Abstract
Leukocyte 8-hydroxydeoxyguanosine (8OHdG) is an indicator of oxidative stress, impaired metabolism, and mitochondrial dysfunction, features that have been implicated in Huntington disease (HD). Increased levels of 8OHdG have been reported in the caudate, parietal cortex, and peripherally in the serum and leukocytes, in patients diagnosed with HD. However, little is known about levels in prodromal patients and changes that might occur as the disease progresses. To address these issues, 8OHdG was tracked over time for a subset of participants enrolled in the PREDICT-HD study. Participants were stratified into four groups based on proximity to HD diagnosis at study entry: Controls (gene-negative individuals), Low (low probability of near future diagnosis), Medium, and High. Blood samples were analyzed using Liquid Chromatography Electrochemical Array, and for comparison purposes, a separate cross-sectional sample was analyzed using liquid chromatography coupled with multiple-reaction-monitoring mass spectrometry. Longitudinal data analysis showed that initial status (at study entry) and annual rate of change varied as a function of proximity group, adjusting for sex, education, age at study entry, and site effects. Overall levels were lowest for the Control group and highest for the High group, and the rate of increase varied in a similar manner. The finding that 8OHdG concentrations increased as a function of proximity to projected disease diagnosis and duration indicates support for the continued assessment of 8OHdG as a robust clinical HD biomarker.
Keywords: 8OHdG, Huntington disease, Biomarker, Oxidative stress, Mitochondrial dysfunction, PREDICT-HD
Introduction
Huntington disease (HD) is an autosomal dominant neurodegenerative disorder of the brain caused by the trinucleotide cytosine– adenine–guanine (CAG) expansion in the gene of the protein huntingtin. There is currently no cure and the few treatments available are for symptom modification. Clinical diagnosis or phenoconversion is often based on the presence of motor symptoms consistent with the disease phenotype and most commonly occur during the fourth decade of life (Langbehn et al., 2004). In addition to motor deterioration, declines in cognition and psychiatric deficits are common (Duff et al., 2007; Paulsen et al., 2008; Stout et al., 2011; Tabrizi et al., 2009).
Identification of the gene responsible for HD (The Huntington's Disease Collaborative Research Group, 1993) has made it possible to identify prodromal research participants who are gene-positive, but are not yet exhibiting clinical motor manifestations sufficient for clinical diagnosis. Research emphasis has since shifted to the development of clinical trials for possible disease-modifying treatments in prodromal persons (Paulsen et al., 2010b), prior to the disease consequences of functional decline, brain atrophy, and adverse clinical symptoms. Investigations with prodromal HD participants allow researchers to gain valuable insight into pathogenesis through phenoconversion and beyond, the understanding of which could potentially lead to intervention measures preventing disease manifestation (Ross and Tabrizi, 2011). Despite ample advances in the characterization of clinical and imaging changes in prodromal HD, few studies have identified possible bio-markers that might be useful in successfully monitoring therapeutic intervention efficacy. Aggressive research in the identification and validation of possible biomarkers for HD is critical to efforts to devise treatments for this relentless brain disease.
One biomarker of interest is leukocyte 8-hydroxydeoxyguanosine (8OHdG), which is an indicator of oxidative stress, impaired metabolism, and mitochondrial dysfunction. Such features have been implicated in the neuronal dysfunction associated with HD (Kikuchi et al., 2002). 8OHdG is a compound formed through hydroxylation of the guanine base by radical oxygen species (ROS). Following oxidation, damaged DNA is repaired by cellular mechanisms, and the hydroxylated guanine is excreted via bodily fluids. Consequently, available levels of 8OHdG in the blood and urine correlate with the degree of internal DNA damage and have been used as markers for such purposes in disease modeling (Valavanidis et al., 2009).
Disruptions to mitochondrial function and metabolism and an increase in oxidative stress through the production of free radical molecules have been implicated in HD (Browne and Beal, 2006; Klepac et al., 2007; Sorolla et al., 2008). Increased levels of 8OHdG have been reported in the caudate and parietal cortex (Browne et al., 1997; Polidori et al., 1999), as well as peripherally in the serum (Hersch et al., 2006) and leukocytes (Chen et al., 2007) of patients diagnosed with HD. Additionally, the R6/2 mouse model, which expresses exon 1 of the human HD gene and an expanded CAG repeat, shows increased levels of 8OHdG in urine, plasma, and striatal microdialysates (Bogdanov et al., 2001). These studies provide support for the utility of 8OHdG measurement in HD and strengthen the rationale for therapeutic strategies that either potentiate antioxidant defenses or avoid oxidative stress generation to delay disease progression. The relationship between prodromal HD individuals and 8OHdG levels, however, has remained unexplored until now.
The PREDICT-HD study is a longitudinal observational study aimed at identifying clinical and biological markers of HD in prodromal and diagnosed individuals who have undergone genetic testing for the HD gene mutation (Paulsen et al., 2006). The study has previously reported the prodromal existence of mild cognitive impairment (Duff et al., 2010), behavioral symptoms (Beglinger et al., 2008), psychiatric symptoms (Duff et al., 2007), motor deficits (Biglan et al., 2009), and neurostructural abnormalities (Nopoulos et al., 2011; Paulsen et al., 2010a). The current study aims to build on these findings by examining possible biochemical changes occurring in HD, specifically those related to mitochondrial dysfunction.
The purpose of the current study is to examine the levels of 8OHdG in plasma samples taken from prodromal HD participants who vary in their disease progression at study entry. The primary goal is to evaluate if 8OHdG is sensitive to change over time and whether such change is a function of disease progression. In addition to change, overall differences among individuals varying in disease progression are of interest. These goals are addressed using two methods of blood specimen analysis.
Methods
Participants
Three separate samples of participants already enrolled in PREDICT-HD were considered for the analysis. Table 1 shows descriptive information of the three samples for a number of demographic variables. Each sample had two types of participants, those who tested positive for the expanded CAG repeat length (cases), and those who tested negative (controls). All participants had undergone genetic testing for the CAG repeat expansion prior to the study. Inclusion criterion for PREDICT-HD and this study was that participants had to be “prodromal” at the first visit, meaning expansion-positive participants did not meet the traditional definition of HD diagnosis as determined by question 17 of the Unified Huntington's Disease Rating Scale (UHDRS). Question 17, also known as “diagnostic confidence level” (DCL), is a 5-point scale on which investigators rate their level of confidence that an individual fulfills the criteria for a clinical motor diagnosis based on the presence of an otherwise unexplained movement disorder. A score of zero indicates that no motor abnormalities are present, whereas a score of 4 corresponds to a confidence level≥99% that an individual is displaying unequivocal signs of HD. Participants with DCL=4 at the first visit were excluded from the analysis. Additional details regarding recruitment procedures in PREDICT-HD are provided by Stout et al. (2011).
Table 1.
Descriptive statistics for the three samples used in the analysis.
| Sample | Variable | Min | Med | Mean | Max | SD |
|---|---|---|---|---|---|---|
| Longitudinal (NL=77, 13 sites) | Duration | 0.00 | 1.03 | 1.05 | 2.72 | 0.85 |
| Entry age | 26.15 | 42.21 | 42.91 | 75.85 | 9.20 | |
| CAG | 16.00 | 41.50 | 36.86 | 47.00 | 10.20 | |
| Education | 8.00 | 16.00 | 15.08 | 20.00 | 2.72 | |
| Sex | 0.55 | |||||
| Cross-sectional (NC=80, 5 sites) | Duration | 0.00 | 2.95 | 2.73 | 4.26 | 1.06 |
| Entry age | 19.15 | 41.73 | 41.83 | 62.59 | 8.39 | |
| CAG | 17.00 | 42.00 | 36.83 | 46.00 | 10.11 | |
| Education | 8.00 | 15.00 | 14.74 | 20.00 | 2.75 | |
| Sex | 0.50 | |||||
| Overlapping (NO=18, 4 sites) | Duration | 1.91 | 1.99 | 2.05 | 2.36 | 0.13 |
| Entry age | 26.84 | 39.02 | 39.10 | 61.17 | 8.42 | |
| CAG | 17.00 | 42.00 | 35.00 | 46.00 | 12.10 | |
| Education | 11.00 | 16.00 | 15.32 | 20.00 | 2.52 | |
| Sex | 0.53 |
Note. Duration=current age-age at entry; sex = proportion of males.
The first sample was longitudinal (L) consisting of NL=77 participants with up to three repeated measures over visits 1–3. The longitudinal participants had their blood specimens processed according to the first method to be discussed below (LCECA analysis). The longitudinal participants were sampled from 13 sites.
The second sample was cross-sectional (C) consisting of NC=80 participants with observations mainly collected over visits 3–6. The cross-sectional participants had their blood specimens processed according to the second method presented below (LC–MS/MS analysis). The cross-sectional participants were sampled from 5 sites.
The third sample was cross-sectional and consisted of eighteen participants who overlapped (O) the two previous samples. That is, NO=18 participants were common among the previous two samples, but only had cross-sectional observations. The overlapping participants had their blood specimens processed by both methods discussed below and were sampled from 4 sites.
The participants were sampled so as to represent various levels of disease progression at time of entry into PREDICT-HD. All expansion-positive participants were classified into groups according to their predicted proximity to HD diagnosis at study entry based on the CAG length/age product (CAP) variable (Zhang et al., 2011). CAP groups are Low, Medium, and High, with the labels reflecting levels of cumulative toxicity of mutant huntingtin at study entry. The Control group consists of the expansion-negative individuals who will never develop HD (all individuals had CAG<31). Table 2 shows the frequency (% of total) of CAP group membership by type of sample. As seen in Table 2, the High group was the largest for all the samples.
Table 2.
CAP group frequency (% of total) by sample.
| Sample | Control | Low | Medium | High |
|---|---|---|---|---|
| Longitudinal | 20 (26) | 8 (10) | 18 (23) | 31 (40) |
| Cross-sectional | 20 (25) | 16 (20) | 6 (8) | 38 (48) |
| Overlapping | 6 (23) | 4 (21) | 2 (11) | 7 (37) |
Validity evidence of the CAP groups for the larger PREDICT-HD data set is provided by Zhang et al. (2011), who show group differences on a large number of clinical and behavioral phenotypes. For present purposes, limited phenotypic information is provided for the longitudinal sample, as this was the sample for the primary analysis (see below). Means and SDs are provided for key phenotypic variables at study entry by CAP group in Table 3, along with the appropriate omnibus test statistic in the last column (F for means and chi-squared for proportions, testing any type of difference among the groups). The variables are striatal volume to total volume×100 (Striatum), UHDRS Total Motor Score (TMS; possible range 0–100 with higher scores reflecting greater symptoms), UHDRS Functional Assessment (FAS; range 0–25 with lower scores reflecting worse functioning), Symbol Digit Modalities Test (SDMT; minimum=0 with lower scores indicating greater impairment), Beck Depression Inventory (BDI; range 0–63 with higher scores indicating greater depression), and the control variables years of education, age at study entry, and proportion of males (Sex). The last row shows the proportion who received a DCL motor diagnosis (i.e., phenoconverted) at some point over the observed duration for the sample (larger values reflect greater disease progression).
Table 3.
Mean (SD) of key variables for the longitudinal sample by disease progression group at the first visit.
| CAP group | |||||
|---|---|---|---|---|---|
|
| |||||
| Variable | Control | Low | Medium | High | Test |
| Striatum | 1.10 (0.13) | 1.02 (0.13) | 0.93 (0.16) | 0.81 (0.14) | F(3,47) = 11.73*** |
| TMS | 2.90 (2.49) | 0.75 (1.49) | 4.16 (4.68) | 9.35 (6.74) | F(3,74) = 10.58*** |
| FAS | 25 (0) | 25 (0) | 24.84 (0.37) | 24.74 (0.68) | F(3,74) = 1.48 |
| SDMT | 56.45 (5.77) | 59.00 (11.74) | 51.89 (13.50) | 44.87 (10.55) | F(3,74) = 6.87*** |
| BDI | 4.60 (5.58) | 4.12 (5.00) | 8.37 (8.78) | 9.52 (10.24) | F(3,74) = 1.86 |
| Education | 16 (2.15) | 14.62 (2.39) | 14.42 (2.97) | 15 (2.92) | F(3,74) = 1.23 |
| Entry age | 42.49 (9.09) | 38.04 (8.55) | 40.79 (9.63) | 45.74 (8.63) | F(3,74) = 2.17 |
| Sex | 0.55 | 0.5 | 0.63 | 0.52 | χ2(3) = 0.74 |
| Diagnosis | 0 | 0 | 0.21 | 0.39 | χ2(3) = 13.52** |
Note. Striatum=ratio of striatal volume to total brain volume×100; TMS = Total Motor Score; FAS = Functional Assessment; SDMT = Symbol Digit Modalities Test; BDI = Beck Depression Inventory; Sex = proportion of males; Diagnosis = proportion who eventually phenoconverted.
p<0.01.
p<0.001.
Table 3 illustrates the baseline progression differences among the CAP groups. Striatal volume, SDMT, and FAS means decreased as disease progression increased, though FAS group differences were not statistically reliable (F(3,74)=1.48, p>0.05). TMS, diagnosis proportion, and BDI means increased as disease progression increased, though BDI group differences were not statistically reliable (F(3,74)=1.86, p>0.05). Additional analyses not presented suggested that differences among the Control and Low groups were attributable to differences in mean age and years of education. For additional CAP group comparisons and additional validation with the larger PREDICT-HD cohort, see Zhang et al. (2011).
Blood specimen collection protocol
Un-fasted blood specimens were obtained from the cubital vein into EDTA-coated tubes, inverted multiple times, and then centrifuged at 4000 rpm at 4 °C for 10 min. Resultant plasma was divided into 1.5–2 mL aliquots, then frozen and stored at −80 °C until analysis.
LCECA analysis
For the longitudinal participants (NL=77), blood specimens were analyzed using a long gradient Liquid Chromatography Electrochemical Array (LCECA) (Bogdanov et al., 1999; Matson et al., 1984, 1987). The LCECA survey method isolates ca. 1500–2000 responses from plasma components to levels of ca. 200 pg/mL. A mixed standard of 60 components of the tyrosine, tryptophan, and purine pathways, sulfur amino acids, and markers of oxidative stress, were used to determine the expression levels of previously reported known compounds.
At the initiation of the study, a pool of all samples was created from sub-aliquots of each sample and was used for both quality assessment and normalization of the analytical data. Samples were randomized for analysis sequence. Samples were run in the sequence order: mixed standard, pool, eight samples, mixed standard, pool, eight samples, etc. All individual samples were time-normalized to a pool in the middle of the study. The data were then exported as digitized values (digital maps) capturing all information from the analytical platform. The digital maps were analyzed in raw form using principle component analysis to determine the extent of any analytical drift over the study, and subsequently normalized to the average pool value for the entire study against the pools bracketing each sample sequence of eight. The digital maps were then trimmed to contain only values below the noise threshold of the instrument.
LC–MS/MS analysis
For the cross-sectional participants (NC=80), 8OHdG was assessed using reverse phase C-18 liquid chromatography coupled to a multiple-reaction-monitoring (MRM) mass spectrometer (LC– MS/MS). This assay was developed and performed by Pharmaceutical Product Development (PPD) and used a stable-heavy-isotope-labeled form of 8OHdG.
Sample processing
The 80 human plasma specimens were received from Coriell Institute in dry ice. The average volume of a specimen was 1.2 mL. A pool produced from human plasma QC samples was obtained from the Stanford University Blood Bank and maintained in a −80 °C freezer before processing. For each sample, 500 μL plasma was mixed with 500 μL water, and 5 μL 15N5-8OHdG was added to the mixture. 150 μL 1 M ammonium acetate (NH4Ac, pH 5.25) was added to the solution and the solution was vigorously vortexed for 10 min on a shaker. The solution was centrifuged at 15,800×g at RT for 2 min, because precipitates were observed in many specimens, which can cause a variety of processing problems. The supernatant was loaded onto Seppak tC18 solid-phase-extraction cartridge (200 mg, Waters, MA) that was preconditioned with 1 mL methanol and 1 mL water. The cartridges were then washed with 3 mL water before the fraction containing 8OHdG was eluted with 1 mL 40% methanol.
The eluent was dried under centrifugation and vacuum and then reconstituted with 100 μL buffer A (10 mM NH4Ac in water, pH 4.75 adjusted by formic acid). Reconstituted solution was filtered with a pre-cleaned 10KD MWCO centrifugal device (wash: 200 μL for 15 min; sample: 100 μL for 12 min, both centrifuged at 15,800×g) and stored in a −80 °C freezer for LC–MS/MS analysis.
Online HPLC–MS/MS (MRM) measurements were carried out chro-matographically on an Agilent capillary 1100 binary pumping system (Agilent Technologies, Santa Clara, CA). Reverse phase-HPLC separations were achieved on a Zorbax® SB C18 capillary HPLC column (150.0.5 mm, particle size: 5 μm, pore size: 8 Å, Agilent Technologies, CA). LC flow rate was 15 μL/min, using 10 mM NH4Ac in water adjusted to pH 4.6 with formic acid (buffer A) and 0.1% formic acid in MeCN (buffer B). Gradient elution was performed as follows: 0–2 min, 0–0% buffer B; 2–10 min, 0–50% buffer B; 10–12 min, 50–50% buffer B; 12–13 min, 50–0% buffer B; 13–20 min, 0–0% buffer B. MS/MS analysis was performed in positive ion mode, using a Q-Trap 4000 triple–quadruple ion trap (ABI-Sciex, Foster City, CA). MRM acquisition mode was used; m/z 168 and 173 (the most abundant product ions resulting from the precursor ions [M+H]+ at m/z 284 and 289, light and heavy) were utilized as quantifier ions with a dwell time 200 ms/cycle per ion.
Statistical analysis
The overall goal of the analysis was to examine CAP group differences in 8OHdG controlling for the covariates of years of education, age at entry, and sex. For the longitudinal participants the CAP group differences were in terms of initial status (intercept) and mean annual rate of change (linear slope). With the cross-sectional participants, the CAP group differences consisted of overall mean differences controlling for the covariates. The secondary goal of the analysis was to compare the 8OHdG processing methods for the NO=18 overlapping participants.
Linear mixed effects regression (LMER) was used for the longitudinal analysis to account for dependency due to repeated measures and to accommodate missing data (Verbeke and Molenberghs, 2000). The time metric for the analysis was duration, defined as the age at observation minus the age at study entry. Thus, initial status reflected the level at duration=0. Site variation was a consideration, as the longitudinal participants were sampled from a relatively large number of sites (see Table 1). The LMER model had duration nested within participants, and participants nested within sites. Random intercepts and random slope terms were specified for duration, along with a random intercept for site. Preliminary statistical and graphical analyses not presented suggested that linear growth curves were adequate.
The first part of the longitudinal analysis consisted of fitting three models: (1) a model with no CAP group differences, i.e., the same initial status and annual rate of increase for all groups; (2) a model with initial status differences among the CAP groups, but equal rates of increase; and (3) a model with initial status and rate of change differences among the CAP groups. All models included the covariates of years of education, age at study entry, and sex. Details of the models are provided in the Appendix A.
Parameters for the LMER analysis were estimated using maximum likelihood methods. The models were compared with the likelihood ratio test (LRT) and Akaike's information criterion (AIC) (Akaike, 1973, 1974) corrected for small sample bias (AICc) (Hurvich and Tsai, 1989). Two scalings of the AICc were also computed, dAICc and wtAICc. dAICc is the AICc for a model minus the smallest AICc of the set. dAICc=0 for the best fitting model and dAICc>0 for the remaining models. Though no statistical testing is performed with dAICc, a rule of thumb is that dAICc>3 represents a “very strong” difference between models (Anderson, 2008). wtAICc is the weight of evidence, which is a probability scaling of the AICc, indicating the probability that a model is the most plausible given the set of models and the data (Burnham and Anderson, 2002; Long, 2012). The weight of evidence has the range 0≤wtAICc≤1, with larger values indicating better fit to the data and higher plausibility.
The second part of the longitudinal analysis focused on specific effects of the best fitting model. The Control group was the reference group for comparison with all the other groups.
The analysis of the cross-sectional sample was similar to that of the longitudinal data in the respect that three models were compared. Though the data were cross-sectional, there was variability in duration due to different individuals having different values (rather individuals having repeated duration observations). Consequently, it was possible to examine the relationship between 8OHdG and duration controlling for the covariates and test the cross-sectional analogs of the three models in the longitudinal analysis.
Preliminary LMER analysis for the cross-sectional data (not presented) indicated zero fitted model variance for sites. Therefore, site variability was not modeled and linear regression (LR) was used for the cross-sectional analysis. The three models were compared using the F-test, AICc, dAICc, and wtAICc.
The size of the overlapping sample was small (NO=18). Therefore, no statistical analysis was performed other than to fit LR models of 8OHdG by duration for descriptive purposes. Graphs of the observed data with the fitted curves are presented below.
Results
Longitudinal sample
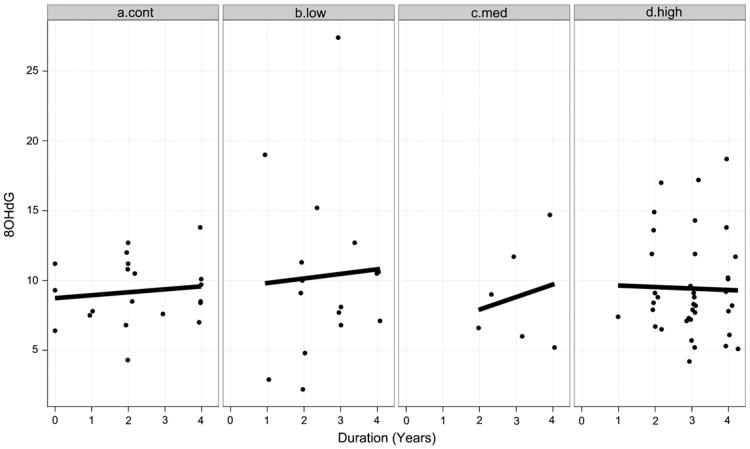
The results of the longitudinal data analysis indicate statistically reliable differences among the CAP groups in terms of initial levels of 8OHdG and differences in the mean rate of 8OHdG increase overtime. The nature of the effects is illustrated in Fig. 1, which is paneled by CAP group. The dashed lines depict observed levels of 8OHdG tracked over time (duration) for individuals, and the solid lines depict linear fits of the mean 8OHdG level over time for the groups. It is the starting point of the solid lines (i.e., the intercepts) and their slopes (i.e., rates of change in 8OHdG per year) that are the focus of the LMER analysis described above.
Fig. 1.
8OHdG levels over time (duration) for individuals (dashed lines) and groups (solid lines). The individual curves are observed 8OHdG levels and the group curves are 8OHdG levels based on linear fits.
As Fig. 1 shows, the slopes of the solid lines varied as a function of CAP group, with the Control group having a slight decrease in 8OHdG level per year, and the High group having the fastest increase in 8OHdG per year. The graph also shows intercept differences in levels of 8OHdG, with the Control group having the lowest mean level at duration=0, and the other groups having higher initial levels.
The results of the global model comparison are shown in Table 4. As seen in the table, Model 3 had substantially better fit than the other two models (wtAICc=0.92). Recall that Model 3 had unequal mean intercepts and unequal mean slopes among the CAP groups, similar to the solid lines in Fig. 1.
Table 4.
Results of the longitudinal sample model comparison.
| Model | K | Chi sq | df | p | AICc | dAICc | wtAICc |
|---|---|---|---|---|---|---|---|
| 1 | 11 | 1281.00 | 30.96 | 0.00 | |||
| 2 | 14 | 32.92 | 3 | <0.0001 | 1254.93 | 4.89 | 0.08 |
| 3 | 17 | 11.96 | 3 | 0.0075 | 1250.04 | 0.00 | 0.92 |
Note. K = number of parameters.
Because Model 3 was the best fitting, additional details are presented. Table 5 shows the estimates of the intercepts (upper portion) and the slopes (lower portion) for each CAP group, adjusting for the covariates. The inferential statistics (SE, z, p) reference the difference between the Control group and the remaining CAP groups (Low– Medium–High), thus they are omitted for the Control group rows in Table 5. The Control group slope estimate (βˆ0C= 1.05) had an associated z=0.40 indicating the slight increase in 8OHdG level after controlling for the covariates was not statistically reliable. The slight increase of the model-based estimate in Table 5 is in conflict with the slight decrease shown in Fig. 1 due to the inclusion of the covariates (Fig. 1 is not adjusted for the covariates). The covariates had relatively small effects compared to CAP group differences, with age at entry having the strongest initial status effect (z=−1.49; younger participants had a weak tendency to start at higher levels), and sex having the strongest slope effect (z=1.04; males had a small tendency to increase at a faster rate).
Table 5.
Estimated mean intercept and mean slope for the 8OHdG regression lines represented by Model 3.
| Effect | CAP group | Estimate | SEa | za | pa |
|---|---|---|---|---|---|
| Intercept | Control | 19.61 | 4.52 | ||
| Low | 22.87 | 2.29 | 1.42 | 0.1543 | |
| Medium | 22.66 | 1.69 | 1.81 | 0.0706 | |
| High | 24.20 | 1.44 | 3.18 | 0.0015 | |
| Slope | Control | 1.05 | 2.66 | ||
| Low | 1.56 | 1.45 | 0.35 | 0.7239 | |
| Medium | 2.40 | 0.98 | 1.38 | 0.1669 | |
| High | 3.99 | 0.87 | 3.38 | 0.0007 |
SE, z, and p refer to the difference between each group and the Control group. Results in this table are adjusted for years of education, age at study entry, and sex.
Focusing first on the CAP group slopes, Table 5 shows that the 8OHdG level increased at a faster rate as the CAP group increased (from Control, βˆ1C = 1.05, to High, βˆ1H = 3:99). The z-ratio column shows that the High group had the largest increase relative to the Control group (z=3.38), followed by Medium versus Control (z=1.38), and Low versus Control (z=0.35).
Turning to the mean intercept differences (at duration=0), the Control group had the smallest estimated initial 8OHdG level (βˆ0C = 19:61) and the High group had the largest estimated level (βˆ0H = 24:20). In terms of effect size, the largest difference was between the Control versus High group (z=3.18), followed by Medium versus Control (z=1.81), and Low versus Control (z=1.42).
Cross-sectional sample
The results indicate no statistically reliable differences among the CAP groups for the cross-sectional sample. The individual observed points and the fitted linear curves are shown in Fig. 2, with the panels representing CAP groups. The reader is reminded that the data in Fig. 2 are cross-sectional as the variability in duration is due to different individuals rather than repeated measures within individuals.
Fig. 2.
Cross-sectional sample observed 8OHdG values (filled circles) and linear fitted curves (solid lines) by CAP group.
The results of the LR analysis are shown in Table 6. The table shows that the model with no CAP group differences was the best fitting model (Model 1; wtAICc=0.93). Parameter estimates for Model 1 not presented showed there was a strong effect for sex (z=3.67), with males having a substantially higher mean 8OHdG level than females.
Table 6.
Results of the cross-sectional sample model comparison.
| Model | K | F | df1 | df2 | p | AICc | dAICc | wtAICc |
|---|---|---|---|---|---|---|---|---|
| 1 | 9 | 442.97 | 0.00 | 0.93 | ||||
| 2 | 12 | 0.82 | 3 | 69 | 0.4900 | 448.21 | 5.24 | 0.07 |
| 3 | 15 | 0.52 | 3 | 66 | 0.6734 | 455.20 | 12.23 | 0.00 |
Note. K = number of parameters.
Overlapping sample
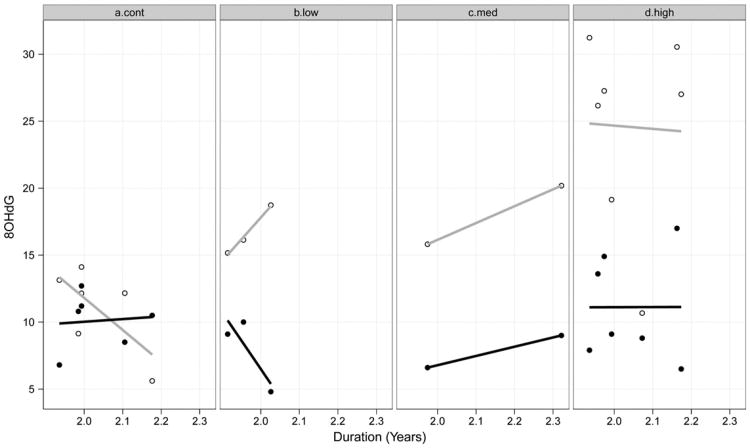
As previously mentioned, there were NO=18 participants whose blood specimens were processed using both methods. The observed points and fitted linear curves are shown in Fig. 3.
Fig. 3.
Observed 8OHdG for two methods of processing by duration and CAP group. Open circles and gray lines indicate LCECA processing; filled circles and solid lines indicate LC– MS/MS processing.
Each vertical cross-section of duration has two points, an open circled and a filled circle, indicating the two types of processing for the same individual. As can be seen in Fig. 3, the fitted curves are similar for the High and Medium groups, but not for the Low and Control groups.
Discussion
The aim of this study was to examine 8OHdG expression levels as a function of HD disease progression. Disease progression was indexed by CAP group (Low, Medium, High), which is based on a proxy variable of time to diagnosis (Zhang et al., 2011) and showed initial differences on several key phenotypic variables for the participants in the primary analysis (see Table 3). Three samples of participants were considered, a longitudinal sample with LCECA blood analysis, a cross-sectional sample with LC–MS/MS analysis, and an overlapping sample that used both methods. The results of the longitudinal sample show that plasma concentrations of 8OHdG varied as a function of CAP group controlling for numerous covariates. Specifically, there were intercept differences (at duration=0) among the groups, with the mutation-negative participants (Control group) having the lowest level, and the participants with the most advanced disease progression (High group) having the highest level. The rate of annual increase in 8OHdG level also varied as a function of disease progression. The mutation-negative participants had no appreciable change over time, similarly for the Low CAP group. The Medium group had a slight increase over time, and the High group had the fastest increase over time (see Fig. 1).
The longitudinal sample results are consistent with previous observations of irregular mitochondrial functioning and increases in DNA damage caused by oxidative stress. Although previous investigators were able to show a similar increase in 8OHdG associated with severity of disease symptoms in diagnosed patients (Chen et al., 2007; Hersch et al., 2006), this is the first study to demonstrate elevated oxidative DNA damage stress with prodromal individuals. 8OHdG concentrations increased as a function of proximity to projected disease diagnosis, indicating support for the continued assessment of 8OHdG as a robust clinical HD biomarker.
This study showed a substantial difference between mutation-negative individuals and mutation-positive individuals who were relatively far in their disease progression. However, this comparison does not establish 8OHdG as a biomarker specific to HD. 8OHdG has been implicated in various forms of cancer (Marnett, 2000), and in different types of neurodegenerative diseases (Sayre et al., 2001). Additional research, possibly with comparison groups representing multiple diseases, is needed to investigate the extent to which 8OHdG is a reliable biomarker for HD.
Due to the cross-sectional nature of the LC–MS/MS data, it was not possible to evaluate if 8OHdG values produced by this method were sensitive to changes over time. Though the 8OHdG by duration relationship was examined by CAP group (see Fig. 2), this provided only indirect information regarding change, and the focus was on overall CAP group differences. The cross-sectional sample did not show CAP group differences in overall level (average level) or in 8OHdG growth rate (see Table 6).
The discrepancy between the results of the longitudinal LCECA analysis and the cross-sectional LC–MS/MS analysis needs to be interpreted in light of participant characteristics and differences in the blood processing. Regarding the first point, the cross-sectional LC– MC/MS sample consisted of individuals who had been in the study longer on average than the longitudinal LCECA sample and were recruited from fewer sites (see Table 1).
Regarding blood processing, there was an increased level of handling, freeze/thawing and transport of specimens to the PPD laboratory for the LC–MS/MS analysis, and all the specimens were processed after the LCECA analysis and nothing was excluded from analysis. The lab used for the LCECA analysis had a larger sample size due to the repeated measures, a more rigorous exclusion criteria, and faster access to samples (less freeze/thaw). The absolute 8OHdG values of the LC–MS/MS analysis were lower than the LCECA analysis and may represent a decreased sensitivity of this assay. The LCECA analysis produced 8OHdG results similar to those previously reported in the literature (Hersch et al., 2006), and the lab used for the LCECA is more familiar with analytical procedures specific to this compound (Bogdanov et al., 1999).
The 18 participants who had their blood specimens analyzed by both methods potentially illustrate the differences in the assay sensitivity. For the Low, Medium, and High groups, the LCECA method produced 8OHdG values larger than the LC–MS/MS method for each individual (see Fig. 3). This was not the case for every individual in the Control group, however. In terms of 8OHdG change over time, the regression lines were very similar for both methods in the High group, which had the largest number of participants (seven). On the other hand, the regression lines for the Control group (six) were dissimilar (see Fig. 3).
One limitation of the current study, as with any research involving prodromal participants, is the classification of groups in regard to proximity to diagnosis. Exclusion criteria for the PREDICT-HD study disqualify individuals clinically diagnosed with HD from enrolling in the study. Therefore, the initial selection of participants for the longitudinal sample was performed following participants' third annual assessment. Subsequently, this retrospective grouping may have inflated the 8OHdG increase seen in participants approaching predicted clinical diagnosis.
The development of robust biomarkers for HD is of the utmost importance. Due the genetic nature of the disease and the ability to identify participants before manifestation of clinical symptoms, there is a unique window for intervention, whereby the course of the disease could theoretically be arrested. Traditional clinical measures, such as the TMS and FAS are often too insensitive to detect subtle changes years prior to phenoconversion. As such, the development of suitable markers of progression, such as 8OHdG, is integral to the formation of earlier therapeutic interventions, as well as assessments of their efficacy.
The findings for the longitudinal sample show that 8OHdG, a biochemical compound easily isolated from multiple bodily fluids, increases with HD disease progression and is sensitive to individual change over time. Though a larger sample is preferred, our results suggest that plasma 8OHdG levels could be employed clinically as a tool to help measure disease progression in gene positive individuals prior to the appearance of overt motor dysfunction. It is unlikely that a single chemical compound would be employed for such purposes, but the current findings could contribute to the development of a combination of robust measures with other biochemical biomarker candidates, as well as complement multiple other marker modalities, such as neuroimaging measures and cognitive and motor assessments to aid in the development and evaluation of interventional HD treatments.
Acknowledgments
Role of the funding sources: The funding sources had no involvement in the study design; in the collection, analysis, or interpretation of the data; in the writing of the report; nor in the decision to submit the article for publication.
This work was supported by the National Institutes of Health, the National Institute of Neurological Disorders and Stroke [NS40068 to J.S.P.] and the CHDI Foundation, Inc. We thank the PREDICT-HD sites, the study participants, and the National Research Roster for Huntington Disease Patients and Families.
PREDICT-HD Investigators, Coordinators, Motor Raters, Cognitive Raters
Thomas Wassink, MD, Stephen Cross, BA, Nicholas Doucette, BA, Mycah Kimble, BA, Patricia Ryan, MSW, LISW, MA, Jessica Wood, MD, PhD, Eric A. Epping, MD, PhD, and Leigh J. Beglinger, PhD (University of Iowa, Iowa City, Iowa, USA);
Edmond Chiu, MD, Olga Yastrubetskaya, PhD, Joy Preston, Anita Goh, D.Psych, Chathushka Fonseka, and Liz Ronsisvalle (St. Vincent's Hospital, The University of Melbourne, Kew, Victoria, Australia);
Phyllis Chua, MD, and Angela Komiti, BS, MA (The University of Melbourne, Royal Melbourne Hospital, Melbourne, Australia);
Lynn Raymond, MD, PhD, Rachelle Dar Santos, BSc, and Joji Decolongon, MSC, CCRP (University of British Columbia, Vancouver, British Columbia, Canada);
Adam Rosenblatt, MD, Christopher A. Ross, MD, PhD, Barnett Shpritz, BS, MA, OD, and Claire Welsh (Johns Hopkins University, Baltimore, Maryland, USA);
William M. Mallonee, MD, Greg Suter, BA, and Judy Addison (Hereditary Neurological Disease Centre, Wichita, Kansas, USA);
Ali Samii, MD, and Alma Macaraeg, BS (University of Washington and VA Puget Sound Health Care System, Seattle, Washington, USA);
Randi Jones, PhD, Cathy Wood-Siverio, MS, Stewart A. Factor, DO, and Claudia Testa, MD, PhD (Emory University School of Medicine, Atlanta, Georgia, USA);
Roger A. Barker, BA, MBBS, MRCP, Sarah Mason, BSC, Anna Goodman, PhD, Rachel Swain, BA, and Anna DiPietro (Cambridge Centre for Brain Repair, Cambridge, UK);
Elizabeth McCusker, MD, Jane Griffith, RN, Clement Loy, MD, David Gunn, BS, and Linda Stewart, RN (Westmead Hospital, Sydney, Australia);
Bernhard G. Landwehrmeyer, MD, Michael Orth MD, PhD, Sigurd Süβmuth, MD, RN, Katrin Barth, RN, and Sonja Trautmann, RN (University of Ulm, Ulm, Germany);
Kimberly Quaid, PhD, Melissa Wesson, MS, and Joanne Wojcieszek, MD (Indiana University School of Medicine, Indianapolis, IN);
Mark Guttman, MD, Alanna Sheinberg, BA, and Irita Karmalkar, BSc (Centre for Addiction and Mental Health, University of Toronto, Markham, Ontario, Canada);
Susan Perlman, MD and Arik Johnson, PsyD (University of California, Los Angeles Medical Center, Los Angeles, California, USA);
Michael D. Geschwind, MD, PhD, Jon Gooblar, BA, and Gail Kang, MD (University of California San Francisco, California, USA);
Tom Warner, MD, PhD, Maggie Burrows, RN, BA, Marianne Novak, MD, Thomasin Andrews, MD, BSC, MRCP, Elisabeth Rosser, MBBS, FRCP, and Sarah Tabrizi, MD, PhD (National Hospital for Neurology and Neurosurgery, London, UK);
Anne Rosser, MD, PhD, MRCP, Kathy Price, RN, and Sarah Hunt, BSc (Cardiff University, Cardiff, Wales, UK);
Frederick Marshall, MD, Amy Chesire, LCSW-R, MSG, Mary Wodarski, BA, and Charlyne Hickey, RN, MS (University of Rochester, Rochester, New York, USA);
Oksana Suchowersky, MD, FRCPC, Sarah Furtado, MD, PhD, FRCPC, and Mary Lou Klimek, RN, BN, MA (University of Calgary, Calgary, Alberta, Canada);
Peter Panegyres, MB, BS, PhD, Elizabeth Vuletich, BSC, Steve Andrew, and Rachel Zombor, MPSYC (Neurosciences Unit, Graylands, Selby-Lemnos & Special Care Health Services, Perth, Australia);
Joel Perlmutter, MD, Stacey Barton, MSW, LCSW, and Amy Schmidt (Washington University, St. Louis, Missouri, USA);
Zosia Miedzybrodzka, MD, PhD, Sheila A. Simpson, MD, Daniela Rae, RN, and Mariella D'Alessandro, PhD (Clinical Genetics Centre, Aberdeen, Scotland, UK);
David Craufurd, MD, Ruth Fullam, BSC, and Elizabeth Howard, MD (University of Manchester, Manchester, UK);
Pietro Mazzoni, MD, PhD, Karen Marder, MD, MPH, and Paula Wasserman, MA (Columbia University Medical Center, New York, New York, USA);
Rajeev Kumar, MD and Diane Erickson, RN (Colorado Neurological Institute, Englewood, Colorado, USA);
Vicki Wheelock, MD, Terry Tempkin, RNC, MSN, Nicole Mans, BA, MS, and Kathleen Baynes, PhD (University of California Davis, Sacramento, California, USA);
Joseph Jankovic, MD, Christine Hunter, RN, CCRC, and William Ondo, MD (Baylor College of Medicine, Houston, Texas, USA);
Justo Garcia de Yebenes, MD, Monica Bascunana Garde, Marta Fatas, BA, and Asuncion Martinez-Descales (Hospital Ramón y Cajal, Madrid, Spain);
Wayne Martin, MD, Pamela King, BScN, RN, and Satwinder Sran, BSC (University of Alberta, Edmonton, Alberta, Canada);
Anwar Ahmed, PhD, Stephen Rao, PhD, Christine Reece, BS, Janice Zimbelman, PhD, PT, Alexandra Bea, BA, Emily Newman, BA, and Alex Bura, BA (Cleveland Clinic Foundation, Cleveland, Ohio, USA).
Steering Committee
Jane Paulsen, PhD, Principal Investigator, Eric A. Epping, MD, PhD, Hans Johnson, PhD, Megan M. Smith, PhD, Janet Williams, PhD, RN, FAAN, Leigh Beglinger, PhD, James Mills, MS (University of Iowa Hospitals and Clinics, Iowa City, IA); Elizabeth Aylward, PhD (Seattle Children's Research Institute, WA); Kevin Biglan, MD (University of Rochester, Rochester, NY); Blair Leavitt, MD (University of British Columbia, Vancouver, BC, Canada); Marcy MacDonald, PhD (Massachusetts General Hospital); Martha Nance, MD (Hennepin County Medical Center, Minneapolis, MN); and Cheryl Erwin, JD, PhD (University of Texas Medical School at Houston).
Scientific Sections
Bio Markers: Blair R. Leavitt, MDCM, FRCPC (Chair) and Michael Hayden, PhD (University of British Columbia); Stefano DiDonato, MD (Neurological Institute “C. Besta,” Italy); Ken Evans, PhD (Ontario Cancer Biomarker Network); Wayne R. Matson, PhD (VA Medical Center, Bedford, MA); Asa Peterson, MD, PhD (Lund University, Sweden), Sarah Tabrizi, MD, PhD (National Hospital for Neurology and Neurology and Neurosurgery, London); Beth Borowsky, PhD (CHDI); Andrew Juhl, BS, James Mills, MS, and David Weir, BSc (University of British Columbia).
Brain: Jean Paul Vonsattell, PhD (Chair), and Carol Moskowitz, ANP, MS (Columbia University Medical Center); Anne Leserman, MSW, LISW, Lynn Schaul, BA, and Stacie Vik, BA (University of Iowa).
Cognitive: Deborah Harrington, PhD (Chair), Gabriel Castillo, BS, Jessica Morison, BS, and Jason Reed, BS (University of California, San Diego), Michael Diaz, PhD, Ian Dobbins, PhD, Tamara Hershey, PhD, Erin Foster, OTD, and Deborah Moore, BA (Washington University Cognitive Science Battery Development); Holly Westervelt, PhD (Chair, Quality Control and Training, Alpert Medical School of Brown University), Jennifer Davis, PhD, and Geoff Tremont, PhD, MS (Scientific Consultants, Alpert Medical School of Brown University); Megan M. Smith, PhD (Chair, Administration), David J. Moser, PhD, Leigh J. Beglinger, PhD, Kelly Rowe, and Danielle Theriault, BS (University of Iowa); Carissa Gehl, PhD (VA Medical Center, Iowa City, IA); Kirsty Matheson (University of Aberdeen); Karen Siedlecki, PhD (Fordham University); Marleen Van Walsem (EHDN); Susan Bonner, BA, Greg Elias, BA, and Melanie Faust, BS (Rhode Island Hospital); Beth Borowski, PhD (CHDI); Noelle Carlozzi (University of Michigan); Kevin Duff, PhD (University of Utah); Nellie Georgiou-Karistianis (St. Vincent's Hospital, The University of Melbourne, Australia); Julie Stout, PhD (Monash University, Melbourne, Australia); Herwig Lange (Air-Rahazentrum); and Kate Papp (University of Connecticut).
Functional: Janet Williams, PhD (Chair), Leigh J. Beglinger, PhD, Anne Leserman, MSW, LISW, Eunyoe Ro, MA, Lee Anna Clark, Nancy Downing, Joan Laing, PhD, Kristine Rees, BA, and Stacie Vik, BA (University of Iowa); Rebecca Ready, PhD (University of Massachusetts); Anthony Vaccarino, PhD (Ontario Cancer Biomarker Network); Sarah Farias, PhD (University of California, Davis); Noelle Carlozzi, PhD (University of Michigan); and Carissa Gehl, PhD (VA Medical Center, Iowa City, IA).
Genetics: Marcy MacDonald, PhD (Co-Chair), Jim Gusella, PhD, and Rick Myers, PhD (Massachusetts General Hospital); Michael Hayden, PhD (University of British Columbia); Tom Wassink, MD (Co-Chair) Eric A. Epping, MD, PhD, Andrew Juhl, BA, James Mills, MS, and Kai Wang, PhD (University of Iowa); Zosia Miedzybrodzka, MD, PhD (University of Aberdeen); and Christopher Ross, MD, PhD (Johns Hopkins University).
Imaging: Administrative: Ron Pierson, PhD (Chair), Kathy Jones, BS, Jacquie Marietta, BS, William McDowell, AA, Greg Harris, BS, Eun Young Kim, MS, Hans Johnson, PhD, and Thomas Wassink, MD (University of Iowa); John Ashburner, PhD (Functional Imaging Lab, London); Steve Potkin, MD (University of California, Irvine); and Arthur Toga, PhD (University of California, Los Angeles). Striatal: Elizabeth Aylward, PhD (Chair, Seattle Children's Research Institute). Surface Analysis: Eric Axelson, BSE (University of Iowa). Shape Analysis: Christopher A. Ross (Chair), MD, PhD, Michael Miller, PhD, and Sarah Reading, MD (Johns Hopkins University); Mirza Faisal Beg, PhD (Simon Fraser University). DTI: Vincent A. Magnotta, PhD (Chair, University of Iowa); Karl Helmer, PhD (Massachusetts General Hospital); Kelvin Lim, MD (University of Ulm, Germany); Mark Lowe, PhD (Cleveland Clinic); Sasumu Mori, PhD (Johns Hopkins University); Allen Song, PhD (Duke University); and Jessica Turner, PhD (University of California, Irvine). fMRI: Steve Rao, PhD (Chair), Erik Beall, PhD, Katherine Koenig, PhD, Michael Phillips, MD, Christine Reece, BS, and Jan Zimbelman, PhD, PT (Cleveland Clinic); and April Bryant (University of Iowa).
Motor: Kevin Biglan, MD (University of Rochester), Karen Marder, MD (Columbia University), and Jody Corey-Bloom, MD, PhD (University of California, San Diego) all Co-Chairs; Michael Geschwind, MD, PhD (University of California, San Francisco); Ralf Reilmann, MD and Zerka Unds (Muenster, Germany); and Andrew Juhl, BS (University of Iowa).
Psychiatric: Eric A. Epping, MD, PhD (Chair), Nancy Downing, RN, MSN, Jess Fiedorowicz, MD, Robert Robinson, MD, Megan M. Smith, PhD, Leigh Beglinger, PhD, James Mills, MS, Kristine Rees, BA, Adam Ruggle, Stacie Vik, BA, Janet Williams, PhD, Dawei Liu, PhD, David Moser, PhD, and Kelly Rowe (University of Iowa); Karen Anderson, MD (University of Maryland); David Craufurd, MD (University of Manchester); Mark Groves, MD (Columbia University); Anthony Vaccarino, PhD and Ken Evans, PhD (Ontario Cancer Biomarker Network); Hugh Rickards, MD (Queen Elizabeth Psychiatric Hospital); Eric van Duijn, MD (Leiden University Medical Center, Netherlands); Irina Antonijevic, MD, PhD, and Joseph Giuliano (CHDI); Phyllis Chua (The University of Melbourne, Royal Melbourne Hospital); and Kimberly Quaid, PhD (Indiana University School of Medicine).
Core Sections
Statistics: James Mills, MS, Dawei Liu, PhD, Jeffrey Long, PhD, Wenjing Lu, Spencer Lourens, and Ying Zhang, PhD (University of Iowa).
Recruitment/Retention: Martha Nance, MD (Chair, University of Minnesota); Anne Leserman, MSW, LISW, Nicholas Doucette, BA, Mycah Kimble, BA, Patricia Ryan, MSW, LISW, MA, Kelli Thumma, BA, Elijah Waterman, BA, and Jeremy Hinkel, BA (University of Iowa).
Ethics: Cheryl Erwin, JD, PhD, (Chair, McGovern Center for Health, Humanities and the Human Spirit); Eric A. Epping, MD, PhD Janet Williams, PhD, Nicholas Doucette, BA, Anne Leserman, MSW, LISW, James Mills, MS, Lynn Schaul, BA, and Stacie Vik, BA (University of Iowa); Martha Nance, MD (University of Minnesota); and Lisa Hughes, MEd (University of Texas Medical School at Houston).
IT/Management: Hans Johnson, PhD (Chair), R.J. Connell, BS, Karen Pease, BS, Ben Rogers, BA, BSCS, Jim Smith, AS, Shuhua Wu, MCS, Roland Zschiegner, Erin Carney, Bill McKirgan, Mark Scully, and Ryan Wyse (University of Iowa); Jeremy Bockholt (AMBIGroup).
Program Management
Administrative: Chris Werling-Witkoske (Chair), Karla Anderson, BS, Jennifer Rapp, BA, Ann Dudler, Jamy Schumacher, Sean Thompson, BA, Leann Davis, Machelle Henneberry, Greg Ennis, MA, and Stacie Vik, BA (University of Iowa).
Financial: Steve Blanchard, MSHA, Kelsey Montross, BA, and Phil Danzer (University of Iowa).
Abbreviations
- 8OHdG
8-hydroxydeoxyguanosine
- HD
Huntington disease
- CAG
cytosine–adenine–guanine
- ROS
radical oxygen species
- UHDRS
Unified Huntington's Disease Rating Scale
- DCL
diagnostic confidence level
- CAP
CAG length/age product
- LCECA
Liquid Chromatography Electrochemical Array
- MRM
multiple-reaction-monitoring
- PPD
pharmaceutical product development
- LMER
linear mixed effects regression
- LRT
likelihood ratio test
- AIC
Akaike's information criterion
- LR
linear regression
Appendix A.
This Appendix provides details of the statistical models considered in the analysis. As mentioned, the LMER analysis considered three models. The most complex model is considered here, as the simpler models omit terms included in the full model.
The time metric for the LMER models was duration (dur), defined as the current age minus the age at study entry. Suppose that agehij is the current age for the ith participant (i=1, …, 77) in the hth site(h=1, …, 13). Then durhij=agehij –agehi1. The most complex Slope model had intercept and slope effects for CAP group and included the same for the covariates of age at entry (agehi1), years of education (educhi), and gender (genhi). Define dummy variables for the CAP groups as capLhi =1 if in the Low group and zero otherwise, capMhi =1 if in the Medium group and zero otherwise, and capHhi if in the High group and zero otherwise. If yhij is the 8OHdG value for the ith participant in the hth site at the jth time point, then Model 3 (see Table 4 and text) is:
The intercept CAP group effects are indexed by β5,β6, and β7, whereas the slope effects are indexed by β11, β12, and β13. The intercept for the Control group is β0C=β0, for the Low group it is β0L=β0+β5, etc. The slope for the Control group is β1C=β1, for the Low group it is β1L=β1+β11, etc. In the model, ah is the random effect for site assumed to be normally distributed with a mean of zero and variance σ2a; b0hi is the random intercept, and b1hi is the random slope. The random intercept and slope are assumed to have zero-means with variance– covariance matrix G, a joint-normal distribution, and orthogonal to ah. Finally, εhij is random error assumed to have zero-mean, a normal distribution, constant variance σ2ε, and orthogonal to the random effects. In the LMER analysis, Model 2 omits all the duration interaction terms for the CAP groups, and Model 1 omits all the CAP group terms. The LR models for the cross-sectional data omit the random effects.
Corrigendum
At the request of CHDI, the authors agree to the corrigendum stated below:
The data generated at PPD/Caprion using the LC-MS/MS-based assay and directed and founded by CHDI Foundation that are reported here were included without the permission of CHDI. Further, CHDI researchers were not consulted about the presentation, analysis, or interpretation of those or any other data included in this article or any conclusions expressed here. Both of these omissions were inadvertent and are regretted.
Footnotes
The authors report no conflicts of interest.
Contributor Information
Jeffrey D. Long, Email: jeffrey-long@uiowa.edu.
Wayne R. Matson, Email: wayne.matson@va.gov.
Andrew R. Juhl, Email: andrew-juhl@uiowa.edu.
Blair R. Leavitt, Email: bleavitt@cmmt.ubc.ca.
Jane S. Paulsen, Email: jane-paulsen@uiowa.edu.
References
- Akaike H. Information theory as an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second International Symposium on Information Theory. Akademia Kiado; Budapest, Hungary: 1973. pp. 267–281. [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control AC. 1974;19:716–723. [Google Scholar]
- Anderson DR. Model-based Inference in the Life Sciences: A Primer on Evidence. Springer; NewYork; London: 2008. [Google Scholar]
- Beglinger LJ, Paulsen JS, Watson DB, Wang C, Duff K, Langbehn DR, et al. Obsessive and compulsive symptoms in prediagnosed Huntington's disease. J Clin Psychiatry. 2008;69:1758–1765. doi: 10.4088/jcp.v69n1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biglan KM, Ross CA, Langbehn DR, Aylward EH, Stout JC, Queller S, et al. Motor abnormalities in premanifest persons with Huntington's disease: the PREDICT-HD study. Mov Disord. 2009;24:1763–1772. doi: 10.1002/mds.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov MB, Beal MF, McCabe DR, Griffin RM, Matson WR. A carbon column-based liquid chromatography electrochemical approach to routine 8-hydroxy-2′-deoxyguanosine measurements in urine and other biologic matrices: a one-year evaluation of methods. Free Radic Biol Med. 1999;27:647–666. doi: 10.1016/s0891-5849(99)00113-6. [DOI] [PubMed] [Google Scholar]
- Bogdanov MB, Andreassen OA, Dedeoglu A, Ferrante RJ, Beal MF. Increased oxidative damage to DNA in a transgenic mouse model of Huntington's disease. J Neurochem. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- Browne SE, Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, et al. Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model Selection and Multimodel Inference. Springer; New York: 2002. [Google Scholar]
- Chen CM, Wu YR, Cheng ML, Liu JL, Lee YM, Lee PW, et al. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington's disease patients. Biochem Biophys Res Commun. 2007;359:335–340. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- Duff K, Paulsen JS, Beglinger LJ, Langbehn DR, Stout JC. Psychiatric symptoms in Huntington's disease before diagnosis: the PREDICT-HD study. Biol Psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Duff K, Paulsen J, Mills J, Beglinger LJ, Moser DJ, Smith MM, et al. Mild cognitive impairment in prediagnosed Huntington disease. Neurology. 2010;75:500–507. doi: 10.1212/WNL.0b013e3181eccfa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, et al. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2′dG. Neurology. 2006;66:250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai C. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- Kikuchi A, Takeda A, Onodera H, Kimpara T, Hisanaga K, Sato N, et al. Systemic increase of oxidative nucleic acid damage in Parkinson's disease and multiple system atrophy. Neurobiol Dis. 2002;9:244–248. doi: 10.1006/nbdi.2002.0466. [DOI] [PubMed] [Google Scholar]
- Klepac N, Relja M, Klepac R, Hecimovic S, Babic T, Trkulja V. Oxidative stress parameters in plasma of Huntington's disease patients, asymptomatic Huntington's disease gene carriers and healthy subjects: a cross-sectional study. J Neurol. 2007;254:1676–1683. doi: 10.1007/s00415-007-0611-y. [DOI] [PubMed] [Google Scholar]
- Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- Long JD. Longitudinal Data Analysis for the Behavioral Sciences Using R. Sage Publications, Inc.; Thousand Oaks, CA: 2012. [Google Scholar]
- Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–371. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- Matson WR, Langlais P, Volicer L, Gamache PH, Bird E, Mark KA. n-Electrode three-dimensional liquid chromatography with electrochemical detection for determination of neurotransmitters. Clin Chem. 1984;30:1477–1488. [PubMed] [Google Scholar]
- Matson WR, Gamache PG, Beal MF, Bird ED. EC array sensor concepts and data. Life Sci. 1987;41:905–908. doi: 10.1016/0024-3205(87)90192-5. [DOI] [PubMed] [Google Scholar]
- Nopoulos PC, Aylward EH, Ross CA, Mills JA, Langbehn DR, Johnson HJ, et al. Smaller intracranial volume in prodromal Huntington's disease: evidence for abnormal neurodevelopment. Brain. 2011;134:137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Hayden M, Stout JC, Langbehn DR, Aylward E, Ross CA, et al. Preparing for preventive clinical trials: the Predict-HD study. Arch Neurol. 2006;63:883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, et al. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J Neurol Neurosurg Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Nopoulos PC, Aylward E, Ross CA, Johnson H, Magnotta VA, et al. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res Bull. 2010a;82:201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Wang C, Duff K, Barker R, Nance M, Beglinger L, et al. Challenges assessing clinical endpoints in early Huntington disease. Mov Disord. 2010b;25:2595–2603. doi: 10.1002/mds.23337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidori MC, Mecocci P, Browne SE, Senin U, Beal MF. Oxidative damage to mitochondrial DNA in Huntington's disease parietal cortex. Neurosci Lett. 1999;272:53–56. doi: 10.1016/s0304-3940(99)00578-9. [DOI] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Sayre LM, Smith MA, Perry G. Chemistry and biochemistry of oxidative stress in neurodegenerative disease. Curr Med Chem. 2001;8:721–738. doi: 10.2174/0929867013372922. [DOI] [PubMed] [Google Scholar]
- Sorolla MA, Reverter Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E. Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic Biol Med. 2008;45:667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Stout JC, Paulsen JS, Queller S, Solomon AC, Whitlock KB, Campbell JC, et al. Neurocognitive signs in prodromal Huntington disease. Neuropsychology. 2011;25:1–14. doi: 10.1037/a0020937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, et al. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. J Environ Sci Health C. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. Springer; New York: 2000. [Google Scholar]
- Zhang Y, Long JD, Mills J, Warner JH, Lu W, Paulsen JS, et al. Indexing disease progression at study entry with individuals at-risk for Huntington disease. Am J Med Genet B Neuropsychiatr Genet. 2011;156:751–763. doi: 10.1002/ajmg.b.31232. [DOI] [PMC free article] [PubMed] [Google Scholar]