Abstract
Objectives
FKBP51 (51 kDa immunophilin) acts as a modulator of the glucocorticoid receptor and a negative regulator of the Akt pathway. Genetic variation in FKBP5 plays a role in antidepressant response. The aim of this study was to comprehensively assess the role of genetic variation in FKBP5, identified by both Sanger and Next Generation DNA resequencing, as well as genome-wide single nucleotide polymorphisms (SNPs) associated with FKBP5 expression in the response to the selective serotonin reuptake inhibitor (SSRI) treatment of major depressive disorder.
Methods
We identified 657 SNPs in FKBP5 by Next Generation sequencing of 96 DNA samples from white patients, and 149 SNPs were selected for the genotyping together with 235 SNPs that were trans-associated with variation in FKBP5 expression in lymphoblastoid cells. A total of 529 DNA samples from the Mayo Clinic PGRN-SSRI Pharmacogenomic trial for which genome-wide SNPs had already been obtained were genotyped for these 384 SNPs, and associations with treatment outcomes were determined. The most significant SNPs were genotyped using 96 DNA samples from white non-Hispanic patients of the NIMH-supported Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study to attempt replication, followed by functional genomic studies.
Results
Genotype–phenotype association analysis indicated that rs352428 was associated with both 8-week treatment response in the Mayo study (odds ratio =0.49; P = 0.003) and 6-week response in the STAR*D replication study (odds ratio = 0.74; P =0.05). The electrophoresis mobility shift assay and the reporter gene assay confirmed the possible role of this SNP in transcription regulation.
Conclusion
This comprehensive FKBP5 sequence study provides insight into the role of common genetic polymorphisms that might influence SSRI treatment outcomes in major depressive disorder patients.
Keywords: FKBP5, genotype–phenotype association, major depressive disorder, Next Generation DNA resequencing, selective serotonin reuptake inhibitor, single nucleotide polymorphism
Introduction
Major depressive disorder (MDD) is a common psychiatric disease with an estimated incidence of 16% in the general population of the USA [1]. Selective serotonin reuptake inhibitors (SSRIs) are one of the most widely prescribed classes of antidepressant drugs [2]. Clinical trials have shown large individual variation in SSRI treatment outcomes, with approximately half of the treated patients failing to benefit from therapy and many developing undesirable drug-related side effects [3]. FKBP5 encodes the FKBP51 protein, a member of the family of large immunophilins [4]. Recently, we reported that FKBP51 acted as a scaffolding protein regulating Akt activity [5]. Activity of Akt has been shown to play a role in a variety of neuronal physiological functions [6–9]. Therefore, alterations in Akt activity might have implications in the development and treatment of psychiatric disorders [10–12].
In addition, it is known that the glucocorticoid receptor (GR) plays a role in stress-related psychiatric disorders, including MDD, probably by affecting the hypothalamic– pituitary–adrenal axis [13–15]. FKBP51 is also a cochaperon for GR maturation, modulating its sensitivity and, thus, playing a role in regulation of the stress response [16]. The GR can increase FKBP5 transcription through intronic GR response elements. An increased FKBP51 level confers elevated GR resistance, completing an ultrashort negative feedback loop on GR sensitivity [17]. Because of the role of FKBP51 in the glucocorticoid pathway and in stress-related disease, previous studies have attempted to assess the role of genetic variation in FKBP5 in MDD and in response to SSRI treatment. These studies reported that sequence variation in the FKBP5 gene may be associated with risk for posttraumatic stress disorder, risk for recurrence of depression, and variation in response to antidepressant therapy [17–23]. FKBP5 has also been reported to be associated with risk for attempted suicide and the occurrence of depressive episodes in bipolar patients [17–23]. Although these studies suggest that variation in the sequence or expression of FKBP5 might be associated with variation in SSRI treatment outcome [5,18,19,24,25], none of them explored the full range of DNA variants present in the gene, and only one study by Binder et al. [18] suggested that one of the potential mechanisms by which those genetic variants might influence FKBP51 function is through their influence on protein levels. Therefore, the aim of the present study was to comprehensively investigate the role of genetic variation in FKBP5, identified by Next Generation DNA sequencing, followed by association studies carried out on depressed patients treated with SSRIs and functional characterization of selected single nucleotide polymorphism (SNPs). SNPs that were associated with SSRI treatment outcome were then genotyped in an independent patient cohort, the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) patient cohort. Our results indicate that SNPs associated with FKBP5 expression may be involved in its transcriptional regulation and, ultimately, modulation of clinical outcomes after SSRI therapy of patients with MDD.
Materials and methods
FKBP5 gene resequencing
Both Sanger and Next Generation sequencing were used to resequence FKBP5 (primers listed in Supplementary Table 1, http://links.lww.com/FPC/A572), as described previously [26]. Sanger sequencing was used to resequence all exons, exon–intron splice junctions, and ~1000 bp of the 5′ and 3′ flanking regions using 96 DNA samples from lymphoblastoid cells generated from white American patients included in the ‘Human Variation Panel’ (HD100CAU; Coriell Institute, Camden, New Jersey, USA) [27]. Deep sequencing using an Illumina Next Generation sequencing platform (Genome Analyzer IIx; Illumina, San Diego, California, USA) was performed with the same DNA sample set to resequence a 160 kb genomic region on chromosome 6p21 that contained FKBP5. There was 99.8% sequence concordance between regions resequenced using both methods. For the small number of discordant genotypes, Sanger sequencing was repeated and discordant genotypes were replaced with Sanger sequencing results. The Next Generation sequencing results were also compared with Illumina and Affymetrix genome-wide SNP (Affymetrix, Santa Clara, California, USA) genotyping data obtained using the same DNA samples, and there was 98.4% concordance between these results. In this case, if there were discordant genotypes, the Illumina or Affymetrix genotype data were used. Sanger sequencing was also used to identify variations in FKBP5 in the two other ethnic groups included in the ‘Human Variation Panel’, specifically DNA samples from 96 African Americans (AA) and 96 Han Chinese Americans (HCA; HD100AA and HD100CHI, respectively, Coriell Institute), with regard to exons, splice junctions, and 1000 bp of 5′ and 3′ flanking regions (Supplementary Table 2, http://links.lww.com/FPC/A572).
Expression quantitative trait loci analysis
We have also generated expression array and genomewide SNP data for 287 of the ‘Human Variation Panel’ lymphoblastoid cell lines (LCLs) [28,29]. The SNPs and expression array data have been deposited under the SuperSeries accession number GSE24277. Association analysis for expression and SNP data was carried out using Pearson’s correlations, as described previously [29].
Study patients
DNA for our initial clinical SSRI study was obtained from 529 MDD patients treated with either citalopram or escitalopram in the Mayo Clinic Pharmacogenomics Research Network-Antidepressant Medication Pharmacogenomic Study (Mayo PGRN-AMPS), a study that has been described elsewhere [30]. Specifically, patients had to meet diagnostic criteria for MDD with a Hamilton Depression Rating Scale (Ham-D) score of 14 or higher at baseline to be enrolled. Fourteen of the patients were not white non-Hispanic (WNH) and were excluded from the analysis and three samples failed genotyping, resulting in 512 WNH patients in the final analyses. The design of the Mayo PGRN-AMPS trial was based on that of the large multicenter NIMH-supported STAR*D study, the largest MDD treatment-response study performed to date [31]. STAR*D was designed to assess which treatment strategies, and in what order, were most effective in depression management, always beginning with an SSRI, citalopram, a drug that was also used in the Mayo study. A total of 960 samples from the STAR*D study were used in our replication study. They were selected because they were from treatment-compliant WNH patients with initial Ham-D scores of 14 or higher at baseline, the entry criteria for our study.
Selective serotonin reuptake inhibitors outcome phenotypes
Treatment outcomes in both the Mayo PGRN-AMPS and the STAR*D studies were assessed using the 16-item Quick Inventory of Depressive Symptomatology-Clinician rating scores. Treatment outcome phenotypes that were analyzed during the association studies included ‘response’ (defined as ≥ 50% reduction in QIDS score from the beginning of treatment to the visit evaluated) and ‘remission’ (defined as a QIDS score of ≤ 5 at the last visit). The ‘response’ and ‘remission’ phenotypes used in this analysis were assessed at both 4 and 8 weeks after starting SSRI therapy for the Mayo PGRN-AMPS and at 4 and 6 weeks for the STAR*D study. The ‘last visit’ for ‘response’ or ‘remission’ phenotypes refers to an analysis of all patients enrolled in the study – that is both those who completed the full 8-week treatment regimen and those who dropped out before the 8-week time point. For ‘last-visit’ analyses, outcomes were defined based on the last observation carried forward. These analyses were not adjusted for time spent in study. Among the patients included in this study, 36% administered citalopram and 64% escitalopram. The drug to be administered was selected by the physician in consultation with the patient. When we compared the outcomes between patients treated with citalopram versus escitalopram, we found no statistically significant differences in remission or response rates between the two groups. Therefore, we did not carry out stratified analyses in the current study.
Single nucleotide polymorphism genotyping
To be included in the analysis of the Mayo PGRN-AMPS samples, SNPs identified from Next Generation sequencing had to have a minor allele frequency (MAF) of 1% or higher and had to pass the Illumina Golden Gate genotyping platform quality control criteria. A total of 149 SNPs of the 657 resequenced in FKBP5 met those criteria and were selected for inclusion in the genotyping panel. We also genotyped 235 SNPs that were transgenome- wide associated with FKBP5 expression [FKBP5 expression Quantitative Trait Locus (eQTL)] in our ‘Human Variation Panel’ LCLs. Similar to the selection of SNPs identified from our resequencing effort, these SNPs also had to pass the Illumina Golden Gate genotyping quality control criteria. This final panel of 384 SNPs was used to genotype the 529 DNA samples using the Illumina BeadXpress platform (Illumina, San Diego, California, USA). These patient samples had also been subjected to genome-wide genotyping using the Illumina 610 BeadChip kit (Illumina) [32]. A TaqMan assay (Applied Biosystems, Foster City, California, USA) was used to perform replication genotyping using STAR*D DNA samples.
Statistical methods
After genotyping of 384 SNPs from our Mayo SSRI patient samples, quality control was also applied to remove SNPs that were significantly deviated from Hardy–Weinberg equilibrium, those with low call rate, which resulted in 340 SNPs remaining in the association analysis. We also removed three samples that failed genotyping. The effect of SNP genotypes on the binary phenotypes of ‘response’ and ‘remission’ in the Mayo PGRN-AMPS trial was assessed using logistic regression models adjusting for possible population stratification using four eigen vectors constructed from the genomewide SNP data [32,33]. The effect of each SNP adjusted for population stratification was tested using a likelihood ratio test. The relationship of the SNPs with percentage change in QIDS from baseline to last visit was assessed using Spearman’s partial correlations and an F-test, in which the phenotype was adjusted for possible population stratification. To replicate Mayo PGRN-AMPS findings, the associations of SNPs of interest were also determined in the STAR*D patient population. Binary phenotypes of ‘response’ and ‘remission’ were tested using likelihood ratio tests. Finally, because association analysis of individual markers can be underpowered for rare markers, we used a novel test, the difference in MAF test, to determine associations of a set of markers with phenotypes. To test the association between groups of SNPs in subregions of FKBP5 and response and remission, variants were analyzed with sliding windows containing between 10 and 50 variants. Detailed statistical methods are described in the Supplementary methods.
Cell culture and transfections
Site-directed mutagenesis was performed to create variant constructs (Arg28, Gln154, and Phe437), using a wildtype (WT) FKBP51 construct as a template, as described previously [34]. Primers are listed in Supplementary Table 3a, (http://links.lww.com/FPC/A572). HEK293T cells [American Type Culture Collection cells were used for transfections with the pIRES-GFP/Flag WT and variant constructs, as well as with the empty vector, using the Lipofectamine2000 protocol (Invitrogen; Life Technology, Grand Island, New York, USA)]. Green fluorescent protein was used to correct for transfection efficiency.
Electrophoresis mobility shift assay
Electrophoresis mobility shift assays (EMSAs) for the rs352428 SNP were performed with nuclear extracts from two glioblastoma cell lines, U-87MG and U251 (ATCC), as well as from a pool of lymphoblastoid cells from healthy individuals (Coriell Institute). Total protein concentrations were assayed using the Bradford method. EMSAs were performed using the LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, Illinois, USA), as described previously [34]. Oligonucleotide sequences (sense and antisense) for WT and variant sequences of the rs352428 (G/A) SNP are listed in Supplementary Table 3b (http://links.lww.com/FPC/A572). For competitive assays, a 400-fold excess of unlabeled probe was added to the reaction mixture.
Reporter gene assay
A 231 bp region surrounding the rs352428 SNP was amplified using DNA isolated from LCLs containing WT or variant genotypes. Primer sequences are listed in Supplementary Table 3c (http://links.lww.com/FPC/A572). The PCR product was cloned into the pGL3-promoter vector (Promega, Madison, Wisconsin, USA). DNA sequences were verified by sequencing both strands. Vector without an insert was used as a control. Specifically, U-87MG and U251 cells were transfected with WT and variant constructs together with a pRL-TK DNA construct encoding Renilla luciferase, as a control for transfection efficiency. Luciferase activity was measured by a dual luciferase activity assay using a TD-20/20 Luminometer (Turner Designs, Sunnyvale, California, USA). Results are expressed as the ratio of firefly luciferase to Renilla luciferase light units, and all values are expressed as a percentage of the pGL3-promoter construct activity. All assays were performed in triplicate.
Results
Single nucleotide polymorphism selection for genotyping
Introduction
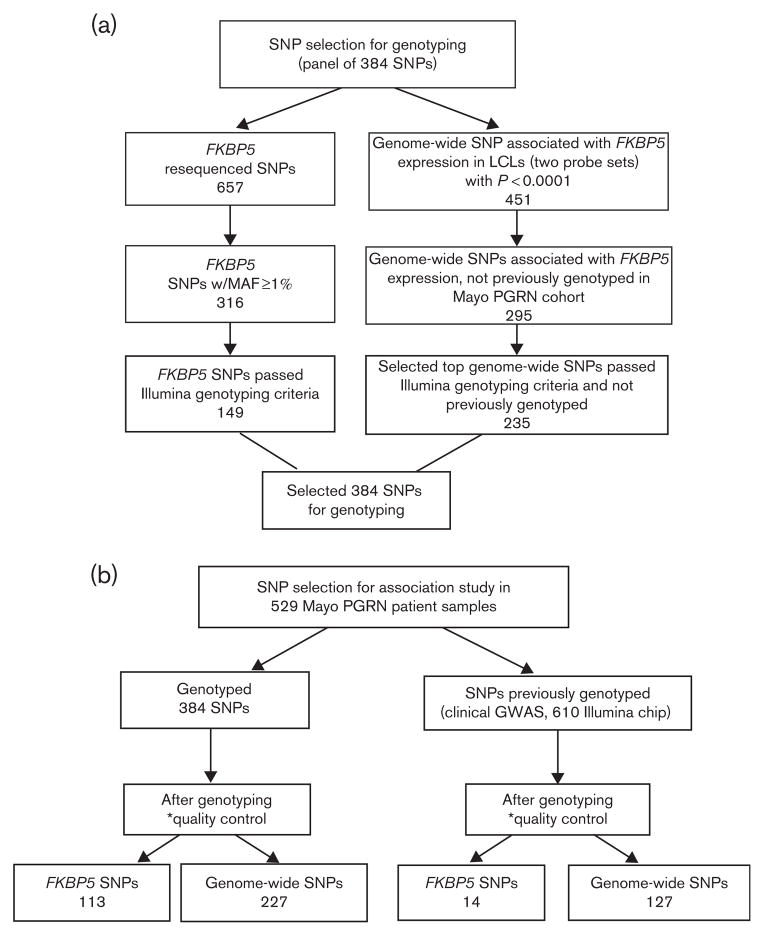
For genotyping, we selected a total of 384 SNPs by combining results from both FKBP5 gene resequencing and FKBP5 eQTL analysis in LCLs. The selection processes are represented in Fig. 1.
Fig. 1.
SNP selection for association analysis. The flow chart outlines the selection criteria for SNPs used for the genotyping and association studies. Genome-wide SNPs are FKBP5 eQTLs. *SNP level quality control. SNPs selected for an association analysis had to meet the quality control measures: minor allele frequency >0.01, per SNP call rate>0.95, and Hardy–Weinberg equilibrium P-value>0.001. eQTLs, expression Quantitative Trait Locus; GWAS, genome-wide association study; LCLs, lymphoblastoid cell lines; MAF, minor allele frequency; Mayo PGRN, Mayo Clinic Pharmacogenomics Research Network; SNPs, single nucleotide polymorphisms.
FKBP5 gene resequencing
Our FKBP5 resequencing covered an area of 160 kb on chromosome 6 and identified 657 SNPs (Supplementary Table 4, http://links.lww.com/FPC/A572), as described in detail by Pelleymounter et al. [26]. The majority of the polymorphisms, including 44 indels (insertions/deletions), were located in introns, flanking regions, and untranslated regions. All but 18 SNPs (indicated in Supplementary Table 4, http://links.lww.com/FPC/A572) were in Hardy–Weinberg equilibrium (P>0.05). In total, 315 SNPs were novel as compared with data from the ‘1000 Genomes Project’ (Phase 1 data [35]) and/or dbSNP. A total of 316 SNPs had MAFs greater or equal to 1%. Sanger sequencing was also used to resequence FKBP5 in 96 additional AAs and HCA DNA samples, respectively (Supplementary Table 2, http://links.lww.com/FPC/A572), resulting in the identification of another 29 novel SNPs, including three nonsynonymous (NS) SNPs (Gly22Arg in HCA, Arg154Gln in AA, and Val437Phe in AA), all with MAFs less than 5% (Supplementary Figure 1, http://links.lww.com/FPC/A572). Only 149 SNPs from FKBP5 resequencing of HCA samples passed the Illumina genotyping criteria and were used for genotyping the Mayo PGRN-AMPS cohort.
FKBP5 expression Quantitative Trait Locus analysis in LCLs
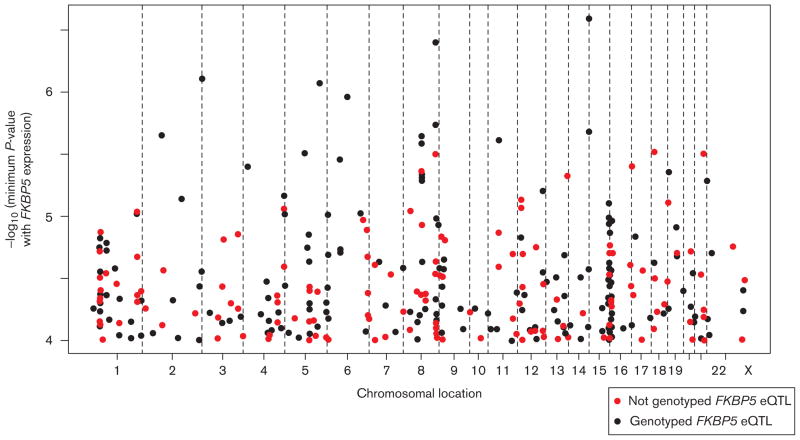
To identify SNPs that might be associated with FKBP5 expression through either cis or trans regulation, we carried out eQTL analysis for all 287 LCLs from all three ethnic groups. A total of 451 SNPs were associated with two of the three FKBP5 Affymetrix expression probe sets (Affymetrix), 224856_at or 224840_at, with P-values less than 0.0001, as illustrated in the expression Manhattan plot in Fig. 2. The third FKBP5 probe set, 224560_at, is not highly correlated with the other two probe sets; therefore, it was excluded from the analysis. Among the SNPs that were associated with FKBP5 expression, 295 had not been genotyped previously in the Mayo PGRN-AMPS cohort using the genome-wide Illumina 610 Beadchip and, therefore, were included in our current genotyping study (black dots in Fig. 2). We selected the top 235 of the 295 genome-wide SNPs that passed the Illumina genotyping criteria to build a panel together with resequenced FKBP5 variants consisting of 384 SNPs for genotyping the Mayo PGRN-AMPS cohort.
Fig. 2.
Graphical representation of the ‘genome-wide’ trans FKBP5 expression Quantitative Trait Locus (eQTL) single nucleotide polymorphism (SNPs; P<10−4). SNPs genotyped in this study are shown as red dots, whereas black dots were obtained from a genome-wide association study of these samples.
Genotype–phenotype analysis of the Mayo Pharmacogenomics Research Network-Antidepressant Medication Pharmacogenomic Study patient samples
The panel consisting of 384 SNPs selected as outlined in Fig. 1a was genotyped in 529 Mayo PGRN-AMPS patient samples. This set represented 149 SNPs from FKBP5 resequencing in addition to 235 FKBP5 eQTL SNPs. In addition, because we had already performed the genome-wide association study (GWAS) genotyping for the Mayo PGRN-AMPS [32], on the basis of our LCL analysis, we were also able to include additional SNPs in FKBP5 as well as those that were associated with FKBP5 expression and were present on the GWAS platform (Fig. 2).
After quality control, genotype–phenotype association analyses for ‘response’ and ‘remission’ during SSRI therapy were carried out with 113 FKBP5 resequenced SNPs, 14 additional FKBP5 SNPs present on the GWAS platform, as well as trans FKBP5 eQTL SNPs (P<10− 4), including 227 genotyped in this study together with 127 SNPs present on the GWAS platform (Fig. 1b). We found that the FKBP5 rs9380524 SNP (A allele) was associated with poor response at both the last visit [P=0.0249; odds ratio (OR)=0.65] and after 8 weeks of treatment (P=0.0175; OR=0.59) and that the 35758265 SNP (genomic location as no rs number has yet been assigned) was associated with percentage change in QIDS-C after the last visit (P=0.042; Table 1). In addition, 22 FKBP5 SNPs, of which 21 were in the same haplotype block, were associated (P<0.05) with better remission at the last visit or after 8 weeks of treatment (OR>1; Table 1). Fifteen trans FKBP5 eQTL SNPs were associated with response at the last visit or after 8 weeks of treatment and/or percentage change in QIDS-C after treatment with P-value less than 0.05 (Table 2), and six SNPs were associated (P<0.05) with remission at the last visit or after 8 weeks of treatment (Table 2). These FKBP5 eQTL SNPs were present in 14 different annotated genes, and three SNPs, rs235317, rs17818663, and rs4964463, had a P-value less than 0.05 for both phenotypes – that is, remission (last visit or 8 weeks) and response (last visit or 8 weeks). None of the associations were significant after correction for multiple testing.
Table 1.
Top FKBP5 SNP association results (P < 0.05) for response, remission, and percentage change in QIDS-C after SSRI treatment (on the basis of logistic regression adjustment for four eigen vectors)
| FKBP5 SNP | Location | Position | Response (last visit)
|
Response (8 weeks)
|
Percentage change in QIDS
|
|||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI)a | P-valuea | OR (95% CI)a | P-valuea | Spearmana | P-valuea | |||
| FKBP5 SNPs associated with the response outcome and percentage change in QIDS-C after SSRI treatment (P < 0.05) | ||||||||
| rs9380524 | Intron 3 | 35589070 | 0.65 (0.44–0.95) | 0.025 | 0.59 (0.38–0.91) | 0.02 | NS | NS |
| 35758265 | Intron 1 | 35650287 | NS | NS | NS | NS | 0.09 | 0.042 |
| FKBP5 SNP | Location | Position | Remission (last visit)
|
Remission (8 weeks only)
|
||||
| OR (95% CI)a | P-valuea | OR (95% CI)a | P-valuea | |||||
|
| ||||||||
| FKBP5 SNPs associated with the remission outcome after SSRI treatment (P < 0.05) | ||||||||
| 35658181 | Intron 8 | 35550203 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| 35660167 | Intron 8 | 35552189 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| 35673999 | Intron 5 | 35566021 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| 35678460 | Intron 5 | 35570482 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| 35686829 | Intron 5 | 35578851 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| 35689604 | Intron 5 | 35581626 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs11966198 | Intron 5 | 35575656 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs16878806 | Intron 5 | 35569119 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs16879318 | Intron 3 | 35590391 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs28675670 | Intron 3 | 35601529 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs34866878 | Exon 10 | 35544942 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs41270080 | 3′UTR | 35542045 | 2.34 (0.99–5.54) | 0.05 | 3.22 (1.13–9.17) | 0.02 | ||
| rs45586932 | Intron 10 | 35543995 | NS | NS | 3 (1.04–8.61) | 0.03 | ||
| rs59320339 | Intron 8 | 35550915 | NS | NS | 3 (1.04–8.61) | 0.03 | ||
| rs16878591 | Intron 8 | 35552627 | NS | NS | 3 (1.04–8.61) | 0.03 | ||
| 35692412 | Intron 5 | 35584434 | NS | NS | 3 (1.04–8.61) | 0.03 | ||
| 35702644 | Intron 3 | 35594666 | NS | NS | 3 (1.04–8.61) | 0.03 | ||
| rs12110366 | Intron 2 | 35610340 | NS | NS | 2.98 (1.04–8.56) | 0.03 | ||
| 35672403 | Intron 6 | 35564425 | NS | NS | 3.21 (1.13–9.14) | 0.03 | ||
| 35751398 | Intron 1 | 35643420 | NS | NS | 3.46 (1.09–11) | 0.04 | ||
| rs2092427 | Intron 1 | 35622207 | NS | NS | 2.99 (1.04–8.61) | 0.05 | ||
| 35696209 | Intron 3 | 35588231 | NS | NS | 3.22 (1.13–9.17) | 0.02 | ||
Highlighted in bold are the SNPs selected for replication study.
CI, confidence interval; OR, odds ratio; QIDS-C, Quick Inventory of Depressive Symptomatology-Clinician Rated; SNP, single nucleotide polymorphism; SSRI, selective serotonin reuptake inhibitor.
Adjusted for population stratification.
Table 2.
Top FKBP5 eQTL (P < 0.05) association results for response, remission, and percentage change in QIDS-C after SSRI treatment (on the basis of logistic regression adjustment for four eigen vectors)
| rs SNP | Chr. | Annotated gene | Position | Response (last visit)
|
Response (8 weeks)
|
% change in QIDS-C
|
|||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | Spearman | P-value | ||||
| FKBP5 eQTLs associated with the response outcome and percent change in QIDS-C after SSRI treatment (P < 0.05) | |||||||||
| rs11045870 | 12 | SLCO1B1 | 21371075 | 0.71 (0.51–0.98) | 0.034 | NS | NS | NA | NS |
| rs11045871 | 12 | SLCO1B1 | 21372224 | 0.71 (0.51–0.98) | 0.034 | NS | NS | NA | NS |
| rs235317 | 21 | PTTG1P | 46275047 | 0.76(0.58–0.99) | 0.041 | 0.64 (0.47–0.87) | 0.005 | NA | NS |
| rs7960384 | 12 | – | 21395908 | 0.72 (0.52–1.00) | 0.049 | NS | NS | NA | NS |
| rs4964463 | 12 | POLR3B | 106789188 | 1.58 (1.07–2.32) | 0.021 | 1.64 (1.02–2.62) | 0.040 | 0.080 | 0.006 |
| rs4603275 | 11 | CNTN5 | 98320413 | 0.73 (0.54–0.99) | 0.041 | NS | NS | NA | NS |
| rs6595125 | 5 | DTWD2 | 117748700 | 1.31 (1.02–1.68) | 0.038 | 1.38 (1.02–1.86) | 0.038 | NA | NS |
| rs7173166 | 15 | BNC1 | 83934748 | NS | NS | 0.68 (0.50–0.94) | 0.021 | NA | NS |
| rs8027193 | 15 | HDGFRP3 | 83879618 | NS | NS | 0.67 (0.51–0.97) | 0.027 | NA | NS |
| rs4341707 | 15 | BNC1 | 83927857 | NS | NS | 0.71 (0.52–0.98) | 0.039 | NA | NS |
| rs17818663 | 9 | – | 13833407 | NS | NS | 0.65 (0.43–0.98) | 0.043 | NA | NS |
| rs352428 | 8 | FZD3 | 28478892 | NS | NS | 0.49 (0.32–0.76) | 0.002 | NA | NS |
| rs10965529 | 9 | DMRTA1 | 22943509 | 0.75 (0.58–0.98) | 0.031 | 0.66 (0.48–0.90) | 0.008 | NA | NS |
| rs11078539 | 17 | PLD2 | 4715977 | NS | NS | NS | NS | − 0.103 | 0.020 |
| rs2784113 | 1 | PTPRC | 198773703 | NS | NS | 0.71 (0.50–0.99) | 0.046 | NA | NS |
| rs SNP | Chr. | Annotated gene | Position | Remission (last visit)
|
Remission (8 weeks)
|
||||
| OR (95% CI) | P-value | OR (95% CI) | P-value | ||||||
|
| |||||||||
| FKBP5 eQTLs associated with the remission outcome after SSRI treatment (P < 0.05) | |||||||||
| rs235317 | 21 | PTTG1P | 46275047 | 0.74 (0.56–0.97) | 0.029 | 0.68 (0.50–0.91) | 0.001 | ||
| rs17818663 | 9 | – | 13833407 | NS | NS | 0.64 (0.43–0.96) | 0.030 | ||
| rs6502015 | 17 | TBCD | 80888164 | NS | NS | 1.41 (1.03–1.94) | 0.033 | ||
| rs4964463 | 12 | POLR3B | 106789188 | 1.69 (1.16–2.45) | 0.006 | 1.67 (1.1–2.53) | 0.016 | ||
| rs11650232 | 17 | DLG4 | 7088923 | 1.29 (1.00–1.67) | 0.047 | NS | NS | ||
| rs1479957 | 3 | MAGI1 | 65228662 | NS | NS | 1.65 (1.06–2.57) | 0.026 | ||
Highlighted in bold are the SNPs selected for replication study.
CI, confidence interval; eQTLs, expression quantitative trait locus; OR, odds ratio; QIDS-C, Quick Inventory of Depressive Symptomatology-Clinician Rated; SNP, single nucleotide polymorphism; SSRI, selective serotonin reuptake inhibitor.
Sequenced Treatment Alternatives to Relieve Depression replication
On the basis of the results of the initial genotype– phenotype association analysis, we selected six SNPs for inclusion in the replication study. These SNPs were genotyped in 960 WNH DNA samples from the STAR*D study after excluding noncompliant and low baseline Hamilton-D patients. Replication genotyping was followed by association analysis that included phenotypes similar to those included in the analysis of the Mayo PGRN-AMPS sample set. The six SNPs included the three SNPs in FKBP5 that had the lowest association P-values for the response or remission phenotypes (Table 1), rs9380524 (P=0.0175 and OR=0.588 for response at 8 weeks), rs34866878 (P=0.0194 and OR=3.22 for remission at 8 weeks), and rs16878591 (P=0.0194 and OR=3.22 for remission at 8 weeks). rs34866878 and rs16878591 represented a large haplotype, within which 21 SNPs were associated with the remission phenotype (P<0.05). We chose two SNPs to represent this haplotype block for the replication genotyping panel because rs34866878 was a coding SNP located in the FKBP5 exon 10 and rs16878591 was in strong linkage disequilibrium with other SNPs within that block (mean r2=0.9186). Three SNPs trans-associated with FKBP5 expression in LCLs with the lowest association P-values for SSRI outcomes were also included in the replication study. These three SNPs were rs4964463 (P=0.006 and OR=1.69) for remission at the last visit, rs352428 (P=0.002 and OR=0.49) for response at 8 weeks, and rs235317 (P=0.001 and OR 0.68) for remission at 8 weeks (Table 2). Among the six replication SNPs, only rs352428 had a replicated P-value of 0.05 (OR=0.74) for response at 6 weeks (Table 3). This SNP also showed a consistent direction of effect with OR less than 1 for both the PGRN-AMPS and STAR*D sample sets (Supplementary Figure 2, http://links.lww.com/FPC/A572). Supplementary Figure 2 (http://links.lww.com/FPC/A572) shows OR comparisons for the six SNPs selected for the replication study in the two samples sets, with an OR less than 1 indicating an association with poor response at 8/6 weeks of SSRI treatment. The sliding window method was also used to assess a combination of rare and common SNPs genotyped in FKBP5 and their association with SSRI treatment outcomes (Supplementary Figure 3, http://links.lww.com/FPC/A572). However, no significant findings were observed (the lowest value was P=0.124 for remission at 8 weeks).
Table 3.
Replication study
| rs SNP | Last visit
|
6 weeks
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF | MAF | |||||||||
|
|
|
|
||||||||
| Nonresponders | Responders | n | OR (95% CI) | P-value | Nonresponders | Responders | n | OR (95% CI) | P-value | |
| SNPs associated with the response outcome after SSRI treatment in STAR*D | ||||||||||
| rs16878591 | 0.04 | 0.04 | 930 | 0.87 (0.58–1.31) | 0.50 | 0.05 | 0.04 | 815 | 0.87 (0.57–1.32) | 0.51 |
| rs235317 | 0.32 | 0.32 | 933 | 0.98 (0.8–1.19) | 0.80 | 0.32 | 0.32 | 816 | 1.00 (0.81–1.24) | 0.97 |
| rs34866878 | 0.03 | 0.02 | 917 | 0.76 (0.44–1.33) | 0.33 | 0.03 | 0.02 | 802 | 0.78 (0.43–1.42) | 0.42 |
| rs352428 | 0.13 | 0.10 | 928 | 0.77 (0.58–1.02) | 0.07 | 0.14 | 0.10 | 812 | 0.74 (0.54–1.00) | 0.05 |
| rs4964463 | 0.13 | 0.14 | 926 | 1.08 (0.82–1.41) | 0.59 | 0.14 | 0.14 | 813 | 1.02 (0.76–1.35) | 0.91 |
| rs9380524 | 0.11 | 0.09 | 923 | 0.83 (0.62–1.11) | 0.21 | 0.11 | 0.09 | 808 | 0.85 (0.62–1.18) | 0.33 |
| rs SNP | Last visit
|
6 weeks
|
||||||||
| MAF | MAF | |||||||||
|
|
|
|
||||||||
| Remitters | Nonremitters | n | OR (95% CI) | P-value | Remitters | Nonremitters | n | OR (95% CI) | P-value | |
|
| ||||||||||
| SNPs associated with the remission outcome after SSRI treatment in STAR*D | ||||||||||
| rs16878591 | 0.04 | 0.04 | 933 | 0.97 (0.64–1.47) | 0.89 | 0.04 | 0.04 | 815 | 0.98 (0.64–1.49) | 0.91 |
| rs235317 | 0.31 | 0.33 | 936 | 1.10 (0.90–1.35) | 0.35 | 0.31 | 0.33 | 816 | 1.11 (0.89–1.37) | 0.36 |
| rs34866878 | 0.03 | 0.02 | 920 | 0.57 (0.31–1.06) | 0.06 | 0.03 | 0.02 | 802 | 0.61 (0.32–1.16) | 0.12 |
| rs352428 | 0.13 | 0.10 | 931 | 0.79 (0.59–1.07) | 0.13 | 0.13 | 0.10 | 812 | 0.74 (0.54–1.02) | 0.06 |
| rs4964463 | 0.14 | 0.14 | 929 | 0.98 (0.75–1.28) | 0.87 | 0.15 | 0.13 | 813 | 0.91 (0.68–1.21) | 0.52 |
| rs9380524 | 0.11 | 0.10 | 926 | 0.89 (0.66–1.21) | 0.45 | 0.10 | 0.10 | 808 | 0.93 (0.67–1.28) | 0.65 |
Highlighted in bold is the SNP that shows the lowest P-value for both last visit and 6 weeks response.
Selected SNPs were genotyped in STAR*D white non-Hispanic patient samples (excluding low baseline Ham-D patients) and associated with SSRI treatment outcomes, including response and remission.
CI, confidence interval; MAF, minor allele frequency; OR, odds ratio; SNP, single nucleotide polymorphism; SSRI, selective serotonin reuptake inhibitor; STAR*D, Sequenced Treatment Alternatives to Relieve Depression.
Functional characterization of rs352428
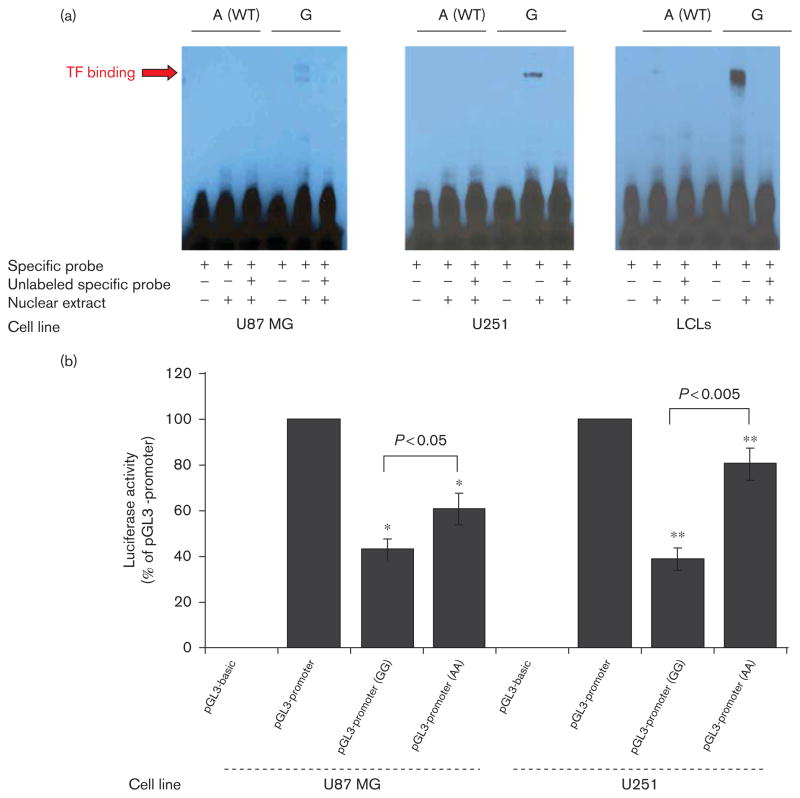
To characterize possible functional consequences of the rs352428 SNP (Table 3 and Supplementary Figure 2, http://links.lww.com/FPC/A572) that was located in an intragenic region on chromosome 8 between FZD3 (~58 kbp) and EXTL3 (~95 kbp), we performed a series of experiments. The TransFast in-silico transcription factor database suggested that transcription factor C/EBPα bound to both A and G alleles for the SNP. However, transcription factor C/EBPβ bound only to the G allele. We then performed EMSA using two human brainderived cell lines, U-87MG and U251, and a pool of non-brain derived cell lines, lymphoblastoid cells. Fig. 3a shows that nuclear extract binding resulted in a ‘shift’ for the rs352428 variant nucleotide. This difference in nuclear protein binding between WT and variant oligonucleotides was present in all three cell lines. We next performed a reporter gene assay to determine the effect of the SNP on transcriptional activity. This region containing the variant SNP sequence resulted in a more than two-fold reduction in luciferase activity as compared with the WT sequence (Fig. 3b). There was also a reduction in luciferase activity when compared with the vector control. A significant decrease in luciferase activity for a construct carrying the rs352428 variant sequence, as compared with WT, might explain the negative correlation of this variant with FKBP5 mRNA expression in LCLs (R= −0.267), observed during our association analysis. These results suggested not only that the region surrounding the rs352428 SNP is transcriptionally active, but also that the SNP could alter binding to transcription factors. In summary, these functional genomic studies identified and validated one region that potentially functions as a ‘silencer’. By affecting the expression level of FKBP5, this regulatory region might also affect physiological and pharmacological function, in this case response to SSRIs.
Fig. 3.
Functional characterization of the rs352428 single nucleotide polymorphism (SNP). (a) Electrophoretic mobility shift assays (EMSAs) with nuclear extract prepared from U87-MG, U251, and a pool from lymphoblastoid cells. A different binding pattern was observed between wild-type (WT) and variant sequences in each case. The arrow indicates the band that was observed with the variant but not the WT sequence. (b) Results from dual luciferase reporter gene assays performed in U87-MG and U251 glioblastoma cell lines. Error bars for each construct represent the average of relative luciferase activity calculated as a % of the pGL3-promoter construct activity obtained during six independent transfections (mean±SEM). * and ** represent T-test P-values for comparing values of pGL3-promoter (AA) and pGL3-promoter (GG) activity. LCLs, lymphoblastoid cell lines; TF, transcription factor.
Functional characterization of Gly22Arg, Arg154Gln, Val437Phe
Only three NS complementary single nucleotide polymorphisms (cSNPs) were observed during our Sanger resequencing study. Arg22 and Phe437 were observed only in HCA samples, with an MAF of 1%, and Gln154 only in the AA group, with a MAF of 2% (Supplementary Table 2, http://links.lww.com/FPC/A572). Because NS cSNPs have the potential to significantly alter protein function [36], we created expression constructs for WT and variant sequences for the NS cSNPs to determine their possible effect on protein function. Quantitative western blot analysis and quantitative real-time-PCR were performed to determine protein and mRNA expression levels (Supplementary Figure 4a and b, http://links.lww.com/FPC/A572). There were no statistically significant differences between WT and variant allozymes for levels of protein or mRNA expression.
Discussion
To our knowledge, this is the first application of in-depth Next Generation resequencing to study response to the treatment of MDD. The treatment of this disease remains challenging. Both environmental and genetic factors can contribute to MDD treatment outcomes. In the present study, we set out to comprehensively study how genetic variation in FKBP5 and genome-wide SNPs associated with FKBP5 expression might play a role in variation in treatment response for MDD. We chose to resequence FKBP5 because of its involvement in the modulation of Akt activity [5], a pathway known to be important in many behavioral phenotypes and physiological functions in the brain [37–41]. In addition, FKBP5 is a modulator of GR sensitivity [42], thus playing a role in the activity of the hypothalamic–pituitary–adrenal axis and, therefore, in the response to stress [43]. Moreover, several previous studies have implicated FKBP5 in stress-related diseases and response to SSRIs [17–20,22, 23,44–52]. However, none of those studies took a comprehensive approach to identify genetic polymorphisms present in the FKBP5 gene. We systematically resequenced FKBP5 by both Next Generation and Sanger sequencing to identify SNPs present in the gene, followed by examining their association with SSRI treatment outcomes in depressed patients enrolled in a large clinical trial. SSRI treatment outcomes were assessed using QIDS and Ham-D scores.
Our resequencing study identified 657 SNPs in FKBP5, 362 of them novel (Supplementary Table 4, http://links.lww.com/FPC/A572). In addition, in AAs and HCAs, we identified 29 novel SNPs, including three NS cSNPs, by Sanger sequencing (Supplementary Table 2, http://links.lww.com/FPC/A572). In our study, we also took advantage of a genomic data-rich panel of 287 LCLs to identify SNPs that were associated with FKBP5 gene expression and combined those SNPs with resequenced FKBP5 SNPs to develop a panel that was used to genotype DNA from Mayo PGRN-AMPS patients treated with SSRIs [32]. Genotype–phenotype association studies for SSRI treatment outcomes in the Mayo PGRN-AMPS cohort patients revealed 24 SNPs within FKBP5 and 19 SNPs trans-associated with FKBP5 expression that were associated with SSRI treatment outcomes with P-values less than 0.05 (Tables 1 and 2). None of these SNPs were significant after correction for multiple testing. We also applied a sliding window analysis, an analysis that takes correction for multiple rare and common SNPs into account to test for association. It is worth mentioning that rs1360780, rs3800373, and rs4713916, SNPs that have previously been reported to be associated with SSRI treatment outcomes in MDD [19,24,25], were not significantly associated with any of the SSRI treatment phenotypes in our study. One of the reasons why we did not observe an association of rs4713916, an SNP reported in a recent meta-analysis to be associated with SSRI response [25], with any response phenotypes in our study could be because of the discrepancies between the previous studies and Mayo PGRN-AMPS, including differences in baseline clinical characteristics of patients, which could contribute to our inability to replicate results from these studies.
In our replication study using STAR*D white non-Hispanic samples, only the FKBP5 eQTL SNP, rs352428, was shown to be associated with response after 6 weeks (P=0.05; Table 3). Although the point of estimate of the effect size in the replication stage is weaker than the effect identified in the discovery stage, which is not unusual as discovery studies tend to have biased effect sizes because of what is known as ‘winner’s curse’, the SNP showed the same OR trend in both studies (OR<1, Supplementary Figure 2, http://links.lww.com/FPC/A572), strongly indicating that it is associated with poor response at 8 or 6 weeks (Mayo PGRN-AMPS or STAR*D sample sets, respectively). rs352428 is an intergenic SNP that maps between FZD3 and EXTL3 on chromosome 8p21. Its chromosomal location has been described as a putative locus for the development of schizophrenia [53–55]. On the basis of the microarray database that is available for our LCLs, this SNP was associated with FKBP5 expression with a P-value of 9.17×10− 6 (R= −0.267) but was not associated with either FZD3 or EXTL3 expression. In addition, the region surrounding this SNP did not encode long noncoding RNA, as evaluated through an in-silico database search (http://www.lncrnadb.org) and through searching a reference catalog of human long noncoding RNA generated by Cabili et al. [56], nor did the region encode a validated miRNA. Our functional EMSA and reporter gene assays indicated that the region surrounding rs352428 was not only transcriptionally active but also showed a striking difference between the A and G allele signals (Fig. 3). The exact mechanisms by which this SNP might influence SSRI response through the regulation of FKBP5 expression will require further studies. However, previous studies have also shown that SNPs resulting in alteration of FKBP5 expression could contribute to the response to treatment in depressed patients, as reported by Binder et al. [18], who found that SNPs that increased the expression of FKBP5 resulted in a good response to treatment. In our case, rs352428 caused a decreased transcriptional activity and low FKBP5 expression and resulted in an association with poor response to SSRIs, an observation that is consistent with a previous finding.
The Mayo PGRN-AMPS trial was designed to mirror the initial phase of the STAR*D study using similar enrollment criteria and using the same SSRI drug (citalopram). However, several factors might contribute to differences between the two studies and, therefore, to the lack of reproducibility of results between the two. Particularly, MDD is often a chronic condition that tends to coexist with substantial psychiatric comorbidity and other medical conditions. Our observations highlight the challenges of performing psychiatric genomic research across studies and also suggest that functional genomic validation might provide a complementary strategy to help validate and characterize the functional consequences of any genomic markers identified [31]. In addition, we also acknowledge that we did not have a placebo arm in our Mayo designed trial; therefore, we could not exclude the possibility that the regulatory SNP is a prognostic factor for outcome. Therefore, to replicate our findings in other independent patient cohorts with similar clinical and demographic characteristics would be desired to better estimate the significance of our initial results.
Conclusion
Our study provides insight into the role of common genetic polymorphisms in FKBP5 that might help predict SSRI treatment response in depression. Further, we evaluated both genetic variants in FKBP5 itself and trans-SNPs associated with FKBP5 expression and their effect on gene transcription. Finally, additional in-depth functional genomic studies are needed to determine their role in SSRI response mechanisms.
Supplementary Material
Acknowledgments
This study was supported by US National Institutes of Health grants K22 CA130828, R01 CA138461, P50 CA102701, U19 GM61388 (The Pharmacogenomics Research Network), R01 GM28157, R01 CA132780, R21 GM86689, as well as by a KL2 Mentored Career Development Award (NCRR Grant KL2TR000136), and a Gerstner Family Mayo Career Development Award in Individualized Medicine. The authors thank the RIKEN Center for Genomic Medicine, Yokohama, Japan, for making it possible for them to use the GWAS data obtained from the Mayo Clinic PGRN-SSRI Pharmacogenomic trial DNA samples. They also thank Luanne Wussow for her help with the preparation of this manuscript.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.pharmacogeneticsandgenomics.com).
Conflicts of interest
Dr Mrazek has developed intellectual property that has been licensed by AssureRx Health and subsequently incorporated into physician decision support software. He has also received research funding from AssureRx Health to create and maintain a bibliographic system designed to monitor the scientific literature. The rest of the authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 2.Preskorn S, Feighner JP, Stanga CY, Ross R. Antidepressants: past, present and future. Berlin; Springer Verlag: 2004. p. 242. [Google Scholar]
- 3.Thase ME, Haight BR, Richard N, Rockett CB, Mitton M, Modell JG, et al. Remission rates following antidepressant therapy with bupropion or selective serotonin reuptake inhibitors: a meta-analysis of original data from 7 randomized controlled trials. J Clin Psychiatry. 2005;66:974–981. doi: 10.4088/jcp.v66n0803. [DOI] [PubMed] [Google Scholar]
- 4.Nair SC, Rimerman RA, Toran EJ, Chen S, Prapapanich V, Butts RN, et al. Molecular cloning of human FKBP51 and comparisons of immunophilin interactions with Hsp90 and progesterone receptor. Mol Cell Biol. 1997;17:594–603. doi: 10.1128/mcb.17.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol. 2004;15:177–182. doi: 10.1016/j.semcdb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3- kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 9.Wymann MP, Zvelebil M, Laffargue M. Phosphoinositide 3-kinase signalling – which way to target? Trends Pharmacol Sci. 2003;24:366–376. doi: 10.1016/S0165-6147(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 10.Beaulieu JM, Gainetdinov RR, Caron MG. Akt/GSK3 signaling in the action of psychotropic drugs. Annu Rev Pharmacol Toxicol. 2009;49:327–347. doi: 10.1146/annurev.pharmtox.011008.145634. [DOI] [PubMed] [Google Scholar]
- 11.Duman RS, Voleti B. Signaling pathways underlying the pathophysiology and treatment of depression: novel mechanisms for rapid-acting agents. Trends Neurosci. 2012;35:47–56. doi: 10.1016/j.tins.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Modulation in activation and expression of phosphatase and tensin homolog on chromosome ten, Akt1, and 3-phosphoinositide-dependent kinase 1: further evidence demonstrating altered phosphoinositide 3-kinase signaling in postmortem brain of suicide subjects. Biol Psychiatry. 2010;67:1017–1025. doi: 10.1016/j.biopsych.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charney DS, Manji HK. Life stress, genes, and depression: multiple pathways lead to increased risk and new opportunities for intervention. Sci STKE. 2004;2004:re5. doi: 10.1126/stke.2252004re5. [DOI] [PubMed] [Google Scholar]
- 14.De Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 15.Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biol Psychiatry. 2001;49:1023–1039. doi: 10.1016/s0006-3223(01)01157-x. [DOI] [PubMed] [Google Scholar]
- 16.Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 17.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34 (Suppl 1):S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 20.Horstmann S, Lucae S, Menke A, Hennings JM, Ising M, Roeske D, et al. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35:727–740. doi: 10.1038/npp.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy A, Gorodetsky E, Yuan Q, Goldman D, Enoch MA. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology. 2010;35:1674–1683. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supriyanto I, Sasada T, Fukutake M, Asano M, Ueno Y, Nagasaki Y, et al. Association of FKBP5 gene haplotypes with completed suicide in the Japanese population. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:252–256. doi: 10.1016/j.pnpbp.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Willour VL, Chen H, Toolan J, Belmonte P, Cutler DJ, Goes FS, et al. Family-based association of FKBP5 in bipolar disorder. Mol Psychiatry. 2009;14:261–268. doi: 10.1038/sj.mp.4002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lekman M, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, et al. The FKBP5-gene in depression and treatment response – an association study in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Cohort. Biol Psychiatry. 2008;63:1103–1110. doi: 10.1016/j.biopsych.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou YF, Wang F, Feng XL, Li WF, Tao JH, Pan FM, et al. Meta-analysis of FKBP5 gene polymorphisms association with treatment response in patients with mood disorders. Neurosci Lett. 2010;484:56–61. doi: 10.1016/j.neulet.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Pelleymounter LL, Moon I, Johnson JA, Laederach A, Halvorsen M, Eckloff B, et al. A novel application of pattern recognition for accurate SNP and indel discovery from high-throughput data: targeted resequencing of the glucocorticoid receptor co-chaperone FKBP5 in a Caucasian population. Mol Genet Metab. 2011;104:457–469. doi: 10.1016/j.ymgme.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Weinshilboum RM. Pharmacogenomics: candidate gene identification, functional validation and mechanisms. Hum Mol Genet. 2008;17 (R2):R174–R179. doi: 10.1093/hmg/ddn270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Fridley BL, Kalari K, Jenkins G, Batzler A, Weinshilboum RM, et al. Gemcitabine and arabinosylcytosin pharmacogenomics: genome-wide association and drug response biomarkers. PLoS One. 2009;4:e7765. doi: 10.1371/journal.pone.0007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20:1482–1492. doi: 10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, et al. Sequenced treatment alternatives to relieve depression (STAR*D): rationale and design. Control Clin Trials. 2004;25:119–142. doi: 10.1016/s0197-2456(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 32.Ji Y, Biernacka JM, Hebbring S, Chai Y, Jenkins GD, Batzler A, et al. Pharmacogenomics of selective serotonin reuptake inhibitor treatment for major depressive disorder: genome-wide associations and functional genomics. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2012.32. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Ellsworth KA, Moon I, Pelleymounter LL, Eckloff BW, Martin YN, et al. Functional genetic polymorphisms in the aromatase gene CYP19 vary the response of breast cancer patients to neoadjuvant therapy with aromatase inhibitors. Cancer Res. 2010;70:319–328. doi: 10.1158/0008-5472.CAN-09-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinshilboum R, Wang L. Pharmacogenetics: inherited variation in amino acid sequence and altered protein quantity. Clin Pharmacol Ther. 2004;75:253–258. doi: 10.1016/j.clpt.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Dummler B, Hemmings BA. Physiological roles of PKB/Akt isoforms in development and disease. Biochem Soc Trans. 2007;35 (Pt 2):231–235. doi: 10.1042/BST0350231. [DOI] [PubMed] [Google Scholar]
- 38.Hsiung SC, Adlersberg M, Arango V, Mann JJ, Tamir H, Liu KP. Attenuated 5-HT1A receptor signaling in brains of suicide victims: involvement of adenylyl cyclase, phosphatidylinositol 3-kinase, Akt and mitogen-activated protein kinase. J Neurochem. 2003;87:182–194. doi: 10.1046/j.1471-4159.2003.01987.x. [DOI] [PubMed] [Google Scholar]
- 39.Karege F, Perroud N, Burkhardt S, Schwald M, Ballmann E, La Harpe R, et al. Alteration in kinase activity but not in protein levels of protein kinase B and glycogen synthase kinase-3beta in ventral prefrontal cortex of depressed suicide victims. Biol Psychiatry. 2007;61:240–245. doi: 10.1016/j.biopsych.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 40.Krishnan V, Han MH, Mazei-Robison M, Iniguez SD, Ables JL, Vialou V, et al. AKT signaling within the ventral tegmental area regulates cellular and behavioral responses to stressful stimuli. Biol Psychiatry. 2008;64:691–700. doi: 10.1016/j.biopsych.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai WS, Xu B, Westphal KG, Paterlini M, Olivier B, Pavlidis P, et al. Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc Natl Acad Sci USA. 2006;103:16906–16911. doi: 10.1073/pnas.0604994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T, et al. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 2003;22:1158–1167. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith SM, Vale WW. The role of the hypothalamic–pituitary–adrenal axis in neuroendocrine responses to stress. Dialogues Clin Neurosci. 2006;8:383–395. doi: 10.31887/DCNS.2006.8.4/ssmith. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brent D, Melhem N, Ferrell R, Emslie G, Wagner KD, Ryan N, et al. Association of FKBP5 polymorphisms with suicidal events in the Treatment of Resistant Depression in Adolescents (TORDIA) study. Am J Psychiatry. 2010;167:190–197. doi: 10.1176/appi.ajp.2009.09040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartmann J, Wagner KV, Liebl C, Scharf SH, Wang XD, Wolf M, et al. The involvement of FK506-binding protein 51 (FKBP5) in the behavioral and neuroendocrine effects of chronic social defeat stress. Neuropharmacology. 2012;62:332–339. doi: 10.1016/j.neuropharm.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 46.Ising M, Depping AM, Siebertz A, Lucae S, Unschuld PG, Kloiber S, et al. Polymorphisms in the FKBP5 gene region modulate recovery from psychosocial stress in healthy controls. Eur J Neurosci. 2008;28:389–398. doi: 10.1111/j.1460-9568.2008.06332.x. [DOI] [PubMed] [Google Scholar]
- 47.Kirchheiner J, Lorch R, Lebedeva E, Seeringer A, Roots I, Sasse J, et al. Genetic variants in FKBP5 affecting response to antidepressant drug treatment. Pharmacogenomics. 2008;9:841–846. doi: 10.2217/14622416.9.7.841. [DOI] [PubMed] [Google Scholar]
- 48.Sarginson JE, Lazzeroni LC, Ryan HS, Schatzberg AF, Murphy GM., Jr FKBP5 polymorphisms and antidepressant response in geriatric depression. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:554–560. doi: 10.1002/ajmg.b.31019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scharf SH, Liebl C, Binder EB, Schmidt MV, Muller MB. Expression and regulation of the Fkbp5 gene in the adult mouse brain. PLoS One. 2011;6:e16883. doi: 10.1371/journal.pone.0016883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatro ET, Everall IP, Kaul M, Achim CL. Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain Res. 2009;1286:1–12. doi: 10.1016/j.brainres.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Touma C, Gassen NC, Herrmann L, Cheung-Flynn J, Bull DR, Ionescu IA, et al. FK506 binding protein 5 shapes stress responsiveness: modulation of neuroendocrine reactivity and coping behavior. Biol Psychiatry. 2011;70:928–936. doi: 10.1016/j.biopsych.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 52.Velders FP, Kuningas M, Kumari M, Dekker MJ, Uitterlinden AG, Kirschbaum C, et al. Genetics of cortisol secretion and depressive symptoms: a candidate gene and genome wide association approach. Psychoneuroendocrinology. 2011;36:1053–1061. doi: 10.1016/j.psyneuen.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, et al. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 54.Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, et al. Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, et al. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3–24 and 20q12.1-11. 23. Am J Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.