Abstract
High-resolution 2D 13C-13C NMR correlation spectra of uniformly 13C-labeled molecules in solution are obtained by homonuclear 13C-decoupling along both dimensions by the application of indirect covariance NMR to constant-time NMR spectra. The spectra are optimally suited for chemical structure elucidation and molecular identification of the components of complex mixtures, such as ones from uniformly 13C-labeled cell cultures.
The characterization of chemical mixtures and their components is an essential task in many areas of Chemistry. NMR spectroscopy is a powerful tool enabling quantitative studies of complex mixtures, e.g. in metabolomics, to monitor changes both in terms of chemistry and concentrations without requiring extensive chromatographic fractionation. The vast majority of applications is based on 1D and 2D 1H NMR taking advantage of the high sensitivity afforded by 100% abundance of protons and their large gyromagnetic ratio. Although the benefits of 2D 13C-1H HSQC correlation spectroscopy of complex mixtures at 13C natural abundance have been demonstrated [1, 2], the absence of correlation information between different C-H pairs impedes identification of entire spin systems and thereby limits applications to compounds catalogued in NMR databases, such as the BMRB [3] and HMDB [4]. To establish correlations between all spins in a molecule or a spin system in an efficient manner, the use of 13C-enriched metabolite samples hold significant promise. Such samples can be produced, for example, by uniformly 13C-labeled cell cultures and organisms. Unfortunately, the presence of large 13C-13C 1J-couplings (>30 Hz) generates broad multiplet structures, which leads to substantial spectral crowding and cross-peak overlap when applied to complex mixtures. 13C constant-time (CT) spectroscopy [5-9] can help overcome the resolution issue along the indirect dimension, but the problem persists along the detection dimension. Here, we demonstrate how the combination of constant-time 13C-13C TOCSY with indirect covariance processing [10-14] produces homonuclear-decoupled high-resolution, high-sensitivity 13C-13C TOCSY spectra suitable for studying complex mixtures. This strategy has recently been demonstrated for the resolution enhancement of pure shift 1H correlation spectra [15, 16].
NMR Samples
A uniformly 13C-labeled algal amino acid mixture, purchased from Sigma-Aldrich, was prepared by dissolving 0.5 mg mixture in 2 ml D2O. The resulting suspension was centrifuged and the supernatant was used for measurements. Uniformly 13C-labeled glucose, purchased from Cambridge Isotope Laboratories, Inc., was prepared as a 1 M solution in D2O.
NMR Experiments and Processing
All 2D 13C-13C CT-TOCSY [9] and 2D 13C-13C TOCSY [17] data sets were collected with 512 N1 and 1024 N2 complex data points, with 56 ms DIPSI-2 mixing [18] for TOCSY and 56 ms FLOPSY-16 mixing [19] for CT-TOCSY. All NMR spectra were collected using a cryogenically cooled TCI probe (from Bruker Biospin) at 700 MHz proton frequency with 110 pm 13C spectral widths at 298 K temperature. The NMR data were zero-filled to 1024 (N1) and 2048 (N2), apodized using shifted sine-bell functions, and Fourier transformed, phase and baseline corrected using NMRPipe [20], and converted to a Matlab-compatible format for subsequent processing and analysis. The experimental time of the CT-TOCSY spectra was 5 hours for glucose and 20 hours for the algal amino acid mixture.
Indirect covariance processing takes the 2D FT NMR spectrum F as input and generates the new spectrum C according to [10, 13]:
| (1) |
In the present context, the method takes advantage of the high resolution of F along the indirect dimension ω1 endowed by the constant-time scheme by mapping it onto the direct dimension. This results in a symmetric 2D spectrum C that is homonuclear decoupled along both dimensions.
Since CT-experiments are susceptible to the appearance of (minor) intermittent extra-peaks [5-9], we apply an exclusion mask to C [21, 22]. For this purpose, a 2D FT CT-TOCSY spectrum F′ is computed from the same 2D time-domain data with zero-filling along ω1 to the same total number of datapoints N2 as along ω2 . Application of Gaussian line broadening (40 Hz) along the ω2 dimension and symmetrization by selecting min{|F′ij|,|F′ji|} from spectral points F′ij and F′ji at positions that are symmetric with respect to the main diagonal yields a medium-resolution spectrum. Line broadening is required, otherwise cross-peaks whose multiplets have no signal at the center frequency will disappear by the symmetrization procedure. Regions in C are then set to zero for which F′ lies below a given threshold. The application of the mask suppresses peaks that are not present in the original 2D FT spectrum while retaining the narrow peak shapes of the indirect covariance spectrum, which is demonstrated below.
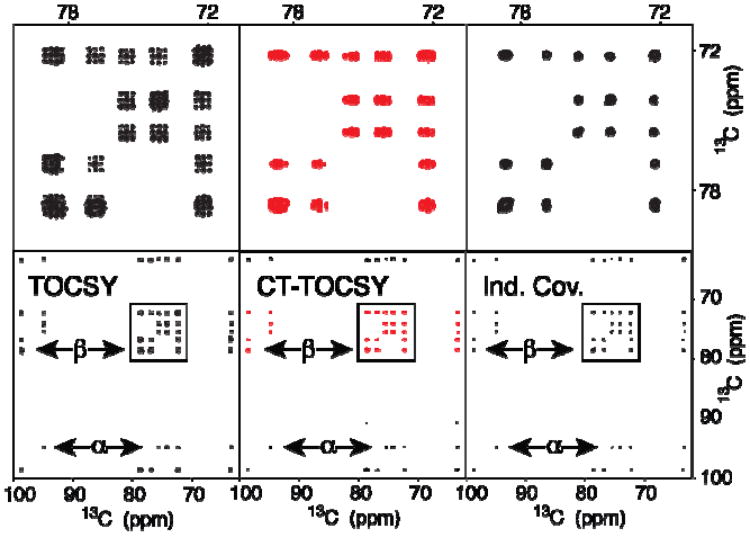
Figure 1 illustrates the method for uniformly 13C-labeled glucose. The two panels on the far left show a standard 13C-13C 2D TOCSY spectrum (bottom) with a zoomed region at the top. The presence of homonuclear 1JCC, 2JCC, 3JCC leads to prominent peak splittings that by far exceed the intrinsic line width. Nuclei that are bonded to two adjacent carbons show characteristic 1:2:1 multiplet patterns that cover a wide spectral range of 2·1JCC ≈ 70-90 Hz, which makes these cross-peaks naturally prone to overlaps. The middle two panels depict the corresponding 13C-13C 2D FT CT-TOCSY spectrum, which shows good decoupling along the ω1 dimension, while the ω2 dimension is fully impacted by 13C-13C J-coupling effects. Application of the covariance method of Eq. (1) to the 13C-13C 2D CT-TOCSY spectrum in the middle leads after masking to the spectrum on the right, which is effectively homonuclear decoupled along both dimensions. Although glucose does not suffer from cross-peak overlaps, Figure 1 illustrates the method.
Figure 1.
2D 13C-13C TOCSY (left), 13C-13C CT-TOCSY (middle), and indirect covariance 13C-13C CT-TOCSY spectra (right) of 13C labeled glucose. The 3 top panels depict expansions of the boxed spectral regions in the lower panels. The double-arrows indicate selected cross-sections that belong to the α and β forms of glucose.
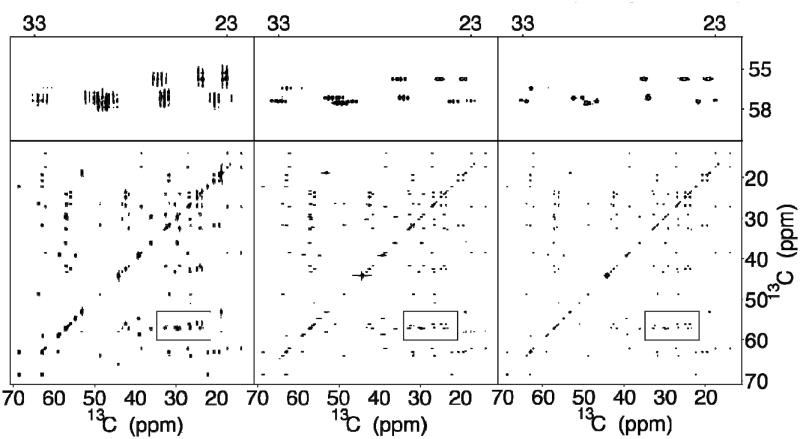
The chance of overlaps increases with the number of cross-peaks present in the spectrum as is the case for the amino-acid mixture shown in Figure 2. This figure is laid out analogously to Figure 1 comparing the standard 13C-13C 2D TOCSY (left panels) with the corresponding 13C-13C 2D CT-TOCSY spectrum (middle panels) and the indirect covariance 13C-13C 2D CT-TOCSY spectrum (right panels). The resolution improvement from left to right is striking as can be seen in the zoomed regions in the three panels at the top. Cross-peak clusters that evade direct analysis in the standard 2D FT spectrum (left) become partially resolved in the CT spectrum (middle) and fully resolved after indirect covariance processing (right). The resolution enhancement over standard 13C-13C 2D FT amounts to a factor 4, improving the average resolution from 70 Hz to 17 Hz along both dimensions. This trait considerably facilitates analysis of cross-peak connectivities, spin system assignment, component identification, or even chemical structure elucidation. Although a skilled NMR spectroscopist can still extract useful information from the standard 13C-13C 2D TOCSY spectrum (left panels), the covariance 13C-13C spectrum (right panels) permits a straightforward interpretation and is directly amenable to automated analysis. Figure 3 illustrates complete 13C-spin system identification for isoleucine taking optimal advantage of the resolution gain afforded by the CT-covariance approach.
Figure 2.
2D 13C-13C TOCSY (far left), 13C-13C CT-TOCSY (middle), and indirect covariance 13C-13C CT-TOCSY spectra (far right) of a uniformly 13C- labeled amino-acid mixture. The 3 top panels depict expansions of the boxed spectral regions in the lower panels.
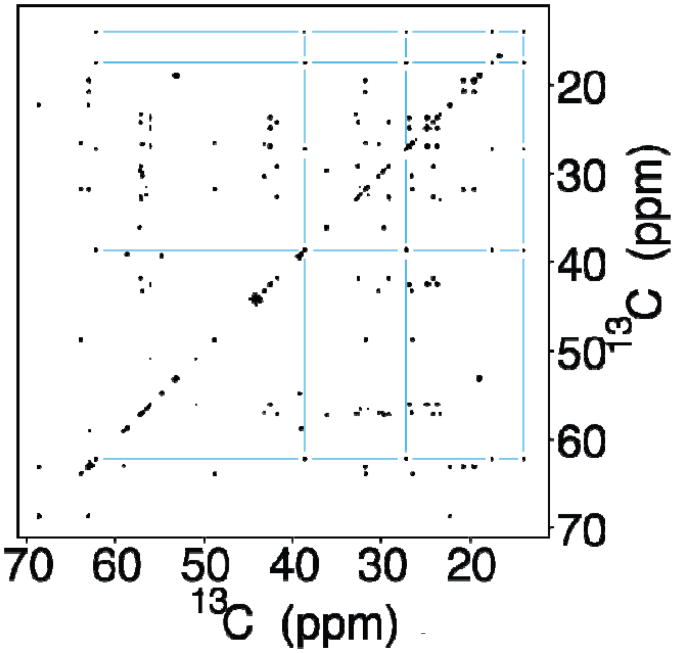
Figure 3.
Indirect covariance 13C-13C CT-TOCSY spectrum of a uniformly 13C-labeled amino acid mixture determined according to Eq. (1). The 13C-spin connectivity network for isoleucine is indicated by blue lines.
Homonuclear decoupling of 2D NMR experiments along both dimensions has been a long-standing challenge, which has elicited both experimental and computational approaches. Composite pulse decoupling [23, 24] and adiabatic decoupling [25, 26] can alter resonance positions and they work best for relatively small frequency ranges. The maximum entropy method [27] has been successfully applied to Cα resonances of amino acids that have a 1J-coupling to Cβs, but its performance for 13C nuclei with more than one coupling partner has not been demonstrated. Spin-state selective methods, such as S3E [28], IPAP [29], and DIPAP [30, 31] are tailored to a fixed number of coupling partners and their performance is sensitive to the exact magnitudes of the involved couplings. A homonuclear decoupling scheme based on pulsed-field gradient slice selection (‘pure shift spectroscopy’) [32] has recently been combined with covariance NMR to improve the resolution of 1H-1H 2D TOCSY, COSY, and NOESY spectra [15, 16]. Although this approach can in principle achieve better decoupling than CT-based methods, the resolution gain is offset by a sensitivity loss caused by slice selection. The CT-covariance spectrum, on the other hand, retains the inherent sensitivity of the 2D FT parent spectrum.
Our results demonstrate that dramatic cross-peak sharpening can be achieved along both 13C dimensions by the CT-covariance method producing high-resolution homonuclear-decoupled 13C-correlation spectra with qualitatively improved spectral behavior. While this property significantly assists the analysis complex organic molecules and mixtures of moderate complexity, as is the case for the amino-acid mixture of Fig. 2, it will be even more beneficial for studying extracts and lysates from 13C-labeled organisms, such as bacteria, yeast, and plants, for metabolic profiling, flux analysis, and de novo structure determination of metabolites [33].
Highlights.
2D TOCSY spectra of 13C-labeled metabolites could be decoupled along both dimensions
Indirect covariance processing of constant-time 2D NMR spectrum provides resolution enhancement
High-resolution 13C-13C TOCSY spectra of complex mixtures display reduced cross-peak overlaps
Acknowledgments
This work was supported by the National Institutes of Health (grant R01 GM066041).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Fax: 850-644-8281; Tel.: 850-644-1768; bruschweiler@magnet.fsu.edu
References
- 1.Keun HC, Beckonert O, Griffin JL, Richter C, Moskau D, Lindon JC, Nicholson JK. Cryogenic probe C-13 NMR spectroscopy of urine for metabonomic studies. Anal Chem. 2002;74:4588–4593. doi: 10.1021/ac025691r. [DOI] [PubMed] [Google Scholar]
- 2.Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, Westler WM, Suissman MR, Markley JL. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional H-1-C-13 NMR spectra. Anal Chem. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Mller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao HY, Markley JL. BioMagResBank. Nucleic Acids Research. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia JG, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong YP, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bax A, Freeman R. Investigation of Complex Networks of Spin-Spin Coupling by Two-Dimensional Nmr. Journal of Magnetic Resonance. 1981;44:542–561. [Google Scholar]
- 6.Rance M, Wagner G, Sorensen OW, Wuthrich K, Ernst RR. Application of Omega-1-Decoupled 2D Correlation Spectra to the Study of Proteins. J Magn Reson. 1984;59:250–261. [Google Scholar]
- 7.Santoro J, King GC. A Constant-Time 2D Overbodenhausen Experiment for Inverse Correlation of Isotopically Enriched Species. J Magn Reson. 1992;97:202–207. [Google Scholar]
- 8.Vuister GW, Bax A. Resolution Enhancement and Spectral Editing of Uniformly C-13-Enriched Proteins by Homonuclear Broad-Band C-13 Decoupling. J Magn Reson. 1992;98:428–435. [Google Scholar]
- 9.Eletsky A, Moreira O, Kovacs H, Pervushin K. A novel strategy for the assignment of side-chain resonances in completely deuterated large proteins using C-13 spectroscopy. J Biomol NMR. 2003;26:167–179. doi: 10.1023/a:1023572320699. [DOI] [PubMed] [Google Scholar]
- 10.Brüschweiler R. Theory of covariance nuclear magnetic resonance spectroscopy. Journal of Chemical Physics. 2004;121:409–414. doi: 10.1063/1.1755652. [DOI] [PubMed] [Google Scholar]
- 11.Brüschweiler R, Zhang F. Covariance nuclear magnetic resonance spectroscopy. Journal of Chemical Physics. 2004;120:5253–5260. doi: 10.1063/1.1647054. [DOI] [PubMed] [Google Scholar]
- 12.Trbovic N, Smirnov S, Zhang F, Brüschweiler R. Covariance NMR spectroscopy by singular value decomposition. Journal of Magnetic Resonance. 2004;171:277–283. doi: 10.1016/j.jmr.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Brüschweiler R. Indirect covariance NMR spectroscopy. Journal of the American Chemical Society. 2004;126:13180–13181. doi: 10.1021/ja047241h. [DOI] [PubMed] [Google Scholar]
- 14.Martin GE, Hilton BD, Blinov KA, Williams AJ. Using indirect covariance spectra to identify artifact responses in unsymmetrical indirect covariance calculated spectra. Magnetic Resonance in Chemistry. 2008;46:138–143. doi: 10.1002/mrc.2141. [DOI] [PubMed] [Google Scholar]
- 15.Morris GA, Aguilar JA, Evans R, Haiber S, Nilsson M. True Chemical Shift Correlation Maps: A TOCSY Experiment with Pure Shifts in Both Dimensions. J Am Chem Soc. 2010;132:12770–12772. doi: 10.1021/ja1039715. [DOI] [PubMed] [Google Scholar]
- 16.Aguilar JA, Colbourne AA, Cassani J, Nilsson M, Morris GA. Decoupling Two-Dimensional NMR Spectroscopy in Both Dimensions: Pure Shift NOESY and COSY. Angew Chem Int Ed Engl. 2012;51:6460–6463. doi: 10.1002/anie.201108888. [DOI] [PubMed] [Google Scholar]
- 17.Ernst R, Bodenhausen G, Wokaun A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Clarendon Press; Oxford: 1987. [Google Scholar]
- 18.Shaka AJ, Lee CJ, Pines A. J Magn Reson. 1988;77:274–293. [Google Scholar]
- 19.Kadkhodaie M, Rivas O, Tan M, Mohebbi A, Shaka AJ. Broad-Band Homonuclear Cross Polarization Using Flip-Flop Spectroscopy. Journal of Magnetic Resonance. 1991;91:437–443. [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Chen YB, Zhang F, Bermel W, Brüschweiler R. Enhanced covariance spectroscopy from minimal datasets. Journal of the American Chemical Society. 2006;128:15564–15565. doi: 10.1021/ja065522e. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser C, Lopez JJ, Bermel W, Glaubitz C. Dual transformation of homonuclear solid-state NMR spectra--an option to decrease measuring time. Biochim Biophys Acta. 2007;1768:3107–3115. doi: 10.1016/j.bbamem.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Shaka AJ, Barker PB, Freeman R. Computer-Optimized Decoupling Scheme for Wideband Applications and Low-Level Operation. J Magn Reson. 1985;64:547–552. [Google Scholar]
- 24.Shaka AJ, Keeler J, Frenkiel T, Freeman R. An Improved Sequence for Broad-Band Decoupling - WALTZ-16. J Magn Reson. 1983;52:335–338. [Google Scholar]
- 25.Kupce E. Nuclear Magnetic Resonance of Biologica Macromolecules Pt A. Academic Press Inc; San Diego: 2001. Applications of adiabatic pulses in biomolecular nuclear magnetic resonance; pp. 82–111. [DOI] [PubMed] [Google Scholar]
- 26.Kupce E, Wagner G. Multisite band-selective decoupling in proteins. J Magn Reson Series B. 1996;110:309–312. doi: 10.1006/jmrb.1996.0048. [DOI] [PubMed] [Google Scholar]
- 27.Shimba N, Stern AS, Craik CS, Hoch JC, Dotsch V. Elimination of C-13 alpha splitting in protein NMR spectra by deconvolution with maximum entropy reconstruction. J Am Chem Soc. 2003;125:2382–2383. doi: 10.1021/ja027973e. [DOI] [PubMed] [Google Scholar]
- 28.Meissner A, Duus JO, Sorensen OW. Integration of spin-state-selective excitation into 2D NMR correlation experiments with heteronuclear ZQ/2Q pi rotations for (1)J(XH)-resolved E.COSY-type measurement of heteronuclear coupling constants in proteins. J Biomol NMR. 1997;10:89–94. doi: 10.1023/a:1018331001961. [DOI] [PubMed] [Google Scholar]
- 29.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 30.Bermel W, Bertini I, Duma L, Felli IC, Emsley L, Pierattelli R, Vasos PR. Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angewandte Chemie-International Edition. 2005;44:3089–3092. doi: 10.1002/anie.200461794. [DOI] [PubMed] [Google Scholar]
- 31.Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R. C-13-detected protonless NMR spectroscopy of proteins in solution. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:25–45. [Google Scholar]
- 32.Zangger K, Sterk H. Homonuclear broadband-decoupled NMR spectra. J Magn Reson. 1997;124:486–489. [Google Scholar]
- 33.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. Carbon backbone topology of the metabolome of a cell. Journal of the American Chemical Society. 2012;134:9006–9011. doi: 10.1021/ja3033058. [DOI] [PMC free article] [PubMed] [Google Scholar]