Abstract
Pathogenic New World hantaviruses cause severe disease in humans characterized by a vascular leak syndrome, leading to pulmonary oedema and respiratory distress with case fatality rates approaching 40%. Hantaviruses infect microvascular endothelial cells without conspicuous cytopathic effects, indicating that destruction of the endothelium is not a mechanism of disease. In humans, high levels of inflammatory cytokines are present in the lungs of patients that succumb to infection. This, along with other observations, suggests that disease has an immunopathogenic component. Currently the only animal model available to study hantavirus disease is the Syrian hamster, where infection with Andes virus (ANDV), the primary agent of disease in South America, results in disease that closely mimics that seen in humans. Conversely, inoculation of hamsters with a passaged Sin Nombre virus (SNV), the virus responsible for most cases of disease in North America, results in persistent infection with high levels of viral replication. We found that ANDV elicited a stronger innate immune response, whereas SNV elicited a more robust adaptive response in the lung. Additionally, ANDV infection resulted in significant changes in the blood lymphocyte populations. To determine whether the adaptive immune response influences infection outcome, we depleted hamsters of CD4+ and CD8+ T cells before infection with hantaviruses. Depletion resulted in inhibition of virus-specific antibody responses, although the pathogenesis and replication of these viruses were unaltered. These data show that neither hantavirus replication, nor pathogenesis caused by these viruses, is influenced by the adaptive immune response in the Syrian hamster.
Keywords: hantavirus, hantavirus cardiopulmonary syndrome, infectious disease, T cells, zoonosis
Introduction
Hantaviruses are members of the family Bunyaviridae and are the aetiological agents of two distinct human diseases; hantavirus cardiopulmonary syndrome (HCPS) and haemorrhagic fever with renal syndrome in humans. Pathogenic New World hantaviruses are responsible for HCPS, resulting in case fatality rates of approximately 30–40%.1,2 Virus infection is zoonotic and transmission occurs from exposure to dried or aerosolized excreta from the small mammal hosts of these viruses, without an intermediate vector.3 The two most important agents of HCPS are Sin Nombre virus (SNV) and Andes virus (ANDV) from North and South America, respectively.4–6 Virus infection of the rodent hosts of HCPS-causing viruses results in a persistent, yet asymptomatic infection, despite high levels of viral replication within many tissue types, especially the lung.7
Rodent hosts infected with hantaviruses mount an adaptive immune response, as evidenced by the presence of high-titre IgG antibodies specific for the virus, indicating that both B-cell and T-cell responses are elicited. Furthermore, deer mice, the natural host of SNV, mount a regulatory T (Treg) cell response that is probably evolutionarily adapted to limit immunopathology, while at the same time allowing for persistence of the virus.8 This has also been studied in Norway rats, the natural reservoir of Seoul virus, where Treg-cell responses are also important for maintaining viral persistence.9–11
The mechanisms of disease in humans are largely undefined. Hantaviruses predominantly infect microvascular endothelial cells of many tissues, with high levels of antigen found in the lungs and heart.1 Despite high levels of replication, there is a general lack of cytopathology associated with infection and endothelial cells are primarily intact, indicating that disease is not caused by the direct effects of infection.1,12,13 Infection does, however, result in a capillary leak syndrome in the lungs with conspicuous oedema, which in turn manifests in cardiac strain and a shock syndrome. Several mechanisms may contribute to disease, including alterations in the permeability of endothelial cells as a function of cellular dysregulation upon infection, and immunopathogenic mechanisms where robust innate or adaptive immune responses lead to a hyperactive inflammatory response, so leading to disease. The latter is evidenced by the observation that patients that succumb to infection possess high levels of inflammatory cytokines and immune cell infiltration in the lungs.12 Additionally, immune system parameters have been correlated with disease severity and outcome, including tumour necrosis factor promoter activity, interferon-γ levels and HLA genotype.14–18 These studies suggest that more robust immune responses are detrimental and that human disease has an immunopathogenic component.
Currently, only one animal model exists that mimics HCPS and is useful to study infection and pathology caused by hantaviruses: the Syrian hamster.19 Upon infection with ANDV, Syrian hamsters develop a disease consistent with several of the hallmarks of HCPS in humans. We have recently expanded upon the hamster model to include SNV, where inoculation with a virus that has been serially passaged in hamsters results in a persistent infection with high levels of replication without pathology, similar to what is observed in the natural rodent host.20 Comparisons between ANDV and SNV in this model allow for the study of mechanisms underlying pathology and persistence. It has recently been shown that CD4+ or CD8+ T cells do not contribute to disease caused by ANDV in the hamster model of HCPS.21 Herein, we expand these findings by comparing the outcome of infection by both ANDV and SNV, and show that virus replication is unaltered by the presence of the adaptive immune response, and that neither pathology nor persistence is dependent on the activity of T cells, despite differences in the immune responses elicited by these two viruses.
Materials and methods
Viruses and Syrian hamster inoculations
Andes virus strain Chile-9717869, originally isolated from an infected rodent,22 was propagated on VeroE6 cells in Dulbecco's modified Eagle's medium (DMEM) containing 2% fetal bovine serum (FBS). SNV was derived from the homogenate of lungs from SNV strain 77734-infected hamsters after five passages in hamsters as described previously.20 Female Syrian hamsters 6–7 weeks of age (Harlan Laboratories, Indianapolis, IN) were inoculated with ANDV (200 focus forming units) diluted in sterile DMEM, or SNVp5 100 μl of lung homogenate, intranasally while under inhalational isoflurane by delivering 50 μl of virus preparation in each nare using a pipette.
Depletion of T cells from hamsters
Syrian hamsters were injected intraperitoneally while under inhalational isoflurane with 500 μl of a solution containing either a mixture of isotype control antibodies (functional grade mouse IgG1 k and rat IgG2b k), or a mixture of anti-mouse CD4 (clone GK1.5) and anti-rat CD8b (clone 341) antibodies (all from eBioscience, San Diego, CA). Antibodies were diluted in sterile PBS and each hamster received 400 μg of each antibody mixture (200 μg of each antibody).
Tissue preparation and antibody staining for flow cytometry
Single-cell suspensions were generated from the spleens of hamsters by placing approximately half of a dissected spleen in a gentleMACs C tube (Miltenyi Biotec, Bergisch Gladbach, Germany) containing 4 ml dissociation buffer (PBS, 0·5% BSA, 2 mm EDTA) on ice. Spleens were dissociated with a gentleMACs dissociator using program m_spleen-2 (Miltenyi Biotec). Cells were then passed through a 70-μm cell strainer (BD, Franklin Lakes, NJ) into 15-ml conical tubes and centrifuged at 350 g for 7 min at 4° before resuspending in 2 ml of room temperature ACK lysis buffer and incubating for 5 min with periodic inversions to lyse red blood cells (Gibco, Grand Island, NY). The cells were then washed twice in dissociation buffer and counted.
For preparing single-cell suspensions of hamster lung tissue, the lower right lung lobe was excised and placed in a gentleMACs C tube containing 5 ml HEPES buffer supplemented with 2 mg/ml collagenase D (Roche, Basel, Switzerland) and 80 U/ml DNase I (Sigma-Aldrich, St Louis, MO) on ice. The tissue was dissociated using a gentleMACs (program m_lung_01), incubated for 30 min at 37° with periodic inversions, then placed on the gentleMACs for further disruption using program m_lung_02. The cells were then passed through a 70-μm cell strainer into 15-ml conical tubes and centrifuged at 350 g for 7 min at 4°, washed twice in 5 ml dissociation buffer and counted.
To prepare cells for flow cytometry, 1 × 106 cells were resuspended in 50 μl blocking buffer (PBS, 2 mm EDTA, 2% FBS, 2% mouse serum, 2% rat serum) in 96-well plates and incubated on ice for 10 min. Cells were then stained using a mixture of anti-mouse CD4-allophycocyanin (clone GK1.5, 1 : 250), anti-rat CD8b-phycoerythrin (clone 341, 1 : 150), and anti-mouse/rat MHC II (I-Ek)-FITC (clone 14-4-4s, 1 : 1000) antibodies in blocking buffer and incubated on ice in the dark for 15 min. As a control, samples of cells were stained with the appropriate isotype control antibodies using matched fluorophores. All antibodies were purchased from eBioscience. The cells were then washed twice in cold PBS containing 2 mm EDTA and 2% FBS before resuspending cells in 250 μl of 4% paraformaldehyde. The cells were incubated overnight at 4°, centrifuged, and the cell pellet was resuspended in fresh 4% paraformaldehyde and transferred to 2-ml microcentrifuge tubes for removal from high containment following standard operating procedures. Following another overnight incubation, cells were washed twice in PBS containing 2 mm EDTA and 2% FBS and resuspended in this buffer for analysis. Flow cytometry analysis was performed using an LSR II (BD Biosciences, San Jose, CA) and data were acquired using FACSDiva software (BD Biosciences). Data were analysed using FlowJo software (Treestar Inc., Ashland, OR) .
Quantitative real-time reverse transcription-PCR
Hamster tissue samples were excised and placed in 2-ml tubes containing 600 μl RNeasy lysis buffer (RLT) and a single stainless steel bead. Approximately 30 mg of tissue was homogenized using a tissue lyser (Qiagen, Hilden, Germany) before total RNA extraction using an RNeasy kit (Qiagen), following the manufacturer's instructions.
For quantification of viral S-segment RNA, 40 ng total RNA was added to the components of a one-step Rotor-Gene RT-PCR kit (Qiagen) according to the manufacturer's instructions, along with forward and reverse primers and a gene-specific probe, in a final reaction volume of 25 μl, as previously reported.23 Cycle thresholds were compared with cycle thresholds of dilutions of in vitro-transcribed S-segment RNA of known copy numbers, and the absolute copies of S-segment RNA per mass total RNA were extrapolated from this standard curve.
The method for the relative quantification of hamster genes and the sequences of the primers and probes used was previously described.24 Briefly, RNA was added to components of a one-step Rotor-Gene RT-PCR kit, along with gene-specific primers and probes and multiplexed with primers and a probe-specific for RPL18, which served as an internal reference gene. The final reaction volume was 25 μl and thermocycling conditions were 50° for 10 min, 95° for 5 min and 40 cycles consisting of 95° for 5 seconds followed by 60° for 10 seconds. The relative template abundance was calculated using the ΔΔCt method and fold changes are calculated relative to RNA from tissues of uninfected control hamsters using the ΔΔCt method.
Histology and haematology
Tissues were fixed in 10% neutral buffered formalin with two changes, for a minimum of 7 days before removal from high containment. Tissues were placed in cassettes and processed with a Sakura VIP-5 Tissue Tek, on a 12-hr automated schedule, using a graded series of ethanol, xylene and ParaPlast Extra. Embedded tissues were sectioned at 5 μm and dried overnight at 42° before staining. For immunohistochemistry, tissues were stained with a rabbit anti-N polyclonal antibody. Haematology was performed on whole EDTA blood samples collected from hamsters via cardiac puncture. A HemaVet 950FS (Drew Scientific, Barrow-in-Furness, UK) was used to count specific cell populations.
ELISA
Serum was prepared from the whole blood of hamsters infected with SNVp5 and euthanized 28 days post-inoculation (dpi). Recombinant SNV N antigen, kindly provided by Dr Hjelle (UNM-HSC), was suspended in PBS at 2 μg/ml and used to coat ELISA plates (Nunc, Rochester, NY) overnight at 4°. The plates were then blocked for 1 hr in diluent (PBS, 5% milk, 0·1% Tween-20) at room temperature. Serum was then twofold serially diluted in diluent and was incubated for 1 hr at 37°. The plates were washed and incubated with peroxidase-labelled anti-Syrian hamster IgG (H+L; KPL) antibodies diluted 1 : 1000 for 1 hr at 37°. After washing, this was followed by the addition of ABTS peroxidase substrate (KPL Inc., Gaithersburg, MD) for 20 min at room temperature. The optical densities of the wells were then measured spectrophotometrically at 405 nm and the dilution in which wells were positive was calculated to be when the optical density was > 3 standard deviations above the same dilution of control sera.
Biosafety and ethical statement
All work with hantavirus-infected hamsters and potentially infectious materials was conducted in the BSL4 facility at the Rocky Mountain Laboratories, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Sample removal from high containment following inactivation was performed according to standard operating protocols approved by the Institutional Biosafety Committee. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Rocky Mountain Laboratories and performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC) by certified staff in an AAALAC-approved facility.
Results
ANDV is pathogenic, whereas SNV causes an apathogenic infection in Syrian hamsters
Reports have previously shown that infection of Syrian hamsters with ANDV results in pathology and death similar to HCPS observed in humans.19,23 To compare the pathogenic process between ANDV and SNV, hamsters were inoculated intranasally with either ANDV, or a hamster-passaged SNV (SNVp5) that persistently infects Syrian hamsters and replicates to high levels in several tissues, including the lung.20 Hamsters inoculated with ANDV displayed signs of respiratory disease and succumbed to infection with an average time to death of 13·2 days, whereas SNV-inoculated hamsters showed no signs of disease up to 28 dpi (Fig. 1a). We then sought to determine whether differences in virus load between ANDV and SNV might be responsible for the disease caused by ANDV. Every other day, starting at 3 dpi and continuing to 11 dpi, groups of hantavirus-infected hamsters were killed and lung tissues were examined for the amount of viral RNA and pathological changes. ANDV RNA was detectable by 3 dpi and increased throughout the course of infection. SNV was first detectable at 5 dpi. Both viruses ultimately produced similar amounts of viral RNA in the lungs at 11 dpi, although the kinetics varied at the earlier time-points (Fig. 1b). Histologically, hamsters infected with ANDV began to develop perivascular oedema in the lungs at 9 dpi. The oedematous tunica adventitia was frequently infiltrated by lymphocytes and macrophages, with fewer plasma cells and neutrophils. At 11 dpi, hamsters generally demonstrated multifocal interstitial pneumonia characterized by thickening of the alveolar septae by oedema fluid and moderate numbers of macrophages, neutrophils and fewer lymphocytes. There were no histological changes observed in the hamsters infected with SNV (Fig. 1c).
Figure 1.
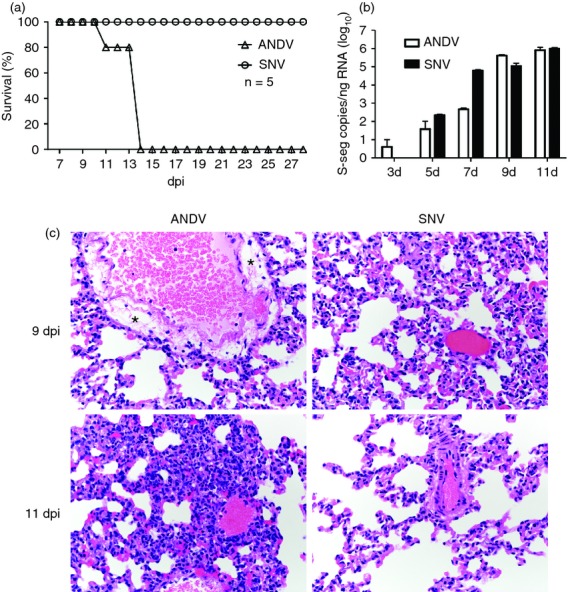
Andes virus (ANDV) is pathogenic, whereas Sin Nombre virus (SNV) causes an apathogenic infection in hamsters. (a) Groups of five hamsters were inoculated intranasally with ANDV or SNV and monitored for signs of disease and survival. (b, c) Hamsters (groups of six) were inoculated with ANDV or SNV and necropsied at the indicated time-points. (b) Lungs of virus-inoculated hamsters were analysed for the presence of viral S-segment RNA using virus-specific quantitative reverse transcription-PCR and in vitro transcribed RNA as standards. Data are represented as the mean and standard deviation of six hamsters per group. (c) Lung tissue was examined histologically after haematoxylin & eosin staining. *Areas of perivascular oedema.
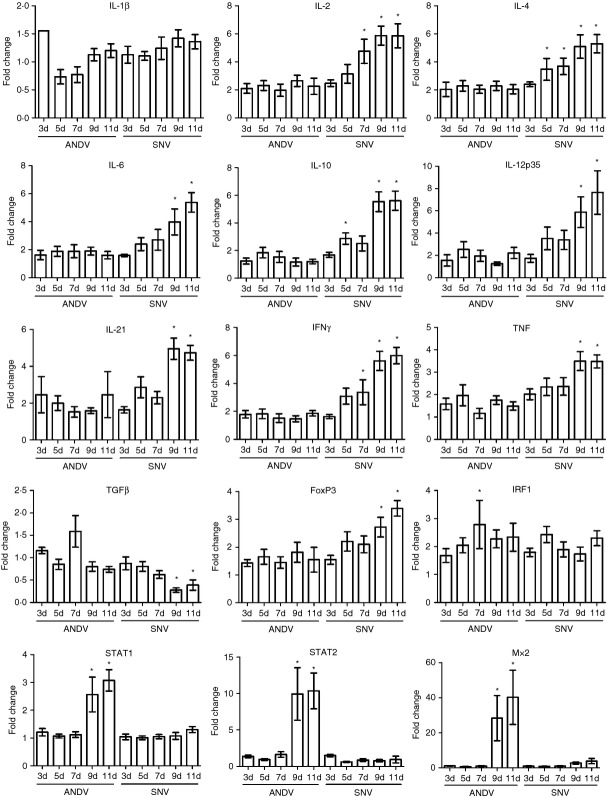
ANDV and SNV differently induce immune gene expression in hamster lungs
It is thought that HCPS is an immune-mediated syndrome. Therefore, we directly compared the expression levels of several immune-related genes by quantitative reverse transcription (qRT-) PCR to elucidate potential mechanistic differences between a pathogenic ANDV infection, and an apathogenic SNV infection. Despite the similar replication kinetics of these viruses in the lung, measured by the accumulation of viral RNA, SNV induced a more robust adaptive immune response, as measured by up-regulation of the RNAs for several interleukins (IL), including IL-2, IL-4, IL-10, IL-12p35 and IL-21, and interferon-γ (Fig. 2). The acute-phase cytokines IL-6, secreted by macrophages and T cells, and tumour necrosis factor, secreted primarily by macrophages, were also up-regulated to a greater extent by SNV at 9 and 11 dpi, corresponding with the dramatic increase in viral RNA accumulation. These genes were only moderately induced by ANDV, despite high levels of viral RNA in the lungs. However, examination of the interferon regulatory factor 1 (IRF1) and signal transducer and activator of transcription-1 (STAT1) and STAT2 transcription factors, as well as the innate antiviral gene myxovirus resistance gene-2 (Mx2), showed that ANDV induced a robust innate immune response at 9 and 11 dpi, corresponding with increased viral replication. SNV induced minimal levels of Mx2 at 9 and 11 dpi, and STAT1 and STAT2 were not induced at any time-point.
Figure 2.
Andes virus (ANDV) and Sin Nombre virus (SNV) differently induce immune gene expression in hamster lungs. Groups of six hamsters were inoculated with SNV or ANDV, or Dulbecco's modified Eagle's medium (DMEM) as a control. Hamsters were killed at the indicated time-points and total RNA was extracted from the lung tissue for quantitative reverse transcription-PCR analysis of host genes. Cycle threshold values were normalized to an internal reference gene (RPL18) and fold changes were calculated relative to control hamsters inoculated with DMEM. Mean and SEM are shown. A one-way analysis of variance with a Dunnett post-test was used to compare the individual groups with control hamsters (*P < 0·05).
Hantaviruses induce immunoreactivity in Syrian hamsters
To determine whether ANDV and SNV differentially induce the proliferation of adaptive immune response cells in hamsters, immune cell populations were measured 11 dpi, a time when hamsters display pathology and soon before they succumb to ANDV-induced disease. The total white blood cell counts in the blood of infected hamsters were similar to those in uninfected control hamsters, and were similar between infection groups. However, ANDV infection resulted in a significant increase in the absolute number of neutrophils (1·9-fold), a decrease in total lymphocytes (1·5-fold), and a decrease in the number of monocytes (3·0-fold) in the blood, compared with control hamsters. Values for SNV-infected hamsters were not different from uninfected controls. Lymphocyte, eosinophil and basophil numbers were similar to control hamsters and did not differ between SNV-infected and ANDV-infected hamsters (Fig. 3a). All hamsters infected with ANDV displayed obvious splenomegaly upon gross examination and this was reflected in the cell counts obtained from this tissue, where spleens from ANDV-infected animals had approximately 30–40% more cells after red blood cell lysis than SNV-infected hamsters, which were only slightly elevated compared with control hamsters (data not shown). The absolute number of cells expressing CD4, CD8 and MHC II were measured by flow cytometry in these samples. Total CD4+ T cells were increased by approximately 1·8-fold in the spleens of hamsters infected with either ANDV or SNV, compared with uninfected controls. Slight elevations in CD8+ T cells were measured in hamsters with both inocula. MHC II-expressing cells were elevated 1·3-fold upon SNV infection and 1·7-fold upon ANDV infection (Fig. 3b).
Figure 3.
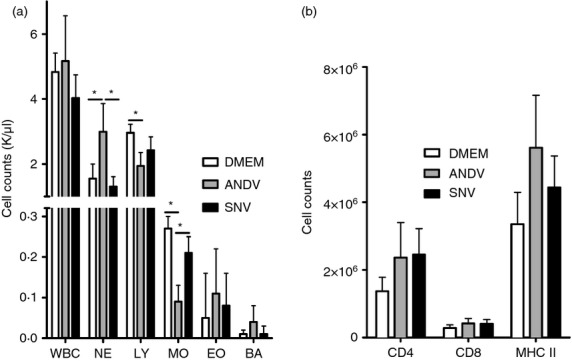
Hantavirus infection results in immune cell expansion. Hamsters inoculated with Sin Nombre virus (SNV) or Andes virus (ANDV), or Dulbecco's modified Eagle's medium (DMEM; control) were euthanized 11 days post-infection (dpi). (a) Whole blood was used to measure immune cell parameters by haematology (WBC, white blood cells; NE, neutrophils; LY, lymphocytes; MO, macrophages; EO, eosinophils; BA, basophils). (b) Single-cell suspensions were prepared from the spleens and cells were counted after red blood cell lysis. The cells were then stained for CD4, CD8 and MHC II expression using fluorescently conjugated antibodies, and absolute numbers of these cell types were measured using flow cytometry. Bars represent the mean and standard deviation of six hamsters per group.
T-cell depletion is efficacious in Syrian hamsters
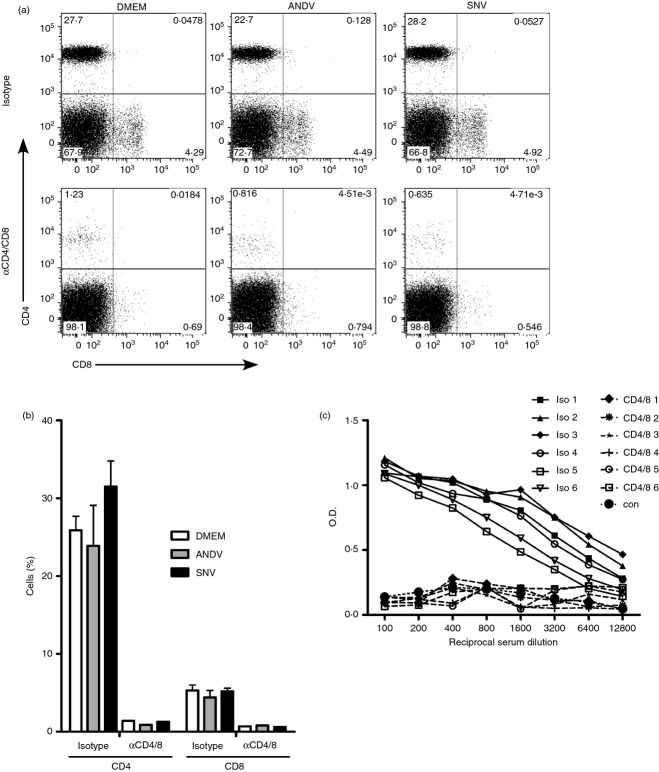
Efficient depletion of T cells from hamsters has previously been reported.21,25 To determine whether T cells play a role in the outcome of hantavirus infection, hamsters were treated with a mixture of antibodies to deplete both CD4+ and CD8+ T cells, or isotype control antibodies. In preliminary experiments, we determined that T-cell depletion in hamsters, measured in blood and spleens, is highly efficient by 2 days after antibody administration (data not shown). Two days after administration of antibodies, hamsters were challenged with ANDV or SNVp5, or mock-infected (DMEM). At 11 dpi, groups of hamsters were killed to measure T-cell depletion efficacy by flow cytometry. Antibody depletion of CD4+ and CD8+ T cells resulted in a reduction of the percentage of these cells by 94·8% and 87·2% in the spleens of control hamsters, respectively (Fig. 4a,b). Hamsters infected with hantaviruses remained depleted by 96·0% and 81·4% for ANDV infection, and 95·5% and 88·4% for SNV infection, for CD4+ and CD8+ cells, respectively.
Figure 4.
T-cell depletion is efficacious and inhibits specific B-cell responses. Groups of six hamsters were given CD4-depleting and CD8-depleting antibodies, or isotype control antibodies 2 days before inoculation with Andes virus (ANDV) or Sin Nombre virus (SNV), or mock infection (Dulbecco's modified Eagle's medium; DMEM). (a, b) Single-cell suspensions were prepared from the spleens 11 days post-infection (dpi) and stained with fluorescently conjugated anti-CD4 and anti-CD8 antibodies and analysed by flow cytometry. (a) Representative panels showing CD4 and CD8 staining for isotype control and T-cell-depleted hamsters 11 dpi. (b) The depletion efficiency was determined by flow cytometry and expressed as the per cent of CD4 or CD8 cells in the spleens of animals 11 dpi (13 days post-depletion). (c) Hamsters were given either isotype control or anti-CD4 and anti-CD8 antibodies 2 days before inoculation with SNV. Hamsters were bled 28 dpi and the sera were tested for the presence of anti-SNV N-specific antibodies by ELISA. Sera from uninfected hamsters were used as controls.
Robust antibody responses require the interaction of T and B cells. To test whether T-cell depletion had a biological effect on the immune response, anti-SNV N-specific antibodies were measured at 28 dpi from hamsters infected with SNVp5. Hamsters receiving T-cell depleting antibodies before infection had no detectable antibody responses, even at a 1 : 100 dilution of serum, demonstrating that T-cell depletion was effective at inhibiting the production of virus-specific antibodies. This was in contrast to hamsters receiving isotype control antibodies, all of which had high titres of anti-SNV N antibodies (Fig. 4c).
T-cell depletion does not influence survival or virus replication
Hamsters that were treated with CD4-depleting and CD8-depleting, or control antibodies, 2 days before inoculation with hantaviruses were monitored for disease progression and survival. Combined depletion of CD4+ and CD8+ T cells from hamsters had no effect on the outcome of either ANDV or SNVp5 infection (Fig. 5a). All ANDV-infected hamsters had to be killed or succumbed to infection regardless of antibody treatment and the mean time to death was not statistically different between these two groups. All hamsters infected with SNVp5 survived to 28 dpi and neither the T-cell-depleted nor the isotype control hamsters displayed any signs of disease. To determine whether T-cell depletion had an effect on virus replication, viral S-segment RNA was assessed in the lungs of hamsters killed at 11 dpi that were inoculated with ANDV or SNVp5, and in hamsters killed at 28 dpi that were inoculated with SNVp5. Viral RNA abundance was not different between the isotype control and the T-cell-depleted groups, regardless of challenge virus or time-point (Fig. 5b).
Figure 5.
T-cell depletion does not influence survival or virus replication. Groups of six hamsters were given CD4-depleting and CD8-depleting antibodies, or isotype control antibodies 2 days before inoculation with Andes virus (ANDV) or Sin Nombre virus (SNV). (a) Hamsters were monitored for signs of disease and survival following depletion and hantavirus infection. (b) Total RNA was extracted from lung tissue of hamsters 11 days post-infection (dpi) (ANDV and SNV) and 28 dpi (SNV only), and virus-specific RNA was quantified by quantitative reverse transcription-PCR. Data are represented as the mean and standard deviation of six animals per group.
Discussion
Pathogenic hantaviruses can cause severe disease in humans through a yet undiscovered mechanism. Although the adaptive immune response is elicited in humans following infection by HCPS-causing hantaviruses, little is known about how the immune response is either inadequate, or possibly immunopathogenic. In the hamster model of HCPS, it has recently been shown that disease caused by ANDV is probably not facilitated by aberrant T-cell responses.21 SNV causes HCPS in humans, similar to the disease observed upon infection with ANDV, but replicates poorly in Syrian hamsters and does not cause disease.26 We have recently generated a passaged SNV (SNVp5) that causes a persistent, apathogenic infection in hamsters despite high levels of viral replication.20 In this study we use ANDV, as well as SNVp5, as tools to examine potential adaptive immune response mechanisms of disease and persistence. We demonstrate that, although elicited, the adaptive immune response does not influence the outcome of infection with pathogenic ANDV, or persistent SNV infection. Furthermore, the presence of T cells does not alter the replication of these viruses in hamsters.
Hantaviruses infect microvascular endothelial cells and this infection is non-cytopathic.27 In hamsters infected with SNVp5, we measured high levels of viral RNA by qRT-PCR, and these levels were similar to that of ANDV. Despite this, no obvious histological changes or immune cell infiltration was observed in the lungs of SNV-infected hamsters. In the case of ANDV infection, immune infiltration was observed histopathologically, and changes, including interstitial pneumonia and oedema, were observed. Virus replication, as measured by the accumulation of viral RNA, was assessed in the lung and we found that SNVp5 and ANDV replicated to a similar extent at 11 dpi, suggesting that direct effects of hantavirus replication within endothelial cells is not in itself a mechanism of pathology. Similar results were previously reported upon infection of hamsters with Choclo virus, a pathogenic HCPS-causing virus in humans.28 Choclo virus replicates efficiently in hamster lung endothelial cells without apparent pathology, again suggesting that virus replication is not the only factor that contributes to disease.
Quantification of immune-related genes in the lung demonstrated that SNVp5 elicits a more pronounced cytokine response than does ANDV. By 9 dpi, increases in transcription of cytokines produced by T cells, as well as cytokines produced by monocytes/macrophages were measured in SNV-infected animals, indicating that an adaptive immune response is elicited. The transcription of IL-21, IL-12p35, interferon-γ, IL-6 and tumour necrosis factor indicates that an inflammatory response is present in the lungs at these time-points, although IL-10 was also up-regulated, suggesting that an anti-inflammatory response is also present and might help to control pathology. Immune infiltration was not observed histologically in the lungs of SNV-infected hamsters, despite moderate increases in CD4+ and CD8+ T cells measured in the spleens of SNV-infected hamsters. In contrast, ANDV induced very little cytokine transcription at any time-point, and even at 11 dpi, a time just before severe pathology and death, most of these cytokines were only slightly up-regulated, despite immune infiltration observed histologically. This is in contrast to the observations in humans that succumb to HCPS, where elevated pro-inflammatory cytokines are detectable in the lungs.12 This discrepancy could be a result of the sensitivity of qRT-PCR, as RNA from non-reactive cell types, which is the vast majority, might effectively mask the up-regulation of specific transcripts produced by a relatively small number of cells.
Immune responses were elicited by ANDV as indicated by the increase in the absolute counts of T cells and MHC II-expressing cells in the spleen, along with conspicuous splenomegaly. High numbers of neutrophils were measured in the blood of ANDV-infected hamsters, which is a hallmark of human disease, as left-shifted neutrophilia is a common observation and a diagnostic tool for HCPS.13,29,30 Blood lymphocytes and monocytes were decreased in ANDV-infected hamsters, possibly because of egress from the blood into tissues. SNVp5 infection resulted in no significant changes in white blood cell counts in the blood, despite higher cytokine expression in the lungs. This could be a result of the activation of small numbers of lymphocytes in the lungs as the fold-changes of these genes were low. There is an apparent discrepancy between the immune gene regulation and the histological observations. Although ANDV infection resulted in pathology and immune cell infiltration in the lung, SNVp5 induced a more robust immune response measured by qRT-PCR. This could be because of the genes selected for analysis, or of stronger activation of resident immune cells by SNVp5 but more robust recruitment of immune cells by ANDV. To assess this, it would be advantageous to analyse expression of chemokines that might influence immune cell egress into the lung, which are currently unavailable.
The innate immune response was strongly elicited by ANDV at 9 and 11 dpi, indicating that ANDV is recognized by the type I interferon response. This up-regulation coincided with a drastic increase in viral RNA in the lung and is probably a response to high amounts of virus replication. Pathogenic hantaviruses have been shown to subvert the innate immune response, allowing for viral replication, whereas non-pathogenic hantaviruses possess less antagonistic potential and replication is controlled by the interferon response.31,32 In contrast, the innate immune response was not measurably activated in hamsters infected with SNVp5, as there was no up-regulation of STAT1 or Mx2, potentially because of interferon antagonism strategies. However, despite high amounts of viral RNA, there was no disease, suggesting that pathogenesis is neither simply a function of virus replication, nor is it a function of innate immune response antagonism. Likewise, the robust innate response measured in response to ANDV infection may actually contribute to, or even be instrumental in, the development of disease.
The dichotomy in infection outcome by two related hantaviruses that replicate similarly in this animal model provides a unique platform to examine whether the adaptive immune response is immunopathogenic or protective. If the immune response is protective, the deletion of the adaptive immune response might render SNVp5 pathogenic. In contrast, if the disease caused by ANDV infection is immunopathogenic in nature, perturbations in the adaptive response might result in a decrease or delay in pathogenesis. Depletion of T cells in Syrian hamsters has been previously established using the same antibodies as we used herein.21,25 In our study, depletion of T cells from hamsters was highly efficient, measured by flow cytometry, and was confirmed functionally, by measuring virus-specific antibody responses. Hamsters that were given isotype control antibodies seroconverted in response to SNVp5 infection by producing high titres of anti-SNV N antibodies, whereas T-cell-depleted hamsters did not produce measurable antibodies. T cells are required for efficient antibody responses as CD4+ T cells are necessary for class switching from IgM to IgG, as well as affinity maturation.33–35 In this respect, depletion of the T-cell response also cripples the specific B-cell response to virus infection. Despite this defect, SNV RNA accumulated to the same levels in the control hamsters that were able to mount an efficient B-cell response, as it did in the T-cell-depleted hamsters at both 11 and 28 dpi. The same was observed for ANDV infection, where T-cell depletion did not affect ANDV replication. This indicates that although these viruses are recognized by the adaptive immune response, the presence of a virus-specific adaptive immune response does not influence virus replication. To have elicited a specific antibody response, the virus was clearly detected, but interestingly, detection has no consequence in the case of SNVp5. It is possible that the presence of virus-specific antibodies has no biological significance after the establishment of an infection, as immunization of hamsters with recombinant ANDV-N renders them resistant to ANDV-induced disease, but anti-N antibodies given to deer mice concomitantly with SNV challenge do not have an effect on virus replication in tissues.36,37 In the former study, it is possible that the mechanism of protection upon immunization is cell-mediated immunity, and the actual presence of antibody might be inconsequential. In the natural reservoirs of hantaviruses, high levels of antibody are detected, despite viral persistence.7,10,38 It is currently unclear how hantaviruses evade antibody responses. However, in human cases of HCPS, it has been shown that high titres of virus-specific antibodies correlate with a less severe disease course.39,40 The observation that vaccination against ANDV can protect hamsters from disease demonstrates that pre-existing immunity can efficiently inhibit pathogenesis. Herein we demonstrate that a naive animal is not able to mount a protective immune response in the case of ANDV infection, and that the immune response that is mounted does not contribute to disease via an immunopathogenic mechanism in hamsters.
It is important to note that several populations of T cells exist that do not express CD4 or CD8, termed double-negative T cells. The studies presented herein use a strategy that specifically depletes CD4-expressing and CD8-expressing T cells and immunopathology elicited by double-negative T cells might not be affected. Despite nearly complete depletion of CD4+ and CD8+ T cells as measured by flow cytometry, populations of T cells might remain after depletion, and these could play a role in the pathogenic process.41 Of the clinical parameters measured, the observed neutrophilia and macrophage/lymphocyte infiltration in the lung most similarly recapitulates findings in humans. Neutrophils are implicated in the pathophysiology of acute respiratory distress syndrome, which in many ways parallels aspects of HCPS.42 In a previous study, cyclophosphamide administration did not alter the time-to-death upon ANDV challenge in hamsters.21 Due to a lack of specific hamster reagents, the neutrophil and cytokine environment in the lungs after cyclophosphamide treatment was not measured.
We demonstrate herein that CD4+ or C8+ T cells do not contribute to protection or the pathogenic process in hamsters challenged with ANDV. Furthermore, we report that a persistent infection by SNV is not influenced by the presence of these T-cell populations and that both ANDV and SNVp5 are not affected by the presence of a specific immune response. The mechanisms of persistence or pathology caused by hantaviruses remains to be elucidated in the hamster infection model. Although T-cell and B-cell responses are elicited in this model, the activation of the adaptive immune response is inconsequential. Further efforts to elucidate the mechanisms of HCPS pathology will prove important as reagents become available for examining the immune response in the hamster model.
Acknowledgments
We would like to thank Dan Long, Tina Thomas and Rebecca Rosenke for histopathology work. We would also like to thank Anita Mora for the preparation of graphics. This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Disease, National Institutes of Health.
Disclosures
The authors declare that they have no conflicts of interests to disclose.
References
- 1.Nolte KB, Feddersen RM, Foucar K, et al. Hantavirus pulmonary syndrome in the United States: a pathological description of a disease caused by a new agent. Hum Pathol. 1995;26:110–20. doi: 10.1016/0046-8177(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 2.Khan AS, Khabbaz RF, Armstrong LR, et al. Hantavirus pulmonary syndrome: the first 100 US cases. J Infect Dis. 1996;173:1297–303. doi: 10.1093/infdis/173.6.1297. [DOI] [PubMed] [Google Scholar]
- 3.Botten J, Mirowsky K, Ye C, Gottlieb K, Saavedra M, Ponce L, Hjelle B. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J Virol. 2002;76:7587–94. doi: 10.1128/JVI.76.15.7587-7594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott LH, Ksiazek TG, Rollin PE, et al. Isolation of the causative agent of hantavirus pulmonary syndrome. Am J Trop Med Hyg. 1994;51:102–8. doi: 10.4269/ajtmh.1994.51.102. [DOI] [PubMed] [Google Scholar]
- 5.Childs JE, Ksiazek TG, Spiropoulou CF, et al. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis. 1994;169:1271–80. doi: 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- 6.Mertz GJ, Hjelle BL, Bryan RT. Hantavirus infection. Adv Intern Med. 1997;42:369–421. [PubMed] [Google Scholar]
- 7.Botten J, Mirowsky K, Kusewitt D, Ye C, Gottlieb K, Prescott J, Hjelle B. Persistent Sin Nombre virus infection in the deer mouse (Peromyscus maniculatus) model: sites of replication and strand-specific expression. J Virol. 2003;77:1540–50. doi: 10.1128/JVI.77.2.1540-1550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schountz T, Prescott J, Cogswell AC, Oko L, Mirowsky-Garcia K, Galvez AP, Hjelle B. Regulatory T cell-like responses in deer mice persistently infected with Sin Nombre virus. Proc Natl Acad Sci USA. 2007;104:15496–501. doi: 10.1073/pnas.0707454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Easterbrook JD, Zink MC, Klein SL. Regulatory T cells enhance persistence of the zoonotic pathogen Seoul virus in its reservoir host. Proc Natl Acad Sci USA. 2007;104:15502–7. doi: 10.1073/pnas.0707453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Easterbrook JD, Klein SL. Seoul virus enhances regulatory and reduces proinflammatory responses in male Norway rats. J Med Virol. 2008;80:1308–18. doi: 10.1002/jmv.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Easterbrook JD, Klein SL. Corticosteroids modulate Seoul virus infection, regulatory T-cell responses and matrix metalloprotease 9 expression in male, but not female, Norway rats. J Gen Virol. 2008;89:2723–30. doi: 10.1099/vir.0.2008/03715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori M, Rothman AL, Kurane I, et al. High levels of cytokine-producing cells in the lung tissues of patients with fatal hantavirus pulmonary syndrome. J Infect Dis. 1999;179:295–302. doi: 10.1086/314597. [DOI] [PubMed] [Google Scholar]
- 13.Zaki SR, Greer PW, Coffield LM, et al. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am J Pathol. 1995;146:552–79. [PMC free article] [PubMed] [Google Scholar]
- 14.Borges AA, Donadi EA, Campos GM, et al. Association of -308G/A polymorphism in the tumor necrosis factor-α gene promoter with susceptibility to development of hantavirus cardiopulmonary syndrome in the Ribeirao Preto region Brazil. Arch Virol. 2010;155:971–5. doi: 10.1007/s00705-010-0655-7. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Wang J, Zhu Y, Xu Z, Yang K, Yang A, Jin B. Cellular immune response to Hantaan virus nucleocapsid protein in the acute phase of hemorrhagic fever with renal syndrome: correlation with disease severity. J Infect Dis. 2009;199:188–95. doi: 10.1086/595834. [DOI] [PubMed] [Google Scholar]
- 16.Kilpatrick ED, Terajima M, Koster FT, Catalina MD, Cruz J, Ennis FA. Role of specific CD8+ T cells in the severity of a fulminant zoonotic viral hemorrhagic fever, hantavirus pulmonary syndrome. J Immunol. 2004;172:3297–304. doi: 10.4049/jimmunol.172.5.3297. [DOI] [PubMed] [Google Scholar]
- 17.Saksida A, Wraber B, Avsic-Zupanc T. Serum levels of inflammatory and regulatory cytokines in patients with hemorrhagic fever with renal syndrome. BMC Infect Dis. 2011;11:142. doi: 10.1186/1471-2334-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makela S, Mustonen J, Ala-Houhala I, Hurme M, Partanen J, Vapalahti O, Vaheri A, Pasternack A. Human leukocyte antigen-B8-DR3 is a more important risk factor for severe Puumala hantavirus infection than the tumor necrosis factor-α(-308) G/A polymorphism. J Infect Dis. 2002;186:843–6. doi: 10.1086/342413. [DOI] [PubMed] [Google Scholar]
- 19.Hooper JW, Larsen T, Custer DM, Schmaljohn CS. A lethal disease model for hantavirus pulmonary syndrome. Virology. 2001;289:6–14. doi: 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- 20.Safronetz D, Prescott J, Haddock E, Scott DP, Feldmann H, Ebihara H. Hamster-adapted Sin Nombre virus caused disseminated infection and efficiently replicates in pulmonary endothelial call without signs of disease. J Virol. 2013;87:4778–82. doi: 10.1128/JVI.03291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerbeck CD, Hooper JW. T cells are not required for pathogenesis in the Syrian hamster model of hantavirus pulmonary syndrome. J Virol. 2011;85:9929–44. doi: 10.1128/JVI.05356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meissner JD, Rowe JE, Borucki MK, St Jeor SC. Complete nucleotide sequence of a Chilean hantavirus. Virus Res. 2002;89:131–43. doi: 10.1016/s0168-1702(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 23.Safronetz D, Zivcec M, Lacasse R, et al. Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus. PLoS Pathog. 2011;7:e1002426. doi: 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zivcec M, Safronetz D, Haddock E, Feldmann H, Ebihara H. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus. J Immunol Methods. 2011;368:24–35. doi: 10.1016/j.jim.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dondji B, Sun T, Bungiro RD, Vermeire JJ, Harrison LM, Bifulco C, Cappello M. CD4 T cells mediate mucosal and systemic immune responses to experimental hookworm infection. Parasite Immunol. 2010;32:406–13. doi: 10.1111/j.1365-3024.2010.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahl-Jensen V, Chapman J, Asher L, Fisher R, Zimmerman M, Larsen T, Hooper JW. Temporal analysis of Andes virus and Sin Nombre virus infections of Syrian hamsters. J Virol. 2007;81:7449–62. doi: 10.1128/JVI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagihara R, Silverman DJ. Experimental infection of human vascular endothelial cells by pathogenic and nonpathogenic hantaviruses. Arch Virol. 1990;111:281–6. doi: 10.1007/BF01311063. [DOI] [PubMed] [Google Scholar]
- 28.Eyzaguirre EJ, Milazzo ML, Koster FT, Fulhorst CF. Choclo virus infection in the Syrian golden hamster. Am J Trop Med Hyg. 2008;78:669–74. [PMC free article] [PubMed] [Google Scholar]
- 29.Koster F, Foucar K, Hjelle B, Scott A, Chong YY, Larson R, McCabe M. Rapid presumptive diagnosis of hantavirus cardiopulmonary syndrome by peripheral blood smear review. Am J Clin Pathol. 2001;116:665–72. doi: 10.1309/CNWF-DC72-QYMR-M8DA. [DOI] [PubMed] [Google Scholar]
- 30.Mertz GJ, Hjelle B, Crowley M, Iwamoto G, Tomicic V, Vial PA. Diagnosis and treatment of new world hantavirus infections. Curr Opin Infect Dis. 2006;19:437–42. doi: 10.1097/01.qco.0000244048.38758.1f. [DOI] [PubMed] [Google Scholar]
- 31.Geimonen E, Neff S, Raymond T, Kocer SS, Gavrilovskaya IN, Mackow ER. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc Natl Acad Sci USA. 2002;99:13837–42. doi: 10.1073/pnas.192298899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiropoulou CF, Albarino CG, Ksiazek TG, Rollin PE. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J Virol. 2007;81:2769–76. doi: 10.1128/JVI.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawabe T, Naka T, Yoshida K, et al. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–78. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 34.Neuberger MS, Ehrenstein MR, Rada C, Sale J, Batista FD, Williams G, et al. Memory in the B-cell compartment: antibody affinity maturation. Philos Trans R Soc Lond B Biol Sci. 2000;355:357–60. doi: 10.1098/rstb.2000.0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–75. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Martinez VP, Padula PJ. Induction of protective immunity in a Syrian hamster model against a cytopathogenic strain of Andes virus. J Med Virol. 2012;84:87–95. doi: 10.1002/jmv.22228. [DOI] [PubMed] [Google Scholar]
- 37.Medina RA, Mirowsky-Garcia K, Hutt J, Hjelle B. Ribavirin, human convalescent plasma and anti-β3 integrin antibody inhibit infection by Sin Nombre virus in the deer mouse model. J Gen Virol. 2007;88:493–505. doi: 10.1099/vir.0.82459-0. [DOI] [PubMed] [Google Scholar]
- 38.Hutchinson KL, Rollin PE, Peters CJ. Pathogenesis of a North American hantavirus, Black Creek Canal virus, in experimentally infected Sigmodon hispidus. Am J Trop Med Hyg. 1998;59:58–65. doi: 10.4269/ajtmh.1998.59.58. [DOI] [PubMed] [Google Scholar]
- 39.Bharadwaj M, Nofchissey R, Goade D, Koster F, Hjelle B. Humoral immune responses in the hantavirus cardiopulmonary syndrome. J Infect Dis. 2000;182:43–8. doi: 10.1086/315657. [DOI] [PubMed] [Google Scholar]
- 40.MacNeil A, Comer JA, Ksiazek TG, Rollin PE. Sin Nombre virus-specific immunoglobulin M and G kinetics in hantavirus pulmonary syndrome and the role played by serologic responses in predicting disease outcome. J Infect Dis. 2010;202:242–6. doi: 10.1086/653482. [DOI] [PubMed] [Google Scholar]
- 41.D'Acquisto F, Crompton T. CD3+CD4–CD8– (double negative) T cells: saviours or villains of the immune response? Biochem Pharmacol. 2011;82:333–40. doi: 10.1016/j.bcp.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Fujishima S, Aikawa N. Neutrophil-mediated tissue injury and its modulation. Intensive Care Med. 1995;21:277–85. doi: 10.1007/BF01701489. [DOI] [PubMed] [Google Scholar]