Abstract
Metabolomics technologies enable the examination and identification of endogenous biochemical reaction products, revealing information on the precise metabolic pathways and processes within a living cell. Metabolism is either directly or indirectly involved with every aspect of cell function, and metabolomics is thus believed to be a reflection of the phenotype of any cell. Metabolomics analysis of cells has many potential applications and advantages compared to currently used methods in the postgenomics era. Cell metabolomics is an emerging field that addresses fundamental biological questions and allows one to observe metabolic phenomena in cells. Cell metabolomics consists of four sequential steps: (a) sample preparation and extraction, (b) metabolic profiles of low-weight metabolites based on MS or NMR spectroscopy techniques, (c) pattern recognition approaches and bioinformatics data analysis, (d) metabolites identification resulting in putative biomarkers and molecular targets. The biomarkers are eventually placed in metabolic networks to provide insight on the cellular biochemical phenomena. This article analyzes the recent developments in use of metabolomics to characterize and interpret the cellular metabolome in a wide range of pathophysiological and clinical contexts, and the putative roles of the endogenous small molecule metabolites in this new frontier of postgenomics biology and systems medicine.
Introduction
Apromising approach to explore the cell phenotype is metabolomics, a rapidly growing field of postgenomics biochemistry, defined as the metabolic complement of functional genomics, and is especially useful in medical and life sciences research (Kinross et al., 2011; Zhang et al., 2012a). Metabolomics enables the characterization of endogenous small molecules (referred to as metabolites) that are the products of biochemical reactions, revealing connections among different pathways that operate within a living cell (Wang et al., 2011). More specifically, the ability to uncover and evaluate biochemical differences within healthy and diseased organisms provides information as to the underlying cause(s) of disease, which in turn suggests targets for pharmacological intervention (Wang et al., 2012a,b). Metabolome analysis could describe qualitatively and quantitatively the final products of cellular regulatory pathways and can be seen as the ultimate response of a biologic system to genetic factors and/or environmental changes (Cuperlović-Cul et al., 2010). Cell metabolome can be defined as the set of all the metabolites present in cells and metabolites can be considered the best indicator of an organism's phenotype (Nomura et al., 2011; Tautenhahn et al., 2012).
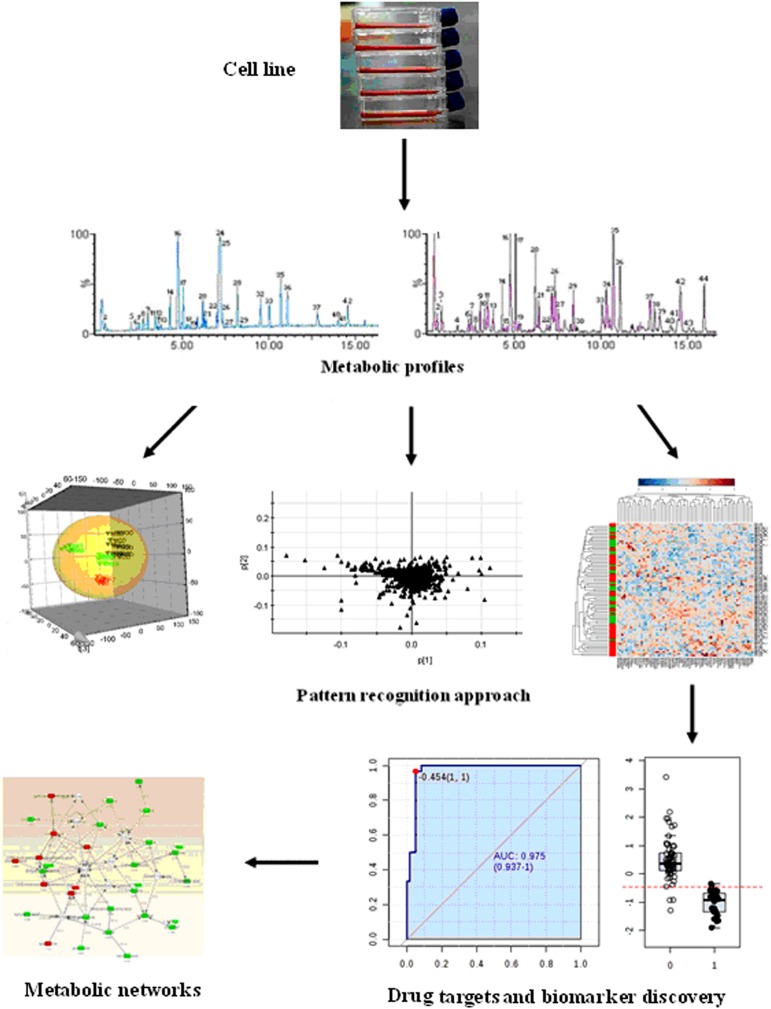
Metabolomics capitalizes on the small molecules in cell to construct a ‘fingerprint’ that can be unique to the individuals. Small-molecule metabolites as primary indicators have an important role in biological systems and represent attractive candidates to understand cell phenotypes (Riedelsheime et al., 2012; Tomita et al., 2012). Cells are used extensively in disease research for understanding the molecular mechanism of disease progression, response, and resistance to therapeutics. Cell applications are easier to control, less expensive and easier to interpret than analysis of both animal models and human subjects. As such, it represents an untapped resource for identification of specific metabolite biomarkers that would help distinguish the normal and abnormal states, as well as response to drugs or stress agents. Metabolomic studies have shown that cellular metabolic networks are robust and the dynamic behavior of biochemical pathways that are governed by a highly interconnected regulatory system (Chen et al., 2012; Finley et al., 2012; Zhang et al., 2011). The general procedures of cell metabolomics are shown in (Figure 1).
FIG. 1.
A scheme of the cell metabolomics workflow. Cell metabolomics consists of four sequential steps: (a) sample preparation and extraction, (b) metabolic profiles of low-weight metabolites based on MS or NMR spectroscopy techniques, (c) pattern recognition approaches and bioinformatic data analysis, (d) metabolites identification resulting in biomarkers and targets. The biomarkers are eventually placed in metabolic networks to provide insight on the biochemical phenomena.
The focus on the metabolic profile of the whole organism provides relevant information about specific cell types under different conditions, which is crucial for a more holistic understanding of cell properties and functions (Cai et al., 2010; Mintz-Oron et al., 2012). By using a combination of feature selection, pattern recognition, and multivariate data analysis approaches, metabolomic profiling aims to provide a comprehensive assessment of the alterations in the metabolite levels in cells (Lange et al., 2011; Stringari et al., 2011; Turnbaugh et al., 2011). Recent technological advancements in nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) have led to wide use of these technologies for precise measurements of metabolites with improved sensitivity, resolution, and mass accuracy (Fischer et al., 2012; Kleiner et al., 2012). At the same time, data provided by metabolic profiling of cells can be complementary to the whole system results. Application of metabolomics in the area of cell is relatively undeveloped and thus the aim of the present review is to provide an insight into the issues pertaining to metabolome analysis, as well as to explore its possible applications in cell culture.
Methodology of Sample Preparation and Extraction
Metabolomic analysis of cells has emerged as an important technique for studying cellular biochemistry. Cell metabolomic experiments can be divided into several general steps: cell culture growth or stimulation, quenching and metabolite extraction, and data processes. With the advent of liquid chromatography-tandem mass spectrometry (LC-MS) and gas chromatography-tandem mass spectrometry (GC-MS) methods, large numbers of metabolites can be quantified to provide detailed insight into the metabolic status of cells (Suhre et al., 2011). The first necessary step, termed ‘quenching,’ is to stop the enzymatic activity or changes in the metabolite levels rapidly. Freezing is generally considered to be the easiest way of stopping enzyme activity, provided that cells are not allowed to partially thaw before extracting metabolites. Apart from the development of sensitive detection systems, a big challenge in cell metabolomics is to cope with the fast changes of intracellular concentrations of metabolites. It imposes stringent requirements on the sample preparation (Blow et al., 2008). The time and method of sampling are important issues to be considered to ensure reproducibility in the bio-samples, especially the biological replicates commonly used. Hence, integrated procedures that allow for sampling, quenching, and extraction to be simplified into a single step have been devised (Kleiner et al., 2012; Sreekumar et al., 2009). Intracellular metabolite concentrations are subsequently determined by subtracting the metabolite content of the cell-free extracellular medium. A simple, fast, and reproducible sample preparation procedure has been developed for relative quantification of metabolites in adherent mammalian cells using the clonal β-cell line INS-1 as a model sample (Lorenz et al., 2011). Maximal recovery can be achieved using a single rapid extraction step. Given the power of the metabolomics technology, the availability of standardized sample preparation methods for cell is critical toward augmenting research in this direction.
Analytical Instrument Platforms
Metabolite concentrations represent sensitive markers of phenotypic changes. Consequently, the development of robust metabolomic platforms will greatly facilitate various applications of cell cultures, and would be invaluable tools for gaining insights into these areas. A variety of analytical platforms has been developed to facilitate these and other types of metabolomics experiments (Locasale et al., 2012). For instance, NMR, GC, and LC have been coupled to MS detection methods in order to perform metabolomic profiling of a variety of biological samples (Qin et al., 2012). MS detector coupled with LC has been effective for characterizing chemical content from cells. There are a number of features of LC that make it a method of choice for such analyses. For example, it is possible to concentrate and separate analytes simultaneously. In addition, because the scaling laws of LC make it amenable to small-volume sampling, it has been used extensively for single cell and even subcellular analyses, and is well suited as a separation method for use in metabolomics experiments (Zhang et al., 2013). In a study by Frezza et al. (2011), LC-MS metabolomics was used to assess the effects of hypoxia on cellular metabolism. It revealed the importance of synchronized and regulated catabolism as a mechanism of adaptation to bioenergetic stress.
An MS and high-throughput analysis will open up the possibility to perform metabolomics on large populations of cells (Cui et al., 2008). In fact, MS has already become the main analytical platform for cell metabolomics. MS has also been used to characterize the contents of individual neurons, with considerable efforts devoted to characterizing their peptide complements. Many of these studies employed direct MS profiling; however, pairing LC to MS often leads to better analyte coverage. Furthermore, electrospray ionization (ESI) coupled with tandem MS/MS allows for the identification and/or characterization of unknown or unexpected compounds, such as metabolites, via their mass-to-charge ratio (m/z) and MS/MS fragmentation pattern, and complements the other detection schemes used for cell measurements. Intracellular metabolites can reflect the physiological state of cells. Hyphenated UPLC-MS is a relatively new technique for the separation of complex samples and shows promise for metabolomics (Bricker et al., 2012; Paglia et al., 2012; Wu et al., 2011). It is a potential technology for the classification of the cells. Cell growth, metabolic activity, and protein productivity measurements, which are currently used to monitor the cellular physiological state, suggested consistency across bioreactors and over the course of the cultivation.
Data Processes
Using pattern recognition approaches such as “unsupervised” principal component analysis (PCA) and “supervised” partial least squares-discriminant analysis (PLS-DA), metabolomics could provide a comprehensive assessment of the metabolite in cells. PCA is a data analysis method that can deal with large volumes of data. Owing to the complexity and volume of the data generated by today's advanced technologies in metabolomics, PCA has become predominant in the medical sciences (Wang et al., 2012c; 2013a). In the PCA scores, each point represents an individual sample. The PCA results are displayed as score plots indicating the scatter of the samples, which indicate similar metabolomic compositions when clustered together and compositionally different metabolomes when dispersed. The purpose of the PLS-DA analysis was to calculate models that differentiate between groups and between model and control subjects. In the PLS-DA modeling, the samples from the different groups were sorted into different classes using score plots, and endogenous metabolites that contribute to the classification were identified in loading plots, which showed the importance of each variable to the classification (Zhang et al., 2012b). Potential markers were extracted from loading-plots constructed following the PLS-DA analysis, and markers were chosen based on their contribution to the variation and correlation within the data set. Because the ultimate goal of biomarker discovery is the translation of those biomarkers to clinical practice, it is clear that the metabolomics needs to generate their receiver operator characteristic (ROC) curves that is generally considered the standard method for performance assessment.
Bringing Metabolomics to the Forefront of Cell Research
It has been noted that the testing of drugs on animals is insufficient in clinical testing, and that the cell cultures may be an alternative for understanding the specific metabolism of drug candidates (Jenkins et al., 2004). Currently, the greatest interest focuses on cell phenotype analysis and molecular classification of disease subtype and drug-testing studies, discovery of markers of sample phenotypes or drug targets (Khoo et al., 2007; Olivier et al., 2012). Metabolic analysis of cell cultures has many potential applications and advantages to currently used methods for cell testing (du Preez et al., 2012). Metabolite concentrations represent sensitive markers of both genomic and phenotypic changes. Metabolomics represents the downstream of systems biology and has drawn significant interest for studying the metabolic networks from cells to organisms (Arakaki et al., 2008). Cell metabolomics is an emerging field that addresses fundamental biological questions and allows observation of metabolic phenomena in cells. Thus, metabolomics has been used for optimizing cell cultures for antibody production (Wu et al., 2012), testing drugs (Sabidó et al., 2012), cell transfections, determinant of apoptosis, and comparing lung cancer cell phenotypes, identifying novel underlying metabolic pathways (Ward et al., 2011).
Current Applications
Cancer cells
There are many possible applications for cell metabolomics in a context of cancer, and some prominent examples are outlined here. Cancer cells have several specific metabolic features, which have been explored for targeted therapies. Agents that promote apoptosis are currently considered as a powerful tool for cancer therapeutics. Metabolomic signatures might be used in the tests of efficacy of agents causing apoptosis in cell culture. HEK and HepG2 cells were searched for metabolic biomarkers of apoptosis differing from that of necrosis (Halama et al., 2011). Several metabolites indicative for apoptotic processes in cell culture including aspartate, glutamate, methionine, alanine, glycine, propionyl carnitine, and malonyl carnitine were observed. These signatures could be obtained in fast high-throughput screening. In a study by Dewar et al. (2010), metabolic differences were to examine between chronic myelogenous leukemic cell, MyL, and MyL-R (Dewar et al., 2010). Specific metabolite identification and quantification were used to examine metabolic differences between the cell types. It demonstrated a clear difference in the metabolite profiles of drug-resistant and sensitive cells, with the biggest difference being an elevation of creatine metabolites in the imatinib-resistant MyL-R cells.
To investigate the mechanisms behind the oncogenic stress response, Bcr-Abl cells were cultivated in presence of imatinib (Dengler et al., 2011). Cell death was preceded by enhanced glycolysis, glutaminolysis, and amino acid metabolism, leading to elevated ATP and protein levels. The enhanced metabolism could be linked to induction of cell death as inhibition of glycolysis or glutaminolysis. It indicated that these metabolic changes are major mediators of oncogenic stress in Bcr-Abl cells. In a report done by Collier et al. (2011), using 832/13 and INS-1E rat insulinoma cells, the apoptosis is unlikely to be the primary pathway underlying β-cell death in response to IL-1β+γ-IFN (Collier et al., 2011). Results demonstrate that pancreatic β-cells undergo apoptosis in response to camptothecin or staurosporine, but not pro-inflammatory cytokines. A study was performed to characterize the global metabolic changes during the process of tumor invasion and metastasis to murine osteosarcoma cell (Hua et al., 2011). Serum metabolic profiling revealed that many key metabolites in glycolysis and tricarboxylic acid (TCA) cycle, as well as most of the amino acids were elevated at rapidly growing stages of tumor, presumably resulting from a high energy demand and turnover of anabolic metabolism during the tumor cell proliferation. Several other excellent examples of the application of quantitative metabolomics in cell culture analysis were presented for the analysis of neuroendocrine cancers and in the analysis of cancer cell metabolic phenotype (Costello et al., 2011). Examples of such analysis include profiling of central metabolism in human cancer cells, identification of metabolic fluxes in hepatic cells (Westra et al., 2011), and several other examples that have been reviewed previously (Prigione et al., 2011).
Cellular response to pathogen infections
Metabolic profiling allowed the differentiation of fatty acid biosynthesis and cholesterol metabolism during viral replication in the A549 and AGS cells (Lin et al., 2010). It revealed the different responses between A549 and AGS to the virus infection. From the pattern recognition results, AGS cell might be more susceptible to influenza A virus. Regarding the fact that AGS is a poorly differentiated gastric adenocarcinoma cell line, whereas A549 is a relatively differentiated lung tumor one, it is speculated that viral replication might be associated with the cell differentiations. A multiplatform analytical approach combining proton nuclear magnetic resonance NMR and MS, together with pattern recognition tools in a metabolomic study was used to investigate the effects of dengue virus infection (Birungi et al., 2010). Distinct effects of infection by each serotype were demonstrated, and these differences were attributed to changes in levels of metabolites, including amino acids, and dicarboxylic acids related to the tricarboxylic acid cycle. These studies demonstrated application of metabolomics to improve understanding of the effect of dengue infection on endothelial cells' metabolome.
Cellular response to toxicity
Toxicity testing is vital to protect human health from exposure to toxic chemicals in the environment. Furthermore, by combining novel cellular models with molecular profiling technologies, metabolomics can add new insights into the molecular basis of toxicity and provide a rich source of biomarkers that are urgently required in a 21st Century approach to toxicology. NMR-based metabolic profiling approach has been used to characterize the metabolome of the RPTEC/TERT1 cells, an immortalized non-tumor human renal epithelial cell line that recapitulates phenotypic characteristics that are absent in other in vitro renal cell models (Ellis et al., 2011). Metabolic profiling of RPTEC/TERT1 cells can report on the effect of chemical exposure on multiple cellular pathways, producing different response profiles for the different compounds tested with a greater number of major metabolic effects observed in the toxin-treated cells. Importantly, compounds with established links to chronic renal toxicity produced more diverse and severe perturbations to the cellular metabolome than nontoxic compounds. As these changes can be rationalized with the different pharmacological and toxicity profiles of the chemicals, it is suggested that metabolic profiling in the RPTEC/TERT1 model would be useful in investigating the mechanism of action of toxins at a low dose.
Cellular response to environmental factors
Hypoxia is one of the features of poorly vascularized areas of solid tumors, but cancer cells can survive in these areas despite the low oxygen tension. The adaptation to hypoxia requires biochemical responses that culminate in a metabolic rearrangement to counter-balance the decrease in energy supply from mitochondrial respiration. The understanding of metabolic adaptations under hypoxia could reveal novel pathways that, if targeted, would lead to specific death of hypoxic regions. In a study done by Frezza et al. (2011), metabolomics was used to assess the effects of hypoxia on cellular metabolism of HCT116 cancer cells, utilizing LC-MS, to determine the metabolic profile of hypoxic cells. It revealed the importance of synchronized and regulated catabolism as a mechanism of adaptation to bioenergetic stress. By investigating the central carbon metabolism that includes most of the energy transfer molecules such as nucleotides, sugar mono- and biphosphates, and cofactors, a conclusion about phenotypes and stress answers in microorganisms is possible (Liebeke et al., 2010). Detection of several remarkable differences (e.g., in nucleotide metabolism and especially cell wall precursor metabolites), indicates a previously unreported importance of serine/threonine kinase/phosphatase on peptidoglycan and wall teichoic acid biosynthesis. These findings may lead to new insights into the regulation of cell wall metabolism.
Other applications
Emerging metabolomic tools can now be used to establish metabolic signatures of specialized circulating hematopoietic cells in physiologic or pathologic conditions and in human hematologic diseases. Metabolomic analysis of young and old normal red blood cells indicates metabolites whose levels are directly related to sickle cell disease (Darghouth et al., 2011). To study the metabolic changes associated with the differentiation program, the exometabolome of differentiating murine B lymphoma cells and primary B cells were compared (Garcia-Manteiga et al., 2011). During proliferation, lactate production increased, together with consumption of essential amino acids; massive Ig secretion was paralleled by alanine and glutamate production, glutamine being used as carbon and energy sources. Notably, ethanol and 5'-methylthioadenosine were produced during the last phase of protein secretion and the proliferative burst, respectively. The high-resolution NMR spectra were applied to present differentiation of Caco-2 (Leeet al., 2009). Results clearly reveal differences in the metabolic profiles over time as the Caco-2 cells differentiate. While a majority of metabolites are present at both the early undifferentiated state and the late differentiated states, the levels of certain metabolites are seen to change dramatically, and in particular, the ratio of myo-inositol and taurine. The appearance of glucose resonances in the differentiated cells spectra suggests that these cells become gluconeogenic. It represents a novel method to analyze the differentiation of Caco-2 cells using a metabolomic approach.
Challenges in Cell Metabolomics
The comprehensive characterization of the cell metabolome, however, is a daunting task, as the endogenous metabolites vary widely in their physical and chemical properties, which in turn, makes their concurrent extraction, separation, and detection a major challenge. Some of these include the fact that the chemical properties of a great number of metabolites are still unknown, adding to the problem of standardizing quenching and extraction methods; standard compounds are not readily available and reference libraries are still incomplete; not all metabolic pathways are known, and this causes difficulties in mapping and biological interpretation of these markers. Limited work has focused on development of sample preparation techniques for metabolomic analysis of adherent mammalian cells. The differences in optimized cell culture growth conditions present another major concern for cell line metabolomics. The standard enhancement of cell culture medium with serum of animal origin can add another level of complexity in cell growth condition optimization. Cultured cells are useful models in biomedical research that characterize metabolic responses to various stimuli and explore the underlying mechanisms (MSI et al., 2007; Selvarasu et al., 2012; Tizian et al., 2009). In addition to the main challenges imposed by cell analysis, sample preparation protocols that do not alter the metabolic status of the cells analyzed must be developed. Yet, these important studies are not sufficient for generating a complete picture of the molecular components regulating cellular function. Without analytical standards and quality control rules, quantification of individual metabolites and validation of the cell metabolomic methods will not be possible. With the rapid developments in powerful analytical technologies, we can expect a plethora of metabolite data. The grand challenges are to validate and make sense of the data, put molecular components and dynamic changes into particular pathways, integrate with other types of data, and connect into molecular networks of cell functions (Rivas-Ubach et al., 2012).
Conclusion and Future Outlook
The purpose of this article was to describe the state of the art of a new emerging discipline, cell metabolomics in the understanding of how the metabolites affect cell behavior and function, and highlight the past successes in applications of metabonomics to contribute to low-molecular-weight metabolites discovery in cell research. In recent years, metabolomic technologies have moved beyond simple cataloging towards large-scale molecular quantification and network analysis. As a vital component of a ‘systems biology’ approach, it is believed to be a good reflection of the phenotype of any cell. A systemic knowledge of how cells work will certainly aid in our effort toward a holistic understanding of cell function. Metabolomics produce highly useful information about cell biology. In addition, future fundamental research should provide a more complete list of metabolites. The detailed experimental design, experimentation, and analysis for metabolomics will provide useful information and become a truly essential analysis for cells. We believe that the availability of high throughput methods for analysis of metabolites in cells will substantially enhance our abilities to gain insight into biochemical reaction networks, to understand mechanistically how the metabolites affect cell behavior, and by extension, health and disease as well individual differences in response to health interventions such as drugs, nutrition, and vaccines.
Acknowledgments
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant Nos. 90709019, 81173500, 81102556, 81202639), National Key Technology Research and Development Program of the Ministry of Science and Technology of China (Grant Nos. 2011BA103B03, 2011BA103B06, 2011BA103B08), Key Science and Technology Program of Heilongjiang Province, China (Grant Nos. GC06C501, GA08C303, GA06C30101), Foundation of Heilongjiang University of Chinese Medicine (Grant No. 201209), and National Key Subject of Drug Innovation (Grant No. 2009ZX09502-005).
Author Disclosure Statement
The authors declare no competing financial interests.
References
- Ajikumar PK. Xiao WH. Tyo KE, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki AK. Skolnick J. McDonald JF. Marker metabolites can be therapeutic targets as well. Nature. 2008;456:443. doi: 10.1038/456443c. [DOI] [PubMed] [Google Scholar]
- Birungi G. Chen SM. Loy BP. Ng ML. Li SF. Metabolomics approach for investigation of effects of dengue virus infection using the EA.hy926 cell line. J Proteome Res. 2010;9:6523–6534. doi: 10.1021/pr100727m. [DOI] [PubMed] [Google Scholar]
- Blow N. Metabolomics: Biochemistry's new look. Nature. 2008;455:697–700. doi: 10.1038/455697a. [DOI] [PubMed] [Google Scholar]
- Bricker DK. Taylor EB. Schell JC, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z. Zhao JS. Li JJ, et al. A combined proteomics and metabolomics profiling of gastric cardia cancer reveals characteristic dysregulations in glucose metabolism. Mol Cell Proteomics. 2010;9:2617–2628. doi: 10.1074/mcp.M110.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. Mias GI. Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier JJ. Burke SJ. Eisenhauer ME, et al. Pancreatic β-cell death in response to pro-inflammatory cytokines is distinct from genuine apoptosis. PLoS One. 2011;6:e22485. doi: 10.1371/journal.pone.0022485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello LC. Franklin RB. Integration of molecular genetics and proteomics with cell metabolism: How to proceed; how not to proceed! Gene. 2011;486:88–93. doi: 10.1016/j.gene.2011.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q. Lewis IA. Hegeman AD, et al. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- Cuperlović-Culf M. Barnett DA. Culf AS. Chute I. Cell culture metabolomics: Applications and future directions. Drug Discov Today. 2010;15:610–621. doi: 10.1016/j.drudis.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Darghouth D. Koehl B. Madalinski G, et al. Pathophysiology of sickle cell disease is mirrored by the red blood cell metabolome. Blood. 2011;117:e57–66. doi: 10.1182/blood-2010-07-299636. [DOI] [PubMed] [Google Scholar]
- Dengler MA. Staiger AM. Gutekunst M, et al. Oncogenic stress induced by acute hyper-activation of Bcr-Abl leads to cell death upon induction of excessive aerobic glycolysis. PLoS One. 2011;6:e25139. doi: 10.1371/journal.pone.0025139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar BJ. Keshari K. Jeffries R. Dzeja P. Graves LM. Macdonald JM. Metabolic assessment of a novel chronic myelogenous leukemic cell line and an imatinib resistant subline by H NMR spectroscopy. Metabolomics. 2010;6:439–450. doi: 10.1007/s11306-010-0204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Preez I. Loots du T. Altered fatty acid metabolism due to rifampicin-resistance conferring mutations in the rpoB gene of Mycobacterium tuberculosis: Mapping the potential of pharmaco-metabolomics for global health and personalized medicine. OMICS. 2012;16:596–603. doi: 10.1089/omi.2012.0028. [DOI] [PubMed] [Google Scholar]
- Ellis JK. Athersuch TJ. Cavill R, et al. Metabolic response to low-level toxicant exposure in a novel renal tubule epithelial cell system. Mol Biosyst. 2011;7:247–257. doi: 10.1039/c0mb00146e. [DOI] [PubMed] [Google Scholar]
- Finley LW. Lee J. Souza A, et al. Skeletal muscle transcriptional coactivator PGC-1α mediates mitochondrial, but not metabolic, changes during calorie restriction. Proc Natl Acad Sci USA. 2012;109:2931–2936. doi: 10.1073/pnas.1115813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R. Trudgian DC. Wright C, et al. Discovery of candidate serum proteomic and metabolomic biomarkers in ankylosing spondylitis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013904. M111.013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C. Zheng L. Tennant DA, et al. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS One. 2011;6:e24411. doi: 10.1371/journal.pone.0024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manteiga JM. Mari S. Godejohann M, et al. Metabolomics of B to plasma cell differentiation. J Proteome Res. 2011;10:4165–4176. doi: 10.1021/pr200328f. [DOI] [PubMed] [Google Scholar]
- Halama A. Möller G. Adamski J. Metabolic signatures in apoptotic human cancer cell lines. OMICS. 2011;15:325–35. doi: 10.1089/omi.2010.0121. [DOI] [PubMed] [Google Scholar]
- Hua Y. Qiu Y. Zhao A, et al. Dynamic metabolic transformation in tumor invasion and metastasis in mice with LM-8 osteosarcoma cell transplantation. J Proteome Res. 2011;10:3513–3521. doi: 10.1021/pr200147g. [DOI] [PubMed] [Google Scholar]
- Jenkins H. Hardy N. Beckmann M, et al. A proposed framework for the description of plant metabolomics experiments and their results. Nat Biotechnol. 2004;22:1601–1606. doi: 10.1038/nbt1041. [DOI] [PubMed] [Google Scholar]
- Khoo SH. Al-Rubeai M. Metabolomics as a complementary tool in cell culture. Biotechnol Appl Biochem. 2007;47:71–84. doi: 10.1042/BA20060221. [DOI] [PubMed] [Google Scholar]
- Kinross JM. Holmes E. Darzi AW. Nicholson JK. Metabolic phenotyping for monitoring surgical patients. Lancet. 2011;377:1817–1819. doi: 10.1016/S0140-6736(11)60171-2. [DOI] [PubMed] [Google Scholar]
- Kleiner M. Wentrup C. Lott C, et al. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci USA. 2012;109:E1173–1182. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM. Mahmoud SS. Wildung MR, et al. Improving peppermint essential oil yield and composition by metabolic engineering. Proc Natl Acad Sci USA. 2011;108:16944–16949. doi: 10.1073/pnas.1111558108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IJ. Hom K. Bai G. Shapiro M. NMR metabolomic analysis of caco-2 cell differentiation. J Proteome Res. 2009;8:4104–4108. doi: 10.1021/pr8010759. [DOI] [PubMed] [Google Scholar]
- Liebeke M. Meyer H. Donat S. Ohlsen K. Lalk M. A metabolomic view of Staphylococcus aureus and its ser/thr kinase and phosphatase deletion mutants: Involvement in cell wall biosynthesis. Chem Biol. 2010;17:820–830. doi: 10.1016/j.chembiol.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Lin S. Liu N. Yang Z, et al. GC/MS-based metabolomics reveals fatty acid biosynthesis and cholesterol metabolism in cell lines infected with influenza A virus. Talanta. 2010;83:262–268. doi: 10.1016/j.talanta.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Locasale JW. Melman T. Song S, et al. Metabolomics of human cerebrospinal fluid identifies signatures of malignant glioma. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014688. M111.014688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MA. Burant CF. Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem. 2011;83:3406–3414. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S. Meir S. Malitsky S. Ruppin E. Aharoni A. Shlomi T. Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci USA. 2011;109:339–344. doi: 10.1073/pnas.1100358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MSI Board Members. Sansone SA. Fan T, et al. The metabolomics standards initiative. Nat Biotechnol. 2007;25:846–848. doi: 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- Nomura DK. Morrison BE. Blankman JL, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011;334:809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier I. Loots du T. A metabolomics approach to characterise and identify various Mycobacterium species. J Microbiol Methods. 2012;88:419–426. doi: 10.1016/j.mimet.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Paglia G. Hrafnsdóttir S. Magnúsdóttir M, et al. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS) Anal Bioanal Chem. 2012;402:1183–1198. doi: 10.1007/s00216-011-5556-4. [DOI] [PubMed] [Google Scholar]
- Prigione A. Lichtner B. Kuhl H, et al. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell-like metabolic reprogramming. Stem Cells. 2011;29:1338–1348. doi: 10.1002/stem.683. [DOI] [PubMed] [Google Scholar]
- Qin XY. Akanuma H. Wei F, et al. Effect of low-dose thalidomide on dopaminergic neuronal differentiation of human neural progenitor cells: A combined study of metabolomics and morphological analysis. Neurotoxicology. 2012;12:00212–00214. doi: 10.1016/j.neuro.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C. Lisec J. Czedik-Eysenberg A, et al. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA. 2012;109:8872–8877. doi: 10.1073/pnas.1120813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Ubach A. Sardans J. Pérez-Trujillo M. Estiarte M. Peñuelas J. Strong relationship between elemental stoichiometry and metabolome in plants. Proc Natl Acad Sci USA. 2012;109:4181–4186. doi: 10.1073/pnas.1116092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabidó E. Quehenberger O. Shen Q, et al. Targeted proteomics of the eicosanoid biosynthetic pathway completes an integrated genomics-proteomics-metabolomics picture of cellular metabolism. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014746. M111.014746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarasu S. Ho YS. Chong WP, et al. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnol Bioeng. 2012;109:1415–1429. doi: 10.1002/bit.24445. [DOI] [PubMed] [Google Scholar]
- Sreekumar A. Poisson LM. Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Stringari C. Cinquin A. Cinquin O. Digman MA. Donovan PJ. Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K. Shin SY. Petersen AK, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y. Yin P. Tang L, et al. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: Potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010694. M111.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R. Cho K. Uritboonthai W. Zhu Z. Patti GJ. Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiziani S. Lodi A. Khanim FL. Viant MR. Bunce CM. Günther UL. Metabolomic profiling of drug responses in acute myeloid leukaemia cell lines. PLoS One. 2009;4:e4251. doi: 10.1371/journal.pone.0004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita M. Kami K. Systems biology, metabolomics, and cancer metabolism. Science. 2012;336:990–991. doi: 10.1126/science.1223066. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ. Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2011;134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Wang TJ. Larson MG. Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Zhang A. Han Y, et al. Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol Cell Proteomics. 2012a;11:370–380. doi: 10.1074/mcp.M111.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Zhang A. Sun H. Future perspectives of Chinese medical formulae: Chinmedomics as an effector. OMICS. 2012b;16:414–421. doi: 10.1089/omi.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Yang B. Sun H. Zhang A. Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal Chem. 2012c;84:428–439. doi: 10.1021/ac202828r. [DOI] [PubMed] [Google Scholar]
- Wang X. Zhang A. Wang P, et al. Metabolomics coupled with proteomics advancing drug discovery towards more agile development of targeted combination therapies. Mol Cell Proteomics. 2013a;12:1226–1238. doi: 10.1074/mcp.M112.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JL. Baker JM. Llewellyn AM. Hawkins ND. Beale MH. Metabolomic analysis of Arabidopsis reveals hemiterpenoid glycosides as products of a nitrate ion-regulated, carbon flux overflow. Proc Natl Acad Sci USA. 2011;108:10762–10767. doi: 10.1073/pnas.1018875108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW. Schlage WK. Frushour BP, et al. Construction of a computable cell proliferation network focused on non-diseased lung cells. BMC Syst Biol. 2011;5:105. doi: 10.1186/1752-0509-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Volponi JV. Oliver AE. Parikh AN. Simmons BA. Singh S. In vivo lipidomics using single-cell Raman spectroscopy. Proc Natl Acad Sci USA. 2011;108:3809–3814. doi: 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. Wu Z. Wang X, et al. Metabolomic analysis reveals that carnitines are key regulatory metabolites in phase transition of the locusts. Proc Natl Acad Sci USA. 2012;109:3259–3263. doi: 10.1073/pnas.1119155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Han Y, et al. Exploratory urinary metabolic biomarkers and pathways using UPLC-Q-TOF-HDMS coupled with pattern recognition approach. Analyst. 2012a;137:4200–4208. doi: 10.1039/c2an35780a. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang P. Han Y. Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012b;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang X. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072–2077. doi: 10.1002/hep.26130. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Filiou MD. Reckow S, et al. Proteomic and metabolomic profiling of a trait anxiety mouse model implicate affected pathways. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.008110. M111.008110. [DOI] [PMC free article] [PubMed] [Google Scholar]