and Chinese populations, though other genes associated with vitiligo susceptibility or protection may be population-specific, particularly TYR, OCA2 and MC1R. Similarly, the Indian–Pakistani GWAS reported association only with the MHC class II gene region, and formal trans-ethnic analysis indicated that association signals in Caucasians and Indian–Pakistani patients in this genomic region likely share the same ancestral origin.103
Table 1.
Confirmed and suggestive vitiligo susceptibility loci identified by genome-wide association or linkage studies
| Chromosome | Candidate gene | Populations | Protein | Function |
|---|---|---|---|---|
| 1p13.2 | PTPN22 | C | LYP protein tyrosine phosphatase | Regulates T-cell signaling |
| 1p31.3 | FOXD3 † | C | Forkhead box D3 | Transcriptional regulator of neural crest; melanoblast differentiation |
| 1p36.23 | RERE | C | Atrophin-1-like protein isoform b | Lymphoid transcriptional co-repressor; apoptotic regulator |
| 2q24.2 | IFIH1 | C | Interferon-induced RNA helicase | Regulates innate antiviral immune responses |
| 2q33.2 | CTLA4 ‡ | C | Cytotoxic T-lymphocyte-associated-4 | Inhibits T cells via interaction with CD80 and CD86 |
| 3p13 | FOXP1 | C | Forkhead box P1 | Transcriptional regulator of B-cell, T-cell, monocyte development |
| 3q13.33 | CD80 | C | B-cell activation antigen B7-1 | T-cell priming by B cells, T cells, dendritic cells; interacts with CTLA-4 |
| 3q28 | LPP | A,C | LIM domain containing preferred translocation | Transcriptional co-activator? |
| 4p16.1 | CLNK | C | Mast cell immunoreceptor signal transducer | Positive regulator of immunoreceptor signaling |
| 5q22.1 | TSLP § | C | Thymic stromal lymphopoietin protein | Cytokine regulator of skin dendritic (Langerhans) cell maturation |
| 6p22.1 | HLA-A | C | Leucocyte antigen A α-chain | Presents peptide antigens |
| 6p22.1 | HLA-B-C | A | Leukocyte antigen B or C α-chain | Presents peptide antigens |
| 6p21.32 | HLA-DRB1-DQA1 | C,I | Major histocompatibility complex class II region | Presents peptide antigens |
| 6q15 | BACH2 | C | BTB and CNC homology 1, basic leucine zipper | B-cell transcriptional repressor |
| 6q27 | CCR6 | A,C | Chemokine (C-C motif) receptor 6 | Regulates differentiation and function of B cells, T cells, dendritic cells |
| 6q27 | SMOC2 ¶ | C | SPARC-related modular calcium-binding protein | Regulate cellextracellular matrix i nteractions |
| 8q24.22 | TG/SLA | C | Thyroglobulin, Src-like adaptor isoform c | Regulates antigen receptor signaling in T cells, B cells, dendritic cells |
| 10p15.1 | IL2RA | A,C | Interleukin 2 receptor α-chain | Regulates interleukin 2-mediated activation of T-cells, regulatory T-cells |
| 10q22.1 | Gene desert | A | ||
| 10q25.3 | CASP7 | A | Caspase 7 | Apoptotic executioner protein |
| 11p13 | CD44 | C | CD44 antigen | T-cell regulator |
| 11q14.3 | TYR | C | Tyrosinase | Melanin biosynthetic enzyme |
| 11q21 | Gene desert | C | None | TYR regulation? |
| 11q23.3 | Gene desert | A | ||
| 12q13.2 | IKZF4 | A,C | Ikaros zinc finger protein, subfamily 1A, 4 | T-cell transcriptional regulator |
| 12q24.12 | SH2B3 | C | LNK adaptor | B-cell, T-cell developmental regulator |
| 14q12 | GZMB | C | Granzyme B | Mediates CTL-induced target cell apoptosis, helper T-cell apoptosis |
| 15q12-13.1 | OCA2 | C | Oculocutaneous albinism II | Melanosomal membrane transporter/pump |
| 16q24.3 | MC1R | C | Melanocortin-1 receptor | Regulates melanogenesis |
| 17p13.2 | NLRP1 | C | NLR family, pyrin domain containing 1 | Regulates IL-1 β innate immune response via NLRP1 inflammasome |
| 19p13.3 | TICAM1 †† | C | Toll-like receptor adaptor molecule 1 | Mediates innate antiviral immune responses |
| 21q22.3 | UBASH3A | C | Ubiquitin associated and SH3 domain containing | Regulates T-cell signaling, apoptosis |
| 22q12.1 | XBP1§ | A,C | X-box binding protein 1 | Transcriptional regulator of MHC class II expression, plasma cells |
| 22q12.3 | C1QTNF6 | A,C | C1q and tumor necrosis factor related protein 6 | Innate immune response to light-induced apoptosis? |
| 22q13.2 | TOB2 | C | Transducer of ERBB2, 2 | Inhibitor of cell cycle progression; involved in T-cell tolerance |
| Xp11.23 | FOXP3 § | C | Forkhead box P3 | Transcriptional regulator of regulatory T-cell function and development |
CTLA4 is only associated with vitiligo in patients with other concomitant autoimmune diseases.74–76 Association of CTLA4 with vitiligo may thus be secondary, driven by primary association with these other diseases.
XBP1, FOXP3 and TSLP are the only vitiligo candidate genes that achieve suggestive association in analysis of genome-wide association study data in Caucasians.75XBP1 was identified as a positional candidate based on linkage in Chinese.70
SMOC2 is located very close to CCR6 and may represent the same association signal.
T/CAM1 achieves highly suggestive association in analysis of GWAS data in Caucasians.101
DNA sequencing studies
Gene-specific DNA sequencing analyses are, in effect, highly detailed candidate gene association studies, typically comparing the frequency of either specific variants or of all observed variation in specific candidate genes in cases versus in controls. Until recently, most such studies have lacked sufficient statistical power or rigor to prove that observed DNA sequence variations are truly causal for the disease versus being rare non-causal polymorphisms.
The first such study was of GCH1,105 and the claimed association of vitiligo with GCH1 mutations was quickly refuted.106 DNA sequences of a number of additional vitiligo candidate genes have subsequently been compared in vitiligo cases versus controls, mostly in relatively small numbers. These include ASIP,107,108MC1R,107–109MYG1/c12orf10110 and POMC.109 None of these studies showed convincing significant differences in cases versus controls after appropriate correction for multiple testing.
More recently, several vitiligo susceptibility genes detected by GWAS have been subjected to NextGeneration DNA re-sequencing to identify sequence variation that is apparently causal for disease. Jin et al.104 sequenced both HLA-A and TYR in Caucasian vitiligo patients, finding that the predominant HLA-A vitiligo-associated susceptibility allele is HLA-A*02:01:01:01, while finding that two common non-synonymous substitutions of TYR, S192Y and R402Q, appear to exert both individual and synergistic protective effects. HLA-A2 presents tyrosinase peptide as an autoimmune antigen, whereas the TYR 192Y and 402Q substitutions likely reduce the amount of tyrosinase peptide available for presentation, suggesting that these loci and variants act via a common pathway. Ferrara et al.111 sequenced GZMB and found that the causal allele involved a common multi-variant haplotype containing three non-synonymous substitutions in strong linkage disequilibrium, Q55R-P94A-Y247H, though the mechanism by which this multi-variant granzyme B affects vitiligo susceptibility is not yet known. Levandowski et al.112 sequenced NLRP1 and found that the common high-risk haplotype contains three non-synonymous substitutions in strong linkage disequilibrium, L155H-V1059M-M1184V, while a less common but even higher risk haplotype contains nine non-synonymous substitutions, L155H-T246S-T782S-T878M-T995I-M1119V-M1184V-V1241L-R1366C. These authors showed that the common multi-variant high-risk haplotype results in 1.8-fold elevation of processing of the inactive interleukin (IL)-1β precursor to biologically active IL-1β cytokine by the NLRP1 inflammasome, presenting a likely mechanism for disease pathogenesis associated with this haplotype.
Gene expression studies
A number of studies have compared expression of genes in normal versus vitiligo skin or melanocytes, either genome-wide or of selected candidate genes. A serious problem in such studies is the difficulty in distinguishing causal changes from those that result from the disease state or from melanocyte stress or senescence. The first such study identified VIT1, a gene downregulated in vitiligo melanocytes113 and later renamed FBXO11, the role of which in vitiligo pathogenesis remains uncertain.114 Analyses comparing genes differentially expressed in vitiligo lesions versus in uninvolved skin principally detect genes that encode melanocyte components,115 which is not surprising given that a greatly reduced number of melanocytes within vitiligo lesions is the hallmark of the disease. Recently, Yu et al.116 carried out transcriptome analyses of vitiligo patients and found evidence of upregulated innate immunity in non-lesional skin, perhaps consistent with implication of these pathways in vitiligo pathogenesis by previous genetic studies.
Other investigators have carried out differential expression studies of a very large number of candidate genes, but it is difficult to have confidence that any of the reported differences are in fact causal for vitiligo, or would even remain statistically significant could appropriate multiple-testing corrections be applied across the numerous such reports. Two such studies89,92 have correlated altered gene expression with genetic association of specific SNPs in the corresponding genes, and thus provide at least some external validation that the candidate gene in question may be relevant to vitiligo pathogenesis.
CURRENT UNDERSTANDING
To date, approximately 36 loci with convincing or strongly suggestive evidence for a role in vitiligo susceptibility have been identified (Table 1). Most of these loci contain or are in close proximity to plausible biological candidate genes. Approximately 90% of these genes encode immunoregulatory proteins, whereas approximately 10% encode melanocyte proteins that likely serve as autoantigens that both stimulate the melanocyte-specific immune response and act as targets for immune recognition and cell killing (or in the case of HLA class I and perhaps class II molecules, present those autoantigens to the immune system). Together, these proteins constitute a dense immunoregulatory network that highlights systems and pathways that mediate vitiligo susceptibility.101 Moreover, DNA sequencing and functional analyses have identified apparent causal variation for TYR,104HLA-A,104NLRP1112 and GZMB,111 yielding deeper insights into the roles played by these proteins in disease pathogenesis.
Several of the genes identified as conferring vitiligo susceptibility, particularly those expressed in melanocytes, have also emerged as genes involved in susceptibility to malignant melanoma, the same SNP having genetically opposite roles in vitiligo versus melanoma susceptibility.81,101 This apparent genetically inverse relationship of melanocyte-specific genes to vitiligo versus melanoma led to the suggestion that vitiligo may represent a dysregulated normal process of immune surveil-lance against malignant melanoma.117
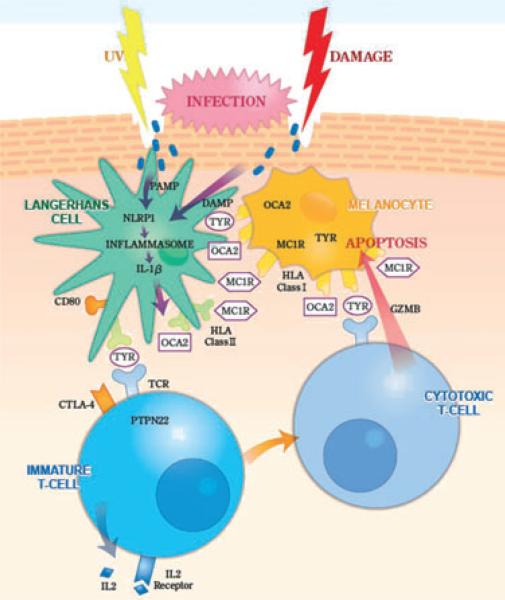
These genetic and functional studies have begun to provide a hypothetical framework for understanding the initial triggering, immune propagation and ultimate cytotoxicity of the anti-melanocyte immune response (Fig. 1). A likely common pathway for initial vitiligo triggering may be Köbnerization, with various types of skin damage resulting in localized cell killing and perhaps localized microinfection. Melanocyte peptide antigens are presented to skin-resident dendritic cells (principally Langerhans cells) by HLA class I molecules on the melanocyte surface. At the same time, Langerhans cells take up infection-derived molecules that display “pathogen-associated molecular patterns” (PAMP) and damage-derived molecules that display “damage-associated molecular patterns” (DAMP), which bind to NLRP1 and thereby induce assembly of the NLRP1 inflammasome; other pathways of inflammasome activation and innate immune induction are also possible. NLRP1 inflamma-some assembly activates caspases that cleave the IL-1β precursor to biologically active secreted IL-1β. IL-1β is a potent pro-inflammatory cytokine, perhaps facilitating presentation of autoantigens that trigger or provide specificity to the immune response. Within the dendritic cells, melanocyte autoantigens are transferred from HLA class I molecules to HLA class II mol ecules, which then present these antigens to immature T cells. Immature T cells then express and secrete IL-2, which binds to IL-2 receptor expressed on the cell surface, inducing their maturation to cytotoxic T-cells (cytotoxic T lymphocytes, CTL) that express T-cell receptor molecules specific to the cognate melanocyte autoantigen, as well as cytolytic molecules such as granzyme B. Melanocyte-specific CTL then recognize the cog-nate melanocyte autoantigens presented on the melanocyte surface by HLA class I molecules, and the target cells are ultimately lysed by granzyme B. This “circuit” of melanocyte triggering, immune propagation, autoantigen programming and target cell killing by immune effector cells thus incorporates many of the genes and proteins that modulate vitiligo susceptibility. While certainly not complete in all details, and perhaps not even substantially correct, this model nevertheless provides for the first time a conceptualization of vitiligo pathogenesis that encompasses advanced biological knowledge of the disease and that may highlight potential new avenues for therapy and even disease prevention in individuals with high genetic susceptibility.
Figure 1.
“Circuit” of melanocyte damage, melanocyte autoantigen presentation, immune triggering and propagation, T-cell programming and melanocyte target cell killing in vitiligo. Skin is damaged by ultraviolet (UV) or other trauma, perhaps facilitating microinfection. Molecules displaying pathogen-associated molecular patterns (PAMP) or damage-associated molecular patterns (DAMP) interact with NLRP1 in the cytoplasm of Langerhans cells, stimulating nucleation of an NLRP1 inflammasome, thereby activating caspases that cleave the interleukin (IL)-1b precursor to biologically active secreted IL-1β. Langerhans cells take up peptide autoantigens presented by human leukocyte antigen (HLA) class I molecules expressed on the surface of nearby melanocytes, including peptides derived from tyrosinase (TYR), OCA2 and the melanocortin-1 receptor (MC1R), and these peptide autoantigens are then transferred to HLA class II molecules expressed on the Langerhans cell surface. Perhaps stimulated by IL-1β and facilitated by interaction of CD80 with CTLA4 and by the action of PTPN22, these melanocyte-derived peptide autoantigens are then presented to immature T cells that express cognate T-cell receptors (TCR), the response of which is regulated by PTPN22. The activated T cells express IL-2, which binds to the IL-2 receptor expressed on their surface, stimulating maturation to cytotoxic T cells that express granzyme B (GZMB). The TCR expressed by these autoreactive cytotoxic T cells binds its cognate autoantigen presented on the surface of target melanocytes by HLA class I molecules, and GZMB is introduced into the target melanocyte, inducing apoptosis.
ACKNOWLEDGMENTS
This study was supported in part by grants R01AR045584, R01AR056292 and P30AR057212 from the National Institutes of Health.
REFERENCES
- 1.Spritz RA, Hearing VJ. Abnormalities of pigmentation. In: Rimoin D, Pyeritz R, Korf B, editors. Principles and Practice of Medical Genetics. 6th edn. Elsevier; Oxford: 2013. (in press) [Google Scholar]
- 2.Le Cat C-N. Traite de la couleur de la peau humaine en général, de celle des negres en particulier, et de la métamorphose d'une des couleurs en l'autre, soit de naissance, soit accidentellement. Amsterdam: 1765. [N.P.] [Google Scholar]
- 3.Addison T. A collection of the published writing of the late Thomas Addison, M.D., physician to Guy's Hospital. Vol. 2. New Sydenham Society; London: 1937. On the constitutional and local effects of disease of the suprarenal capsules. pp. 244–93. 1868. 1855. Reprinted in Med Classics. [Google Scholar]
- 4.Schmidt M. Eine biglanduiare Erkrankung (Nebennieren und Schilddruse) bei Morbus Addisonii. Verh Dtsch Ges Pathol. 1926;21:212–221. [Google Scholar]
- 5.Neufeld M, Blizzard RM. Polyglandular autoimmune diseases. In: Pinchera A, Doniach D, Fenzi GF, et al., editors. Symposium on Auto-immune Aspects of Endocrine Disorders. Academic Press; New York: 1980. pp. 357–365. [Google Scholar]
- 6.Hebra F, Kaposi M. In: On Diseases of the Skin, including the Exanthemata. Tay W, editor. III. New Sydenham Society; London: 1874. [Google Scholar]
- 7.Köbner H. Zur Aetiologie der Psoriasis. Vjschr Dermatol. 1876;8:559–561. [Google Scholar]
- 8.Kaposi M. Pathologie und Therapie der Hautkrankheiten. Urban und Schwartzenberg; Vienna: 1879. [Google Scholar]
- 9.Hu F, Fosnaugh RP, Lesney PF. In vitro studies on vitiligo. J Invest Dermatol. 1959;33:267–280. doi: 10.1038/jid.1959.150. [DOI] [PubMed] [Google Scholar]
- 10.Breathnach AS, Bor S, Wyllie LM-A. Electron microscopy of peripheral nerve terminals and marginal melanocytes in vitiligo. J Invest Dermatol. 1966;47:125–140. doi: 10.1038/jid.1966.117. [DOI] [PubMed] [Google Scholar]
- 11.Lerner AB. Vitiligo. J Invest Dermatol. 1959;32:285–310. [PubMed] [Google Scholar]
- 12.Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–214. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 13.Behl PN, Bhatia RK. 400 cases of vitiligo: a clinic-therapeutic analysis. Indian J Dermatol. 1972;17:51–56. [PubMed] [Google Scholar]
- 14.Boisseau-Garsaud A-M, Garsaud P, Cales-Quist D, Hélénon R0, Quénéhervé C. Charles Sainte Claire R. Epidemiology of vitiligo in the French West Indies (Isle of Martinique). Int J Dermatol. 2000;39:18–20. doi: 10.1046/j.1365-4362.2000.00880.x. [DOI] [PubMed] [Google Scholar]
- 15.Dutta AK, Mondal SB, Dutta SB. ABO blood group and secretory status in vitiligo. J Indian Med Assoc. 1969;53:186–189. [PubMed] [Google Scholar]
- 16.Hamada T. 5 years’ survey of patients with vitiligo at the Osaka City University Hospital during 1960-1964. Hifuka Kiyo. 1966;61:92–99. [PubMed] [Google Scholar]
- 17.Howitz J, Brodthagen H, Schwartz M, Thomsen K. Prevalence of vitiligo. Epidemiological survey on the Isle of Bornholm, Denmark. Arch Dermatol. 1977;113:47–52. doi: 10.1001/archderm.113.1.47. [DOI] [PubMed] [Google Scholar]
- 18.Khoo OT. Vitiligo. A review and report of treatment of 60 cases in the General Hospital, Singapore from 1954-1958. Singapore Med J. 1962;3:157. [PubMed] [Google Scholar]
- 19.Mehta NR, Shah KC, Theodore C, Vyas VP, Patel AB. Epidemio-logical study of vitiligo in Surat area, south Gujarat. Indian J Med Res. 1973;61:145–154. [PubMed] [Google Scholar]
- 20.Obe WK. Vitiligo in Zimbabwe. Cent Afr J Med. 1984;30:259–264. [PubMed] [Google Scholar]
- 21.Ortonne JP, Perrot H, Thivolet J. Clinical and statistical study of 100 patients with vitiligo. II. Associated lesions. Sem Hop. 1976;52:679–686. [PubMed] [Google Scholar]
- 22.Stuttgen G. Die Vitiligo in erbbiologischer Betrachtung. Z Haut Geschlechtskr. 1950;9:451–457. [PubMed] [Google Scholar]
- 23.Teindel H. Familiäare Vitiligo. Z Haut Geschlechtskr. 1950;9:457–462. [PubMed] [Google Scholar]
- 24.Das SK, Majumder PP, Chakraborty R, Majumdar TK, Haldar B. Studies on vitiligo. I. Epidemiological profile in Calcutta, India. Genet Epidemiol. 1985;2:71–78. doi: 10.1002/gepi.1370020107. [DOI] [PubMed] [Google Scholar]
- 25.Das SK, Majumder PP, Majumdar TK, Haldar B. Studies on vitiligo. II. Familial aggregation and genetics. Genet Epidemiol. 1985;2:255–262. doi: 10.1002/gepi.1370020303. [DOI] [PubMed] [Google Scholar]
- 26.Bhatia PS, Mohan L, Pandey ON, Singh KK, Arora SK, Mukhija RD. Genetic nature of vitiligo. J Dermatol Sci. 1992;4:180–184. doi: 10.1016/0923-1811(92)90017-6. [DOI] [PubMed] [Google Scholar]
- 27.Majumder PP, Nordlund JJ, Nath SK. Pattern of familial aggregation of vitiligo. Arch Dermatol. 1993;129:994–998. [PubMed] [Google Scholar]
- 28.Majumder PP, Das SK, Li CC. A genetical model for vitiligo. Am J Hum Genet. 1988;43:119–125. [PMC free article] [PubMed] [Google Scholar]
- 29.Nath SK, Majumder PP, Nordlund JJ. Genetic epidemiology of vitiligo: multilocus recessivity cross-validated. Am J Hum Genet. 1994;55:981–990. [PMC free article] [PubMed] [Google Scholar]
- 30.Arcos-Burgos M, Parodi E, Salgar M, et al. Vitiligo: complex segregation and linkage disequilibrium analyses with respect to microsatellite loci spanning the HLA. Hum Genet. 2002;110:334–342. doi: 10.1007/s00439-002-0687-5. [DOI] [PubMed] [Google Scholar]
- 31.Kim SM, Chung HS, Hann S-K. The genetics of vitiligo in Korean patients. Int J Dermatol. 1998;38:908–910. doi: 10.1046/j.1365-4362.1998.00549.x. [DOI] [PubMed] [Google Scholar]
- 32.Ramaiah A, Mojamdar M, Amarnath VM. Vitiligo in the SSK community of Bangalore. Indian J Dermatol Venereol Leprol. 1988;54:251–254. [PubMed] [Google Scholar]
- 33.Sun XK, Xu AE, Meng W, et al. Study on genetic epidemiology on 815 patients with vitiligo in the Zhejiang area. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26:911–914. [PubMed] [Google Scholar]
- 34.Boisseau-Garsaud A-M, Saint-Cyr I, Quist D, Arveiler B, Garsaud P. Familial aggregation of vitiligo in the French West Indies (Isle of Martinique). Eur J Dermatol. 2001;11:554–556. [PubMed] [Google Scholar]
- 35.Laberge G, Mailloux CM, Gowan K, et al. Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res. 2005;18:300–305. doi: 10.1111/j.1600-0749.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- 36.El-Hefnawi H, Mohieddin O, Rasheed A. ABO blood groups in various skin diseases. J Egypt med Ass. 1963;47:1097–1101. [PubMed] [Google Scholar]
- 37.Kareemullah L, Taneja V, Begum S, Sarma PK, Baig HA. Association of ABO blood groups and vitiligo. J Med Genet. 1977;14:211–213. doi: 10.1136/jmg.14.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oriente Biondi C, Ruocco V. Vitiligo and blood groups. Rass Int Clin Ter. 1969;49:1395–1399. [PubMed] [Google Scholar]
- 39.Seghal VN, Dube B. ABO blood groups and vitiligo. J Med Genet. 1968;5:308–309. doi: 10.1136/jmg.5.4.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh G, Shanker P. Vitiligo and blood groups. Preliminary report. Br J Dermatol. 1966;78:91–92. doi: 10.1111/j.1365-2133.1966.tb12180.x. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava GN, Shukla RC. ABO blood groups in vitiligo. Indian J Med Res. 1965;53:221–225. [PubMed] [Google Scholar]
- 42.Wasfi AI, Saha N, El Munshid HA, El Sheikh FS, Ahmed MA. Genetic association in vitiligo: ABO, MNSs, Rhesus, Kell and Duffy blood groups. Clin Genet. 1980;17:415–417. doi: 10.1111/j.1399-0004.1980.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 43.Sehgal VN, Dube B. Secretion of blood group specific substances in saliva of vitiligo patients. A preliminary report. Br J Dermatol. 1967;79:704–705. doi: 10.1111/j.1365-2133.1967.tb11441.x. [DOI] [PubMed] [Google Scholar]
- 44.Mujahid Ali M, Banu M, Waheed MA, Quadri GS, Habibullah CM. Serum alpha 1-antitrypsin and haptoglobin phenotypes in vitiligo. Arch Dermatol Res. 1990;282:206–207. doi: 10.1007/BF00372625. [DOI] [PubMed] [Google Scholar]
- 45.Nath RL, Debnath H, Chatterji A, Ghosh NK. Paper electrophoresis of serum proteins in diseases. Bull Calcutta Sch Trop Med. 1967;15:60–61. [PubMed] [Google Scholar]
- 46.Abanmi A, Harthi FA, Baqami RA, et al. Association of HLA loci alleles and antigens in Saudi patients with vitiligo. Arch Dermatol Res. 2007;298:347–352. doi: 10.1007/s00403-006-0699-4. [DOI] [PubMed] [Google Scholar]
- 47.Dunston GM, Halder RM. Vitiligo is associated with HLA-DR4 in black patients. A preliminary report. Arch Dermatol. 1990;126:56–60. [PubMed] [Google Scholar]
- 48.Finco O, Cuccia M, Martinetti M, Ruberto G, Orecchia G, Rabbiosi G. Age of onset in vitiligo: relationship with HLA supratypes. Clin Genet. 1991;39:48–54. doi: 10.1111/j.1399-0004.1991.tb02984.x. [DOI] [PubMed] [Google Scholar]
- 49.Foley LM, Lowe NJ, Misheloff E. Association of HLA-DR4 with vitiligo. J Am Acad Dermatol. 1983;8:39–40. doi: 10.1016/s0190-9622(83)80279-5. [DOI] [PubMed] [Google Scholar]
- 50.Kachru RB, Telischi M, Mittal KK. HLA antigens and vitiligo in an American black population. Tissue Antigens. 1978;12:396–397. doi: 10.1111/j.1399-0039.1978.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 51.Metzker A, Zamir R, Gazit E, David M, Feuerman EJ. Vitiligo and the HLA system. Dermatologica. 1980;160:100–105. doi: 10.1159/000250480. [DOI] [PubMed] [Google Scholar]
- 52.Minev M, Tonkin N, Martinova F. Association of the HLA system with vitiligo. Vestn Dermatol Venerol. 1985;5:41–42. [PubMed] [Google Scholar]
- 53.Nakagawa H, Otuka F, Kukita A, Mizoguchi M, Ito H, Juji T. Histo-compatible antigens in vitiligo vulgaris II. Nihon Hifuka Gakkai Zasshi. 1980;90:939–941. [PubMed] [Google Scholar]
- 54.Retornaz G, Betuel H, Ortonne JP, Thivolet J. HL-A antigens and vitiligo. Br J Dermatol. 1976;95:173–175. doi: 10.1111/j.1365-2133.1976.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 55.Tastan HB, Akar A, Orkunoglu FE, Arca E, Inal A. Association of HLA class I antigens and HLA class II alleles with vitiligo in a Turkish population. Pigment Cell Res. 2004;17:181–184. doi: 10.1111/j.1600-0749.2004.00141.x. [DOI] [PubMed] [Google Scholar]
- 56.Liu J-B, Li M, Chen H, et al. Association of vitligo with HLA-A2: a meta analysis. J Eur Acad Dermatol Venereol. 2007;21:205–213. doi: 10.1111/j.1468-3083.2006.01899.x. [DOI] [PubMed] [Google Scholar]
- 57.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 58.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 59.Butler MG, Hodes ME, Conneally PM, Biegel AA, Wright JC. Linkage analysis in a large kindred with autosomal dominant transmission of polyglandular autoimmune disease type II (Schmidt syndrome). Am J Med Genet. 1984;18:61–65. doi: 10.1002/ajmg.1320180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tripathi RK, Flanders DJ, Young TL, et al. Micropthalmia-Associated Transcription Factor (MITF) locus lacks linkage to human vitiligo or osteopetrosis: an evaluation. Pigment Cell Res. 1999;12:187–192. doi: 10.1111/j.1600-0749.1999.tb00512.x. [DOI] [PubMed] [Google Scholar]
- 61.Liang Y, Yang S, Zhou Y, et al. Evidence for two susceptibility loci on chromosomes 22q12 and 6p22 in Chinese generalized vitiligo families. J Investig Dermatol. 2007;127:2552–2557. doi: 10.1038/sj.jid.5700904. [DOI] [PubMed] [Google Scholar]
- 62.Zamani M, Spaepen M, Sghar SS, et al. Linkage and association of HLA class II genes with vitiligo in a Dutch population. Br J Dermatol. 2001;145:90–94. doi: 10.1046/j.1365-2133.2001.04288.x. [DOI] [PubMed] [Google Scholar]
- 63.Nath SK, Kelly JA, Namjou B, et al. Evidence for a susceptibility gene, SLEV1, on chromosome 17p13 in families with vitiligo-related systemic lupus erythematosus. Am J Hum Genet. 2001;69:1401–1406. doi: 10.1086/324470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spritz RA, Gowan K, Bennett DC, Fain PR. Novel vitiligo susceptibility loci on chromosomes 7 (AIS2) and 8 (AIS3), confirmation of SLEV1 on chromosome 17, and their roles in an autoimmune diathesis. Am J Hum Genet. 2004;74:188–191. doi: 10.1086/381134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin Y, Mailloux CM, Gowan K, et al. NALP1 in vitiligo-associated multiple autoimmune disease. New Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 66.Alkhateeb A, Stetler GL, Old W, et al. Mapping of an autoimmunity susceptibility locus (AIS1) to chromosome 1p31.3-32.2. Hum Mol Genet. 2002;11:661–667. doi: 10.1093/hmg/11.6.661. [DOI] [PubMed] [Google Scholar]
- 67.Alkhateeb A, Fain PR, Spritz RA. Candidate functional promoter variant in the FOXD3 melanoblast developmental regulator gene in autosomal dominant vitiligo. J Invest Dermatol. 2005;125:388–391. doi: 10.1111/j.0022-202X.2005.23822.x. [DOI] [PubMed] [Google Scholar]
- 68.Fain PR, Gowan K, LaBerge GS, et al. A genomewide screen for generalized vitiligo: confirmation of AIS1 on chromosome 1p31 and evidence for additional susceptibility loci. Am J Hum Genet. 2003;72:1560–1564. doi: 10.1086/375451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J-J, Huang W, Gui J-P, et al. A novel linkage to generalized vitiligo on 4q13-q21 identified in a genomewide linkage analysis of Chinese families. Am J Hum Genet. 2005;76:1057–1065. doi: 10.1086/430279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren Y, Yang S, Xu S, et al. Genetic variation of promoter sequence modulates XBP1 expression and genetic risk for vitiligo. PLoS Genet. 2009;5:e1000523. doi: 10.1371/journal.pgen.1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu S, Zhou Y, Yang S, et al. Platelet-derived growth factor receptor alpha mutations in vitiligo vulgaris. Acta Derm Venereol. 2010;90:131–135. doi: 10.2340/00015555-0820. [DOI] [PubMed] [Google Scholar]
- 72.Kemp EH, Ajjan RA, Waterman EA, et al. Analysis of a microsatellite polymorphism of the cytotoxic T-lymphocyte antigen-4 gene in patients with vitiligo. Br J Dermatol. 1999;140:73–78. doi: 10.1046/j.1365-2133.1999.02610.x. [DOI] [PubMed] [Google Scholar]
- 73.Gough SCL, Walker LSK, Sansom DM. CTLA4 gene polymorphism and autoimmunity. Immunol Rev. 2005;204:102–115. doi: 10.1111/j.0105-2896.2005.00249.x. [DOI] [PubMed] [Google Scholar]
- 74.Birlea SA, LaBerge GS, Procopciuc LM, Fain PR, Spritz RA. CTLA4 and generalized vitiligo: two genetic association studies and a meta-analysis of published data. Pigment Cell Melanoma Res. 2009;22:230–234. doi: 10.1111/j.1755-148X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Birlea SA, Jin Y, Bennett DC, et al. Comprehensive association analysis of candidate genes for generalized vitiligo supports XBP1, FOXP3, and TSLP. J Invest Dermatol. 2011;131:371–381. doi: 10.1038/jid.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blomhoff A, Kemp EH, Gawkrodger DJ, et al. CTLA4 polymorphisms are associated with vitiligo, in patients with concomitant autoimmune diseases. Pigment Cell Res. 2004;18:55–58. doi: 10.1111/j.1600-0749.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 77.Canton I, Akhtar S, Gavalas NG, et al. A single-nucleotide polymorphism in the gene encoding lymphoid protein tyrosine phosphatase (PTPN22) confers susceptibility to generalised vitiligo. Genes Immun. 2005;6:584–587. doi: 10.1038/sj.gene.6364243. [DOI] [PubMed] [Google Scholar]
- 78.Brand O, Gough S, Heward J. HLA, CTLA-4 and PTPN22: the shared genetic master-key to autoimmunity? Expert Rev Mol Med. 2005;7:1–15. doi: 10.1017/S1462399405009981. [DOI] [PubMed] [Google Scholar]
- 79.LaBerge GS, Bennett DC, Fain PR, Spritz RA. PTPN22 is genetically associated with risk of generalized vitiligo, but CTLA4 is not. J Investig Dermatol. 2008;128:1757–1762. doi: 10.1038/sj.jid.5701233. [DOI] [PubMed] [Google Scholar]
- 80.Laddha NC, Dwivedi M, Shajil EM, Prajapati H, Marfatia YS, Begum R. Association of PTPN22 1858C/T polymorphism with vitiligo susceptibility in Gujarat population. J Dermatol Sci. 2008;49:260–262. doi: 10.1016/j.jdermsci.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 81.Jin Y, Birlea SA, Fain PR, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. New Engl J Med. 2010;362:1686–1697. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Casp CB, She J-X, McCormack WT. Genes of the LMP/TAP cluster are associated with the human autoimmune disease vitiligo. Genes Immun. 2003;4:492–499. doi: 10.1038/sj.gene.6364016. [DOI] [PubMed] [Google Scholar]
- 83.Yang S, Wang J-Y, Gao M, et al. Association of HLA-DQA1 and DQB1 genes with vitiligo in Chinese Hans. Int J Dermatol. 2005;44:1022–1027. doi: 10.1111/j.1365-4632.2004.02389.x. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X-J, Liu H-S, Liang Y-H, et al. Association of HLA class I alleles with vitiligo in Chinese Hans. J Dermatol Sci. 2004;35:165–168. doi: 10.1016/j.jdermsci.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 85.Fain PR, Babu SR, Bennett DC, Spritz RA. HLA class II haplotype DRB1*04-DQB1*0301 contributes to risk of familial generalized vitiligo and early disease onset. Pigment Cell Res. 2006;19:51–57. doi: 10.1111/j.1600-0749.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 86.Wang XW, Li K, Guo S, et al. The association of functional polymorphisms in the aryl hydrocarbon receptor (AHR) gene with the risk of vitiligo in Han Chinese populations. Br J Dermatol. 2012;166:1081–1087. doi: 10.1111/j.1365-2133.2011.10798.x. [DOI] [PubMed] [Google Scholar]
- 87.Li M, Gao Y, Li C, et al. Association of COX2 functional polymorphisms and the risk of vitiligo in Chinese populations. J Dermatol Sci. 2009;53:176–181. doi: 10.1016/j.jdermsci.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 88.Minashkin MM, Salnikova LE, Lomonosov KM, Korobko IV, Tatarenko AO. Possible contribution of GSTP1 and other xenobiotic metabolizing genes to vitiligo susceptibility. Arch Dermatol Res. 2012 doi: 10.1007/s00403-012-1301-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 89.Imran M, Laddha NC, Dwivedi M, et al. Interleukin-4 genetic variants correlate with its transcript and protein levels in patients with vitiligo. Br J Dermatol. 2012;167:314–323. doi: 10.1111/j.1365-2133.2012.11000.x. [DOI] [PubMed] [Google Scholar]
- 90.Kingo K, Reimann E, Karelson M, et al. Association analysis of genes of the IL19 cluster and their receptors in vitiligo patients. Dermatology. 2010;221:261–266. doi: 10.1159/000317526. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Li C, Li K, Liu I, Jian Z, Gao T. Analysis of inducible nitric oxide synthase gene polymorphisms in vitiligo in Han Chinese people. PLoS ONE. 2011;6:e27077. doi: 10.1371/journal.pone.0027077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Douroudis K, Kingo K, Karelson M, et al. The PRO2268 gene as a novel susceptibility locus for vitiligo. Acta Derm Venereol. 2011;91:189–190. doi: 10.2340/00015555-0999. [DOI] [PubMed] [Google Scholar]
- 93.Karaca N, Ozturk G, Gerceker BT, Turkmen M, Berdeli A. TLR2 and TLR4 gene polymorphisms in Turkish vitiligo patients. J Eur Acad Dermatol Venereol. 2013;27:e85–e90. doi: 10.1111/j.1468-3083.2012.04514.x. [DOI] [PubMed] [Google Scholar]
- 94.Shin MK, Im SH, Kim SK, Yim SV, Chung J-H, Lee M-Y. Association study between polymorphisms of CD28, CTLA4, and ICOS and non-segmental vitiligo in a Korean population. Exp Ther Med. 2011;2:1145–1149. doi: 10.3892/etm.2011.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yasar A, Gunduz K, Onur E, Calkan M. Serum homocysteine, vita-min B12, folic acid levels and methylenetetrahydrofolate reductase (MTHFR) gene polymorphism in vitiligo. Dis Markers. 2012;33:85–89. doi: 10.3233/DMA-2012-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alkhateeb A, AI-Dain Marzouka N, Qarqaz F. SMOC2 gene variant and the risk of vitiligo in Jordanian Arabs. Eur J Dermatol. 2010;20:701–704. doi: 10.1684/ejd.2010.1095. [DOI] [PubMed] [Google Scholar]
- 97.Birlea SA, Fain PR, Spritz RA. A Romanian population isolate with high frequency of vitiligo and associated autoimmune diseases. Arch Dermatol. 2008;144:310–316. doi: 10.1001/archderm.144.3.310. [DOI] [PubMed] [Google Scholar]
- 98.Birlea SA, Gowan K, Fain PR, Spritz RA. Genome-wide association study of generalized vitiligo in an isolated European founder population identifies SMOC2, in close proximity to IDDM8. J Invest Dermatol. 2010;130:798–803. doi: 10.1038/jid.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quan C, Ren YQ, Xiang LH, et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat Genet. 2010;42:614–618. doi: 10.1038/ng.603. [DOI] [PubMed] [Google Scholar]
- 100.Jin Y, Birlea SA, Fain PR, et al. Common variants in FOXP1 are associated with generalized vitiligo. Nat Genet. 2010;42:576–578. doi: 10.1038/ng.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jin Y, Birlea SA, Fain PR, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tang XF, Zhang Z, Hu DY, et al. Association analyses identify three susceptibility Loci for vitiligo in the chinese han population. J Invest Dermatol. 2013;133:403–410. doi: 10.1038/jid.2012.320. [DOI] [PubMed] [Google Scholar]
- 103.Birlea SA, Ahmad FJ, Uddin RM, et al. Association of generalized vitiligo with MHC class II loci in patients from the Indian subcontinent. J Invest Dermatol. 2013 doi: 10.1038/jid.2012.501. doi: 10.1038/jid.2012.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jin Y, Ferrara T, Gowan K, et al. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J Investig Dermatol. 2012;132:1730–1733. doi: 10.1038/jid.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De la Fuente-Fernandez R. Mutations in GTP-cyclohydrolase I gene and vitiligo. Lancet. 1997;350:640. doi: 10.1016/S0140-6736(05)63329-6. [DOI] [PubMed] [Google Scholar]
- 106.Bandyopadhyay D, Lawrence E, Majumder PP, Ferrell RE. Vitiligo is not caused by mutations in GTP-cyclohydrolase I gene. Exp Dermatol. 2000;25:152–153. doi: 10.1046/j.1365-2230.2000.00599.x. [DOI] [PubMed] [Google Scholar]
- 107.Na GY, Lee KH, Kim MK, Lee SJ, Kim DW, Kim JC. Polymorphisms in the melanocortin-1 receptor (MC1R) and agouti signaling protein (ASIP) genes in Korean vitiligo patients. Pigment Cell Res. 2003;16:383–387. doi: 10.1034/j.1600-0749.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 108.Széll M, Baltás E, Bodai L, et al. The Arg160Trp allele of melanocortin-1 receptor gene might protect against vitiligo. Photochem Photobiol. 2008;84:565–571. doi: 10.1111/j.1751-1097.2008.00296.x. [DOI] [PubMed] [Google Scholar]
- 109.Eskandani M, Hasannia S, Golchai J. Assessment of MC1R and α-MSH genes sequences in Iranian vitilgio patients. Indian J Dermatol. 2010;55:325–328. doi: 10.4103/0019-5154.74530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Philips MA, Kingo K, Karelson M, et al. Promoter polymorphism - 119C/G in MYG1 (C12orf10) gene is related to vitiligo susceptibility and Arg4Gln affects mitochondrial entrance of Myg1. BMC Med Genet. 2010;11:56. doi: 10.1186/1471-2350-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferrara TM, Jin Y, Gowan K, Fain PR, Spritz RA. Risk of generalized vitiligo is associated with the common 55R-94A-247H variant haplotype of GZMB (encoding Granzyme B). J Invest Dermatol. 2013 doi: 10.1038/jid.2013.5. doi: 10.1038/jid.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Levandowski CB, Mailloux CM, Ferrara TM, et al. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1β processing via the NLRP1 inflammasome. Proc Natl Acad Sci USA. 2013;110:2952–2956. doi: 10.1073/pnas.1222808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Le Poole IC, Sarangarajan R, Zhao Y, et al. ‘VIT1’, a novel gene associated with vitiligo. Pigment Cell Res. 2001;14:475–484. doi: 10.1034/j.1600-0749.2001.140608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guan C, Lin F, Zhou M, et al. The role of VIT1/FBXO11 in the regulation of apoptosis and tyrosinase export from endoplasmic reticulum in cultured melanocytes. Int J Mol Med. 2010;26:57–65. doi: 10.3892/ijmm_00000435. [DOI] [PubMed] [Google Scholar]
- 115.Ströomberg S, Björklund MG, Asplund A, et al. Transcriptional profiling of melanocytes from patients with vitligo vulgaris. Pigment Cell Melanoma Res. 2007;21:162–171. doi: 10.1111/j.1755-148X.2007.00429.x. [DOI] [PubMed] [Google Scholar]
- 116.Yu R, Broady R, Huang Y, et al. Transcriptome analysis reveals markers of aberrantly activated innate immunity in vitiligo lesional and non-lesional skin. PLoS ONE. 2012;7(12):e51040. doi: 10.1371/journal.pone.0051040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Spritz RA. The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med. 2010;2:78. doi: 10.1186/gm199. [DOI] [PMC free article] [PubMed] [Google Scholar]