Abstract
A customized metabolomics NMR database, TOCCATA, is introduced, which uses 13C chemical shift information for the reliable identification of metabolites, their spin systems and isomeric states. TOCCATA, whose information was derived from information of the BMRB and HMDB databases and the literature, currently contains 463 compounds and 801 spin systems and it can be used through a publicly accessible web server at http://spinportal.magnet.fsu.edu/toccata/webquery.html. TOCCATA allows the identification of metabolites in the sub-mM concentration range from 13C-13C TOCSY experiments of complex mixtures, which is demonstrated for an E.coli cell lysate, a carbohydrate mixture, and an amino acid mixture, all of which were uniformly 13C-labeled.
Introduction
NMR spectroscopy is one of the main analytical tools for the characterization of metabolomic samples.1 This is largely due to its high resolution power, which permits the analysis of complex mixtures without the need for extensive physical separation, e.g., by chromatographic techniques.2 Since NMR spectra of complex mixtures correspond to the linear superposition of the NMR spectra of individual components, the use of NMR database information of single metabolites can considerably facilitate spectral analysis. A primary goal of metabolomics is the identification of all mixture components with high accuracy. Retrieval of such information from 1D NMR spectra alone is often challenging.3 This is because of two factors: (1) the high frequency of peak overlaps impairs compound identification from individual peaks, and (2) the lack of connectivity information between peaks that belong to the same compound limits the combined use of NMR information from multiple nuclei that belong to the same molecule. As a consequence, even minor changes in chemical shifts between the mixture and database spectra can cause ambiguities in component annotation.
The use of 2D NMR spectra can overcome some of these issues, thereby outweighing the generally longer measurement times required. For the matching of 2D NMR spectra against database information a number of different strategies have been proposed. 2D 1H-13C HSQC spectra can be matched cross-peak by cross-peak against database entries.4–7 Although the resolution is increased by the introduction of the indirect 13C dimension, the lack of connectivity information between the different 1H,13C pairs belonging to the same molecule causes similar types of challenges for peak annotation and metabolite identification as in 1D NMR. Connectivity information between resonances from different parts of a molecule is available in 1H-1H TOCSY spectra collected at long mixing times.8 In this case, a cross section through the 2D spectrum along ω1 or ω2 represents the 1D spectrum of a whole spin system,9 which is equivalent to the 1D spectrum obtained after selective excitation of a resonance followed by TOCSY transfer.10 In the case of cross-peak overlap, consensus trace clustering followed by clustering permits the extraction of the 'clean' 1D spectrum of the spin system by taking advantage of the redundancy of connectivity information of TOCSY spectra.11
Recently, we expanded this strategy to uniformly 13C-labeled metabolites by the use of 13C-13C constant-time-TOCSY12 (CT-TOCSY) spectroscopy. Application to uniformly labeled E.coli extracts allowed the determination of carbon topologies of all detectable metabolites in the mM and sub-mM range.13 The analysis also used consensus trace clustering for the extraction of the spectra of individual spin systems. Because the topologies could be determined without any database information, this approach is not limited to the characterization of mixture components that are already catalogued. On the other hand, for those mixture components that are present in metabolite databases, the ability to directly identify them from 13C-13C CT-TOCSY would further enhance the utility of this approach. Since TOCSY traces only correlate resonances that belong to the same spin system, for molecules with multiple spin systems or multiple isomeric forms that are in slow exchange, they yield only part of the entire 1D 13C spectrum. Therefore, query against NMR databases that consist of the full 1D NMR spectra of metabolites leads to matches that are imperfect carrying the risk of false interpretations. Moreover, depending on the matching algorithm used, often molecules with a large a number of resonances are returned since they have a higher chance to match the resonances of the query trace. Presently, none of the current public NMR databases sorts spins into individual spin systems or slowly exchanging isomers for separate queries. To meet this demand, a metabolite database is introduced here, which is specifically geared toward the query of 13C TOCSY traces with the goal to optimize the matching accuracy.
Results and Discussion
1. Generation of TOCCATA database
The new database, which is termed TOCCATA (for TOCSY Customized Carbon Trace Archive), was primarily derived from the BMRB4 and HMDB6 metabolomics databases and presently contains 463 compounds. Out of these 463 compounds, 263 contain a single spin system and adopt a single isomeric state. Therefore only for this subset of compounds, there is a perfect match possible between a 1D 13C TOCSY trace and the 1D 13C spectrum. 163 compounds consist of more than one spin system in a single (isomeric) state, 29 compounds consist of a single spin system in multiple states, and 8 compounds consist of multiple states and multiple spin systems.
The TOCCATA database is structured as follows. First, the chemical shifts of the 463 compounds were subdivided into their individual isomeric states, which were then further subdivided into individual spin systems. Groups of 13C spins are considered to belong to separate spin systems, if they are separated by at least one non-carbon atom, such as an oxygen (e.g. O-glycosidic bond in lactose), a sulfur (e.g. methionine), or a nitrogen (e.g. N-glycosidic bond in adenosine). While this does not exclude the possible existence of small 13C-13C J-couplings between spins that belong to neighboring spin systems, magnetization transfer in 13C-13C TOCSY experiments at mixing times used here (<50 ms) is essentially negligible. This definition of the spin systems yielded a total of 801 different spin systems. A specifically designed web portal at http://spinportal.magnet.fsu.edu/toccata/webquery.html allows querying of the database either by using a list of 13C chemical shifts of a given spin system or by uploading a 13C trace, such as a 13C consensus TOCSY trace. The trace can be peak-picked interactively and subsequently queried against the database.
The BMRB metabolomics database includes NMR spectra of compounds at different pH and solvents. Since pH and solvent may result in NMR peak shifts, only NMR data of compounds dissolved in H2O/D2O at pH 7.4 were included in TOCCATA. For some metabolites not present in the BMRB, such as ribose, the 13C NMR chemical shifts were extracted from the HMDB (it should be noted that the HMDB spectra had been recorded at a slightly lower pH (pH 7.0); no pH correction was applied to the resulting chemical shifts).
The chemical shift assignments of all compounds of TOCCATA were performed manually by extracting spectral information from the BMRB, HMDB, and the literature 14–21. After the assignment of all resonances of a given compound, they were grouped into the different spin systems. In addition, the assignments were also used to determine the peak multiplet patterns for every carbon resonance in the databank. In a uniformly 13C-labeled compound, with all protons decoupled, a 13C multiplet directly reports on the number of directly bonded 13C atoms. A primary, secondary, tertiary, or quaternary carbon possesses a multiplet with intensity ratios of 1:1, 1:2:1 (or 1:1:1:1), 1:3:3:1 and 1:4:6:4:1, respectively. Inspection of multiplet patterns along the ω2-detection dimension in the CT-TOCSY spectrum has proven useful for the independent validation of the top matches returned by database query.13
2. TOCCATA query and web server
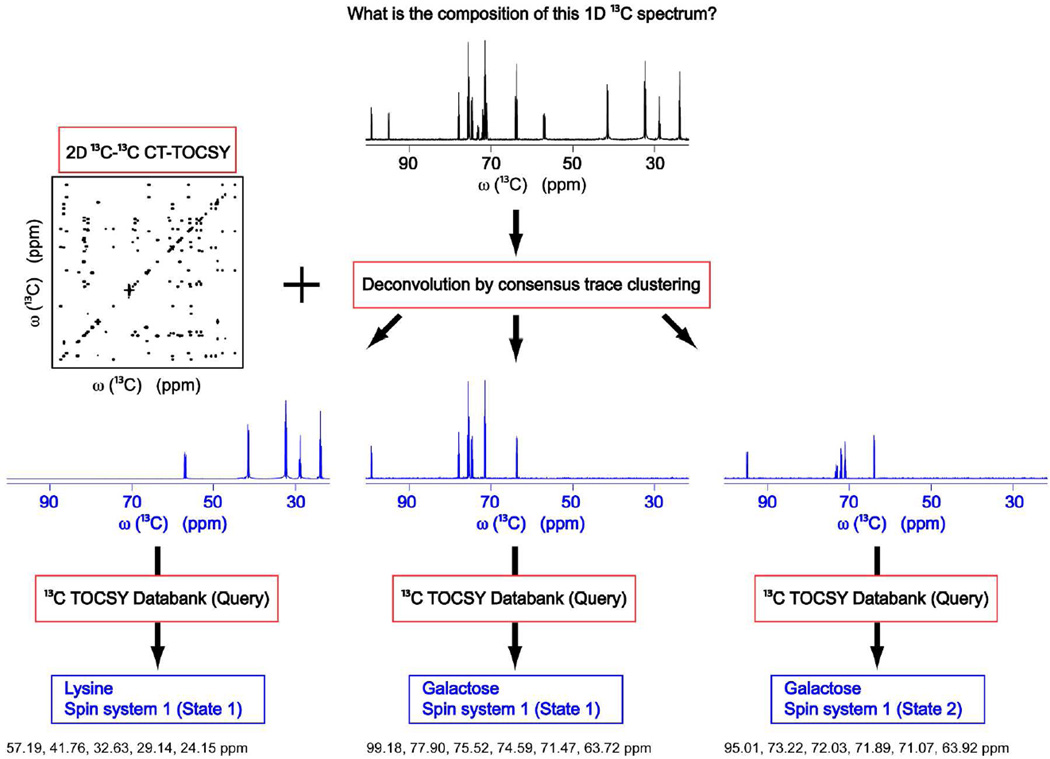
The idea of complex mixture analysis by 13C-13C CT-TOCSY NMR and subsequent database searching using TOCCATA is depicted in Figure 1. In order to identify the components of a mixture consisting of uniformly 13C-labeled metabolites (lysine, galactose β-pyranose, and galactose α-pyranose in the case of Figure 1) giving rise to the 1D 13C NMR spectrum depicted at the top of the figure, a 13C-13C CT-TOCSY spectrum at a sufficiently long TOCSY mixing time is collected (e.g. τm = 47 ms), which is deconvoluted into consensus traces (blue spectra) that represent individual 13C spin systems as described previously13 and is summarized in the Supporting Information. When queried against TOCCATA, the identities of the underlying metabolites are returned. Figure 2 shows a screenshot of the user interface of the TOCCATA web server. Users can manually enter (e.g. by copy and paste) a chemical shift list into the 'Peak List' text box, and submit it for TOCCATA query. Alternatively, a user can directly upload a TOCSY (consensus) trace as a file in a two-column ASCII format (where the first column represents the frequencies in ppm and the 2nd column the spectral intensities in arbitrary units). The trace is then displayed in the web server so that it can be manually peak-picked using the peak-picking button. For each peak multiplet, the center frequency should picked (even in case it has zero intensity, such as in the case of a doublet or quartet). The selected peak list can be interactively edited by clicking on the spectrum: a peak is added to the list if it is not yet in the peak list and a peak is removed if the peak is already contained in the peak list.
Figure 1.
Schematic representation of the deconvolution and TOCCATA database querying based on 2D 13C-13C CT-TOCSY spectrum of complex metabolite mixtures. The method is illustrated in the figure for a model mixture composed of lysine and galactose. The resulting deconvoluted 1D 13C TOCSY consensus traces belong to (from left to right): lysine, galactose β-pyranose, and galactose α-pyranose. They are identified by querying each of these traces against the 13C TOCSY databank TOCCATA.
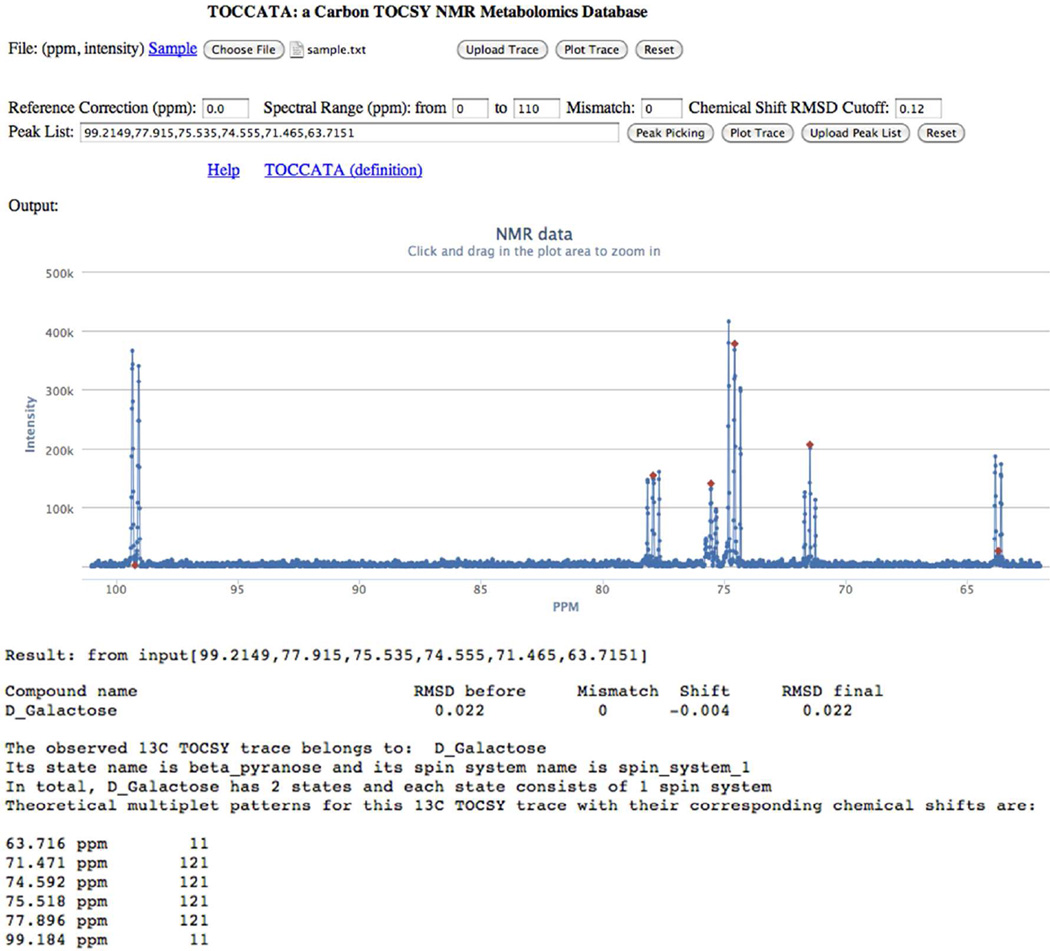
Figure 2.
Screenshot of TOCCATA web server user interface. A 1D 13C trace of interest can be uploaded and interactively peak-picked (red diamonds) with the corresponding chemical shifts displayed in the "Peak List". Querying of this peak list against TOCCATA returns the best matching compound (in this case D-galactose) with the chemical shift RMSD before and after a uniform shift of −0.004 ppm was applied. A mismatch number M = 0 indicates that the number of query peaks and database peaks for D-galactose were the same. The 13C chemical shifts of D-galactose are listed together with their multiplet fine structure (11 = 1:1 doublet, 121 = 1:2:1 triplet, 1111 = 1:1:1:1 quartet, 1331 = 1:3:3:1 quartet, etc) for validation.
2D CT-TOCSY spectra are typically performed on a finite spectral range, e.g. between 0 and 110 ppm in order to minimize off-resonance effects on TOCSY transfers. To take this into account during TOCCATA query, the web server allows users to specify the spectral width on which the database query should be performed by specifying the most downfield and most upfield ppm values. This eliminates potential mismatches arising from 13C resonances not detected in the TOCSY experiment, but which are present in the database. Ideally, the number of query peaks is identical to the number of resonances of the best matching spin system. However, this is not always the case, e.g., because a peak was missed in the query trace or two chemical shifts were assigned to different multiplet components of the same resonance. To facilitate the analysis of mismatches the web server allows the user to specify a maximally tolerated mismatch difference (Mmax) between the number of query peaks and the number of resonances of all possible matches. If the user is confident that all query peaks were correctly identified, then a mismatch parameter Mmax = 0 should be entered (default value). As a general rule, if a mismatch is detected, the user is advised to inspect the NMR raw data to identify the origin of the mismatch.
An important property of NMR chemical shifts is their proper referencing. Ideally, the chemical shifts are referenced against standard compounds, such as 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) or tetramethylsilane (TMS). In case that no standard was used, the web server permits the user to enter a chemical shift value (default 0.00 ppm) in order to reference a spectrum by uniformly increasing or decreasing the chemical shifts of all metabolite signals in the spectrum by the entered ppm value. To find the minimum root-mean-square-deviation (RMSD) for every metabolite, the TOCCATA matching algorithm performs an automated alignment within an interval of +/−0.5 ppm and then applies a weighted matching algorithm22 to find the best matching peak pairs from the query list and the database. Finally, the average chemical shift RMSD between input and database peak pairs is calculated and used as a criterion to select the best match, which is displayed on the screen (Figure 2).
In our experience the database query is most accurate when Mmax = 0 and RMSD < 0.12 ppm (default values). If no database entries satisfy the above criteria, the query returns a “no match” statement. With remarkably few exceptions, the chemical shift lists extracted from the TOCSY traces of the sugar mixture, the amino acid mixture, and the E.coli cell lysate have only one match satisfying these criteria, which are the correct ones. When multiple matches are returned, they are rank-ordered according to increasing RMSDs and displayed in groups with identical mismatch. Concise information about the number of isomeric states and spin systems of a compound is displayed for the top (currently 4) returns (Figure 2). In addition, their expected multiplet patterns are displayed for direct comparison with the multiplet patterns of the experimental input data. In our experience, the use of the multiplet pattern as a 'tie-breaker' resolves the vast majority of ambiguities.
3. Application to carbohydrate model mixture and E.coli cell extract
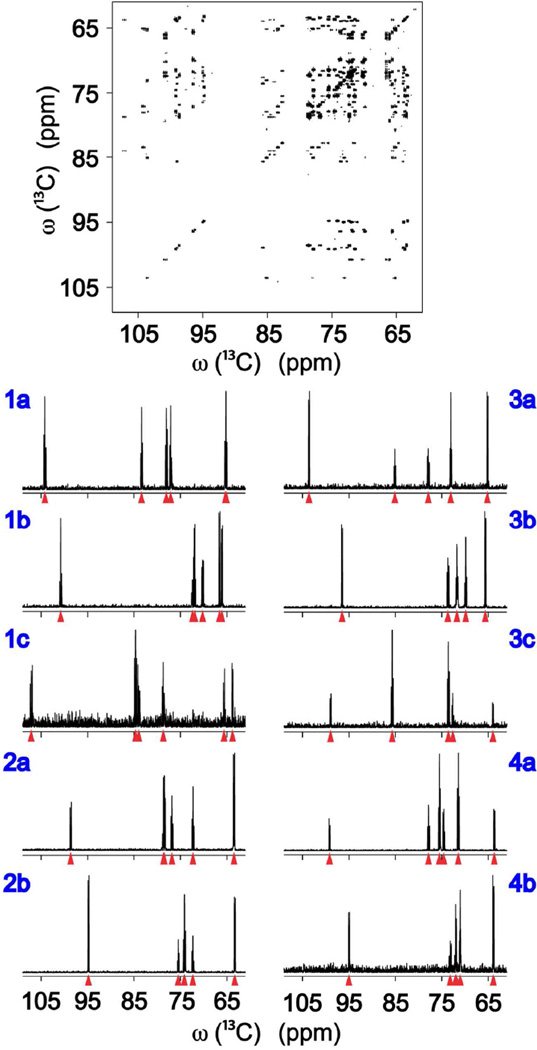
The first application shows the performance of TOCCATA for a carbohydrate mixture consisting of uniformly 13C-labeled fructose, glucose, ribose, and galactose in D2O. Each of these carbohydrates was present in solution either in two (glucose, galactose) or three (fructose, ribose) isomeric forms whereby each isomer constitutes a single 13C spin system. Consensus trace clustering of the 13C-13C CT-TOCSY spectrum yielded the traces shown in Figure 3. Each of these traces were peak-picked as shown by red triangles and queried against TOCCATA, which resulted in the correct identification of each of the 10 isomers present in the mixture with the query results compiled in Table 1. For each hit (see Table 1), the mismatch parameter (M) is returned (whereby M ≤ Mmax where Mmax can be entered on the web server) as well as the RMSD between the query and the database chemical shifts. For the carbohydrate mixture of Figure 3, the RMSD values are all < 0.12 ppm and the mismatch parameters (M) are all zero, reflecting that the number of input peaks queried was equal to the number of matched database peaks. With the selection criteria Mmax = 0 and RMSD < 0.12 ppm, unique and correct matches were found for all the carbohydrate traces. The column “Shift” shows how much the input data were shifted by the TOCCATA matching algorithm to find the minimum RMSD between input and database peaks. The Shift values show small variations indicating that there is no optimal universal shift for all traces. This is not unexpected as each metabolite responds individually to the specific conditions of the mixture.
Figure 3.
Deconvolution and TOCCATA database querying of 2D 13C-13C CT-TOCSY spectrum of carbohydrate mixture. The resulting deconvoluted 1D 13C TOCSY traces belong to: fructose β-furanose (1a), fructose β-pyranose (1b), fructose α-furanose (1c), glucose β-pyranose (2a), glucose α-pyranose (2b), ribose β-furanose (3a), ribose β-pyranose (3b), ribose α-furanose (3c), galactose β-pyranose (4a) and galactose α-pyranose (4b).
Table 1.
TOCCATA query results of deconvoluted 1D 13C TOCSY traces of carbohydrate mixture shown in Figure 3.
| RMSDa | Mb | Shiftc | RMSDa | Mb | Shiftc | ||
|---|---|---|---|---|---|---|---|
| fructose β-furanose | 0.016 | 0 | 0.019 | ribose β-furanose | 0.080 | 0 | 0.060 |
| fructose β-pyranose | 0.020 | 0 | 0.019 | ribose β-pyranose | 0.030 | 0 | 0.015 |
| fructose α-furanose | 0.015 | 0 | 0.024 | ribose α-furanose | 0.119 | 0 | 0.033 |
| glucose β-pyranose | 0.013 | 0 | 0.023 | galactose β-pyranose | 0.011 | 0 | 0.019 |
| glucose α-pyranose | 0.010 | 0 | 0.022 | galactose α-pyranose | 0.018 | 0 | 0.013 |
Root-mean square difference (in units of ppm) between the input and databank chemical shifts.
Integer mismatch parameter between the number of input and databank chemical shifts.
Amount by which the input chemical shifts were uniformly moved (in ppm) so that the RMSD with respect to the databank chemical shifts is minimized.
Application of consensus trace clustering to the 13C-13C CT-TOCSY of uniformly 13C-labeled E.coli cell lysate yielded 112 metabolite carbon topologies.13 We queried each of these traces against TOCCATA using the criteria Mmax = 0 and RMSD < 0.12 ppm, which led to the identification of 36 metabolites corresponding to 43 spin systems or isomeric states (Table 2). Out of the 36 metabolites, 34 have at least one topology for which a single match was returned. For the other 2 metabolites, the query returned 2 matches where one of the returns could be safely discarded: in one case because the expected multiplet pattern did not agree with the experimental one and in the other case because of a large RMSD difference between the best and second best hit. In addition, if a molecule consists of multiple spin systems, one expects to detect all other spin systems of the same molecule, which should be used as an additional criterion for the verification of the identity of a compound. For instance, spermidine consists of two 13C spin systems containing 4 and 3 spins, which were detected independently in the CT-TOCSY and turned out to be best hits in TOCCATA query.
Table 2.
Metabolites identified in E.coli cell lysate by querying against the TOCCATA database. The numbers in parentheses correspond to the query results of different metabolite states or spin systems.
| RMSDa | Mb | Shiftc | RMSDa | Mb | Shiftc | ||
|---|---|---|---|---|---|---|---|
| Valine | 0.044 | 0 | 0.085 | Lysine | 0.016 | 0 | 0.089 |
| Glutathione red. (1) | 0.016 | 0 | 0.084 | Aspartate | 0.041 | 0 | 0.093 |
| Glutathione red. (2) | 0.042 | 0 | 0.037 | Glucose | 0.011 | 0 | 0.104 |
| Glutathione ox. (1) | 0.006 | 0 | 0.079 | Cysteine | 0.052 | 0 | 0.159 |
| Glutathione ox. (2) | 0.008 | 0 | 0.085 | Isoleucine | 0.027 | 0 | 0.074 |
| Coenzyme A (1) | 0.014 | 0 | 0.053 | α-Glycerol phosphate | 0.026 | 0 | 0.047 |
| Coenzyme A (2) | 0.031 | 0 | 0.061 | Inosine | 0.026 | 0 | 0.107 |
| Coenzyme A (3) | 0.072 | 0 | 0.090 | Threonine | 0.034 | 0 | 0.100 |
| Glutamate | 0.016 | 0 | 0.137 | N(α)-Acetyl ornithine | 0.110 | 0 | 0.103 |
| Malate | 0.006 | 0 | 0.089 | N-Acetyl-glutamate | 0.072 | 0 | 0.057 |
| Maltose (1) | 0.066 | 0 | 0.070 | N(α)-Acetyl-lysine | 0.086 | 0 | 0.144 |
| Maltose (2) | 0.090 | 0 | 0.039 | 2-Aminoadipic acid | 0.016 | 0 | 0.089 |
| Maltose (3) | 0.090 | 0 | 0.088 | N-Acetyl-alanine | 0.003 | 0 | −0.011 |
| Maltose (4) | 0.088 | 0 | 0.081 | 2-Aminobutyric acid | 0.041 | 0 | 0.069 |
| Proline | 0.031 | 0 | 0.091 | Gluconate | 0.014 | 0 | 0.062 |
| Adenosine | 0.045 | 0 | 0.144 | NAD+ (1) | 0.047 | 0 | 0.110 |
| Leucine | 0.037 | 0 | 0.066 | NAD+ (2) | 0.069 | 0 | 0.181 |
| UDP_GlcNAc | 0.035 | 0 | 0.053 | NADP+ (1) | 0.118 | 0 | 0.216 |
| Ethanolamine | 0.024 | 0 | 0.037 | NADP+ (2) | 0.101 | 0 | 0.056 |
| Phenylalanine | 0.015 | 0 | 0.110 | Spermidine (1) | 0.078 | 0 | 0.143 |
| Serine | 0.030 | 0 | 0.096 | Spermidine (2) | 0.039 | 0 | 0.164 |
| Galactose | 0.013 | 0 | 0.101 | Uridine | 0.046 | 0 | 0.105 |
| Methionine | 0.025 | 0 | 0.081 | Putrescine | 0.026 | 0 | 0.110 |
Root-mean square difference (in units of ppm) between the input and databank chemical shifts.
Integer mismatch parameter between the number of input and databank chemical shifts.
Amount by which the input chemical shifts were uniformly moved (in ppm) so that the RMSD with respect to the databank chemical shifts is minimized.
Some metabolites in Table 2 demonstrate the capability of TOCCATA to differentiate and detect metabolites with very similar chemical shifts. For example, maltose exists in both α- and β-isomers with each state consisting of 2 glucose spin systems connected by a α(1→4) bond. While the first glucose populates the α state, the second glucose can adopt both the α and the β state (α- and β-maltose). This renders the chemical shifts of the first glucose very similar in the two states. CT-TOCSY analysis yields 3 unique consensus traces for maltose. TOCCATA query showed that one trace belongs to the first glucose of both α- and β-maltose, while the other two traces correspond to the second glucose of α- and β-maltose. NAD+ and NADP+ represent another example. These two metabolites are structurally and chemical shift-wise very similar (the only difference is that in NADP+ the ribose group attached to the adenine base is phosphorylated at the 2' position). Three consensus traces could be identified for NAD+ and NADP+ within the spectral range of the CT-TOCSY. According to TOCCATA query one of them corresponds to the ribose rings attached to the nicotinamide group in both NAD+ and NADP+. The other two traces correspond to the ribose rings attached to the adenine groups of NAD+ and NADP+, respectively. These examples illustrate that by treating the chemical shifts of different spin systems separately, the capability to distinguish between different molecules and their isomeric states can be enhanced, by identifying them through their most distinct (unique) spin systems.
To compare the TOCCATA with other current 13C chemical shift web servers, we submitted the 43 E.coli cell lysate 13C consensus TOCSY traces to the BMRB,4 COLMAR22,23 and the HMDB6 for 1D 13C database querying. The correct hit rates for BMRB (with a “C Range” parameter of 0.15) and COLMAR were 71% and 76%, respectively, and the one for HMDB (with a “13C Shift Tolerance” parameter of 0.15) was significantly lower. It should be noted that unlike the HMDB, the BMRB and COLMAR databases were derived from the same experimental spectra. Overall, this comparison illustrates how the accuracy of compound identification is substantially enhanced when using the customized TOCSY trace database TOCCATA.
Application of TOCCATA query to 13C-TOCSY traces of an amino-acid mixture is shown in the Supporting Information. Consensus trace clustering of the 13C-13C CT-TOCSY spectrum yields the traces shown in Figure S-1. The query results are summarized in Table S-1, which shows that TOCCATA query always identified the correct compound as a clear top match, when using the same criteria as above (Mmax = 0 and RMSD < 0.12 ppm). The relatively strict selection criteria together with multiplicity and completeness analysis prevented the occurrence of false positive hits in all applications reported here.
Conclusions
Unambiguous identification of the components of complex metabolite mixtures is a key step for their biological interpretation. We introduced here the TOCCATA database, which is customized for the identification of spin systems and isomeric states of metabolites from 13C TOCSY spectra. Consensus trace clustering of 2D 13C-13C CT-TOCSY spectra, which minimize the effects of peak overlaps, produces 13C traces that yield database query results at an unprecedented accuracy. As metabolomics databases continue to grow the chances that two entries have very similar NMR properties will also increase. This requires highly accurate database query tools, such as TOCCATA, for the unambiguous identification of mixture components. While metabolomics studies with uniformly 13C-labeled samples are not yet wide-spread, the ease and reliability of interpretation should provide additional motivation for this type of approach. As 13C-labeling of whole organisms, such as bacteria, yeast, and plants, is becoming increasingly common, the emergence of a wealth of new chemical and biological information including both natural product chemistry and metabolomics can be expected. While TOCCATA presently contains >800 spin systems, there is ample room for expansion, as the cell lysate example clearly demonstrates, by adding 13C chemical shift information from a wide range of sources, including existing NMR databases, the chemical literature, and NMR experiments of new compounds.
In addition to the analysis of 2D 13C-13C TOCSY spectra, TOCCATA can also be used to analyze 2D 13C-13C COSY spectra after the user has established complete chemical shift lists of each spin system from a 'COSY-walk' between direct-neighboring 13C spins. Application of TOCCATA to 2D 13C-1H HSQC-TOCSY spectra works only in exceptional cases: because TOCSY transfer in 2D 13C-1H HSQC-TOCSY experiments is mediated by 1H spins, the presence of non-protonated carbons leads to qualitatively different 1H-TOCSY and 13C-TOCSY transfer traces. Therefore, for the analysis of 2D 13C-1H HSQC-TOCSY and 2D 1H-1H TOCSY spectra, a 1H TOCSY database derived in a fashion analogous to TOCCATA will be needed.
Materials and Methods
The carbohydrate mixture of uniformly 13C-labeled glucose was purchased from Sigma-Aldrich, and fructose, galactose, and ribose were purchased from Cambridge Isotope Laboratories, Inc. An NMR sample was prepared by dissolving these carbohydrates in D2O each with a 10 mM final concentration. The E.coli NMR sample was prepared from an extract of the hydrophilic components from E. coli BL21(DE3) strain obtained from cells cultured in M9 medium as described recently13 with 13C-labeled glucose added.
2D 13C-13C CT-TOCSY12 data sets of the E.coli cell lysate and the carbohydrate mixture were collected with 576 N1 and 2048 N2 complex data points for 47 ms FLOPSY-16 mixing.24 All NMR spectra were collected at 800 MHz proton frequency with 110 pm 13C spectral width at 25°C. The NMR data were zero-filled to 2048 (N1) and 8192 (N2), apodized using shifted sine-bell windows, Fourier transformed, phase and baseline corrected using NMRPipe,25 and converted to a Matlab-compatible format for subsequent processing and analysis.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (grant R01 GM 066041).
References
- 1.Lindon JC, Nicholson JK, Holmes E. The Handbook of Metabonomics and Metabolomics. Amsterdam: Elsevier; 2007. [Google Scholar]
- 2.Robinette SL, Bruschweiler R, Schroeder FC, Edison AS. Acc. Chem. Res. 2012;45:288–297. doi: 10.1021/ar2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenz EM, Wilson ID. J. Proteome Res. 2007;6:443–458. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- 4.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao HY, Markley JL. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Nat. Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 6.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia JG, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong YP, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikayama E, Sekiyama Y, Okamoto M, Nakanishi Y, Tsuboi Y, Akiyama K, Saito K, Shinozaki K, Kikuchi J. Anal. Chem. 2010;82:1653–1658. doi: 10.1021/ac9022023. [DOI] [PubMed] [Google Scholar]
- 8.Braunschweiler L, Ernst RR. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 9.Zhang F, Bruschweiler R. Angew. Chem. Int. Ed. 2007;46:2639–2642. doi: 10.1002/anie.200604599. [DOI] [PubMed] [Google Scholar]
- 10.Sandusky P, Raftery D. Anal. Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 11.Bingol K, Bruschweiler R. Anal. Chem. 2011;83:7412–7417. doi: 10.1021/ac201464y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eletsky A, Moreira O, Kovacs H, Pervushin K. J. Biomol. NMR. 2003;26:167–179. doi: 10.1023/a:1023572320699. [DOI] [PubMed] [Google Scholar]
- 13.Bingol K, Zhang F, Bruschweiler-Li L, Bruschweiler R. J. Am. Chem. Soc. 2012;134:9006–9011. doi: 10.1021/ja3033058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitmaier E, Voelter W. Eur. J. Biochem. 1972;31:234–238. doi: 10.1111/j.1432-1033.1972.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 15.Witherup TH, Abbott EH. J. Org. Chem. 1975;40:2229–2234. doi: 10.1021/jo00903a021. [DOI] [PubMed] [Google Scholar]
- 16.Hesbain-Frisque AM, Van Schaftingen E, Hers HG. Eur. J. Biochem. 1981;117:325–327. doi: 10.1111/j.1432-1033.1981.tb06340.x. [DOI] [PubMed] [Google Scholar]
- 17.Bock K, Pedersen C. Adv. Carbohydr. Chem. Biochem. 1983;41:27–66. [Google Scholar]
- 18.Bock K, Pedersen C, Pedersen H. Adv. Carbohydr. Chem. Biochem. 1984;42:193–225. [Google Scholar]
- 19.Rossi C, Donati A, Ulgiati S, Sansoni MR. Bull. Magn. Reson. 1992;14:181–185. [Google Scholar]
- 20.D'Ordine RL, Paneth P, Anderson VE. Bioorg. Chem. 1995;23:169–181. [Google Scholar]
- 21.Kustermann E, Seelig J, Kunnecke B. Am. J. Physiol. 1998;274:E65–E71. doi: 10.1152/ajpendo.1998.274.1.E65. [DOI] [PubMed] [Google Scholar]
- 22.Robinette SL, Zhang FL, Bruschweiler-Li L, Bruschweiler R. Anal. Chem. 2008;80:3606–3611. doi: 10.1021/ac702530t. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Robinette SL, Bruschweiler-Li L, Bruschweiler R. Magn. Reson. Chem. 2009;47:S118–S122. doi: 10.1002/mrc.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadkhodaie M, Rivas O, Tan M, Mohebbi A, Shaka AJ. J. Magn. Reson. 1991;91:437–443. [Google Scholar]
- 25.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.