Abstract
Following recent technological revolutions, the investigation of massive biomedical data with growing scale, diversity, and complexity has taken a center stage in modern data analysis. Although complex, the underlying representations of many biomedical data are often sparse. For example, for a certain disease such as leukemia, even though humans have tens of thousands of genes, only a few genes are relevant to the disease; a gene network is sparse since a regulatory pathway involves only a small number of genes; many biomedical signals are sparse or compressible in the sense that they have concise representations when expressed in a proper basis. Therefore, finding sparse representations is fundamentally important for scientific discovery. Sparse methods based on the norm have attracted a great amount of research efforts in the past decade due to its sparsity-inducing property, convenient convexity, and strong theoretical guarantees. They have achieved great success in various applications such as biomarker selection, biological network construction, and magnetic resonance imaging. In this paper, we review state-of-the-art sparse methods and their applications to biomedical data.
Keywords: Sparse learning, structured sparsity, Gaussian graphical model, magnetic resonance imaging
1. INTRODUCTION
Recent technological revolutions have unleashed a torrent of biomedical data with growing scale, diversity, and complexity [24; 27; 77; 86; 101]. The wealth of data confronts scientists with an urgent need for new methods and tools that can intelligently and automatically extract useful information from data and synthesize knowledge [17; 32; 56; 74]. Although complex, the underlying representations of many real-world data are often sparse [32; 38; 41]. For example, for a certain disease such as leukemia, even though humans have tens of thousands of genes, only a small number of them are relevant to the disease; a gene network is sparse since a regulatory pathway involves only a small number of genes; the neural representation of sounds in the auditory cortex of unanesthetized animals is sparse, since the fraction of neurons active at a given instant is small; many biomedical signals have sparse representations when expressed in a proper basis. Therefore, finding sparse representations is fundamentally important for scientific discovery. The last decade has witnessed a growing interest in the search for sparse representations of data.
The quest for sparsity is further motivated for various reasons. First, sparse representations enhance the interpretability of the model. For example, in many biological applications, the selection of genes or proteins which are related to the study, is crucial to facilitate the biological interpretation [18; 38]. In addition, the resulting gene/protein selection might enable a feasible biological validation with a reduced experimental cost. Second, sparseness is one way to measure the complexity of the learning model [84]. Regularization is commonly employed to penalize the complexity of a learning model and alleviate overfitting. Regularization based on the norm maximizes sparseness, which, however, leads to an NP-hard problem. As a computationally efficient alternative, the norm regularization, which also leads to a sparse model, is widely used in many areas including signal processing, statistics, and machine learning [13; 23; 52; 93; 98; 124; 127]. Finally, finding sparse representations has recently received increasing attention due to the current burst of research in Compressed Sensing (CS) [4; 6; 16; 25; 26; 102]. CS is a technique for acquiring and reconstructing a signal utilizing the prior knowledge that it is sparse or compressible. It encodes a large sparse signal using a relatively small number of linear measurements, and minimizing the norm in order to decode the signal. Recent theories [13; 14; 15; 16; 25] assert that one can recover certain signals and images from far fewer samples or measurements than traditional methods.
In this paper, we review sparse methods for (1) incorporating a priori knowledge on feature structures for feature selection, (2) constructing undirected Gaussian graphical models, and (3) parallel magnetic resonance imaging.
Structured Feature Selection
Although sparse learning models based on the norm such as the Lasso [98] have achieved great success in many applications, they do not take the existing feature structure into consideration. Specifically, these models yield the same solution after randomly reshuffling the features. However, in many applications, the features exhibit certain intrinsic structures, e.g., spatial or temporal smoothness, disjoint/overlapping groups, trees, and graphs [42; 45; 51; 65; 116]. The a priori structure information may significantly improve the classification/regression performance and help identify the important features. For example, in the study of arrayCGH [99; 100], the features—the DNA copy numbers along the genome—have the natural spatial order, and the fused Lasso, which incorporates the structure information using an extension of the , outperforms the Lasso in both classification and feature selection. In this paper, we review various structured sparse learning models including group Lasso, sparse group Lasso, overlapping group Lasso, tree Lasso, fused Lasso, and graph Lasso.
Sparse Undirected Gaussian Graphical Models
Undirected graphical models explore the relationships among a set of random variables through their joint distribution. The estimation of undirected graphical models has applications in many domains, such as computer vision, biology, and medicine. An instance is the analysis of gene expression data. As shown in many biological studies, genes tend to work in groups based on their biological functions, and there exist some regulatory relationships between genes [19]. Such biological knowledge can be represented as a graph, where nodes are the genes, and edges describe the regulatory relationships. Graphical models provide a useful tool for modeling these relationships, and can be used to explore gene activities. One of the popular graphical models is the Gaussian graphical model (GGM), which assumes the variables to be Gaussian distributed [5]. In GGM, the problem of learning a graph is equivalent to estimating the inverse of the covariance matrix (precision matrix), since the nonzero off-diagonal elements of the precision matrix represent edges in the graph [5]. In some applications, we need to estimate multiple related precision matrices. For example, in the modeling of brain networks for Alzheimer's disease using neuroimaging data [43], we want to estimate graphical models for three groups: normal controls (NC), patients of mild cognitive impairment (MCI), and Alzheimer's patients (AD). These graphs are expected to share some common connections, but they are not identical. It is thus desirable to jointly estimate the three graphs. In this paper, we review sparse methods for estimating a single undirected graphical model and for estimating multiple related undirected graphical models and discuss their properties.
Parallel Magnetic Resonance Imaging
Parallel imaging has been the single biggest innovation in magnetic resonance imaging in the last decade. It exploits the difference in sensitivities between individual coil elements in a receive array to reduce the number of gradient encodings required for imaging, and the increase in speed comes at a time when other approaches to acquisition time reduction were reaching engineering and human limits [59]. In the SENSE-type reconstruction approach, researchers have taken advantage of the sparsity promoting penalties (e.g., wavelets and total variations) to reduce the acquisition time while maintaining the image quality. Key components of sparse learning include the estimation of the coil sensitivity profiles, the design of the sparsity promoting regularization, the development of the sampling pattern that takes advantage of sparse learning, and the efficient optimization of the non-smooth inverse problem. In this paper, we review different components of sparse learning in magnetic resonance imaging.
The rest of the paper is organized as follows. We review structured sparse learning for feature selection in Section 2. The estimation of sparse undirected Gaussian graphical models is presented in Section 3. We discuss sparse learning in parallel magnetic resonance imaging in Section 4. Finally, we conclude the paper in Section 5.
2. STRUCTURED FEATURE SELECTION
We are given a set of training samples , where denotes the p-dimensional features for the i-th sample, and is its response (numeric for regression, and categorical for classification). In addition, we are given a feature structure, e.g., a group structure, a tree structure, or a graph structure, as part of the input data. We focus on a linear model with h(a) = xTa, where is the vector of model parameters. To fit the model with the training samples, we learn the model parameter vector x by solving the following optimization problem:
| (1) |
where L(x) is a loss function, Ω(x) is a regularization term encoding the prior knowledge on the input features, and λ > 0 is the regularization parameter controlling the trade-off between the loss L(·) and the penalty Ω(·).
The formulation in (1) can be applied for regression, classification, and longitudinal data analysis:
Regression: The outcome b is a continuous value, e.g., the hippocampus volume or the minimental state examination (MMSE) score of a subject in the study of Alzheimer's disease. The least squares loss is commonly used for regression.
Classification: The outcome b is a discrete value, e.g., disease status, including normal controls and disease patients. The logistic loss is commonly used for classification.
Longitudinal Data Analysis: The outcome b is the observed failure/censoring time. If an event occurs at time t, then the subject has a failure time t. If a patient drops from the study at time t, we consider he/she is censored at time t. The Cox model is a popular approach for longitudinal data analysis, in which the negative log-likelihood function of the proportional hazard is used as the loss function [21].
The regularization term Ω(x) in (1) is commonly employed to penalize the complexity of a learning model and alleviate overfitting, e.g., the regularization used in ridge regression. However, the commonly used regularization leads to a dense model, i.e., almost all model parameters in x are non-zero. To enhance the interpretability of the model, a sparse model is desired. One popular sparse model, known as the Lasso, is based on the penalty:
| (2) |
The Lasso has been applied widely in many biomedical applications [91; 94; 107; 111; 123]. In many applications, the features exhibit certain intrinsic structures, e.g., spatial or temporal smoothness, graphs, trees, and disjoint/overlapping groups. The a priori structure information may significantly improve the classification/regression performance and help to identify the important features.
2.1 Group Lasso and Sparse Group Lasso
In many applications, the features form a natural group structure. For example, the voxels of the positron emission tomography (PET) images in the Alzheimer's Disease study can be divided into a set of non-overlapping groups according to the brain regions [43]; in the multi-factor ANOVA problem, each factor may have several levels and can be represented using a group of dummy variables [117]. The selection of group structures has recently received increasing attention in the literature [3; 44; 45; 64; 78; 117; 120]. The pioneer work [117] focused on the non-overlapping group Lasso, i.e., the groups are disjoint. Assume the features are partitioned into k disjoint groups {G1, · · · , Gk}. The group Lasso formulation uses the penalty on the model parameters:
| (3) |
where ∥ · ∥q is the with q > 1 (most existing work focus on q = 2 or ∞) [68], and wi is the weight for the i-th group. The group selection distinguishes the group Lasso from the Lasso which does not take group information into account and does not support group selection. The group Lasso has been applied for regression [55; 80; 117], classification [78], joint covariate selection for grouped classification [85], and multi-task learning [2; 62; 89].
The group Lasso does not perform feature selection within each feature group. For certain applications, it is desirable to perform simultaneous group selection and feature selection. The sparse group Lasso (sgLasso) incorporates the strengthens from both Lasso and group Lasso, and it yields a solution with simultaneous between- and within- group sparsity [30; 87]. The sparse group Lasso penalty is based on a composition of the and the :
| (4) |
where α ∈ [0, 1], the first term controls the sparsity in the feature level, and the second term controls the sparsity in the group level. The sparse group Lasso has been applied to analyze multiple types of high dimensional genomic data for biomarker discovery [87].
Figure 1 illustrates Lasso, group Lasso, and sparse group Lasso; we use four types of data sources including Proteomics, GWAS (genome-wide association study), MRI (magnetic resonance imaging), and PET from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database1. We construct four feature groups, one for each each data source. As shown in the figure, the Lasso does not consider the group (source) information and selects a subset of features from all four groups; the group Lasso selects a subset of the groups (3 in this example) and all features from these 3 groups are selected; the sparse group Lasso simultaneously selects a subset of the groups and a subset of the features within each selected group.
Figure 1.
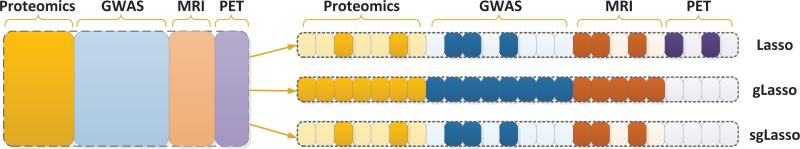
Illustration of Lasso, group Lasso (gLasso), and sparse group Lasso (sgLasso). Four types of data sources, including Proteomics, GWAS (genome-wide association study), MRI (magnetic resonance imaging), and PET from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database are used for illustration. There are four feature groups, one for each data source. The features selected by each algorithm are highlighted.
2.2 Overlapping Group Lasso and Tree Lasso
In group Lasso [117], the groups are disjoint. Some recent work [44; 45; 46; 51; 69; 120] studied the more general case where the groups may overlap. One motivating example is the use of biologically meaningful gene/protein sets (groups). The proteins/genes in the same groups are related if they either appear in the same pathway, or are semantically related in terms of Gene Ontology (GO) hierarchy, or are related from gene set enrichment analysis (GSEA) [97]. The canonical pathway in MSigDB, for example, has provided 639 groups of genes [97]. It has been shown that the group (of proteins/genes) markers are more reproducible than individual protein/gene markers and the use of such group information improves classification performance [19]. Groups may overlap - one protein/gene may belong to multiple groups - and the group Lasso formulation is not applicable. For the general overlapping group patterns, we can make use of the following overlapping group Lasso penalty [120]:
| (5) |
where α ∈ [0, 1], wi > 0 (i = 1, 2, . . . , k), and Gi consists of the indices from the i-th group of features. The k groups of features are pre-specified, and they may overlap. A different overlapping group Lasso formulation was proposed in [44]. In some applications, the features follow a tree structure. For example, an image can be represented using a tree structure where each leaf node corresponds to a feature (pixel) and each internal node corresponds to a group of features (pixels) based on the spatial locality [69]. In such a case, we can make use of the tree structured group Lasso penalty [46; 51; 69; 120]:
| (6) |
where is a constant weight, and , a node at the depth i, consists of all features in the subtree. Note that any parent node is a superset of its children. Thus, if a specific node is not selected (i.e., its corresponding model coefficient is zero), then all its children will not be selected. It is clear that the tree structured group Lasso is a special case of the overlapping group Lasso with a specific tree structure.
2.3 Fused Lasso
In many applications, the features enjoy certain smoothness properties. For example, the adjacent features in the arrayCGH data are close to each other along the genome. Therefore, it is desirable to enforce the model parameters in x to have the structure of smoothness. Such a structure can be induced by the fused Lasso penalty [28; 99]:
| (7) |
where α ∈ [0, 1]. The fused Lasso penalty in (7) shall induce a solution that xi tends to be close or identical to xi+1 for i = 1, · · · , p – 1. The smoothness structure can also be revealed from the fused Lasso signal approximator [28]:
| (8) |
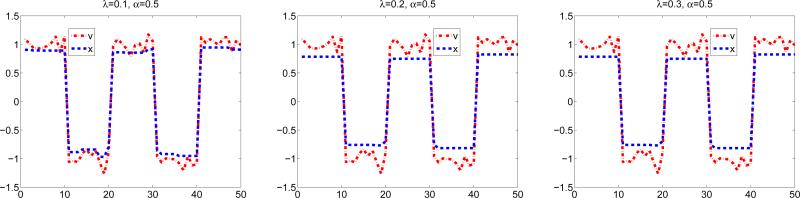
Figure 2 illustrates the fused Lasso signal approximator (8) under different values of λ with α = 0.5. We can observe that the solution is piecewise constant.
Figure 2.
Illustration of the solution of (8), the fused Lasso signal approximator.
2.4 Graph Lasso
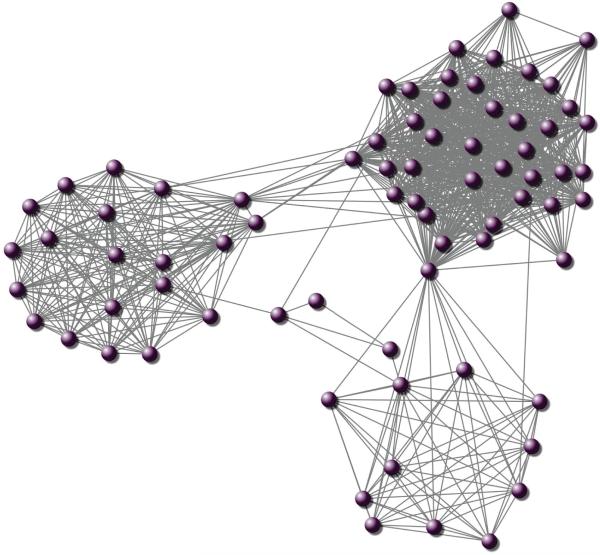
In certain applications, the features form an undirected graph structure, in which two features connected by an edge in the graph are more likely to be selected together. As an example, many biological studies have suggested that genes tend to work in groups according to their biological functions, and there are some regulatory relationships between genes [60]. This biological knowledge can be represented as a graph, where the nodes represent the genes, and the edges imply the regulatory relationships between genes. Figure 3 shows a subgraph consisting of 80 nodes (genes) of the network described in [19]. Several recent studies have shown that the estimation accuracy can be improved using dependency information encoded as a graph. Let (N, E) be a given graph, where N = {1, 2, · · · , p} is a set of nodes, and E is a set of edges. Node i corresponds to the i-th feature. If nodes i and j are connected by an edge in E, then the i-th feature and the j-th feature tend to be grouped.
Figure 3.
Illustration of a subgraph of the network consisting of 80 nodes.
The fussed Lasso penalty in (7) can be extended to a general graph structure; we call it the graph Lasso:
| (9) |
where the second regularization term penalizes a large deviation between two model parameters whose corresponding nodes are connected in the graph. Intuitively, if two genes/proteins are connected in a network, their model parameters are likely to be close to each other, satisfying the so-called smoothness property on a graph. The graph Lasso formulation is computationally expensive to solve. The graph Lasso, or the Laplacian Lasso, is an efficient alternative, which uses the following penalty:
| (10) |
where L is the Laplacian matrix [7; 20] constructed from the graph. It is known that the Laplacian matrix is positive semi-definite, and captures the underlying local geometric structure of the data. When L is an identity matrix, (10) reduces to the elastic net penalty [126]. Existing efficient algorithms for solving the Lasso can be applied to solve the graph Lasso by grouping the loss term L(x) and the Laplacian regularization λ(1 – α)xTLx together, as the latter is both convex and differentiable.
Both and graph Lasso encourage positive correlation between the values of coefficients for the features connected by an edge in the graph. However, in certain applications, two features connected may be negatively correlated. To overcome this limitation, GFlasso employs a different regularization over a graph:
| (11) |
where rij is the sample correlation between two features [50]. The penalty in (11) encourages the coefficients xi, xj for features i, j connected by an edge in the graph to be similar when rij > 0, but dissimilar when rij < 0. GFlasso would introduce additional estimation bias due to possible graph misspecification. For example, additional bias may occur when the sign of rij is inaccurate.
Another alternative is the so-called graph OSCAR (GOSCAR) penalty given by [110]:
| (12) |
where a pairwise regularizer is used to encourage the coefficients to be equal [9], but the grouping constraints are imposed on the nodes connected over the given graph. The regularizer encourages sparseness. The pairwise regularizer puts more penalty on the larger coefficients. Note that max {|xi|, |xj|} can be decomposed as
The GOSCAR formulation is closely related to OSCAR [9]. The penalty of OSCAR is
| (13) |
The regularizer leads to a sparse solution, and the regularizer encourages the coefficients to be equal. OSCAR can be efficiently solved by accelerated gradient methods, whose key projection can be solved by a simple iterative group merging algorithm [121]. However, OSCAR assumes each node is connected to all the other nodes, which is not sufficient for many applications. Note that OSCAR is a special case of GOSCAR when the graph is complete. GOSCAR, incorporating an arbitrary undirected graph, is much more challenging to solve [110].
The penalty in GOSCAR overcomes the limitation of the Laplacian Lasso that the different signs of coefficients can introduce additional penalty. However, under the regularizer, even if |xi| and |xj| are close to each other, the penalty on this pair may still be large due to the property of the max operator, resulting in the coefficient xi or xj being over penalized. The additional penalty would result in biased estimation, especially for large coefficients, as in the Lasso case [98]. In GFlasso, when the pairwise sample correlation wrongly estimates the sign between xi and xj, an additional penalty on xi and xj would occur, introducing estimation bias. This motivates the following non-convex feature grouping and selection penalty:
| (14) |
which shrinks only small differences in absolutes values [110; 125]. As a result, estimation bias is reduced as compared to those convex grouping penalties. Note that the non-convex penalty does not assume the sign of an edge is given; it only relies on the graph structure.
3. SPARSE UNDIRECTED GAUSSIAN GRAPHICAL MODELS
Undirected graphical models are commonly used to describe and explain the relationships among a set of variables based on a collection of observations. In the Gaussian case, the graphical Lasso [29] is a popular approach for learning the structure in an undirected Gaussian graphical model [5]. The basic model for continuous data assumes that the observations have a multivariate Gaussian distribution with mean μ and covariance matrix Σ. If the ijth entry of Θ = Σ–1 is zero, then variables i and j are conditionally independent, given the other variables. Here, Θ is called the precision matrix. Thus, the problem of identifying the structure of the undirected Gaussian graphical model is equivalent to finding the nonzero entries of Θ. In [5], the penalty is imposed on the precision matrix to increase its sparsity. The sparse undirected graphical model has been applied to construct biological networks [5] and brain networks [43].
3.1 Graphical Lasso
Suppose we have n samples independently drawn from a multivariate Gaussian distribution, and these samples are denoted as , where yi is a p dimensional vector, is the mean, and is the covariance matrix. Let be the inverse covariance matrix. The empirical mean is denoted as , and the empirical covariance is denoted as S:
It can be shown that under a multivariate Gaussian model, the maximum likelihood estimate of Θ = Σ–1 can be obtained by solving the following maximization problem:
| (15) |
where tr (SΘ) is the trace of SΘ, given by the summation of the diagonal entries of SΘ. Assume that S is nonsingular. The maximum likelihood estimate of the inverse covariance Θ is Θ = S–1. If the dimensionality is larger than the sample size, i.e., p > n, S is singular. In such a case, regularization is commonly applied, and we estimate Θ = Σ–1 by maximizing the following objective function:
| (16) |
where J(Θ) is a penalty function. The graphical Lasso employs the penalty and solves the following optimization problem [5]:
| (17) |
It is known that a larger value of λ leads to a sparser Θ that fits the data less well, while a smaller value of λ leads to a less sparse Θ that fits the data well. Thus, the choice of λ is an important issue in practical application of the graphical Lasso [63; 79].
Banerjee et al. [5] employed the interior point method to solve the optimization problem in (17). Friedman et al. [29] developed the graphical Lasso (GLasso) which applied the blockwise coordinate descent method to solve (17). The GLasso fails to converge with warm-starts. To resolve this issue, Mazumder and Hastie [76] proposed a new algorithm called DP-GLasso, each step of which is a box-constrained QP problem. The main challenge of estimating a sparse precision matrix is its high computational complexity. Witten et al. [106] and Mazumder and Hastie [75] independently derived a screening rule, which dramatically reduced the computational cost especially for large regularization parameter values.
3.2 The Monotone Property
Huang et al. [43] derived the monotone property of the graphical Lasso. We first introduce the following definition.
Definition 1. In the graphical representation of the inverse covariance, if node i is connected to node j by an arc, then node i is called a “neighbor” of node j. If node i is connected to node k though some chain of arcs, then node i is called a “connectivity component” of node k.
Intuitively, two nodes are neighbors if they are directly connected, whereas two nodes belong to the same connectivity component if they are indirectly connected, i.e., the connection is mediated through other nodes. In other words, if two nodes do not belong to the same connectivity component (i.e., two nodes completely separated in the graph), then they are completely independent of each other. Huang et al. [43] showed that the connectivity components have the following monotone property:
Proposition 1. Let Ck(λ1) and Ck(λ2) be the sets of all the connectivity components of node k with λ = λ1 and λ = λ2, respectively. If λ1 < λ2, then Ck(λ2) ⊆ Ck(λ1).
Intuitively, if two nodes are connected (either directly or indirectly) at one level of sparseness, they will be connected at all lower levels of sparseness. This monotone property can be used to identify how strongly connected each node k is to its connectivity components [43].
3.3 Simultaneous Estimation of Multiple Graphs
In some applications, we need to estimate multiple related precision matrices. A motivating example is the modeling of brain networks for Alzheimer's disease using neuroimaging data such as PET, in which, we want to estimate graphical models for three groups: normal controls (NC), patients of mild cognitive impairment (MCI), and Alzheimer's patients (AD). These graphs are expected to share some common connections, but they are not identical. Furthermore, the graphs are expected to evolve over time, in the order of disease severity from NC to MCI to AD. Estimating the graphical models separately fails to exploit the common structures among them. It is thus beneficial to jointly estimate the three graphs, especially when the number of subjects in each group is small. There is some recent work on the estimation of multiple precision matrices. Guo et al. [36] proposed to jointly estimate multiple graphical models using a hierarchical penalty. The time-varying graphical models were studied by Zhu et al. [122], and Kolar et al. [53; 54]. Danaher et al. [22] estimated multiple precision matrices simultaneously using a pairwise fused penalty and grouping penalty.
Assume we are given K data sets, , k = 1, · · · , K with K ≥ 2, where nk is the number of samples of the ith dataset, and p is the number of features. The p features are common for all K data sets, and all samples are independent. Furthermore, the samples within each data set X(k) are identically distributed with a p-variate Gaussian distribution with zero mean and covariance matrix Σ(k). We assume that there are many conditionally independent pairs of features, i.e., the precision matrix Θ(k) = (Σ(k))–1 is sparse. Denote the sample covariance matrix for each data set X(k) as S(k) and Θ = {Θ(1), . . . , Θ(k)}. We can learn multiple precision matrices together by solving the following optimization problem [22; 109]:
| (18) |
where ,
and λ1 and λ2 are nonnegative regularization parameters. The regularization leads to a sparse solution, and the fused penalty encourages Θ(k) to be similar to its neighbors. The optimization in (18) is computationally expensive to solve. Danaher et al. [22] developed a screening rule for the two graph case to speed up the computation. The screening rule was recently extended to the more general case with more than two graphs in [109]. Specifically, Yang et al. [109] considered the problem of estimating multiple graphical models by maximizing a penalized log likelihood with and fused regularization as in [22]. The regularization yields a sparse solution, and the fused regularization encourages adjacent graphs to be similar. The block-wise coordinate descent method was employed to solve the fused multiple graphical Lasso (FMGL), where each step was solved by the accelerated gradient method [83]. In addition, a screening rule was developed which enabled the efficient estimation of multiple large precision matrices. Specifically, a set of necessary conditions were derived for the solution of FMGL to be block diagonal. These conditions were shown to be sufficient when K ≤ 3. Yang et al. also performed exten sive simulation studies; results indicate that these conditions are likely sufficient for any K > 3 as well.
4. PARALLEL MAGNETIC RESONANCE IMAGING
Magnetic resonance imaging (MRI) [39; 105] is a medical imaging technique used in radiology to visualize internal structures of the body in detail. As a non-invasive imaging technique, MRI makes use of the property of nuclear magnetic resonance to image nuclei of atoms inside the body. MRI has been applied to image the brain, muscles, the heart, cancers, etc.
4.1 Undersampled k-space
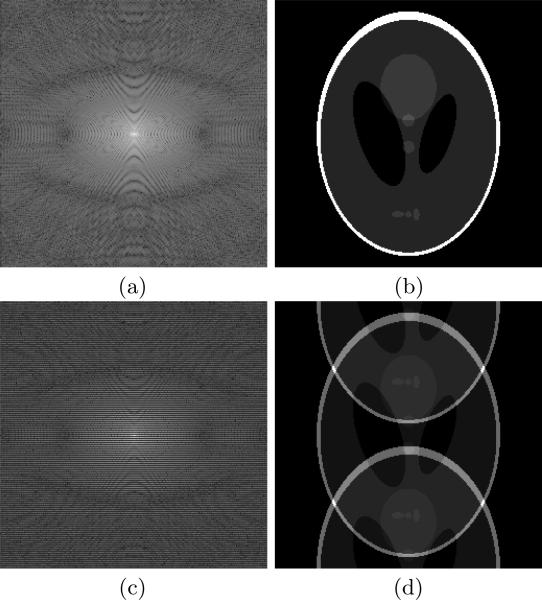
The acquired raw data by an MR scanner are the Fourier coefficients, or the so-called k-space data (see Figure 4 (a) for illustration). The k-space data are typically acquired by a series of phase encodings (each phase encoding covers a given amount of k-space data that are related to the trajectory, e.g., Cartesian sampling, radial sampling). For example, with Cartesian sampling, we need 256 frequency encodings to cover the full k-space of one 256 × 256 image. The time between the repetitions of the sequence is called the repetition time (TR) and it measures the time for acquiring one phase encoding. If TR=50 ms, it takes about 12.8 seconds to acquire the full k-space data of one 256 × 256 image with the Cartesian trajectory. With the same TR, it takes about 15.4 minutes to acquire the full k-space of a 256 × 256 × 72 volume. With higher spatial resolution, the time for acquiring the full k-space can be even longer. In addition, in dynamic cine imaging, we are interested in the study of the motion of the object (heart, blood, etc) over time. This leads to an increased number of phase encodings and increased acquisition time, and one usually has to compromise between spatial resolution and temporal resolution. To save the acquisition time, one has to undersample the k-space, i.e., reducing the number of acquired phase encodings. For example, if the k-space data are acquired every other line, as shown in Figure 4 (c), half of the acquisition time can be saved. The relationship between the acquired k-space data and the image to be reconstructed can be written as
| (19) |
where Fu is a given undersampled Fourier transform operator, f denotes the MR image, y is the acquired k-space data, and n depicts the noise introduced in the acquisition. Unlike the full k-space scenario, one cannot directly apply the inverse Fourier transform to the undersampled data acquired in Figure 4 (c), since otherwise an aliased image shown in Figure 4 (d) will be obtained.
Figure 4.
Illustration of MR image and the k-space data: (a) the full k-space data (displayed in logarithmic scale), (b) the image obtained by applying inverse Fourier transform to (a), (c) the undersampled k-space (displayed in logarithmic scale), and (d) the image obtained by applying inverse Fourier transform to (c).
4.2 Parallel MR Imaging
Parallel imaging [34; 47; 88; 95] has been proven effective for reducing the acquisition time. It exploits the difference in sensitivities between individual coil elements in a receive array to reduce the number of gradient encodings required for imaging. Figure 5 illustrates parallel imaging with 8 coils. Specifically, the first two rows show the coil images seen by the individual coil/channel, and the last two rows show the coil profiles of these 8 coils. It can be observed that the 8 coils have different sensitivities. Parallel imaging tries to reconstruct the target image with the undersampled k-space data.
Figure 5.
Illustration of the coil images and the coil sensitivity profiles (coil images of 8 channels are shown in the first two rows, and the corresponding coil profiles are shown in the last two rows).
Based on how the coil sensitivities are used, parallel imaging can be roughly divided into the following two main categories: 1) the approaches that implicitly make use of the coil sensitivities, represented by GRAPPA [34], and 2) the approaches that explicitly make use of the coil sensitivities, represented by SENSE [88]. In the GRAPPA type approaches, one usually estimates the missing phase encoding lines with the kernels that are estimated by implicitly using the coil sensitivities. In the SENSE type approach, one models the relationship between the target image and the acquired k-space data as:
| (20) |
where yi is the acquired undersampled k-space data by the i-th coil, and Si is the coil sensitivity maps (see the last two rows of Figure 5). The relationships between GRAPPA and SENSE have been studied in the literature [8; 35; 47], and several recent work [57; 58; 72; 73] have shown that GRAPPA and SENSE can be combined to give improved reconstruction performance.
4.3 Coil Profile Estimation
The most common way to determine the sensitivity maps is to obtain low-resolution pre-scans. However, when the object is not static, the sensitivity functions are different between pre-scan and under-sampled scans, and this could lead to reconstruction errors. To compensate for this, joint estimation approaches [103; 113] have been proposed. However, these approaches usually have high computation cost and are restricted to the SENSE type reconstruction.
The eigen-vector approach proposed in [72] is a very promising approach for sensitivity maps estimation. It tried to build a connection between GRAPPA and SENSE-type approaches, by showing that the Coil Profile used in SENSE can be computed with the GRAPPA-type calibration. Such idea was also used in [57; 58]. It was shown in [72] that the coil sensitivities can be computed as the eigen-vector of a given matrix in the image space corresponding to eigenvalues “1”s.
4.4 Sampling Pattern and Fourier Transform
Cartesian sampling is the most natural scheme which under-samples the k-space by skipping some lines. In cardiac MR imaging, TSENSE [37; 48] is a well-known approach that is based on time interleaving of k-space lines in sequential images, and there are studies that makes use of variable density to optimize the sampling scheme, e.g., [12]. The Fourier transform associated with the Cartesian sampling can be efficiently computed.
Spiral and projection (radial) are the most widely used non-Cartesian sampling patterns, among many others. It was observed in several works (e.g., [40]) that the radial sampling exhibits advantages over Cartesian Sampling. The Fourier transform in the non-Cartesian case is much more challenging than the Cartesian one, and gridding is usually employed for performing Non-Uniform FFT [33].
4.5 Incorporating Prior Knowledge and Optimization
To recover f from (19), it is important to note that our target f has certain structures, with which we can better reconstruct f from the undersampled data y. This is where sparse learning can play a role. Typically, we are interested in computing f by solving the following problem
| (21) |
where loss(y, Fuf) depicts the data fidelity, and ϕ(f) incorporates our prior knowledge about the image to be reconstructed.
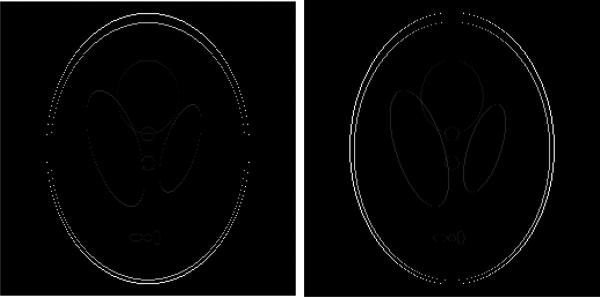
For the data fidelity term, a commonly used one is the squared distance between the acquired data and the prediction: . Recent studies have that the usage of self-consistency [57; 58; 73] can benefit reconstruction. For ϕ(f), one needs to take advantage of the structure in the target image f. Figure 6 shows the gradient of the phantom, and it is easy to observe that such gradient is sparse. Candès et al. [14] proposed to set ϕ(f) = ∥f∥TV, showed the effectiveness of the sparsity promoting penalty in the scenario of single coil, and proved the exact recovery under the so-called Robust Uncertainty Principles (RIP). Later on, compressed sensing was used widely in the reconstruction of MR images, e.g., [1; 57; 61; 71; 112]. When applying sparse learning to parallel MR imaging, one key task is to develop a suitable ϕ(·) that adapts the structure of the image(s) to be reconstructed. Group sparsity [117] has been used for accelerating dynamic MRI [104], and total variation and wavelet transformation have also been used for parallel MR imaging [14; 66; 67; 90; 103; 112]. An important and hot research topic is to develop better sparsity promoting penalties that adapt to the images to be reconstructed.
Figure 6.
Illustration of the gradient of the phantom (shown in Figure 4) along the vertical direction (left) and horizonal direction (right), respectively.
The efficient optimization of problem (21) is crucial for parallel imaging. Several popular approaches include conjugate gradient [40], Newton-type methods [103], Nesterov-type approaches [81; 82; 66; 49], and the alternating direction method of multipliers [1; 10; 31; 112].
5. CONCLUSIONS
In this paper, we review sparse methods for biomedical data in three specific applications. Sparse methods have also been applied to many other applications, e.g., incomplete multi-source data fusion [114] and biological image annotation and retrieval [115]. As with many other data mining and machine learning techniques, the selection of the appropriate sparse method and proper tuning of the associated parameters are critical for finding meaningful and useful results. To this end, one needs to understand the data in a domain specific context and understand the strengths and weaknesses of various sparse methods.
Most existing work on sparse learning focus on prediction, parameter estimation, and variable selection. Very few work address the problem of assigning statistical significance or confidence [11; 118]. However, such significance or confidence measures are crucial in biomedical applications where interpretation of parameters and variables is very important [11]. Most sparse methods in the literature are based on a convex regularizer. Sparse methods based on a non-convex regularizer have recently been proposed and efficient methods based on the difference of convex functions (DC) have been developed [92; 119]. However, their theoretical properties have not been well understood yet, although some recent work demonstrate the advantage of non-convex methods over their convex counterparts [92; 108; 119]. Finally, missing data is ubiquitous in biomedical applications. One important issue that has not been well addressed is how to adapt sparse methods to deal with missing data [70; 96].
ACKNOWLEDGEMENTS
This work was supported in part by NSF (IIS-0953662, MCB-1026710, CCF-1025177) and NIH (R01LM010730).
Footnotes
Contributor Information
Jieping Ye, Arizona State University Tempe, AZ 85287 jieping.ye@asu.edu.
Jun Liu, Siemens Corporate Research Princeton, NJ 08540 jun-liu@siemens.com.
REFERENCES
- 1.Afonso M, Bioucas-Dias J, Figueiredo M. An augmented lagrangian approach to the constrained optimization formulation of imaging inverse problems. IEEE Transactions on Image Processing. 2011;20:681–695. doi: 10.1109/TIP.2010.2076294. [DOI] [PubMed] [Google Scholar]
- 2.Argyriou A, Evgeniou T, Pontil M. Convex multi-task feature learning. Machine Learning. 2008;73(3):243–272. [Google Scholar]
- 3.Bach FR. Consistency of the group lasso and multiple kernel learning. Journal of Machine Learning Research. 2008;9:1179–1225. [Google Scholar]
- 4.Bajwa W, Haupt J, Sayeed A, Nowak R. Compressive wireless sensing. International Conference on Information Processing in Sensor Networks. 2006 [Google Scholar]
- 5.Banerjee O, El Ghaoui L, d'Aspremont A. Model selection through sparse maximum likelihood estimation for multivariate gaussian or binary data. The Journal of Machine Learning Research. 2008;9:485–516. [Google Scholar]
- 6.Baraniuk R. Compressive sensing. IEEE Signal Processing Magazine. 2007;24(4):118–121. [Google Scholar]
- 7.Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Computation. 2003;15:1373–1396. [Google Scholar]
- 8.Blaimer M, Breuer F, Muller M, Heidemann R, Griswold MA, Jakob PM. SMASH, SENSE, PILS, GRAPPA. Top Magn Reson Imaging. 2004;15:223–236. doi: 10.1097/01.rmr.0000136558.09801.dd. [DOI] [PubMed] [Google Scholar]
- 9.Bondell H, Reich B. Simultaneous regression shrinkage, variable selection, and supervised clustering of predictors with oscar. Biometrics. 2008;64(1):115–123. doi: 10.1111/j.1541-0420.2007.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Foundations and Trends in Machine Learning. 2011;3:1–122. [Google Scholar]
- 11.Bühlmann P. Statistical significance in high-dimensional linear models. Arxiv preprint arXiv:1202.1377v1. 2012 [Google Scholar]
- 12.Busse R, Wang K, Holmes J, Brittain J, Korosec F. Optimization of variable-density cartesian sampling for time-resolved imaging. International Society for Magnetic Resonance in Medicine. 2009 [Google Scholar]
- 13.Candès E, Romberg. J. Quantitative robust uncertainty principles and optimally sparse decompositions. Foundations of Computational Mathematics. 2006;6(2):227–254. [Google Scholar]
- 14.Candès E, Romberg J, Tao T. Robust uncertainty principles: Exact signal reconstruction from highly incomplete frequency information. IEEE Transactions on Information Theory. 2006;52(2):489–509. [Google Scholar]
- 15.Candès E, Tao T. Near optimal signal recovery from random projections: Universal encoding strategies? IEEE Transactions on Information Theory. 2006;52(12):5406–5425. [Google Scholar]
- 16.Candès E, Wakin M. An introduction to compressive sampling. IEEE Signal Processing Magazine. 2008;25(2):21–30. [Google Scholar]
- 17.Carroll S, Grenier J, Weatherbee S. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2nd edition. Blackwell Pub; Malden, MA: 2005. [Google Scholar]
- 18.Chu W, Ghahramani Z, Falciani F, Wild D. Biomarker discovery in microarray gene expression data with gaussian processes. Bioinformatics. 2005;21(16):3385–3393. doi: 10.1093/bioinformatics/bti526. [DOI] [PubMed] [Google Scholar]
- 19.Chuang H, Lee E, Liu Y, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular systems biology. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung F. Spectral Graph Theory. American Mathematical Society. 1997 [Google Scholar]
- 21.Cox D. Regression models and life-tables. Journal of the Royal Statistical Society. Series B (Methodological) 1972;34(2):187–220. [Google Scholar]
- 22.Danaher P, Wang P, Daniela D. The joint graphical lasso for inverse covariance estimation across multiple classes. Arxiv preprint arXiv:1111.0324. :2011. doi: 10.1111/rssb.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.d'Aspremont A, El Ghaoui L, Jordan M, Lanckriet GRG. A direct formulation for sparse PCA using semidefinite programming. NIPS. 2005 [Google Scholar]
- 24.Donoho D. High-dimensional data analysis: The curses and blessings of dimensionality. 2000 [Google Scholar]
- 25.Donoho D. Compressed sensing. IEEE Transactions on Information Theory. 2006;52:1289–1306. [Google Scholar]
- 26.Duarte M, Davenport M, Wakin M, Baraniuk R. Sparse signal detection from incoherent projections. ICASSP. 2006 [Google Scholar]
- 27.Fan J, Lv J. A selective overview of variable selection in high dimensional feature space. Statistica Sinica. 2010;20:101–148. [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman J, Hastie T, Höfling H, Tibshirani R. Pathwise coordinate optimization. Annals of Applied Statistics. 2007;1(2):302–332. [Google Scholar]
- 29.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman J, Hastie T, Tibshirani R. A note on the group lasso and a sparse group lasso. Technical report. Department of Statistics; Stanford University: 2010. [Google Scholar]
- 31.Goldstein T, Osher S. The split bregman method for l1-regularized problems. SIAM Journal on Imaging Sciences. 2009;2:323–343. [Google Scholar]
- 32.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 33.Greengard L, Lee J. Accelerating the nonuniform fast fourier transform. SIAM Review. 2004;46:443–454. [Google Scholar]
- 34.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, K. B., Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magnetic Resonance in Medicine. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 35.Griswold MA, Kannengiesser S, Heidemann RM, Wang J, Jakob PM. Field-of-view limitations in parallel imaging. Magnetic Resonance in Medicine. 2004;52:1118–1126. doi: 10.1002/mrm.20249. [DOI] [PubMed] [Google Scholar]
- 36.Guo J, Levina E, Michailidis G, Zhu J. Joint estimation of multiple graphical models. Biometrika. 2011;98(1):1–15. doi: 10.1093/biomet/asq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman MA, Kellman P, Dick AJ, Lederman RJ, McVeigh ER. Real-time accelerated interactive MRI with adaptive TSENSE and UNFOLD. Magnetic Resonance in Medicine. 2003;50:315–321. doi: 10.1002/mrm.10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46(1-3):389–422. [Google Scholar]
- 39.Haacke EM, Brown RW, Thompson MR, Venkatesan R, editors. Magnetic Resonance Imaging: Physical Principles and Sequence Design. Wiley-Liss; 1999. [Google Scholar]
- 40.Hansen MS, Baltes C, Tsao J, Kozerke S, Pruessmann KP, Eggers H. k-t BLAST reconstruction from non-cartesian k-t space sampling. Magnetic Resonance in Medicine. 2006;55:85–91. doi: 10.1002/mrm.20734. [DOI] [PubMed] [Google Scholar]
- 41.Hromádka T, DeWeese M, Zador A. Sparse representation of sounds in the unanesthetized auditory cortex. PLoS Biol. 2008;6(1):e16. doi: 10.1371/journal.pbio.0060016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Zhang T, Metaxas D. Learning with structured sparsity. Journal of Machine Learning Research. 2011;12:3371–3412. [Google Scholar]
- 43.Huang S, Li J, Sun L, Liu J, Wu T, Chen K, Fleisher A, Reiman E, Ye J. Learning brain connectivity of alzheimer's disease from neuroimaging data. NIPS, pages. 2009:808–816. [Google Scholar]
- 44.Jacob L, Obozinski G, Vert J. Group lasso with overlap and graph lasso. ICML. 2009 [Google Scholar]
- 45.Jenatton R, Audibert J-Y, Bach F. Structured variable selection with sparsity-inducing norms. Journal of Machine Learning Research. 2011;12:2777–2824. [Google Scholar]
- 46.Jenatton R, Mairal J, Obozinski G, Bach F. Proximal methods for sparse hierarchical dictionary learning. ICML. 2010 [Google Scholar]
- 47.Kellman P. Parallel imaging: the basics. ISMRM Educational Course: MR Physics for Physicists. 2004 [Google Scholar]
- 48.Kellman P, Epstein FH, McVeigh ER. Adaptive sensitivity encoding incorporating temporal filtering (tsense). Magnetic Resonance in Medicine. 2001;45:846–852. doi: 10.1002/mrm.1113. [DOI] [PubMed] [Google Scholar]
- 49.Khare K, Hardy CJ, King KF, Turski PA, Marinelli L. Accelerated MR imaging using compressive sensing with no free parameters. Magnetic Resonance in Medicine. 2012 doi: 10.1002/mrm.24143. [DOI] [PubMed] [Google Scholar]
- 50.Kim S, Xing E. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS genetics. 2009;5(8):e1000587. doi: 10.1371/journal.pgen.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S, Xing EP. Tree-guided group lasso for multi-task regression with structured sparsity. ICML. 2010 [Google Scholar]
- 52.Koh K, Kim S, Boyd S. An interior-point method for large-scale l1-regularized logistic regression. Journal of Machine Learning Research. 2007;8:1519–1555. [Google Scholar]
- 53.Kolar M, Song L, Ahmed A, Xing E. Estimating time-varying networks. The Annals of Applied Statistics. 2010;4(1):94–123. [Google Scholar]
- 54.Kolar M, Xing E. On time varying undirected graphs. AISTAT. 2011 [Google Scholar]
- 55.Kowalski M. Sparse regression using mixed norms. Applied and Computational Harmonic Analysis. 2009;27(3):303–324. [Google Scholar]
- 56.Kumar S, Jayaraman K, Panchanathan S, Gurunathan R, Marti-Subirana A, Newfeld S. BEST: A novel computational approach for comparing gene expression patterns from early stages of Drosophila melanogaster development. Genetics. 2002;162(4):2037–2047. doi: 10.1093/genetics/162.4.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai P, Lustig M, B. A. C., V. S. S., B. P. J., A. M. Efficient L1SPIRiT reconstruction (ESPIRiT) for highly accelerated 3d volumetric MRI with parallel imaging and compressed sensing. ISMRM. 2010 [Google Scholar]
- 58.Lai P, Lustig M, V. S. S., B. A. C. ESPIRiT (efficient eigenvector-based l1spirit) for compressed sensing parallel imaging - theoretical interpretation and improved robustness for overlapped FOV prescription. ISMRM. 2011 [Google Scholar]
- 59.Larkman DJ, Nunes RG. Parallel magnetic resonance imaging. Physics in Medicine and Biology. 2007;52:R15–55. doi: 10.1088/0031-9155/52/7/R01. [DOI] [PubMed] [Google Scholar]
- 60.Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24(9):1175–1182. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- 61.Liang D, Liu B, Wang J, Ying L. Accelerating SENSE using compressed sensing. Magnetic Resonance in Medicine. 2009;62:1574–1584. doi: 10.1002/mrm.22161. [DOI] [PubMed] [Google Scholar]
- 62.Liu H, Palatucci M, Zhang J. Blockwise coordinate descent procedures for the multi-task lasso, with applications to neural semantic basis discovery. ICML. 2009 [Google Scholar]
- 63.Liu H, Roeder K, Wasserman L. Stability approach to regularization selection (StARS) for high dimensional graphical models. NIPS. 2011 [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Ji S, Ye J. Multi-task feature learning via efficient ℓ2, 1-norm minimization. UAI. 2009 [Google Scholar]
- 65.Liu J, Ji S, Ye J. SLEP: Sparse Learning with Efficient Projections. Arizona State University. 2009 [Google Scholar]
- 66.Liu J, Rapin J, Chang T, Lefebvre A, Zenge M, Mueller E, Nadar MS. Dynamic cardiac MRI reconstruction with weighted redundant haar wavelets. ISMRM. 2012 [Google Scholar]
- 67.Liu J, Rapin J, Chang T, Schmitt P, Bi X, Lefebvre A, Zenge M, Mueller E, Nadar MS. Regularized reconstruction using redundant haar wavelets: A means to achieve high under-sampling factors in non-contrast-enhanced 4D MRA. ISMRM. 2012 [Google Scholar]
- 68.Liu J, Ye J. Efficient ℓ1 / ℓq norm regularization. Arxiv preprint arXiv:1009.4766v1. 2010 [Google Scholar]
- 69.Liu J, Ye J. Moreau-Yosida regularization for grouped tree structure learning. NIPS. 2010 [Google Scholar]
- 70.Loh P, Wainwright M. High-dimension regression with noisy and missing data: Provable guarantees with non-convexity. NIPS. 2011 [Google Scholar]
- 71.Lustig M, Donoho DL, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 72.Lustig M, Lai P, Murphy M, Vasanawala S, Elad M, Zhang J, Pauly J. An eigen-vector approach to autocalibrating parallel MRI, where SENSE meets GRAPPA. ISMRM. 2011 doi: 10.1002/mrm.24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magnetic Resonance in Medicine. 2010;64:457–471. doi: 10.1002/mrm.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marton M, et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nature Medicine. 1998;4(11):1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 75.Mazumder R, Hastie T. Exact covariance thresholding into connected components for large-scale graphical lasso. Arxiv preprint arXiv:1108.3829. :2011. [PMC free article] [PubMed] [Google Scholar]
- 76.Mazumder R, Hastie T. The graphical lasso: New insights and alternatives. Arxiv preprint arXiv:1111.5479. :2011. doi: 10.1214/12-EJS740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Megason S, Fraser S. Imaging in systems biology. Cell. 2007;130(5):784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 78.Meier L, Geer S, Bühlmann P. The group lasso for logistic regression. Journal of the Royal Statistical Society: Series B. 2008;70:53–71. [Google Scholar]
- 79.Meinshausen N, Bühlmann P. Stability selection. Journal of the Royal Statistical Society: Series B. 2010;72:417–473. [Google Scholar]
- 80.Negahban S, Wainwright M. Joint support recovery under high-dimensional scaling: Benefits and perils of ℓ1, ∞-regularization. NIPS. 2008:1161–1168. [Google Scholar]
- 81.Nemirovski A. Efficient methods in convex programming. Lecture Notes; 1994. [Google Scholar]
- 82.Nesterov Y. Introductory Lectures on Convex Optimization: A Basic Course. Kluwer Academic Publishers; 2004. [Google Scholar]
- 83.Nesterov Y. Gradient methods for minimizing composite objective function. CORE; 2007. [Google Scholar]
- 84.Ng A. Feature selection, ℓ1 vs. ℓ2 regularization, and rotational invariance. ICML. 2004 [Google Scholar]
- 85.Obozinski G, Taskar B, Jordan MI. Joint covariate selection for grouped classification. Technical report. Statistics Department; UC Berkeley: 2007. [Google Scholar]
- 86.Peng H. Bioimage informatics: a new area of engineering biology. Bioinformatics. 2008;24(17):1827–1836. doi: 10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng J, Zhu J, B. A., Han W, Noh D-Y, Pol-lack JR, Wang P. Regularized multivariate regression for identifying master predictors with application to integrative genomics study of breast cancer. Annals of Applied Statistics. 2010;4(1):53–77. doi: 10.1214/09-AOAS271SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pruessmann K, Weiger M, Scheidegger M, Boesiger P. SENSE: sensitivity endcoding for fast MRI. Magnetic Resonance in Medicine. 1999;42:952–962. [PubMed] [Google Scholar]
- 89.Quattoni A, Carreras X, Collins M, Darrell T. An efficient projection for ℓ1, ∞ regularization. ICML. 2009 [Google Scholar]
- 90.Ramani S, Fessler JA. Parallel MR image reconstruction using augmented lagrangian methods. IEEE Transactions on Medical Imaging. 2011;30:694–706. doi: 10.1109/TMI.2010.2093536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ryali S, Supekar K, Abrams D, Menon V. Sparse logistic regression for whole-brain classification of fMRI data. Neuroimage. 2010;51(2):752–764. doi: 10.1016/j.neuroimage.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen X, Pan W, Zhu Y. Likelihood-based selection and sharp parameter estimation. Journal of American Statistical Association. 2012;107:223–232. doi: 10.1080/01621459.2011.645783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi J, Yin W, Osher S, Sajda P. A fast algorithm for large scale ℓ1-regularized logistic regression. Technical report, CAAM TR08-07. 2008.
- 94.Shi W, Lee K, Wahba G. Detecting disease-causing genes by lasso-patternsearch algorithm. BMC Proceedings. 2007;1(Suppl 1):S60. doi: 10.1186/1753-6561-1-s1-s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sodickson D, Manning W. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magnetic Resonance in Medicine. 1997;38:591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- 96.Stäadler N, Bühlmann P. Missing values: sparse inverse covariance estimation and an extension to sparse regression. Statistics and Computing. 2012;22:219–235. [Google Scholar]
- 97.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. 1996;58(1):267–288. [Google Scholar]
- 99.Tibshirani R, Saunders M, Rosset S, Zhu J, Knight K. Sparsity and smoothness via the fused lasso. Journal Of The Royal Statistical Society Series B. 2005;67(1):91–108. [Google Scholar]
- 100.Tibshirani R, Wang P. Spatial smoothing and hot spot detection for cgh data using the fused lasso. Biostatistics. 2008;9(1):18–29. doi: 10.1093/biostatistics/kxm013. [DOI] [PubMed] [Google Scholar]
- 101.Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biology. 2002;3(12):1–14. doi: 10.1186/gb-2002-3-12-research0088. research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tropp A, Gilbert A, Strauss M. Algorithms for simultaneous sparse approximation: part I: Greedy pursuit. Signal Processing. 2006;86(3):572–588. [Google Scholar]
- 103.Uecker M, Hohage T, Block KT, Frahm J. Image reconstruction by regularized nonlinear inversion-joint estimation of coil sensitivities and image content. Magnetic Resonance in Medicine. 2008;60:674–682. doi: 10.1002/mrm.21691. [DOI] [PubMed] [Google Scholar]
- 104.Usman M, Prieto C, Schaeffter T, Batchelor PG. k-t group sparse: A method for accelerating dynamic MRI. Magnetic Resonance in Medicine. 2011;66:1163–1176. doi: 10.1002/mrm.22883. [DOI] [PubMed] [Google Scholar]
- 105.Vlaardingerbroek MT, Boer JA, editors. Magnetic Resonance Imaging. Spinger; 2004. [DOI] [PubMed] [Google Scholar]
- 106.Witten D, Friedman J, Simon N. New insights and faster computations for the graphical lasso. Journal of Computational and Graphical Statistics. 2011;20(4):892–900. [Google Scholar]
- 107.Wu T, Chen Y, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25(6):714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiang S, Shen X, Ye J. Efficient sparse group feature selection via nonconvex optimization. Arxiv preprint arXiv:1205.5075. 2012 [Google Scholar]
- 109.Yang S, Pan Z, Shen X, Wonka P, Ye J. Fused multiple graphical lasso. Technical Report. Arizona State University; 2012. [Google Scholar]
- 110.Yang S, Yuan L, Lai Y-C, Shen X, Wonka P, Ye J. Feature grouping and selection over an undirected graph. KDD. 2012 doi: 10.1145/2339530.2339675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ye J, Farnum M, Yang E, Verbeeck R, Lobanov V, Raghavan N, Novak G, DiBernardo A, Narayan V. Sparse learning and stability selection for predicting MCI to AD conversion using baseline ADNI data. BMC Neurology. 2012;12:46. doi: 10.1186/1471-2377-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ye X, Chen Y, Huang F. Computational acceleration for MR image reconstruction in partially parallel imaging. IEEE Transactions on Medical Imaging. 2011;30:1055–1063. doi: 10.1109/TMI.2010.2073717. [DOI] [PubMed] [Google Scholar]
- 113.Ying L, Sheng J. Joint image reconstruction and sensitivity estimation in sense (JSENSE). Magnetic Resonance in Medicine. 2007;57:1196–1202. doi: 10.1002/mrm.21245. [DOI] [PubMed] [Google Scholar]
- 114.Yuan L, Wang Y, Thompson P, Narayand V, Ye J. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. NeuroImage. 2012;61(3):622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yuan L, Woodard A, Ji S, Jiang Y, Zhou Z-H, Kumar S, Ye J. Learning sparse representations for fruit-fly gene expression pattern image annotation and retrieval. BMC Bioinformatics. 2012;13:107. doi: 10.1186/1471-2105-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuan M, Joseph VR, Zou H. Structured variable selection and estimation. Annals of Applied Statistics. 2009;3:1738–1757. [Google Scholar]
- 117.Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal Of The Royal Statistical Society Series B. 2006;68(1):49–67. [Google Scholar]
- 118.Zhang C-H, Zhang S. Confidence intervals for low-dimensional parameters with high-dimensional data. Arxiv preprint arXiv:1110.2563v1. 2011 [Google Scholar]
- 119.Zhang T. Analysis of multi-stage convex relaxation for sparse regularization. Journal of Machine Learning Research. 2010;11:1081–1107. [Google Scholar]
- 120.Zhao P, Rocha G, Yu B. The composite absolute penalties family for grouped and hierarchical variable selection. Annals of Statistics. 2009;37(6A):3468–3497. [Google Scholar]
- 121.Zhong L, Kwok J. Efficient sparse modeling with automatic feature grouping. ICML. 2011 doi: 10.1109/TNNLS.2012.2200262. [DOI] [PubMed] [Google Scholar]
- 122.Zhou S, Lafferty J, Wasserman L. Time varying undirected graphs. COLT. 2008 [Google Scholar]
- 123.Zhu J, Hastie T. Classification of gene microar-rays by penalized logistic regression. Biostatistics. 2004;5(3):427–443. doi: 10.1093/biostatistics/5.3.427. [DOI] [PubMed] [Google Scholar]
- 124.Zhu J, Rosset S, Hastie T, Tibshirani R. 1-norm support vector machines. Neural Information Processing Systems. 2003:49–56. [Google Scholar]
- 125.Zhu Y, Shen X, Pan W. Simultaneous grouping pursuit and feature selection in regression over an undirected graph. Preprint. 2012 doi: 10.1080/01621459.2013.770704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of The Royal Statistical Society Series B. 2005;67(2):301–320. [Google Scholar]
- 127.Zou H, Hastie T, Tibshirani R. Sparse principle component analysis. Journal of Computational and Graphical Statistics. 2006;15(2):262–286. [Google Scholar]