Abstract
Eviction or destabilization of nucleosomes from chromatin is a hallmark of functional regulatory elements of the eukaryotic genome. Historically identified by nuclease hypersensitivity, these regulatory elements are typically bound by transcription factors or other regulatory proteins. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) is an alternative approach to identify these genomic regions and has proven successful in a multitude of eukaryotic cell and tissue types. Cells or dissociated tissues are crosslinked briefly with formaldehyde, lysed, and sonicated. Sheared chromatin is subjected to phenol-chloroform extraction and the isolated DNA, typically encompassing 1–3% of the human genome, is purified. We provide guidelines for quantitative analysis by PCR, microarrays, or next-generation sequencing. Regulatory elements enriched by FAIRE display high concordance with those identified by nuclease hypersensitivity or ChIP, and the entire procedure can be completed in three days. FAIRE exhibits low technical variability, which allows its use in large-scale studies of chromatin from normal or diseased tissues.
Keywords: FAIRE, open chromatin, nucleosome, next-generation sequencing
Introduction
Understanding the regulation of transcription by sequence-specific regulatory factors and subsequent remodeling of chromatin is central to studies of health and disease. The activities of regulatory factors at promoters, enhancers, silencers, and insulators typically cause nucleosomes to be evicted from chromatin in eukaryotic cells1. Therefore, one of the most effective means of discovering transcriptional regulatory elements is through the identification of nucleosome-depleted regions (“open chromatin”). Historically, this was accomplished by exploiting regional hypersensitivity to nucleases such as DNase I2–9. More recently, we demonstrated an alternative methodology for the detection of open chromatin, which we termed FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements)10–13. FAIRE was first characterized in yeast and subsequently applied to human cells and tissues13–16. The technique has proven useful for a wide range of eukaryotes, from Plasmodium17 to maize18. Here, we present recent methodological enhancements that improve the utility and reliability of FAIRE, especially for use on tissues or lipid-laden cells such as adipocytes.
Overview
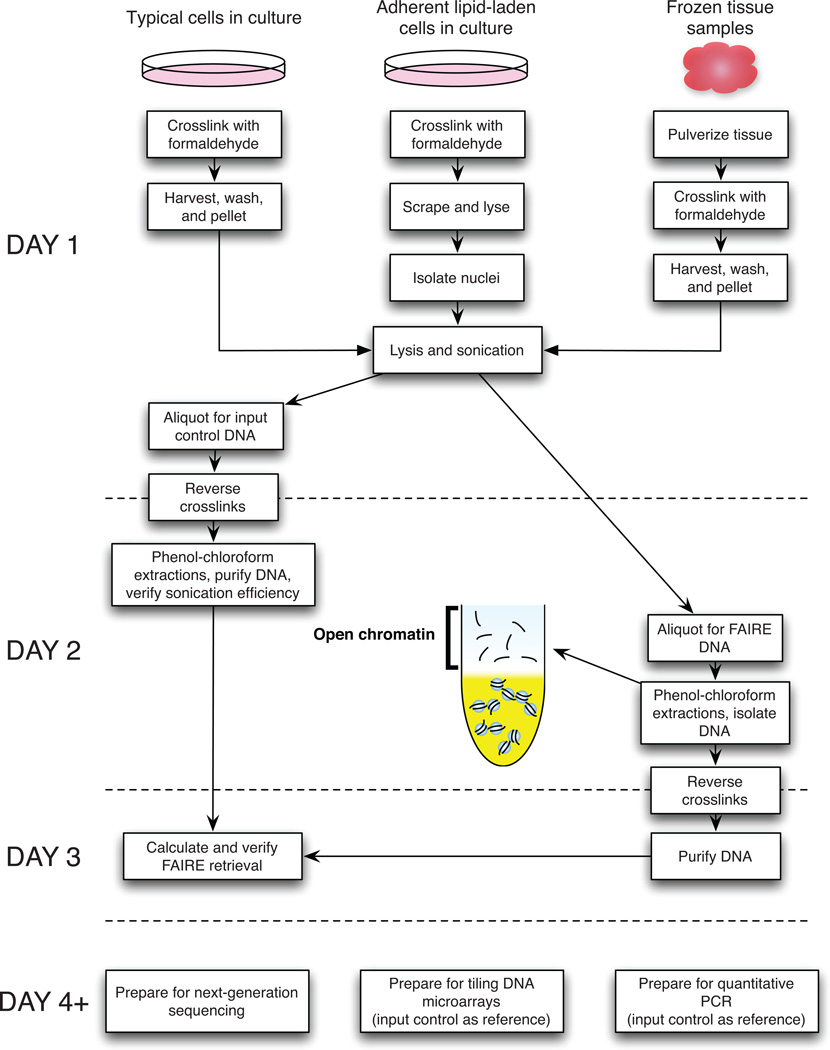
FAIRE does not rely on the use of antibodies or enzymes, and is based on differences in crosslinking efficiencies between DNA and nucleosomes or sequence-specific DNA-binding proteins. DNA in nucleosome depleted regions of chromatin (for example through the activity of a sequence-specific regulatory factor) is much less efficiently crosslinked to protein12. DNA not crosslinked to protein will segregate to the aqueous phase during phenol-chloroform extraction. In contrast, DNA covalently linked to proteins will demonstrate hydrophilic properties, and will become trapped between the organic and aqueous phase. To perform FAIRE (Figure 1), cells or dissociated tissues are cross-linked briefly with formaldehyde, lysed, and sonicated. Sheared chromatin is then subjected to phenol/chloroform extraction. The DNA in the aqueous phase is then purified and assayed. FAIRE-enriched chromatin is detected using one of several quantitative approaches. Options include quantitative amplification by PCR (FAIRE-qPCR)13, hybridization to a tiling DNA microarray (FAIRE-chip)11,13, or sequencing via next-generation sequencing technologies (FAIRE-seq)13,16. Due to declining costs of sequencing and higher quality and resolution of sequencing-based data, FAIRE-seq has now nearly fully supplanted FAIRE-chip and FAIRE-qPCR, especially for larger genomes, but also for smaller genomes through multiplexing. Analysis by next-generation sequencing requires alignment of high-quality reads to a reference genome (e.g. Bowtie19) followed by detection of regions of significant enrichment (we recommend ZINBA20). Bowtie and ZINBA are both freely available.
Figure 1.
Example timeline for FAIRE protocol. Steps are grouped by day for the typical timeline, but utilizing Pause Points will extend the duration.
Applications
Our lab has used FAIRE extensively to characterize active regulatory elements of several human cell lines as part of the ENCODE consortium21, as well as different cell, tissue, and tumor samples from humans, mice, and other eukaryotes. FAIRE has been used to create catalogs of regulatory elements in normal or diseased cells13,14,16, narrow the search space for causal sequence variants in human disease13,22, and understand the interactions between transcription factors and chromatin remodeling23,24. When coupled with high-throughput sequencing, FAIRE can also be used to identify both large- and small-scale structural variations such as copy number variants (CNV)20.
Comparison with other methods
We have previously shown that regions in the yeast genome enriched by FAIRE were anti-correlated with occupancy of histones H3 and H410, and that FAIRE regions encompass promoters, enhancers, insulators, and other regulatory elements, most of which are also captured by DNase I hypersensitivity assays10,12–14,16. An in-depth comparison between regulatory elements captured by FAIRE, DNase I hypersensitivity, and ChIP-seq found that while a large set of elements were identified by all methods, each assay also identified a unique set of features16. FAIRE was able to detect some distal regulatory elements, such as transcriptional enhancers, that DNase-seq could not, whereas DNase-seq identified some promoters that FAIRE did not.
Advantages of FAIRE
Antibody and enzyme independency
In contrast to ChIP, which is highly subject to antibody reliability and variability issues25, FAIRE offers the consistency of a chemical based isolation. Moreover, FAIRE does not require enzymes, such as DNase or MNase, which are commonly used in analogous methods for detecting nucleosome-free regions. Avoiding the optimization and extra steps necessary for enzymatic processing or immunoprecipitations eliminates a major source of variation, and thus makes it a much more reliable and robust method.
Enhancer detection
As described in Comparison with other methods and in Song et al16, FAIRE may identify additional transcriptional enhancers and other distal regulatory elements in comparison to other methods such as DNase-seq.
Sequenced input control not required
As discussed in Rashid et al20, a sequenced input control is not required for proper analysis of FAIRE-enriched regions. This reduces next-generation sequencing costs as well as the cost of reagents.
Applicability to tissue samples
Since FAIRE does not require a single-cell suspension or nuclear isolation, it is easily adapted for use on tissue samples. The only additional step needed is pulverization of frozen tissue into a coarse powder prior to fixation.
Limitations
Promoter detection
As described in Comparison with other methods and in Song et al16, other methods, such as DNase-seq, may be better at identifying nucleosome-depleted promoters of highly expressed genes.
Analysis
As noted below in Experimental design, although FAIRE is relatively straightforward experimentally, an extensive amount of computational processing and analysis are required for comprehensive interpretation of genome-wide results. Groups without access to bioinformatics specialists and computers with sufficient memory, computing power, and storage capacity may experience challenges in interpreting their results. Quantification of FAIRE signal by qPCR or microarrays may be more straightforward.
Absence of transcription factor footprinting
Transcription factor motifs can be identified in regions of open chromatin identified by FAIRE. However, the higher resolution and increased signal-to-noise of DNase-seq permits detection of specific transcription factor footprints in very deeply sequenced data1.
Low signal-to-noise
Relative to ChIP-seq or DNase-seq experiments, FAIRE has a lower signal-to-noise ratio. Therefore, the sites detected by FAIRE can, at times, be only marginally enriched above the background signal. This leads to a reduced confidence in the sites identified. This effect can be exacerbated when using non-sequencing based detection methods. Consequently, primer and array design as well as the selection of control regions are critical. Despite the low signal-to-noise ratio, we note that FAIRE is remarkably reproducible from experiment to experiment.
Fixation variation among tissues
Fixation efficiency can vary drastically due to many reasons, including differences in cellularity, permeability, purity, fat content, and surface area. Although dissociation by pulverization seems to make fixation slightly more consistent compared to mincing or other methods, this variability can still lead to inconsistent results; optimization is thus recommended.
Experimental design
Replicates
Studies utilizing FAIRE, like many other genome-wide assays, should include biological replicates. This entails the use of multiple independently grown batches of cells or tissues treated in the same fashion. Several methods have been developed for the assessment of concordance among replicates, such as Irreproducible Discovery Rate (IDR)26, which is currently employed by the ENCODE consortium. Methods like IDR often require a ranked set of statistically enriched regions, which can be obtained by most peak-calling algorithms, including ZINBA20 (see Analysis below).
Control sample
For sequencing-based detection of FAIRE enrichment, we have found that a control sample, such as genomic or input DNA, while always better to have, is not strictly necessary for samples that have been sequenced to sufficient depth and coverage20. When detecting enrichment by qPCR or tiling DNA microarrays, a genomic or input DNA sample is necessary for use as a reference.
Analysis
Although FAIRE is a relatively straightforward experimental protocol that can be completed in three days, extensive computational processing and analysis are required for interpretation of the results. This includes quality assessment of the sequencing library and the sequencing reactions themselves, reference genome alignment, detection of enrichment, and assessment of replicate concordance. We recommend a combination of the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and TagDust27 for quality control of the sequencing reactions and libraries, respectively. Although we typically use Bowtie19 for reference genome alignment, other similar algorithms such as BWA28 are equally suitable. To detect regions of significant FAIRE enrichment (“peaks”), we found that methods such as MACS29 and Fseq30, though commonly used successfully for ChIP-seq or DNase-seq data, do not perform well on FAIRE-seq data, likely due to its relatively lower inherent signal-to-noise ratio. We thus developed a novel statistical algorithm called ZINBA20. The regions identified by ZINBA can then be used to assess concordance among replicates using algorithms such as IDR26. If possible, the data should be compared to existing maps of open chromatin, such as DNase-seq and FAIRE-seq data made available by the ENCODE consortium21, or with gene expression data. FAIRE enrichment at gene promoters is strongly linked to gene expression. Therefore, strong FAIRE enrichment is expected around genes known to be highly expressed. A large fraction (~30–50%) of the regions enriched by FAIRE are in intergenic regions of the genome Typically only ~5–15% of all FAIRE sites are at proximal promoters13,16. To determine if an experiment was successful, we often examine the pattern from a locus on human chromosome 19 that produces a remarkably consistent level of FAIRE enrichment across cell types (see Anticipated Results).
Detection method
In cases where a reference genome assembly is available, FAIRE coupled with high-throughput sequencing is likely the most cost-effective option, especially if multiplexing is applicable. In smaller eukaryotes or for very targeted experiments, detection by microarray or quantitative PCR may be preferable, but array and primer design will play a key role in the overall success of the experiment (see FAIRE-chip Microarray design and FAIRE-qPCR Primer design below).
Fixation
The most common reason for a failed FAIRE experiment is under-fixation of the cells. We have found that for a majority of mammalian cells in culture, fixation for five minutes with formaldehyde is both adequate and ideal. The protocol below includes quantification of both input control and FAIRE DNA, and we describe a diagnostic for determining if the sample has not been fixed sufficiently. For tissues, samples must first be pulverized into a course powder and then fixed for 7–9 minutes. The adequacy of fixation will depend heavily on the tissue size and composition and thus may need to be optimized. Other techniques or adaptations for fixation may be required for plants or fungi, such as significantly increased fixation time10 or modified fixation solutions31. For lipid-laden cells, it may be beneficial to perform both fixation and cell lysis (to extract nuclei) prior to attempting to harvest the cells, as outlined below in step 1B.
Sonication
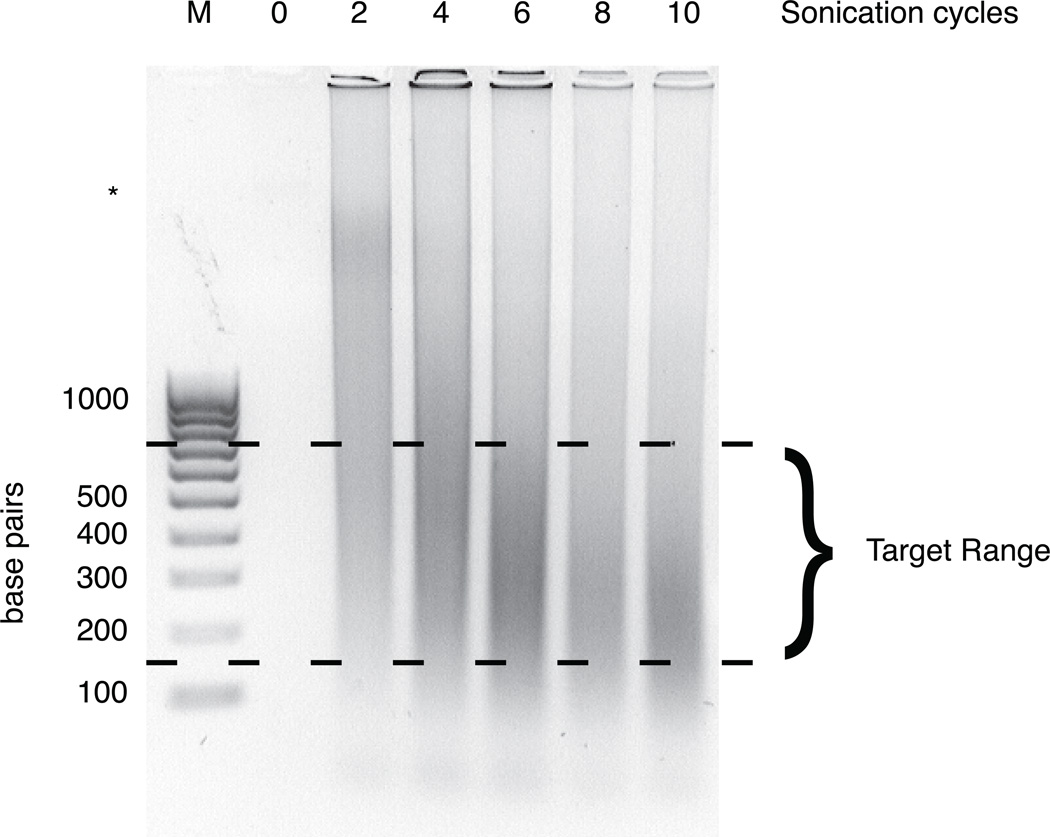
Sonication parameters must be optimized for each experiment due to variation in cell number, composition, sonicator and probe type, and fixation. In Figure 2, we present a representative agarose gel that provides examples of over-, under-, and sufficiently sonicated chromatin. Ideally, chromatin is sheared to a range of about 150–750 bp with an average fragment length around 300–400 bp. Sonication yielding average fragment sizes smaller than this can result in reduced detection of highly nucleosome-depleted regions. High molecular weight bands may be visible especially when beginning with frozen tissue, but their presence in lieu of a distribution of smaller fragments is indicative of under-sonication or poor cell lysis.
Figure 2.
Representative gel image showing varying degrees of sonication. NIH3T3 cells were fixed and lysed as described above. Chromatin was then sheared by sonication for 0, 2, 4, 6, 8, and 10 cycles using the parameters outlined in step 2A. After clearing cell debris, crosslinks were reversed, and purified DNA was run on a 1% agarose gel. A 100 bp ladder (lane marked M) is included for reference. The target range for fragment sizes is shown. Six cycles yields an ideal distribution of fragment lengths; fewer than six cycles of sonication is insufficient for solubilization and shearing of chromatin, whereas sonication beyond six cycles leads to oversonication. A high molecular weight band is slightly visible and marked with an asterisk.
FAIRE-chip Microarray design
The two main considerations for microarray design are the resolution (or spacing) of the probes throughout the genome and the set of genomic loci covered by the probes. Resolution is the genomic distance from one probe to the next and must be sufficiently dense to capture the physiologically relevant size of the DNA fragments recovered by FAIRE (~200 bp). Probe spacing should allow a minimum of 3 probes per FAIRE DNA fragment or ~65 bp resolution. The set of genomic regions represented on the array is important as it provides a relative interpretation of the results. This is due to all the measurements being expressed as a ratio of the FAIRE signal over a reference sample, which is normalized by centering based on the mean ratio. The majority of probes should span regions that correspond to background (not open) chromatin. There are a number of published protocols that address specific aspects of array design and include recommendations for reliable detection32–41.
FAIRE-qPCR Primer design
When detecting FAIRE enrichment via quantitative PCR, careful consideration of experimental design will maximize the chance of success. In addition to the methodology for quantification of the results, selection of an appropriate set of control regions and locations of primers play an important role in calculating relative enrichment. This is often difficult due to the lack of a priori knowledge of both true FAIRE-positive and -negative sites for most cell or tissue types or growth conditions. The data made available by the ENCODE consortium may be helpful in this regard21. We often employ a tiling approach for detection of open chromatin sites using qPCR, such that primer pairs are designed so the amplicons are either directly overlapping or closely spaced across the assayed genomic regions. As a control, we recommend using primer sets that flank the regions isolated by FAIRE. Since primers spanning or near the edges of sonication breakpoints of FAIRE fragments are unlikely to properly amplify, primer pairs should be designed such that they amplify 60–100 bp within the center of the region of interest. Primer sets should be validated on a dilution series of input DNA to confirm consistent and proportionate amplification characteristics. For these and other reasons, FAIRE-chip and FAIRE-seq are strongly preferred over FAIRE-qPCR.
Materials
Reagents
Formaldehyde 37% w/v (Fisher Scientific F79-500). CAUTION: Formaldehyde is toxic by inhalation or if swallowed; is irritating to the skin, eyes, and respiratory system; and may be carcinogenic. Formaldehyde should be used with appropriate safety measures such as protective gloves, glasses, clothing, and sufficient ventilation. All waste should be handled according to hazardous waste regulations.
2.5 M glycine (Fisher Scientific BP381-500).
1 × Dulbecco’s phosphate buffered saline (Cellgro 21-031).
Tris-HCl pH 8.0 (Fisher Scientific BP152-500).
Tris HCl pH 7.4 (Fisher Scientific BP152-500).
Triton X-100 (Fisher Scientific BP151-500).
Sodium dodecyl sulfate (Fisher Scientific BP166-500).
NaCl (Mallinkrodt 7581).
EDTA (Fisher Scientific BP120-500).
KCl (Fisher Scientific BP366-500)
NP-40 (Sigma I8896-100)
Sucrose (Gibco 15503-014)
Protease inhibitors (Roche 11836153001)
DNase-free RNaseA (10ug/uL) (Roche 11119915001).
Proteinase K (20mg/mL) (Roche 03115836001).
Phenol/Chloroform/Isoamyl Alcohol 25:24:1 (Sigma P3803). CAUTION: Phenol/chloroform is harmful if swallowed or in contact with skin, causes severe skin burns and eye damage, is fatal if inhaled, and is potentially carcinogenic. It should be used with appropriate safety measures such as protective gloves, glasses, clothing, and sufficient ventilation. All waste should be handled according to hazardous waste regulations.
Chloroform/Isoamyl Alcohol 24:1 (Sigma C0549). CAUTION: Chloroform/isoamyl alcohol is harmful if swallowed, causes skin and eye irritation, and is potentially carcinogenic. It should be used with appropriate safety measures such as protective gloves, glasses, clothing, and sufficient ventilation. All waste should be handled according to hazardous waste regulations.
95% ethanol (Decon 2801)
Glycogen (20mg/mL) (Roche 901393).
3 M sodium acetate pH 5.2 (Mallinkrodt 7364).
70% ethanol (ice cold, diluted from 95% ethanol)
Double-distilled water
Lysis buffer A (see REAGENT SETUP)
Lysis buffer B (see REAGENT SETUP)
Sucrose pad (see REAGENT SETUP)
SYBR green master mix (Applied Biosystems 4309155)
Equipment
Lab equipment
Cell scrapers (such as Corning 3008)
Liquid nitrogen and appropriate container
Tissue pulverizer (such as Biospec 59012N)
Nutator (such as BD/Clay Adams 421105)
Temperature controlled swinging bucket centrifuge (such as Fisher Scientific Accuspin 1R)
Dounce (such as Kimble-Chase 885300-0000)
Bead-beater (Biospec Mini-BeadBeater-8)
Tubes with metal beads, 2.38 mm (MoBio 13117-50). CRITICAL STEP: We have found that the specified metal beads dissociate tissues or cell clumps better than other materials
Sonicator (such as Branson Sonifier 450D equipped with microtip)
Zymo-I spin columns (C1003-250)
Zymo DNA binding buffer (D4004-1-L)
Zymo Wash buffer (D4003-2-4)
Fluorometer with DNA quantification reagents and standards (such as Invitrogen Q32866) or NanoDrop ND-1000. Quantification of DNA is necessary, and a fluorescence-based system is recommended as it is much more accurate.
MicroAmp™ Optical 96-Well Reaction Plate (applied Biosystems 4306737).
Computer and software
A computer with at least 8 GB RAM and 100 GB hard drive, preferably with a UNIX subsystem, such as Mac OS X or LINUX, and multiple processing cores.
Bowtie19, for reference genome alignment, ZINBA20 for peak-calling, and TagDust27 and FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) for quality control. All are freely available and run easily in a command-line context.
Reagent Setup
Lysis buffer A (10 mM Tris-HCl pH 8.0, 2% Triton X-100, 1% SDS, 100 mM NaCl, 1 mM EDTA). Store at 4°C for a maximum of 6 months.
Lysis buffer B (10 mM Tris-HCl pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.1% NP-40, 5% sucrose, 1X protease inhibitors). Prepare stock solution excluding protease inhibitors and store at 4 °C for a maximum of 6 months.
Sucrose pad (10mM Tris pH 7.4, 15 mM NaCl, 60 mM KCl, 10% sucrose, 1X protease inhibitors). Prepare stock solution excluding protease inhibitors, sterile-filter, and store at 4 °C for a maximum of 6 months.
Equipment Setup
Tissue preparation: To pulverize tissues, pre-chill both the mortar and the pestle of a tissue pulverizer in liquid nitrogen. Tissues should be pulverized until a coarse powder is achieved, with a granularity similar to that of cornmeal or granulated sugar. Samples should only be allowed to begin to thaw after pulverization is completed and before PBS is added, since the PBS will otherwise freeze.
Cell dissociation: For dissociation of cells and tissues we use the Mini-BeadBeater-8, set to homogenize. Although settings will depend on cell or tissue type, we begin with five 2-minute cycles at 4 °C, allowing the sample to cool on ice for 2 minutes between each cycle. We have found that the specified metal beads dissociate tissues or cell clumps better than other materials
Sonication: We typically use a Branson Sonifier 450D, equipped with a microtip. Although the settings will depend on cell or tissue type, growth conditions, and crosslinking, we begin with 30% amplitude and 6 cycles (where each cycle has a 1.0 s burst followed by a 0.5 s pause and a total length of 30 s) and allow the sample to cool in an ice-water bath during and/or between cycles
Procedure
Formaldehyde crosslinking and cell lysis. TIMING Day 1, 4–6 hours
-
1For formaldehyde-crosslinking of cells, chose one of the following three procedures depending on whether you are using (A) typical adherent or suspension cells, (B) adherent lipid-laden cells such as adipocytes (or if a bead-beater is unavailable), or (C) frozen tissues. The procedure described in C has been tested on human and mouse samples. Alterations such as increased fixation length may be necessary for non-mammalian eukaryotes such as plants or fungi; we recommend reviewing existing publications performing chromatin immunoprecipitations for such necessary adaptations. Other modifications may also be necessary, such as those made for pancreatic islets42.
- For typical adherent or suspension cells:
- Culture 1 × 106 – 5 × 107 cells for each experiment. If available, 1 × 107 cells is ideal for most applications.
- Add 37% formaldehyde directly to the media to a final concentration of 1%.
- Fix for 5 minutes while rocking at room temperature (25 °C).
- Add 2.5 M glycine to a final concentration of 125 mM to quench formaldehyde.
- Rock an additional 5 minutes at room temperature.
- Scrape cells if needed and pool in 50mL conical tubes
- Spin at 300–500g for 5 minutes at 4 °C to collect fixed cells. Decant or pipette supernatant into formaldehyde waste.
- Wash pellet with 10 mL 1× PBS.
- Repeat steps 1Avii – viii twice for a total of three washes. During the final wash, transfer cells in PBS to 15 mL conical tubes.
-
Flash-freeze the fixed cell pellets in liquid nitrogen.PAUSE POINT: Once cells have been frozen, they can be stored at −80 °C indefinitely.
- Resuspend fixed cells in 1 mL cold lysis buffer A and transfer to the tubes containing the metal beads.
- Dissociate and lyse cells by bead beating (see Equipment Setup). For most cell and tissue types, five two-minute cycles followed by two minutes of rest (on ice) is sufficient, but additional cycles may be required. CRITICAL STEP: This step must be performed in a 4 °C room.
- Transfer lysate to 15 mL conical tube. Wash the beads that are left behind with an additional 1 mL of cold lysis buffer A. Add this wash to the 15 mL tube for a final lysate volume of 2 mL.
- For adherent lipid-laden cells (or if bead-beater is unavailable):
- Begin with 1 × 106 – 5 × 107 cells for each experiment. If available, 1 × 107 cells is ideal for most applications.
- Add 37% formaldehyde directly to the media to a final concentration of 1%.
- Fix for 5 minutes while rocking at room temperature.
- Add 2.5 M glycine to a final concentration of 125 mM to quench formaldehyde.
- Rock an additional 5 minutes at room temperature.
- Pipette and remove the formaldehyde, glycine, and media mixture without disturbing the cells.
- Scrape cells in 7 mL lysis buffer B added directly to the plate. Multiple plates can be pooled by scraping cells from each plate in the same 7mL of lysis buffer B such that the final volume does not exceed 7mL.
- Dounce cells with 5–10 smooth strokes using a tight fitting pestle.
- Transfer to a 15 mL conical tube and add 2 mL sucrose pad slowly to the bottom of the tube using a Pasteur pipet.
- Centrifuge at 2,100g for 20–25 minutes at 4 °C to collect nuclei
- Aspirate to remove supernatant.
- Resuspend nuclear pellet in 2 mL lysis buffer A.
- For frozen tissues
- Begin with 20 – 200 mg frozen tissue.
- Pulverize frozen tissue into a coarse powder (see Equipment Setup). Allow sample to begin to thaw. CRITICAL STEP: Pulverization of the tissue to a coarse powder, rather than mincing or douncing, allows for a more uniform fixation.
- Add 5 mL PBS to resuspend powder, avoiding freezing of PBS. Transfer to 15 mL conical tube.
- Wash pulverizer mortar and pestle with additional 5 mL PBS and add to conical tube for a total volume of 10 mL.
- Add 37% formaldehyde to a final concentration of 1%.
- Fix for 7–9 minutes while rocking at room temperature.
- Add 2.5 M glycine to a final concentration of 125 mM to quench formaldehyde.
- Rock an additional 5 minutes at room temperature.
- Spin at 300–500g for 5 minutes at 4 °C to collect fixed tissue. Pipette supernatant into formaldehyde waste.
- Wash pellet with 10 mL 1× PBS.
- Repeat steps 1Cvii – viii twice for a total of three washes. During the final wash, transfer tissue in PBS to 15 mL conical tubes.
- Flash-freeze fixed tissue in liquid nitrogen. PAUSE POINT: Once tissues have been frozen, they can be stored at −80°C indefinitely.
- Resuspend fixed tissue in 1 mL cold lysis buffer A and transfer to the tubes containing the metal beads.
- Dissociate tissue and lyse cells by bead beating (see Equipment Setup). For most cell and tissue types, five two-minute cycles followed by two minutes of rest (on ice) is sufficient, but additional cycles may be required. CRITICAL STEP: This must be performed in a 4°C room.
- Transfer lysate to 15 mL conical tube. Wash the beads that are left behind with an additional 1 mL of cold lysis buffer A. Add this wash to the 15 mL tube for a final lysate volume of 2 mL. Note that some clumps or cloudiness may remain, especially if the tissue was particularly vascular.
Sonication. TIMING: Day 1, 1–2 hours
-
2
Sonicate cell lysate to achieve an average DNA fragment size of approximately 300–400 bp. For most cell and tissue types, six 30-second cycles with 1s bursts followed by 0.5s rest at 30% amplitude using the Branson Sonifier 450D is sufficient, but optimization may be required for some cell or tissue types or for other sonicators. Samples should be allowed to cool in an ice-water bath during and/or between sonication cycles or for at least 1–2 minutes.
CRITICAL STEP: Foaming should be avoided, as this likely decreases sonication efficiency. If foaming occurs, let sample settle on ice until bubbles have subsided or centrifuge briefly and gently resuspend all material. Probe positioning heavily influences both sonication efficiency and whether or not sample will foam. In most cases, the probe should be placed in the center of the tube approximately one-quarter to one-half an inch from the bottom.
Preparation of input control DNA. Day 1, 1.5 hours and overnight incubation
-
3
Remove a 100 uL aliquot of cell lysate to check efficiency of sonication. Remaining lysate can be stored temporarily at 4 °C.
-
4
Centrifuge aliquot of lysate at 15,000 – 20,000g for 5 minutes at 4 °C to pellet cell debris.
-
5
Transfer supernatant to fresh 1.5 mL tube
-
6
Add 1uL DNase-free RNaseA. Incubate for 30 minutes at 37°C. CRITICAL STEP: RNaseA must be DNase-free.
-
7
Add 1ul proteinase K, incubate at 55°C for 1 hr, then incubate overnight at 65°C to reverse crosslinks.
Purification and assessment of input control DNA. Day 2, 3–4 hours
-
8
If needed, collect sample by brief centrifugation in a microfuge or tabletop centrifuge. Add 200 uL 10 mM Tris-HCl pH 7.4 to a final volume of 300 uL and ensure all materials are fully resuspended.
-
9
Add 300 uL phenol/chloroform/isoamyl alcohol. CAUTION: See REAGENTS for precautions when using phenol and chloroform
-
10
Vortex for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
11
Transfer aqueous (top) layer to a fresh 1.5 mL tube
-
12
To ensure complete retrieval of aqueous material, add 150 uL 10 mM Tris-HCl pH 7.4 to the tube containing the interphase and organic layer
-
13
Vortex for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge.
-
14
Transfer aqueous (top) layer and combine with previously isolated aqueous material.
-
15
Add 1 volume phenol/chloroform/isoamyl alcohol. CAUTION: See REAGENTS for precautions when using phenol and chloroform
-
16
Vortex for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
17
Transfer aqueous (top) layer to a fresh 1.5 mL tube
-
18
Add 200 ul chloroform/isoamyl alcohol to remove traces of phenol. CAUTION: See REAGENTS for precautions when using phenol and chloroform.
-
19
Vortex for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
20
Transfer aqueous layer to a fresh 1.5 mL tube.
-
21
Add 1/10 volume 3 M Sodium Acetate (pH 5.2), 2 volumes 95% ethanol, and 1 uL 20 mg/mL glycogen
-
22
Incubate at −80 °C for 30 min or longer. PAUSE POINT: Precipitations can be left at −80 °C overnight or longer.
-
23
Centrifuge at 12,000g for 15 minutes at 4 °C to precipitate DNA
-
24
Carefully aspirate supernatant without disturbing DNA pellet.
-
25
Wash pellet with 500 uL ice cold 70% ethanol
-
26
Centrifuge at 12,000g for 5 minutes at 4°C
-
27
Carefully aspirate supernatant without disturbing DNA pellet.
-
28
Dry pellet with a speedvac or by leaving tubes open for 10–20 minutes, then resuspend pellet in 20uL 10mM Tris-HCl pH 7.4. CRITICAL STEP: Remaining traces of ethanol can affect downstream steps. Make sure pellets are completely dry before resuspending.
-
29
Quantify 1 uL of input control DNA with fluorometer or NanoDrop. We recommend fluorometry-based quantification due to its increased accuracy. For experiments beginning with around 1 × 107 cells or 150 mg of tissue, input control yields should be around 50–100 ng/uL with a total volume of 20 uL.
TROUBLESHOOTING.
-
30
Run 500 ng or half of input control DNA on 1% agarose gel and visualize with ethidium bromide. CRITICAL STEP: Sufficient sonication has been achieved if fragments range in size from 100 bp and 1,000 bp, with an approximate average fragment length of between 200bp and 500bp. If only a large molecular weight band is detected or if the average fragment size is significantly larger than 500 bp, additional rounds of sonication are necessary. Retrieve lysate from 4 °C storage and repeat steps 2–31 until this optimal range of fragment sizes has been achieved. PAUSE POINT: Input control DNA can be frozen and stored indefinitely at −80 °C. Cell lysates can be stored at 4 °C for up to several days, or frozen and stored for several weeks at −80 °C. TROUBLESHOOTING
Preparation of FAIRE DNA. Day 2, 3–4 hours and overnight incubation
-
31
Aliquot remaining cell lysate into 1.5 mL tubes each with no more than 500 uL. One aliquot can be stored at −80 °C indefinitely as backup.
-
32
Centrifuge aliquots of lysate at 15,000 – 20,000g for 5 minutes at 4 °C to pellet cell debris.
-
33
Transfer supernatants to fresh 1.5 mL tubes
-
34
Add 1 volume phenol/chloroform/isoamyl alcohol to each aliquot. CAUTION: See REAGENTS for precautions when using phenol and chloroform
-
35
Vortex each tube for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
36
Transfer aqueous (top) layers to fresh 1.5 mL tubes. TROUBLESHOOTING.
-
37
To ensure complete retrieval of aqueous material, add 150 uL 10 mM Tris-HCl pH 7.4 to the tubes containing the interphase and organic layers
-
38
Vortex each tube for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
39
Transfer aqueous (top) layers and combine with previously isolated aqueous material.
-
40
Add 1 volume phenol/chloroform/isoamyl alcohol to each tube. CAUTION: See REAGENTS for precautions when using phenol and chloroform
-
41
Vortex each tube for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
42
Transfer aqueous (top) layers to fresh 1.5mL tubes
-
43
Add 200ul chloroform/isoamyl alcohol to each tube to remove traces of phenol. CAUTION: See REAGENTS for precautions when using phenol and chloroform.
-
44
Vortex for 10 seconds. Centrifuge at 12,000g for 5 minutes in a tabletop centrifuge
-
45
Transfer aqueous layer to fresh 1.5mL tubes.
-
46
Add 1/10 volume 3 M Sodium Acetate (pH 5.2), 2 volumes 95% ethanol, and 1 uL 20 mg/mL glycogen to each tube
-
47
Incubate at −80 °C for 30 min or longer. PAUSE POINT: Precipitations can be left at −80 °C overnight or longer.
-
48
Centrifuge at 12,000g for 15 minutes at 4°C to precipitate DNA
-
49
Carefully aspirate supernatants without disturbing DNA pellets.
-
50
Wash pellets with 500 uL ice cold 70% ethanol
-
51
Centrifuge at 12,000g for 5 minutes at 4°C
-
52
Carefully aspirate supernatants without disturbing DNA pellet.
-
53
Dry pellets with a SpeedVac or by leaving tubes open for 10–20 minutes, then resuspend pellet in 50uL 10mM Tris-HCl pH 7.4. CRITICAL STEP: Remaining traces of ethanol can affect downstream steps. Make sure pellets are completely dry before resuspending.
-
54
Add 1 uL DNase-free RNaseA. Incubate for 30 minutes at 37 °C. CRITICAL STEP: RNaseA must be DNase-free.
-
55
Add 1 ul proteinase K, incubate at 55 °C for 1 hr, then incubate overnight at 65 °C to reverse any DNA-DNA crosslinks.
Purification and assessment of FAIRE DNA. Day 3, 1 hour
-
56
If needed, collect sample with brief centrifugation in a microfuge or tabletop centrifuge
-
57
Purify with Zymo-I spin columns using 2 volumes of DNA binding buffer and using 200 uL wash buffer for each washing step. Elute twice with 10 uL 10mM Tris-HCl pH 7.4, allowing buffer to sit on the column at room temperature for 1–2 minutes.
-
58
Quantify 1uL of FAIRE DNA with fluorometer or NanoDrop. We recommend fluorometry-based quantification due to its increased accuracy. For experiments beginning with around 1 × 107 cells or 150 mg of tissue, FAIRE yields should be around 6–12 ng/uL with a total volume of 20 uL.
CRITICAL STEP: To test if the FAIRE yield is within an acceptable range, we recommend dividing the total FAIRE yield (in nanograms) by the volume of cell lysate used for FAIRE (in uL, the number of lysate aliquots multiplied by the aliquot volume). A similar value should be calculated for the input control (total yield in nanograms over lysate aliquot volume). The volume-normalized ratio of FAIRE DNA isolated with respect to input control DNA isolated should not exceed 5% and will ideally fall in the 1–3% range. A retrieval ratio significantly higher than 5% is often indicative of under-fixation and may predict experimental failure due to poor signal enrichment.
PAUSE POINT: FAIRE DNA can be frozen and stored indefinitely at −80°C.
TROUBLESHOOTING.
Detection of FAIRE enrichment and basic data analysis
-
59For detection of FAIRE enrichment, one of three procedures can be used depending on whether you are utilizing next-generation sequencing (FAIRE-seq)13,16 (A), tiling DNA microarrays (FAIRE-chip)11,13 (B), or quantitative PCR (FAIRE-qPCR)13 (C). The procedure described in A has been tested on the Illumina sequencing platform and thus optimization may be required for other platforms. The procedure described in B has been tested on multiple human tiling DNA microarray platforms including Nimblegen (Roche) and Agilent, but optimization may be required for certain platforms or array types.
- Detection and analysis by next-generation sequencing – FAIRE-seq
- Prepare sequencing libraries using manufacturer’s protocols. We recommend beginning with 100–200 ng of FAIRE DNA. We typically incorporate two rounds of purification with Agencourt AMPure XP beads prior to 18 cycles of amplification by PCR, and size-select the final library to 200–500 bp, avoiding adapter bands, which typically run under 100 bp. CRITICAL STEP: We have found that sufficient depth and coverage on the human genome is typically achieved with no less than 3 × 107 aligned reads.
- Remove sequencing reads with significant adapter contribution using an algorithm such as TagDust27. We typically set the False Discovery Rate (FDR) parameter to 0.001. CRITICAL STEP: For clean libraries, an average of about 0.1–0.2% of reads are filtered at this step. Significantly higher fractions (>10%) are indicative of poor library quality.
- Assess sequencing quality, including confidence scores and nucleotide distributions, using algorithms such as those in the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). CRITICAL STEP: It is important that a relatively even nucleotide distribution is observed for all nucleotides across every read sequence. An over-representation of “N”, abundance of a specific sequence, or wide variability in sequence quality is indicative of poor library complexity or sequencing errors.
- Align high-quality reads to reference genome using an algorithm such as Bowtie19 with default parameters, except restrict the maximum number of allowable alignments to 4 and force Bowtie to pick the highest-scoring alignment when multiple possibilities exist. For most genomes and experiments, ~75–85% of sequencing reads are successfully aligned.
- Create files for visualization and detect regions of significant enrichment with respect to local background using ZINBA20. TROUBLESHOOTING.
- Assess cross-replicate correlation using an algorithm such as IDR26.
- Detection by tiling DNA microarrays – FAIRE-chip.
- Follow manufacturer’s recommended protocols and refer to Lee et al (2006)32 for sample labeling, hybridization and image acquisition procedures.
- For dual channel platforms (such as NimbleGen) data for each probe on the microarray is expressed as a log2 ratio, which is normalized by calculating the Z-score for each probe.
- Identify regions enriched by FAIRE; this can be accomplished using most peak-finding algorithms used for ChIP-chip. We recommend ChIPOTle44 or Mixer45. If applicable, the window size should be sufficiently large enough to contain approximately 10 probes and the step size should be set to equal the resolution of the microarray.
- Detection by quantitative PCR – FAIRE-qPCR
- In each well of a MicroAmp™ Optical 96-Well reaction plate, add 12.5 uL of 2X SYBR green master mix, 5 uL of DNA (2.5 to 25 ng/uL, ideal is 5 ng/uL), 1 uL primers (20 uM concentration) and 6.5 uL dH20.
- Seal plate with optical adhesive film
- Cycling parameters will vary, but typically 50 °C for 2 min, 95 °C for 10 min and the 40 cycles of 95 °C for 15 sec and 60 °C for 1 min and finally 60 °C for 1 min.
- Calculate the relative enrichment for each amplicon using the comparative Ct method46, such that a ratio is calculated for the signal from the FAIRE sample relative to the signal from input control DNA.
Timing
Day 1: Steps 1–8 (approximately 7–8 hours and overnight incubation)
Day 2: Steps 9–31 (approximately 3–4 hours)
Day 2: Steps 32–56 (approximately 3–4 hours and overnight incubation)
Day 3: Steps 57–59 (approximately 1 hour)
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 29 | Low input control yield | Low starting cell number | Start experiment with more cells or larger tissue (step 1) |
| Poor cell lysis | Vary dissociation and cell lysis conditions (step 1) | ||
| 30 | Sheared chromatin has incorrect average fragment length | Solution foamed | Make sure sonicator tip is centered and located ¼ to ½ an inch from bottom of tube and that sample has been cooled in ice-water bath (step 2) |
| Under-sonicated | Increase number of sonication cycles (step 2) | ||
| Under-fixation | Insufficiently crosslinked chromatin will lead to production of very small fragments. Increase fixation time or vary fixation conditions (step 1) | ||
| 36 | Aqueous layer is cloudy | Phenol may be overloaded due to high cell number | Start experiment with fewer cells or smaller tissue (step 1) |
| 58 | High DNA yield | Under-fixation | Insufficiently crosslinked chromatin will lead to high DNA yields with respect to input control. Increase fixation time or vary fixation conditions (step 1) |
| 58 | Low DNA yield | Low starting cell number | Start experiment with more cells or larger tissue (step 1) |
| Over-fixation | Over-crosslinking will reduce recovery of nucleosome-depleted regions. Reduce fixation time or vary fixation conditions (step 1) | ||
| 59 | Poor signal-to-noise | Under-fixation | Insufficiently crosslinked chromatin will lead to decreased enrichment by FAIRE. Increase fixation time or vary fixation conditions (step 1) |
Anticipated Results
Visualize FAIRE-seq or FAIRE-chip data in a browser such as the UCSC Genome Browser47. For data from human cells or tissues, we expect to see enrichment similar to that presented in Figure 3a. This genomic locus on chromosome 19 contains several genes that each contain a nucleosome-depleted promoter detectable in nearly every cell or tissue type assayed to date, including all Tier-1 and Tier-2 cell types assayed by ENCODE (a total of 19 cell types to date)21. Additionally, there are some cell-type-selective regions of open chromatin, such as the region immediately upstream of CNOT3, which is selective for embryonic stem cells and HepG2. The aggregated FAIRE signal around all transcription start sites (TSS) ranked by their gene expression should be similar to that presented in Figure 3b, showing a strong nucleosome-free region approximately 125 bp upstream of TSS and depletion (representing a well-positioned nucleosome) immediately downstream of TSS. The average signal across all genes is presented in Figure 3c. The number of regions of the genome enriched by FAIRE should be approximately 100,000 in any given cell or tissue type. FAIRE additionally detects distal regulatory regions, such as those marked by CTCF (Figure 3d).
Figure 3.
Expected results from FAIRE-seq experiments. A. Genomic locus residing on chromosome 19 as visualized with the UCSC Genome Browser47 shows consistent FAIRE enrichment at transcriptional start sites (TSS) across seven ENCODE cell lines16. Data are presented as number of aligned, in silico extended reads per base, on a scale of 0 to 50 reads. Pink coloring atop tall peaks of enrichment represent where signal exceeded this range. B. Heatmap of normalized GM12878 FAIRE signal ±3kb around TSS ranked by gene expression in GM12878 cells. Color was assigned on a log2 scale of −6 (background) to −2 (enriched). C. Average GM12878 FAIRE signal ±3kb around TSS across all genes. Enrichment peaks around −125bp. D. Average GM12878 FAIRE signal ±3kb around GM12878 CTCF sites, representing a class of distal regulatory elements.
Acknowledgments
We would like to acknowledge members of the Lieb and Davis labs for their constructive feedback. Support for this work was provided by grants from the NHGRI.
Footnotes
Key references:
Hogan, G.J., Lee, C.-K., and Lieb, J.D., PLoS Genet 2 (9), e158 (2006).
Giresi, P.G., Kim, J., McDaniell, R.M. et al., Genome Res 17 (6), 877 (2007).
Giresi, P.G. and Lieb, J.D., Methods 48 (3), 233 (2009).
Author Contributions
The work presented here was carried out in collaboration between all authors. PG and JMS designed and improved the method. JMS, PG, IJD, and JDL wrote the manuscript. All authors have contributed to, seen, and approved of the manuscript.
Competing Interests
The authors declare that they have no competing financial interests.
Contributor Information
Jeremy M. Simon, Email: jmsimon@unc.edu.
Paul G. Giresi, Email: paulg@email.unc.edu.
Ian J. Davis, Email: ian_davis@med.unc.edu.
Jason D. Lieb, Email: jlieb@bio.unc.edu.
References
- 1.Boyle AP, Song L, Lee B-K, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Research. 2011;21(3):456. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford GE, Holt IE, Whittle J, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Research. 2006;16(1):123. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle AP, Davis S, Shulha HP, et al. High-Resolution Mapping and†Characterization of Open Chromatin across the Genome. Cell. 2008;132(2):311. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L, Crawford GE. DNase-seq: A High-Resolution Technique for Mapping Active Gene Regulatory Elements across the Genome from Mammalian Cells. Cold Spring Harb Protoc. 2010;2010(2) doi: 10.1101/pdb.prot5384. pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keene MA, Corces V, Lowenhaupt K, et al. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proceedings of the National Academy of Sciences. 1981;78(1):143. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGhee JD, Wood WI, Dolan M, et al. A 200 base pair region at the 5' end of the chicken adult [beta]-globin gene is accessible to nuclease digestion. Cell. 1981;27(1, Part 2):45. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- 7.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421(6921):448. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 8.Gross DS, Garrard WT. Nuclease Hypersensitive Sites in Chromatin. Annual Review of Biochemistry. 1988;57(1):159. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 9.Stalder J, Larsen A, Engel JD, et al. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980;20(2):451. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- 10.Hogan GJ, Lee C-K, Lieb JD. Cell Cycle-Specified Fluctuation of Nucleosome Occupancy at Gene Promoters. PLoS Genet. 2006;2(9):e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giresi PG, Kim J, McDaniell RM, et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48(3):233. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giresi PG, Kim J, McDaniell RM, et al. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Research. 2007;17(6):877. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy PL, Cleary ML, Brown PO, et al. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6364. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, Zhang Z, Grasfeder LL, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Research. 2011 doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponts N, Harris EY, Prudhomme J, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Research. 2010;20(2):228. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louwers M, Bader R, Haring M, et al. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell. 2009;21(3):832. doi: 10.1105/tpc.108.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashid N, Giresi PG, Ibrahim JG, et al. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12(7):R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium T.E.P. A User's Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E, Lieb JD, Furey TS, et al. Allele-specific and heritable chromatin signatures in humans. Human Molecular Genetics. 2010;19(R2):R204. doi: 10.1093/hmg/ddq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurtado A, Holmes KA, Ross-Innes CS, et al. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eeckhoute J, Lupien M, Meyer C, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Research. 2009 doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egelhofer TA, Minoda A, Klugman S, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18(1):91. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, BJB, Huang H, Bickel P. Measuring reproducibility of high-throughput experiments. Annals of Applied Statistics. 2011 in press. [Google Scholar]
- 27.Lassmann T, Hayashizaki Y, Daub CO. TagDust--a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25(21):2839. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows, ÄìWheeler transform. Bioinformatics. 2009;25(14):1754. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle AP, Guinney J, Crawford GE, et al. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24(21):2537. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haring M, Offermann S, Danker T, et al. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3(1):11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protocols. 2006;1(2):729. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren B, Dynlacht BD. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 2004;376:304. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- 34.Oberley MJ, Tsao J, Yau P, et al. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 35.Oberley MJ, Farnham PJ. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- 36.Lieb JD. Genome-wide mapping of protein-DNA interactions by chromatin immunoprecipitation and DNA microarray hybridization. Methods Mol. Biol. 2003;224:99. doi: 10.1385/1-59259-364-X:99. [DOI] [PubMed] [Google Scholar]
- 37.Ciccone DN, Morshead KB, Oettinger MA. Chromatin immunoprecipitation in the analysis of large chromatin domains across murine antigen receptor loci. Methods Enzymol. 2004;376:334. doi: 10.1016/S0076-6879(03)76022-4. [DOI] [PubMed] [Google Scholar]
- 38.Chaya D, Zaret KS. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol. 2004;376:361. doi: 10.1016/S0076-6879(03)76024-8. [DOI] [PubMed] [Google Scholar]
- 39.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein BE, Humphrey EL, Liu CL, et al. The use of chromatin immunoprecipitation assays in genome-wide analyses of histone modifications. Methods Enzymol. 2004;376:349. doi: 10.1016/S0076-6879(03)76023-6. [DOI] [PubMed] [Google Scholar]
- 41.Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004;376:269. doi: 10.1016/S0076-6879(03)76018-2. [DOI] [PubMed] [Google Scholar]
- 42.Nammo T, Rodriguez-Segui SA, Ferrer J. Mapping open chromatin with formaldehyde-assisted isolation of regulatory elements. Methods Mol Biol. 2011;791(1940-6029 (Electronic)):287. doi: 10.1007/978-1-61779-316-5_21. [DOI] [PubMed] [Google Scholar]
- 43.Ren B, Robert F, Wyrick JJ, et al. Genome-Wide Location and Function of DNA Binding Proteins. Science. 2000;290(5500):2306. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 44.Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6(11):R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun W, Buck M, Patel M, et al. Improved ChIP-chip analysis by a mixture model approach. BMC Bioinformatics. 2009;10(1):173. doi: 10.1186/1471-2105-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protocols. 2008;3(6):1101. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 47.Fujita PA, Rhead B, Zweig AS, et al. The UCSC Genome Browser database: update 2011. Nucleic acids research. 2011;39(Database issue):D876. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]