Abstract
A report in this issue of Molecular Cell provides evidence that a translocating SWI/SNF-nucleosome complex efficiently displaces neighboring nucleosomes in vitro and may account for SWI/SNF-dependent nucleosome eviction in vivo.
Ever since the first biochemical characterization of the SWI/SNF complex over 15 years ago, researchers have elucidated aspects of the mechanism by which this and related enzymes utilize energy derived from ATP hydrolysis to increase the accessibility of nucleosomal DNA in chromatin. Indeed, a long list of biochemical activities for the SWI/SNF complex have been described (Cairns, 2007), and references therein) However, the activities most relevant to SWI/SNF's in vivo function as a pleiotropic effector of gene expression remain unclear. Now, a report in this issue of Molecular Cell provides evidence that nucleosome-dependent SWI/SNF translocation resulting in displacement of neighboring nucleosomes might be a primary function of this enzyme in vivo.
Previous work has shown that SWI/SNF is required to evict nucleosomes upon activation of specific promoters in vivo, in collaboration with histone chaperones such as Asf-1 (Adkins et al., 2004; Boeger et al., 2003; Reinke and Horz, 2003). However, the mechanism of eviction is poorly understood. In vitro studies show that the SWI/SNF complex or paralogs such as the RSC complex, can catalyze transfer of H2A/H2B dimers from nucleosomes to acceptor H3/H4-DNA complexes (Bruno et al., 2003). In addition, total disruption of nucleosomes has been reported resulting in naked DNA after remodeling activity (Lorch et al., 2006). However, these activities occur with relatively low efficiency with mononucleosomes – substrates usually used in such studies – and require large amounts of acceptor or competitor species. (Dechassa et al., 2010).
In the current work, Bartholomew and colleagues noticed that remodeling of dior trinucleosomes resulted in significant amounts of species with reduced histone content. These substrates have an advantage over large arrays of nucleosomes, another commonly used substrate for remodeling studies, in that discrete products are easily characterized on nucleoprotein gels. Some may have assumed that the more rapidly migrating products simply represented alteration of nucleosome translational positions, a well-characterized outcome of SWI/SNF remodeling. However, using a quantitative double-label technique, Bartholomew and colleagues demonstrated that a fraction of the core histones of are lost in the products of dinucleosome remodeling, whereas no loss is observed with mononucleosomes. Specifically, they found that one H2A/H2B dimer is lost from an initial product, while an entire histone octamer is lost in a second major product. Interestingly, histone displacement occurs in the absence of chaperones or naked DNA as acceptors. Thus, in contrast to the remodeling of mononucleosomes, the results indicate SWI/SNF remodeling of dinucleosomes results in efficient eviction of one of the two original nucleosomes.
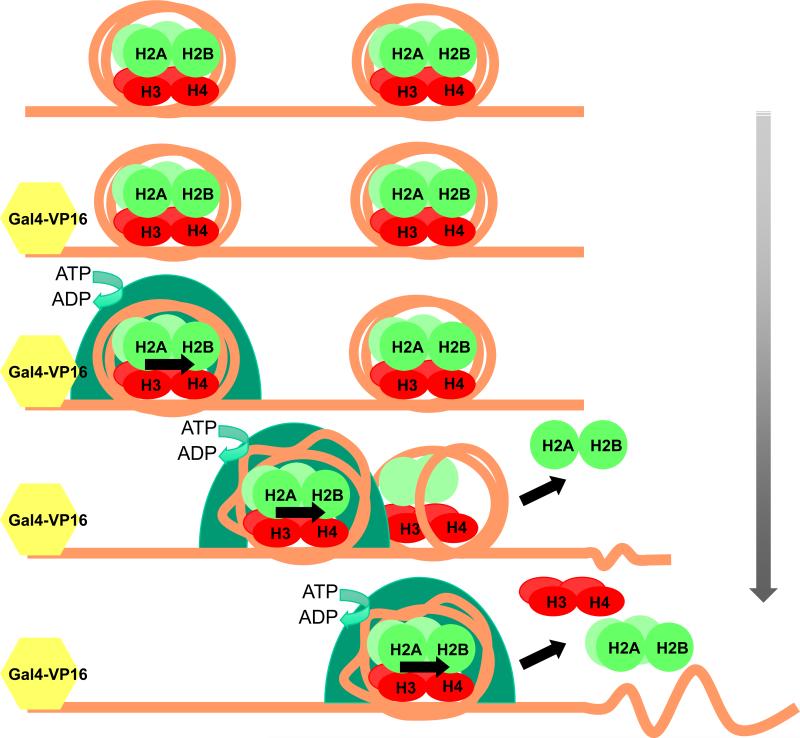
The researchers then precisely mapped histone-DNA interactions using a site-directed crosslinking technique and a DNA methylase protection assay known as MapIT that provides single-molecule information (Dechassa et al., 2010 and references therein). Results from these studies indicate nucleosomes are primarily moved to one end of the DNA fragment and that one of the two nucleosomes is evicted during the process, consistent with the histone content data. Moreover, by specifically recruiting SWI/SNF to one end of the dinucleosome template via the binding of the transcriptional activator Gal4-VP16, the mapping evidence suggests that the nearby (proximal) nucleosome is moved further downstream and eventually the distal nucleosome is disrupted and evicted (see Figure).
Figure.
Proposed mechanism of SWI/SNF nucleosome eviction. Top of figure shows unperturbed chromatin with two nucleosomes. SWI/SNF is recruited by the transcription factor (Gal4-VP16, yellow hexagon) and binds the proximal nucleosome. SWI/SNF remodels the nucleosome in its active site and translocates in one direction along the DNA. Upon encountering the downstream nucleosome, an H2A dimer is first displaced, followed by eviction of the entire neighboring nucleosome from the DNA template.
These results correlate well with a recent report from the Owen-Hughes laboratory that showed remodeling can result in one nucleosome invading the region of DNA inhabited by a neighbor (Engeholm et al., 2009). However, the Bartholomew group finds that remodeling is inhibited if the length of linker DNA between two nucleosomes gets too short. Rather than “pushing” the invader into the neighbor, SWI/SNF appears to contact DNA ahead of the advancing nucleosome and bump into the soon-to-be evicted nucleosome itself. A model then emerges whereby a nucleosome occupies a large pocket on the surface of the SWI/SNF complex and stimulates its ATPase-driven DNA translocase activity. The nucleosome in the pocket retains all of its histones, although its structure may be drastically altered, while the nucleosome in the path of the complex is evicted from the DNA (see Figure).
Several questions immediately arise from this work. First, it will be important to determine whether eviction would occur with the same efficiency in situations (as in native chromatin) where no free DNA ends are available. Nevertheless, the nucleosome-dependent SWI/SNF translocation likely would not be affected by nearby DNA ends. Second, how does the binding of the nucleosome in the pocket increase the processivity of translocation? Also how many nucleosomes can be displaced by the translocating complex and what, if anything, regulates this process?
It is also worth noting that Dechassa et al. detected a large group of remodeled products that exhibited protection of more than one nucleosome's worth of DNA. This may be due to bona fide remodeled nucleosome structures harboring internal loops, as has been recently been reported for RSC remodeling of mononucleosomes (Shukla et al., 2010). Moreover, this work suggests that a directionality is imparted by Gal4-VP16 recruitment of the remodeling complex, perhaps related to the observation of directional nucleosome movement away from positioning sequences in vivo by related ATP-dependent chromatin remodeling complexes (Whitehouse and Tsukiyama). The authors of the current study also point out that SWI/SNF eviction of nucleosomes may have some characteristics in common with how RNA Polymerase II disrupts histone-DNA interactions during transcription as recently detailed (Kulaeva et al., 2009). Interestingly, the Pol II studies were done with the same high-affinity nucleosome positioning elements used in the current work. The ability of Pol II to negotiate these nucleosomes varies greatly depending on the orientation of these sequences, giving insights into the mechanism by which histone-DNA interactions are disrupted. It would therefore be interesting to investigate the efficiency of SWI/SNF nucleosome eviction when the downstream nucleosome positioning sequence is inverted to determine if the Pol II mechanism is perhaps more generally employed. It would also be interesting to see whether distinct subunits in the SWI/SNF complex are necessary for the nucleosome eviction activities and whether histone acetylation stimulates nucleosome eviction in this system.
References
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 2010 doi: 10.1016/j.molcel.2010.02.040. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc Natl Acad Sci U S A. 2010;107:1936–1941. doi: 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]