Abstract
The Human Connectome Project (HCP) seeks to map the structural and functional connections between network elements in the human brain. Magnetoencephalography (MEG) provides a temporally rich source of information on brain network dynamics and represents one source of functional connectivity data to be provided by the HCP. High quality MEG data will be collected from 50 twin pairs both in the resting state and during performance of motor, working memory and language tasks. These data will be available to the general community. Additionally, using the cortical parcellation scheme common to all imaging modalities, the HCP will provide processing pipelines for calculating connection matrices as a function of time and frequency. Together with structural and functional data generated using magnetic resonance imaging methods, these data represent a unique opportunity to investigate brain network connectivity in a large cohort of normal adult human subjects. The analysis pipeline software and the dynamic connectivity matrices that it generates will all be made freely available to the research community.
Keywords: Magnetoencephalography (MEG), connectome, processing pipeline, functional connectivity
1. INTRODUCTION
The connectome was conceived as a comprehensive structural description of the network elements and connections comprising the human brain (Sporns et al., 2005). This connectome theoretically constitutes the structural support for brain function. While such a connectome could be conceived at multiple scales, the Human Connectome Project (HCP) has chosen to work at the macroscale level, in which distinct brain regions comprised of large neuronal populations are defined as network nodes (whether based on individual voxels or on voxel clusters derived from functional imaging studies) between which both structural and functional connections are defined (Van Essen et al., 2013; Van Essen et al., 2012).
For macroscopic connectome representation, the primary source of structural connection data lies in diffusion weighted magnetic resonance imaging (dMRI) methods (Sporns, 2011) which return a static map of resolvable anatomical connections between brain regions. While brain activity is underpinned by anatomical connectivity, it cannot be understood in those terms alone. Thus, for example, functional connectivity may be seen in the absence of direct anatomical connections (Deco et al., 2011; Honey et al., 2009; Vincent et al., 2007). The value of correlating structural and functional connectivity has been recognized for many years (Rubinov et al., 2009; Sporns et al., 2005). Functional information is provided to the HCP from two sources, which differ in their spatial and temporal resolution as well as in the basis of their signal generation (see Table 1). While fMRI provides a vascular surrogate for neural activity that broadly correlates with anatomical connectivity (Honey et al., 2010; Rubinov et al., 2009; Skudlarski et al., 2008), MEG represents population neuronal activity, which is likely to have a less straightforward correspondence to dMRI-derived anatomical mappings due to the rapid transit of information through indirect pathways (Rubinov et al., 2009). Inclusion of MEG data provides a measure of brain network connectivity at time scales used in neural communication, an important adjunct to that provided by the more static maps provided by anatomical and BOLD functional connectivity data.
Table 1.
Connectivity characteristics of imaging modalities
| Dynamic structure |
Imaging basis |
Imaging characteri stics |
Variety of connecti vity metrics |
|
|---|---|---|---|---|
|
Anatomical
connectivity |
static | Anisotropic diffusion of water |
Recoverabl e with DTI/DSI (~1 mm resolution) |
One |
|
fMRI
connectivity |
Weakly nonstation ary, frequency range nominally <0.1 Hz |
Indirect measure of neural population activity (Hemodyna mic response to synchronized neural population activity, BOLD) |
Recoverabl e with fMRI (~3 mm resolution) |
A few |
|
Electrophysiol
ogical connectivity |
Strongly nonstation ary, frequency range DC to ~1,000 Hz |
Direct measure of synchronized neural population activity (magnetic/el ectric fields generated by synchronized synaptic current) |
Model dependent (~ 4mm precision for sparse cortical activations, ~ 10 mm spatial resolution) |
Many |
As a macroscale measure, MEG represents the activity of neuronal populations in which communication has been shown to be accomplished in part via synchronized oscillatory activity which has been related to binding of relevant, or inhibition of irrelevant, information during cognitive activity (Buzsaki, 2009; Fries, 2009; Singer, 1993; Stanley, 2013; Varela et al., 2001). The goal of the MEG component of the HCP is to integrate the temporal information provided by electrophysiological methods with the structural data inherent in anatomical connectome and fMRI functional connectivity studies to better understand the way in which brain networks transmit and process information.
Owing to substantial differences in spatial resolution, temporal resolution and coverage, relating electromagnetic brain activity to the anatomical and fMRI connectome is challenging (Table 1). In the spatial domain, the anatomical connectome is resolvable to submillimeter precision (Calamante et al., 2011; Calamante et al., 2010; Ugerbil et al., 2013). The spatial resolution of fMRI in the HCP will be 2 millimeter isotropic voxels (see (Ugerbil et al., 2013)). In contrast, the spatial resolution of electrophysiological methods is, at best, 10 mm at the cortical surface (Lin et al., 2006). In the temporal domain, the anatomical connectome is, by definition, static. For fMRI, the upper frequency limit of physiologically meaningful signal plausibly is ~0.1 Hz (Hathout et al., 1999) while that for MEG could be up to 1 kHz (Xiang et al., 2009). This difference in spectral content translates to a substantial difference in information acquired over a typical recording epoch. Thus, MEG supports a wide variety of analytic strategies for characterizing signal interactions between regional brain pairs that are not accessible to fMRI.
The HCP will deliver multimodal neuroimaging and behavioral data on a large cohort of subjects with the goal of characterizing normal brain connectivity patterns in healthy adult human subjects (Barch et al., 2013; Van Essen et al., 2012). In keeping with initial definitions of the human connectome, a major focus of the HCP will lie in characterization of structural brain connections. Expansion of the concept of connectome to investigation of functional networks will include both fMRI and MEG. Here we present the first full description of the MEG component of the HCP. While the expectation is that this effort will lead to substantial new contributions in the study of dynamic brain network connectivity, the following will focus primarily on the fundamental approach taken in creating an electrophysiological complement to the human connectome.
2. THE ELECTROPHYSIOLOGICAL APPROACH TO CONNECTOMICS
A major goal of the HCP, in addition to data acquisition, is to provide automated pipelines for processing of the data made publicly available via Connectome DB, with visualization tools provided by the Connectome Workbench (Glasser et al., 2013; Marcus et al., 2013; Marcus et al., 2011). The MEG component of the HCP will provide one set of utility pipelines and two connectivity pipelines (see 2.2-2.5 below).
2.1. Data collection
MEG data is collected on a whole head MAGNES 3600 (4D Neuroimaging, San Diego, CA) system housed in a magnetically shielded room, located at the Saint Louis University medical campus (Figure 1). Data collection follows best practices for MEG data acquisition as detailed by Gross and colleagues (Gross et al., 2013). One hundred subjects (50 same-sex monozygotic twin pairs) will be studied using both resting and task evoked MEG. The use of twin pairs allows evaluation of heritable traits, and recent studies have shown that MEG responses to somatosensory stimuli are partly heritable (Van’t Ent et al., 2010). These 100 subjects will also participate in the full HCP study design including structural and functional magnetic resonance image studies (MRI, fMRI), diffusion tensor imaging (dMRI) and behavioral and genetic testing (see (Van Essen et al., 2012)). Significant effort has been made to ensure that MEG and fMRI are performed on the same subjects using the same tasks performed, as closely as possible, with the same timing. Together with behavioral and genetic information in these subjects, these data will facilitate the study of individual variability across a large population.
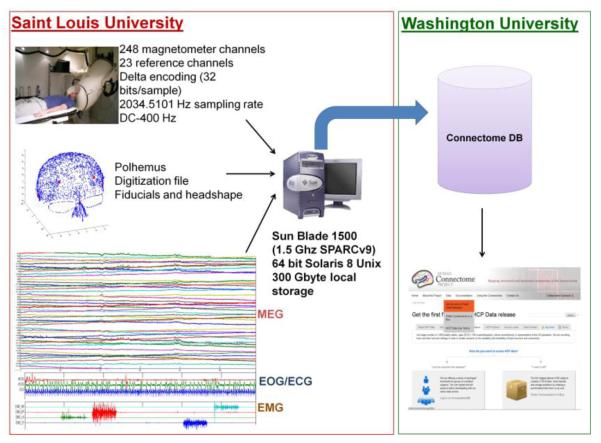
Figure 1.
Data Acquisition. Data will be collected on a whole head 4D Neuroimaging system housed in a magnetically shielded room at Saint Louis University (SLU). The MEG system includes 248 magnetometer channels together with 23 reference channels. Data will be sampled at 2 kHz and delta encoding is used to increase readout resolution, providing a quantization of ~0.3 fT. Spatial digitization of head shape and fiducials will be accomplished using a Polhemus FASTRAK-III system. A Sun Blade running 64 bit Solaris 8 Unix and communicating with a dedicated UltraAX-12 system is used for data collection and sensor configuration. Following data acquisition, data will be uploaded via https to the Washington University in St. Louis (WashU) internal HCP database. Public data releases will be accomplished through the external ConnectomeDB interface.
2.2. Utility pipelines: Quality control and artifact identification
Empty room measurements (5 min) are acquired daily and used to monitor the MEG system for hardware malfunctions or excessive environmental noise. These data are compared to a set of reference scans collected to estimate baseline noise levels and corrective action will be taken appropriately.
During pre-processing of human data, one quality assessment (QA) module detects noisy channels by examining the signal similarity between each sensor and its neighbors. Channels exhibiting poor correlation or high variance ratio to neighboring channels (Winter et al., 2007) will be flagged as bad and removed from further analysis. A second QA module based on an iterative independent components analysis (ICA) algorithm removes bad channels and bad segments using spatial and temporal criteria (Mantini et al., 2011).
Physiological artifacts are identified using ICA (Mantini et al., 2011). Signal decomposition is done iteratively starting from different initial guesses. For each decomposition, independent components (ICs) are classified as ‘Brain’ or ‘Noise’ using six parameters derived from a large number of recordings. The values of three parameters (correlation between IC signals, correlation between power time courses, and correlation between spectra) are thresholded based on receiver operating characteristics (ROC) obtained from a large number of subject’s MEG data. Three additional parameters derived from both spectral and temporal properties are added to aid in classification of system or environmental noise (see (Mantini et al., 2011). The final classification is made automatically by selecting the iteration accounting for the highest brain component subspace dimensionality and the lowest residual artifact contamination. Identified noise components usually consist of magneto- and electro-cardiogram, eye movements, power supply bursting and 1/f –like environmental noise.
2.3. Utility pipelines: Anatomy processing and source reconstruction
The anatomy pipeline links MEG to structural MRI (sMRI) and dMRI. This pipeline begins with co-registration of the MEG sensors to the anatomical coordinate system of the sMRI data. The sMRI data is then used to generate volume conduction models of the head and provide anatomical constraints for source localization analyses.
Accurate source reconstruction of MEG data is critical, since it is a prerequisite for comparison of the electrophysiological results to those obtained from other imaging modalities. Working in source space is necessary to avoid errors in connectivity analyses (Schoffelen and Gross, 2009) and provides a more straightforward physiological interpretation of the results (Schoffelen and Gross, 2009). Three source reconstruction strategies are implemented. For resting state data (Figure 2) weighted minimum-norm estimates (wMNE) are used to reconstruct source space current density maps from sensor-space ICs (Hyvarinen and Oja, 2000). In this approach, based on the work of de Pasquale and colleagues (de Pasquale et al., 2010; de Pasquale et al., 2012; Mantini et al., 2011), the regularization for the minimum norm estimation is tuned to each IC and obtains computationally efficient and reliable projections of resting activity into source space. For task data, two beamformer reconstruction approaches will be used. Linear constrained minimum variance beamformers (LCMV) reconstruct source space data in the time domain and have proven useful for detection of connectivity in oscillatory brain activity (Brookes et al., 2011a; Schoffelen and Gross, 2009). Dynamic imaging of coherent source (DICS) reconstructs source-space data in the frequency domain (Gross et al., 2001; Van Veen et al., 1997). Following source reconstruction, seed-based or data-driven group-ICA methods can be used to process data for dynamic connectivity.
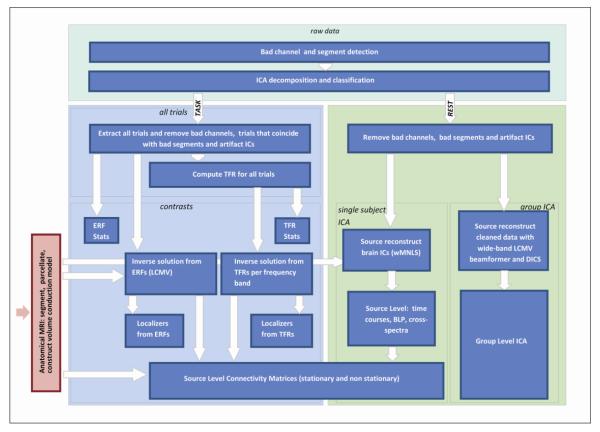
Figure 2.
MEG Analysis Pipelines. The upper part of the pipeline labelled as “raw data” operates on the entire data set and is shared between the task and rest pipelines. For TASK (left panel), the box labelled as “all trials” operates on the concatenated epochs from all conditions; the box labelled “contrasts” operates on epochs of individual conditions and contrasts between conditions. For REST (right panel), the box labelled “single subject ICA” operates on temporal ICs at the individual level and the box labelled “group level ICA” operates on spatial ICA at the group level.
The advantage of beamformers is that they are adaptive, data-driven methods for deriving the inverse solution from empirical evidence (sensor-space covariance or cross-spectral density). Beamformers solve the inverse problem for each source independently. In contrast, minimum norm solutions are model-driven methods for computing the inverse solution. Minimum norm inverse estimates are affected by the topology of the source model. The FieldTrip toolbox (Oostenveld et al., 2011) supports wMNE, LCMV and DICS.
Additional processing is performed to map the electrophysiological connectivity matrices onto the anatomical representation of the connectome. An important aspect of this mapping is the use of a scheme by which network nodes are defined on a common anatomical parcellation. MEG source reconstructions may consist of up to ~8000 nodes with electrophysiological connections between 64 million node pairs. Dense connectivity matrices produced using fMRI or dMRI consist of several orders of magnitude greater numbers of nodes, which are typically grouped into functionally or anatomically consistent regions. For visualization, the electrophysiological connections are mapped onto this anatomically parcellated representation.
2.4. Resting state MEG (rMEG)
Evidence from multiple imaging studies shows that the brain at rest is organized as a complex set of networks (Beckmann et al., 2005; Biswal et al., 1995; Brookes et al., 2011a; Damoiseaux et al., 2006; de Pasquale et al., 2010; de Pasquale et al., 2012; Fiecas et al., 2012; Fox and Raichle, 2007; Fox et al., 2005) in which the two fundamental properties of functional segregation and dynamic integration (Friston, 2002) can be implemented. Three successive rMEG runs are collected at the beginning of each experimental session during which subjects are instructed to lie quietly with eyes fixed on a central cross.
Recent source level analyses of resting state MEG networks indicate that stationary and non-stationary interactions occur within and/or across networks and vary according to oscillation frequency (Brookes et al., 2011b; Brookes et al., 2011c; de Pasquale et al., 2010; de Pasquale et al., 2012; Guggisberg et al., 2008; Hipp et al., 2012; Martino et al., 2011; Marzetti et al., 2013; Sekihara et al., 2011). Two data-driven approaches will be used to study this phenomenology. The first utilizes seed-based functional connectivity in which signal stationarity is not assumed a priori (de Pasquale et al., 2010; de Pasquale et al., 2012). The second approach uses a data-driven group-ICA for resting state network (RSN) identification (Brookes et al., 2011b; Brookes et al., 2011c).
2.4.1 Seed based functional connectivity
The approach developed by de Pasquale et al. (2010, 2012) is based on identifying temporal epochs during which source space band limited power (BLP) correlations within RSNs are maximal. These epochs are termed maximal correlation windows (MCWs). The spatial topography of RSNs is derived from independent fMRI experiments. Critically, the temporal frequencies over which BLP correlations are defined are in the infraslow range (<0.1 Hz), i.e., comparable to frequencies accessed by fMRI.
In addition to correlation, alternative connectivity metrics applicable to faster time scales will be computed. One such alternative is the imaginary part of coherence obtained by the cross-spectrum normalized by the power (Nolte et al., 2004), as is done for task-related data where it is used to map frequency specific lagged interactions. Due to potential mismatching in functional to structural image co-registration, together with the potential to include currents oriented in multiple directions due to the size of voxel patches used in analysis, local activity cannot be assumed to occur along one principal source orientation and an alternative strategy is needed. The Multivariate Interaction Measure (MIM) is a frequency-specific connectivity measure (Ewald et al., 2011) that allows characterization of interactions between multi-dimensional subspaces. Electrophysiological activity is generated by current flow, which inherently is a vector quantity while the blood oxygen level dependent (BOLD) signal (the physiological basis of fMRI) is a scalar quantity. MIM is based on finding the vector weights that maximize the imaginary part of coherence between two brain loci. In a seed-based approach, MIM provides seed- and frequency-specific maps of lagged interactions. The MIM measure excludes non-lagged (i.e. instantaneous) correlations, producing results indicative of lagged interactions that are physiologically meaningful (Marzetti et al., 2013). MIM, therefore, cannot be used to study zero-lagged interactions. However, MIM can be used to study coupling between networks or parcels at electrophysiological frequencies (1-800 Hz range).
2.4.2 Data-driven group-ICA approach
Source space group-ICA analysis is a technique that identifies RSNs in MEG data (Brookes et al., 2011c). Resting state fMRI data is typically analyzed using spatial ICA (Beckmann et al., 2005) to derive RSNs of functional (Smith et al., 2009) and clinical relevance (Filippini et al., 2009). The HCP will apply temporal ICA to concatenated source-space MEG data represented as BLP time series (Brookes et al., 2011c; de Pasquale et al., 2010; de Pasquale et al., 2012; Hipp et al., 2012)(see 2.4.1 above). Importantly, BLP time series are low pass filtered using a moving average with a time window of 2 seconds to enhance the detection of functional connectivity (Luckhoo et al., 2012) at low frequencies consistent with those inherent to fMRI. In contrast to seed-based MEG functional connectivity, ICA analyses are independent of fMRI-derived spatial priors. The comparison of MEG RSN spatial topography with that defined by fMRI is a crucial step, since the original observations of RSN came from fMRI.
2.5. Task MEG (tMEG)
HCP task paradigms to be studies with MEG are a subset of those used in tfMRI (Barch et al., 2013). The MEG paradigms are matched in temporal structure to fMRI tasks to maximize the scientific value of the multi-modal design of the HCP (Van Essen et al., 2012). Adapting the paradigms for MEG requires increasing the total number of trials to overcome the lower signal-to-noise properties of non-invasive electrophysiology.
2.5.1 Experimental paradigms for tMEG
Three experimental paradigms will be used to provide data on sensory-motor, working memory and language processing.
(1) In the sensory-motor task, participants execute a simple hand or foot movement. Which limb on which side is instructed by a visual cue, which serves to pace the movement (Figure 3a). The paradigm includes task and rest blocks (Figure 3b). Motor tasks modulate power in sensorimotor cortex, mainly in the alpha (mu), beta and gamma bands (Crone et al., 1998; Miller et al., 2009; Pfurtscheller and Lopes da Silva, 1999) and motor preparation and execution have been shown to be dependent on corticospinal gamma-band coherence (Schoffelen et al., 2005). Open questions remain concerning the sensorimotor areas in which these frequency specific modulations occur (Jurkiewicz et al., 2006) and how the different oscillatory modes interact.
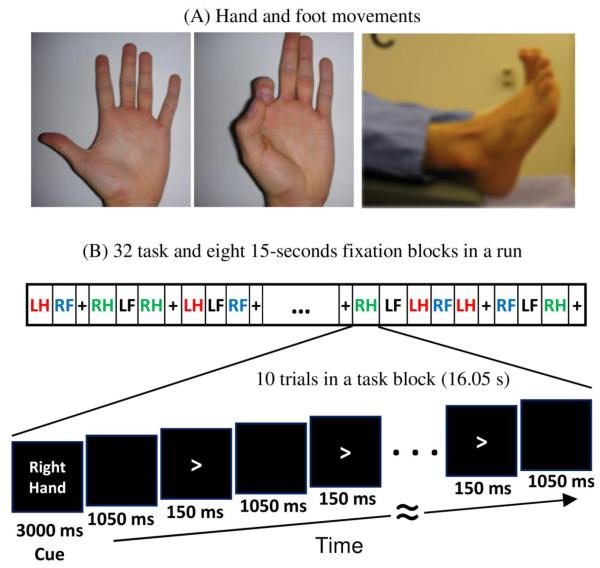
Figure 3.
The motor task paradigm consists of a set of hand and foot movements (A) performed with both right (RH/RF) and left (LH/LF) limbs. A block design matching that used in task fMRI paces each movement (B). Each block begins with a 3 sec cue telling the subject which appendage to move in that trial.
(2) In the working memory task, participants are presented with pictures of tools or faces (Figure 4). Working memory load is controlled by alternating 0-back and 2-back conditions (Brookes et al., 2011b). Match and no-match responses are recorded by right index and right middle finger button presses respectively. Working memory tasks provide an interface between perception, long term memory and action (Baddeley, 2003). fMRI studies indicate that working memory tasks activate mainly prefrontal and parietal cortical areas (Collette et al., 2006; Curtis and D’Esposito, 2003). Electrophysiological studies of working memory have demonstrated that power and coherence in theta (Brookes et al., 2011b; Klimesch, 2006) and alpha bands (Jensen et al., 2002; Khader et al., 2010) increase with memory load. The role of gamma band is still unclear (Brookes et al., 2011c; Howard et al., 2003; Roux et al., 2012).
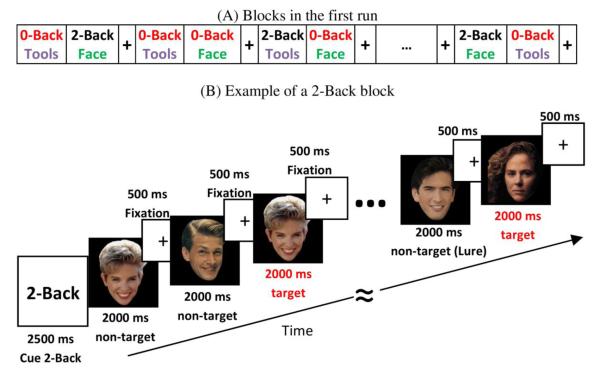
Figure 4.
The working memory paradigm is designed to match the N-back task performed during task fMRI. For MEG studies, only two categories are presented (A) in a block design of 16 task blocks in each of two runs. (B) illustrates the design for a 2-back face task.
(3) The language processing task will be the same as that used in the tfMRI component of the HCP (Barch et al., 2013). Subjects will listen either to auditory narratives (30 sec duration) or matched-duration simple arithmetic problems (Binder et al., 2011) followed by a 2-alternative forced choice question. Subjects will respond by right hand button press (index or middle finger). While this is a somewhat unusual task for MEG studies, it has been shown to activate a large cortically distributed language network (Barch et al., 2013). It is reported that slow temporal modulation of speech is synchronized with low-frequency (δ and θ) cortical activity during speech recognition (Ding and Simon, 2013); the precision of the speech recognition can be predicted from this synchronization. Phase locking of cortical oscillation to the low-frequency (4-8 Hz) envelope of speech is predictive of important speech events (Peelle and Davis, 2012). Use of the same task paradigm in MEG and fMRI will facilitate integrated analysis of the combined datasets (Hari and Salmelin, 2012; Pulvermueller, 2010; Renvall et al., 2012).
2.5.2 T-MEG processing pipeline
The tMEG processing pipeline is illustrated in Figure 2. For each task data set, all trials for all conditions are extracted over a predefined pre- and post-stimulus time interval. Bad channels, noisy time segments and artifact ICs are removed to produce a “clean” set of all trials in the time domain (event related field, ERF). Each trial is then analyzed in the time-frequency domain (spectral modulations as a function of latency relative to task events) using the multitaper method (Mitra and Pesaran, 1999; Oostenveld et al., 2011). Sensor-level time frequency analysis is computed for a typical range of frequencies (1 to 100 Hz) to produce a set of all trials in the time-frequency domain (TFR).
Following the separation of trials into conditions, response averages are computed and contrasts between conditions are defined. The first level of analysis is computation of basic statistics in sensor space. Sensor-level analysis provides early insight into whether expected effects of the task on brain activity are observed. The LCMV beamformer is used to compute TFR contrasts between conditions based on sensory covariance. DICS is a beamformer in the frequency domain. DIC operates on the sensor-space cross-spectral density matrix to compute band-limited power and coherence in source-space (see section 2.3). We will apply this method to compute BLP contrasts across conditions in all experimental tasks. Complete 1 Hz spectral resolution representation of responses in source space is not feasible owing to memory and storage limitations. Therefore, the spectrum is divided into frequency bands known to play distinct roles in brain function under various conditions (δ(1-4), θ(4-7), α(8-13), β-lower(13 20), β-upper(20 30), γ-lower(30-50), γ-upper(50-90)).
The final step in the tMEG pipeline is computation of connectivity metrics. These metrics are based on TFR source level representations (see section 2.5.3 below).
2.5.3 Connectivity metrics in tMEG
Numerous functional and effective connectivity metrics are available to assess brain networks as participants perform cognitive tasks. In general, these can be computed both in sensor and source space. For task based data analysis, connectivity will be initially assessed with a set of established metrics selected for computational efficiency. This set of measures includes coherence, imaginary coherence, MIM, phase locking value (PLV), phase slope index (PSI), bivariate Granger causality and power envelope correlation. Each of these metrics is available in the FieldTrip toolbox (Oostenveld et al., 2011).
Coherence (correlation in the frequency domain) is the most basic measure of synchrony between two oscillatory signals. However, simple coherence suffers from bias resulting from field spread. MIM (see section 2.4.1) avoids this problem. PLV (Lachaux et al., 1999) is similar to coherence but with the effect of amplitude in synchrony estimates removed. However it still depends on the SNR of the data and significant power changes can lead to confounded PLV differences(Muthukumaraswamy and Singh, 2011). The PSI (Nolte et al., 2008) is a metric tailored to provide robust estimates of the direction of information flow in the presence of independent background activity. The theoretical basis of PSI is that a causal relationship between two signals at a consistent time lag manifests as a constant proportionality between cross-spectral phase and frequency. Granger causality (Geweke, 1984; Granger, 1969) is a technique for estimating causal relations between time-series. The method fundamentally tests for the ability of one time series to forecast the behavior of a second time series, under the assumption that the first causally drives the second. Bivariate Granger causality is implemented is based on the derivation of the transfer function of the system from its time-frequency cross-spectra (Chen et al., 2009). Finally, instantaneous power envelope correlations have been used (Hipp et al., 2012) to quantify concurrent variations in symmetrical brain sub-networks in resting state data. This method removes field spread effects through a joint orthogonalization and can be used to infer functional connectivity even between spatially proximate areas.
3. HCP MEG DELIVERABLES
The ConnectomeDB is the public face of the HCP (Marcus et al., 2013). The ConnectomeDB will manage all MEG data and analysis results and be the primary site from which raw and processed data may be downloaded (Marcus et al., 2011). All data will be shared in standardized formats or well-defined custom formats should standard formats not be readily available. Imaging data will be shared in the original DICOM or in NIfTI format, with dense and sparse connectivity matrices shared in the CIFTI format (Glasser et al., 2013; Marcus et al., 2011). Since there is no commonly accepted standard format for MEG data, both MEG and EEG data will be shared in the original 4D Magnes file format. Open Source reading routines for the 4D data format are provided in FieldTrip and in compiled form, which allow conversion of the data to other formats, including plain ASCII or binary. Parts of the processed data will be shared in native MATLAB format, such as the volume conduction models. Small files, such as lists of electrode positions and the specification of bad channels and time segments, will be shared in ASCII format. The provided compiled software will allow for conversion of all native MATLAB files into an ASCII representation provide MATLAB-independent users full access to the data.
The ConnectomeDB will also manage provenance information for HCP data. Data and results will be released in versioned release packages. Results produced by HCP processing pipelines will be linked to the version of the processing software used to create that set of results. The specific processing parameters will be included in processing scripts that are released with the software. The goal is to completely document all processing used to produce intermediate and final results. This provenance information will permit the research community to not only explore and use HCP generated results via Connectome Workbench but to move data sets into their own environment and re-analyze them starting at arbitrary points in the pipeline.
3.1 MEG Data
MEG data and associated metadata (e.g., sampling rate, event timing, bad channels and data segments with excessive motion artifacts) will be managed in the ConnectomeDB. These data will be linked to descriptions of the experimental protocols and results of quality control processes. The utility pipeline identifies bad channels and segments for each run and each subject. These results will be shared in the form of ASCII files containing the bad channel names and the start and end times of each of the bad temporal segments in the respective run. These will be accompanied by figures (bitmap image files) presenting the topology and characteristics of the bad segments.
One utility pipeline step consists of multiple fastICA iterations used in automatic classification of signal into Brain and Noise components. The results of the best iteration will be provided to the user in the form of a mixing and unmixing matrix with the classification of each IC noted. Additionally, figures characterizing each component will be provided that include the IC sensor map and its corresponding time-course and power spectral density.
3.2 Software pipelines
In addition to providing access to raw MEG data, the HCP will also share the software that comprises the analysis pipelines. This will allow users of the HCP to reanalyze the shared data, optionally using different parameters with the various algorithms. Additionally, the software allows for the same analyses to be performed on MEG data that is or will be acquired outside the HCP.
All MEG analysis software will be implemented in MATLAB (Mathworks, Natick, MA), using the FieldTrip toolbox (Oostenveld et al., 2011). Most of the analyses will be implemented in scripts that call FieldTrip functions and custom written HCP functions. The scripts comprise the HCP MEG analysis pipelines and extend the FieldTrip toolbox according to the requirements of this project. The FieldTrip toolbox is freely available as Open Source under the General Public License (GPLv2) and analysis pipelines will be available for download.
Although all analysis scripts and functions are to be made available free of charge, the MATLAB environment on which they are built is a commercial software package that might not be available to all external HCP users. To enable all researchers to use the software, we will provide compiled versions of all pipelines using the MATLAB Compiler toolbox. The compiled executables use the MATLAB Compiler Runtime (MCR), which can be shared with the compiled executables and is available royalty-free from Mathworks. The executable and MCR will be provided for the 64 bit versions of the Linux, Mac OS X and Windows operating systems. The Linux version of these compiled applications, running on the Washington University Center for High Performance Computing (CHPC) system (Marcus et al., 2011), are the instantiation of the MEG pipelines used by the HCP team.
3.3 Anatomical data
Anatomical models are used to define the locations in the brain at which electrical source activity are estimated. Because HCP MRI data has been processed to remove facial features for subject privacy (Milchenko and Marcus, 2012) pre-computed head models will be made available for each subject.
The anatomy pipeline will generate three types of output: head models, source models and coregistration information. For forward modeling, a single shell model will be provided for MEG (Nolte, 2003). The BEM system matrix, computed with OpenMEEG (Gramfort et al., 2011) will be provided for source modeling. The models for distributed sources in the HCP are based on high-quality extracted cortical sheets. The volumetric source models for beamformer reconstruction are based on regular 3-D grids in MNI space, constructed from a template brain. Individual anatomies are (non-linearly) warped to this template brain, and we apply the inverse of this warp to the template source model. The individually warped 3-D volumetric grid facilitates group analysis, as it allows for computationally efficient averaging in normalized space without the need for interpolation. Coregistration information is provided to link subject-specific head coordinate systems to the standard coordinate system provided by the HCP. Co-registration is accomplished using FSL’s linear registration tool [FLIRT;(Greve and Fischl, 2009; Jenkinson et al., 2002; Jenkinson and Smith, 2001)].
3.4 rMEG results
The processing pipeline for rMEG data is modular, allowing for the application of different source projection algorithms. Sensor level analyses are used to identify independent components of brain origin, which are then projected onto source space using the source model provided by the Anatomy Pipeline. The single subject branch of the rMEG pipeline provides source level network information based on a wMNE algorithm (see section 2.5.2). As noted in section 2, these data are then analyzed for connectivity using either a correlation approach, or MIM (Ewald et al., 2011).
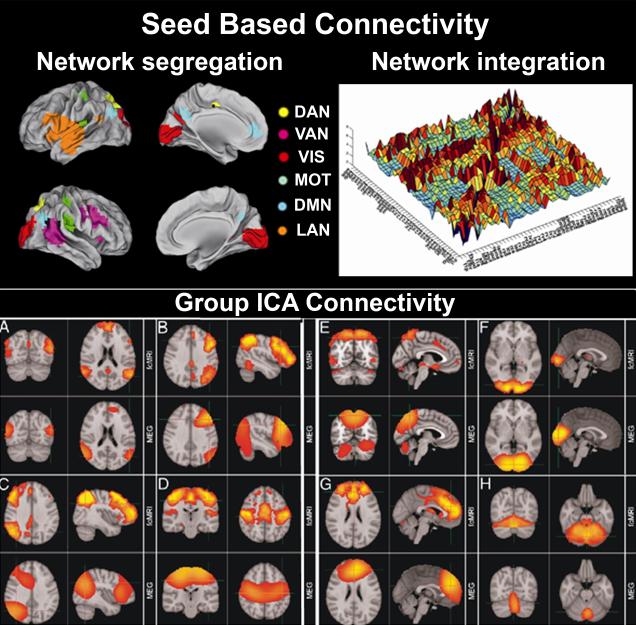
Band limited power in the classic frequency bands will be provided for each voxel/parcel in source space. At this stage, time-series data consists of matrices of dimension K × N, where K is the number of voxels/parcels in the brain and N is the number of time points. This data representation supports seed-based or a data-driven analyses. For the seed based approach, the MCW algorithm (see section 2.4.1) identifies epochs in which the contrast between within-network vs. external-to-network correlation is maximal, while the external-to-network correlation is minimal (de Pasquale et al., 2010; de Pasquale et al., 2012). The outputs provided by this approach are: i) a set of time points of the observed maximum correlation windows (MCWs); ii) seed-based connectivity matrices of every defined network (Figure 5, upper left); iii) cross-correlation matrices among different RSNs at the node level (Figure 5, upper right); iv) cross-correlation matrices among different RSNs averaged across nodes at the network level; v) temporal coverage of internal coupling of every network; vi) temporal overlap of MCWs from different RSNs.
Figure 5.
Resting State Network Analysis. Upper: Seed-based approach allowing the study of network segregation/integration. Upper left: segregated RSNs obtained in the related MCWs; upper right: 3D plot of covariance across RSN nodes showing the central role of DMN. Lower: Comparison of brain networks obtained using ICA independently on MEG [Lower] and fMRI [Upper] data. (A) DMN; (B) left lateral frontoparietal network; (C) right lateral frontoparietal network; (D) sensorimotor network; (E) medial parietal regions; (F) visual network; (G) frontal lobes including anterior cingulate cortex; (H) cerebellum.
Similarly, MIM (see section 2.4.1) for each frequency bin will be provided for each voxel/parcel in source space. The output of this connectivity pipeline is a connectome of size Nsources × Nsources × Nfrequency bins, where a source can be a voxel, a parcel or a network.
The group branch of the rMEG pipeline provides source level network information based on a LCMV beamformer algorithm (Schoffelen and Gross, 2009), and computation of RSN dynamics is run separately for each frequency band. The input data is BLP time courses over all K voxel/parcels for each of the J subjects in the group. Following low pass filtering, the BLP time courses are concatenated over subjects along the time dimension, to give an (NJ × K) data matrix, where N is the number of time points. This matrix is then fed into temporal ICA, using the fastICA algorithm. The output from this is the (Q × NJ) matrix of component time courses for each subject, and the (Q × K) matrix of component spatial maps, where Q is the dimensionality of the ICA. These Q component spatial maps correspond to the MEG resting state networks (Figure 5, lower). Group-ICA analyses depend on the selection of subjects and so cannot be precomputed. However, the Connectome Workbench will provide online selection of subjects based on user-specified criteria. The group-ICA computation will be submitted to the CHPC-HCP compute cluster and made available to the user when the job completes.
3.5 tMEG results
The tMEG pipeline input divides each pre-processed dataset into trials, providing a clean set of epochs in the time domain, which are subsequently transformed to time-frequency representations. The final output of the initial pipeline includes the sensor-level average ERFs and TFRs for all conditions, which will be provided to the user in native MATLAB format.
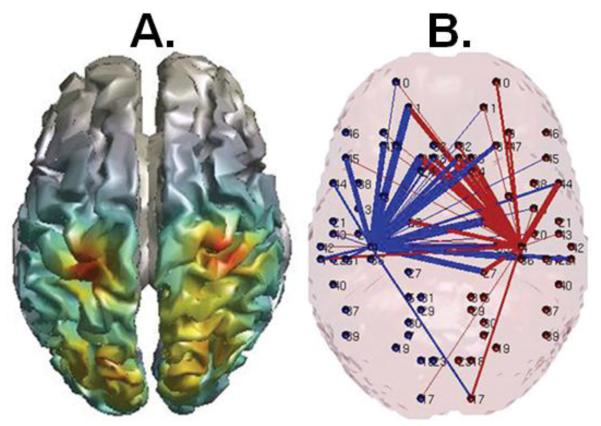
The next step of the pipeline is analysis of BLP contrasts in source space (see section 2.5.2). The results of these computations will be available to users in CIFTI format. An example of DICS source localization for the left hand motor task for the period prior to movement onset is shown in Figure 6A. The frequency range for which the inverse solution is illustrated is the upper beta band, which can clearly be seen to peak in motor and posterior areas.
Figure 6.
Frequency specific source localization (A) and connectivity map based on Talairach atlas labeling scheme (Lancaster et al., 2000). (A) DICS source modeling in the beta band for LH movement in the motor task (−0.5-0 sec before movement onset). Beta band power is greatest in motor and more posterior regions. (B) Seed regions of interest were placed bilaterally in Brodmann’s area 4, and imaginary coherence calculated in the beta band −0.5-0 sec before movement onset. Note high levels of connectivity cross-hemispherically to homolateral motor cortex and medial prefrontal regions. Line thickness is proportional to the value of imaginary coherence, blue lines represent connections originating in the left hemisphere, and red lines originate in the right hemisphere.
The final step of the tMEG pipeline is the computation of a set of connectivity metrics (see section 2.5.3). Connectivity maps will be available to the users in CIFTI format. Figure 6B illustrates the result of analysis of motor seed regions using imaginary coherence in the beta band as the connectivity metric of interest. The final pipeline outputs will include all-to-all node connectivity matrices for each metric together with the ability to project resultant data to the HCP defined cortical surface parcellation.
3.6 Exploring the connectome
The Connectome Workbench (Marcus et al., 2013; Marcus et al., 2011) provides interactive tools for visualizing HCP results and performing on-line experiments primarily using data that has been pre-computed by the HCP team. While dMRI and fMRI derived connectomes may be represented by parcel × parcel connectivity matrices, MEG derived connectomes have the additional dimensions of frequency and time given the greater temporal resolution inherent in this modality. Thus basic visualization tools in the Connectome Workbench will be extended to permit researchers to explore the additional dimensionality of MEG results.
By necessity the HCP team must make specific choices from the wide array of analytic techniques. These choices will be well documented in ConnectomeDB and in the open source pipeline software released with the MEG data. ConnectomeDB will enable the global research community to extract data at multiple levels in the processing pipeline for alternative processing and analysis by investigators using their own computational environments.
Preliminary estimates indicate that the raw MEG data, metadata and preprocessing information for each quarterly data release will total to approximately 328 GB. The source-space time series and connection matrices are not included in this calculation. ConnectomeDB provides a user interface for identifying and downloading via the Internet each data release. The download mechanism is based on multi-threaded TCP. Total download time depends on the round-trip latency between the ConnectomeDB servers and requester and is throttled by the lowest bandwidth link between sender and receiver. Based on measurements conducted between ConnectomeDB and requesters at the University of Minnesota and Oxford University, we estimate that a complete quarterly MEG raw data release can be downloaded in approximately 3-3.5 hours. It is also possible to request the data release be shipped on removable media (Connectome in a Box) (Marcus et al., 2011).
4. DISCUSSION
4.1 Methodological considerations
Inverse MEG source modeling is fundamentally an ill-posed problem (Gramfort et al., 2012; Helmholtz, 1853). The relative advantages and disadvantages of the various methods for computing source-space activity are discussed above (section 2.3). Each method is associated with a point-spread function (PSF) that varies across the brain (Hauk et al., 2011). It is important to emphasize that these PSFs limit the capacity to correctly identify true interactions, since they might determine possible crosstalk between both nearby and distant regions. But, there is no theoretical limit to the precision with which the center coordinates of an extended source can be localized; although in practice, mean squared localization error does vary across methods (Hauk et al., 2011). The MEG component of the HCP will implement wMNE and two varieties of beamforming spatial filters. These approaches have provided meaningful results in resting state (Brookes et al., 2011c; de Pasquale et al., 2010; de Pasquale et al., 2012; Hipp et al., 2012) and task (Brookes et al., 2012; Brookes et al., 2011b) MEG. Compared to other inverse modeling techniques, e.g., sLORETA and dSPM, wMNE has the smallest PSF (Hauk et al., 2011). However, regardless of which inverse modeling technique is used, the spatial resolution of MEG is much lower than tfMRI and dMRI. Hence, spurious interactions in connectivity estimates cannot be entirely avoided (Schoffelen and Gross, 2009). Results from both source reconstructions will be made available in the ConnectomeDB as they complement each other by highlighting different aspects of MEG connectivity.
Two contrasting analytic approaches, specifically, seed-based correlation mapping (Biswal et al., 1995) and spatial ICA (Beckmann et al., 2005) currently dominate the field of resting state fMRI. A similar situation obtains in the field of resting state MEG, as exemplified by the work of de Pasquale and colleagues (de Pasquale et al., 2010; de Pasquale et al., 2012) and Brookes and colleagues (Brookes et al., 2011a) respectively. The MEG component of the HCP will support both approaches. The main advantage of a seed based approach is that connectivity matrices derived by different modalities can be directly compared. The main drawbacks are reliance on prior knowledge of seed locations based on fMRI, the assumption that ROIs are correctly chosen and are concordant between modalities, and the dependence on accurate inverse source modeling together with information on neurovascular coupling to register BOLD data with electrophysiological sources. In contrast, ICA directly estimates RSNs from the MEG data without prior specification of ROIs. However, the over-arching design of the HCP is formulated in terms of parcel-to-parcel connectivity. Presently, it is unclear how best to map spatial components derived by MEG ICA onto parcel parcel connectivity matrices
4.2 Linking MEG large-scale connectivity to the human connectome
Strong evidence links fMRI RSNs to task-evoked activity (Smith et al., 2009). In addition, resting state functional connectivity based on fMRI is linked to anatomical connectivity, both in humans (Honey et al., 2009; Honey et al., 2010; Skudlarski et al., 2008) and in animals (Schwarz et al., 2013). However, the correspondence between functional and anatomical connectivity is not one-to-one (for discussion see Deco 2011; Vincent 2007)). This correspondence is even less clear at the higher frequencies of MEG data (Honey et al., 2010). Further, while structure constrains the dynamics of complex systems such as the brain, plasticity (Rubinov et al., 2009) can produce different functional network configurations on the same structural backbone. At present, few studies have examined the relationship between structural connectivity at the macroscopic level and functional connectivity derived from MEG. Initial resting state studies have been directed at showing that spatial structures involved in complex brain networks derived from MEG are similar to those derived from fMRI where these networks were initially defined (Biswal et al., 1995; de Pasquale et al., 2010; Fox et al., 2005). Having shown that similar spatial structures are involved in these complex networks at rest, MEG techniques can then be used to add valuable temporal and frequency information not available from fMRI. In addition to illuminating the question of information transfer at higher frequencies, on cross-frequency interactions or multiplexing (Marzetti et al., 2013), MEG rules out the hypothesis that these networks are a vascular phenomena. Further, MEG directly reflects the dynamics of neural populations at behaviorally meaningful temporal scales. Thus, integration of MEG functional connectivity with the fMRI and dMRI components of the HCP offers the potential for increased understanding of human brain functions.
Because MEG and fMRI have complementary spatiotemporal characteristics, integration of these two modalities theoretically should increase our understanding of the functional organization of brain network dynamics (see (Rosa et al., 2010; Snyder and Raichle, 2010) for reviews). However, integration of MEG and fMRI data remains elusive, despite considerable efforts by several laboratories (Babajani and Soltanian-Zadeh, 2006; Dale et al., 2000; Debener et al., 2006; Larson-Prior et al., 2011; Rosa et al., 2010; Sotero and Trujillo-Barreto, 2008). A fully integrated analysis of MEG and fMRI requires a single biophysical model that describes data from both modalities (Babajani and Soltanian-Zadeh, 2006; Riera et al., 2006; Sotero and Trujillo-Barreto, 2008). While such a bottom-up approach symmetrically utilizes data from both modalities, it is mathematically complicated. More commonly, an asymmetric approach is taken in which data from one modality are used as prior information for the other. For example, several studies have used fMRI-driven spatial priors to inform the MEG inverse problem (Ahlfors and Simpson, 2004; Dale et al., 2000; Henson et al., 2009). Alternatively, MEG temporal information or spectral power has been used as a predictor in fMRI (Debener et al., 2006; Kobayashi et al., 2007).
The relation between the hemodynamic response and neuronal activity is still not fully understood (Logothetis, 2008; Magri et al., 2012). Recent evidence indicates that astrocytic processes govern the dynamics of “neuro-vascular coupling” (Schummers et al., 2008). Various studies have shown that trial-by-trial BOLD fluctuations during task performance correlate negatively with power in the alpha and beta frequency bands and positively in the gamma band (Feige et al., 2005; Moosmann et al., 2003; Scheeringa et al., 2011; Yuan et al., 2010). Nevertheless BOLD activation maps with their high spatial resolution are used in many studies as priors for MEG inverse solutions.
The first part of this paper discussed the very different spatial and temporal scales that characterize the domains of MEG, fMRI and dMRI. Hence, integrating connectivity across these domains is challenging. The HCP approach to meeting this challenge is parcellation and measurement of parcel parcel connectivity matrices. Anatomical parcellations might provide the spatial smoothing and integration required to confidently fuse the different modalities. Atlases have been used in MEG analysis (Hillebrand et al., 2012) to parcellate and examine connectivity of the entire cortex. Source level group-ICA can be used to produce MEG data-derived network parcellations, which can be used to interrogate different subpopulations of the HCP database. Alternatively, parcels may be defined from ICA, resting-state fMRI, or task-based fMRI, EEG or MEG data and used as seed regions for connectivity analyses. It might be argued that high spatial resolution modalities, e.g., resting-state fMRI, should be used to define the parcels used in MEG functional connectivity analyses. However, MEG derived parcellations are adapted to the characteristics of the MEG data, and as such may be more appropriate to use in subsequent analyses within this modality.
4.3 Future directions
The availability of resting and task MEG data in the ConnectomeDB will enable the exploration of multiple features of the data using both existing and yet to be developed analysis techniques. In the future, more elaborate connectivity metrics are likely to become available. Metrics such as those based on multivariate autoregressive models, e.g. Partial Directed Coherence (PDC) (Baccala and Sameshima, 2001) and Directed Transfer Function (DTF) (Kaminski et al., 2001), adopt a semi arbitrary normalization (making connectivity comparisons meaningful only with the same starting or ending voxel) and are generally sensitive to noise (Baccala and Sameshima, 2001; Michalareas et al., 2012). Metrics based on non-parametric spectral factorization can be adopted to study directionality, such as spectrally resolved Granger causality (Bosman et al., 2012). These metrics are planned for implementation in the HCP at a later stage, after initial evaluation. Other methods of inferring effective connectivity such Dynamic Causal Modeling (DCM) (Friston et al., 2003; Kiebel et al., 2008) or Transfer Entropy(TE) (Wibral et al., 2009) are computationally limited to a few nodes. These methods are best suited to studies where clear data- or theory-driven hypotheses define the regions participating in a functional network. DCM may be implemented if indicated by initial results.
HIGHLIGHTS.
Dynamic functional connectivity at millisecond resolution will be provided to the Human Connectome Project using MEG
MEG data will be collected both at rest and during performance of motor and cognitive tasks
Task paradigms are matched to tfMRI designs to provide cross-modal information
A wide array of connectivity metrics will be implemented for MEG data at both sensor and source levels
Automated processing pipelines will be made publically available
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahlfors SP, Simpson GV. Geometrical interpretation of fMRI-guided MEG/EEG inverse estimates. Neuroimage. 2004;22:323–332. doi: 10.1016/j.neuroimage.2003.12.044. [DOI] [PubMed] [Google Scholar]
- Babajani A, Soltanian-Zadeh H. Integrated MEG/EEG and fMRI model based on neural masses. Biomedical Engineering, IEEE Transactions on. 2006;53:1794–1801. doi: 10.1109/TBME.2006.873748. [DOI] [PubMed] [Google Scholar]
- Baccala LA, Sameshima K. Partial directed coherence: a new concept in neural structure determination. Biological cybernetics. 2001;84:463–474. doi: 10.1007/PL00007990. [DOI] [PubMed] [Google Scholar]
- Baddeley A. Working memory: Looking back and looking forward. Nature reviews neuroscience. 2003;4:829–839. doi: 10.1038/nrn1201. [DOI] [PubMed] [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss S, Dixit S, Feldt C, Nolan D, Bryant E, Hartley T, Footer O, Bjork JM, Poldrack R, Smith S, Johansen-Berg H, Snyder AZ, Van Essen D, for the WU-Minn HCP Consortium Function in the Human Connectome: Task-fMRI and Individual Differences in Behavior. 2013 doi: 10.1016/j.neuroimage.2013.05.033. In press, NeuroImage Special Issue “Mapping the Connectome”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder JR, Gross WL, Allendorfer JB, Bonilha L, Chapin J, Edwards JC, Grabowski TJ, Langfitt JT, Loring DW, Lowe MJ, Koenig K, Morgan PS, Ojemann JG, Rorden C, Szaflarski JP, Tivarus ME, Weaver KE. Mapping anterior temporal lobe language areas with fMRI: a multicenter normative study. Neuroimage. 2011;54:1465–1475. doi: 10.1016/j.neuroimage.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bosman C, Schoffelen J-M, Brunet N, Oostenveld R, Bastos A, Womelsdorf T, Rubehn B, Stieglitz T, De Weerd P, Fries P. Attentional Stimulus Selection through Selective Synchronization between Monkey Visual Areas. Neuron. 2012;75:875–888. doi: 10.1016/j.neuron.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes M, Liddle E, Hale J, Woolrich M, Luckhoo H, Liddle P, Morris P. Task Induced Modulation of Neural Oscillations in Electrophysiological Brain Networks. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Hale JR, Zumer JM, Stevenson CM, Francis ST, Barnes GR, Owen JP, Morris PG, Nagarajan SS. Measuring functional connectivity using MEG: Methodology and comparison with fcMRI. Neuroimage. 2011a;56:1082–1104. doi: 10.1016/j.neuroimage.2011.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes MJ, Wood JR, Stevenson CM, Zumer JM, White TP, Liddle PF, Morris PG. Changes in brain network activity during working memory tasks: A magnetoencephalography study. Neuroimage. 2011b;55:1804–1815. doi: 10.1016/j.neuroimage.2010.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes MJ, Woolrich M, Luckhoo H, Price D, Hale JR, Stephenson MC, Barnes GR, Smith SM, Morris PG. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc Natl Acad Sci U S A. 2011c;108:16783–16788. doi: 10.1073/pnas.1112685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G. Rhythms of the Brain. Oxford University Press; USA: 2009. [Google Scholar]
- Calamante F, Tournier J-D, Heidemann RM, Anwander A, Jackson GD, Connelly A. Track density imaging (TDI): validation of super resolution property. Neuroimage. 2011;56:1259–1266. doi: 10.1016/j.neuroimage.2011.02.059. [DOI] [PubMed] [Google Scholar]
- Calamante F, Tournier J-D, Jackson GD, Connelly A. Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. Neuroimage. 2010;53:1233–1243. doi: 10.1016/j.neuroimage.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bressler SL, Ding M. Dynamics on networks: assessing functional connectivity with Granger causality. Computational & Mathematical Organization Theory. 2009;15:329–350. [Google Scholar]
- Collette F, Hogge M, Salmon E, Van der Linden M. Exploration of the neural substrates of executive functioning by functional neuroimaging. Neuroscience. 2006;139:209–221. doi: 10.1016/j.neuroscience.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Crone NE, Miglioretti DL, Gordon B, Sieracki JM, Wilson MT, Uematsu S, Lesser RP. Functional mapping of human sensorimotor cortex with electrocorticographic spectral analysis. I. Alpha and beta event-related desynchronization. Brain. 1998;121:2271–2299. doi: 10.1093/brain/121.12.2271. [DOI] [PubMed] [Google Scholar]
- Curtis CE, D’Esposito M. Persistent activity in the prefrontal cortex during working memory. Trends in cognitive sciences. 2003;7:415–423. doi: 10.1016/s1364-6613(03)00197-9. [DOI] [PubMed] [Google Scholar]
- Dale AM, Liu AK, Fischl BR, Buckner RL, Belliveau JW, Lewine JD, Halgren E. Dynamic statistical parametric neurotechnique mapping: combining fMRI and MEG for high-resolution imaging of cortical activity. Neuron. 2000;26:55–67. doi: 10.1016/s0896-6273(00)81138-1. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder AZ, Lewis C, Mantini D, Marzetti L, Belardinelli P, Ciancetta L, Pizzella V, Romani GL, Corbetta M. Temporal dynamics of spontaneous MEG activity in brain networks. Proceedings of the National Academy of Sciences. 2010;107:6040–6045. doi: 10.1073/pnas.0913863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder AZ, Marzetti L, Pizzella V, Romani GL, Corbetta M. A Cortical Core for Dynamic Integration of Functional Networks in the Resting Human Brain. Neuron. 2012;74:753–764. doi: 10.1016/j.neuron.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debener S, Ullsperger M, Siegel M, Engel AK. Single-trial EEG-fMRI reveals the dynamics of cognitive function. Trends in cognitive sciences. 2006;10:558–563. doi: 10.1016/j.tics.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Ding N, Simon JZ. Adaptive Temporal Encoding Leads to a Background Insensitive Cortical Representation of Speech. The Journal of Neuroscience. 2013;33:5728–5735. doi: 10.1523/JNEUROSCI.5297-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A, Marzetti L, Zappasodi F, Meinecke FC, Nolte G. Estimating true brain connectivity from EEG/MEG data invariant to linear and static transformations in sensor space. Neuroimage. 2011;60:476–488. doi: 10.1016/j.neuroimage.2011.11.084. [DOI] [PubMed] [Google Scholar]
- Feige B, Scheffler K, Esposito F, Di Salle F, Hennig J, Seifritz E. Cortical and subcortical correlates of electroencephalographic alpha rhythm modulation. Journal of neurophysiology. 2005;93:2864–2872. doi: 10.1152/jn.00721.2004. [DOI] [PubMed] [Google Scholar]
- Fiecas M, Ombao H, van Lunen D, Baumgartner R, Coimbra A, Feng D. Quantifying temporal correlations: A test-retest evaluation of functional connectivity in resting-state fMRI. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.09.052. [DOI] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-e4 allele. Proceedings of the National Academy of Sciences. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries P. Neuronal gamma-band synchronization as a fundamental process in cortical computation. Annual review of neuroscience. 2009;32:209–224. doi: 10.1146/annurev.neuro.051508.135603. [DOI] [PubMed] [Google Scholar]
- Friston K. Beyond phrenology: what can neuroimaging tell us about distributed circuitry? Annual review of neuroscience. 2002;25:221–250. doi: 10.1146/annurev.neuro.25.112701.142846. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Geweke JF. Measures of conditional linear dependence and feedback between time series. Journal of the American Statistical Association. 1984;79:907–915. [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, for the WU-Minn HCP Consortium The Minimal Preprocessing Pipelines for the Human Connectome Project. 2013 doi: 10.1016/j.neuroimage.2013.04.127. In press, NeuroImage Special Issue “Mapping the Connectome”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A, Kowalski M, Hämäläinen M. Mixed-norm estimates for the M/EEG inverse problem using accelerated gradient methods. Physics in medicine and biology. 2012;57:1937. doi: 10.1088/0031-9155/57/7/1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A, Papadopoulo T, Olivi E, Clerc M. Forward field computation with OpenMEEG. Computational intelligence and neuroscience. 2011:2011. doi: 10.1155/2011/923703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37:424–438. [Google Scholar]
- Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48:63. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Baillet S, Barnes G, Henson R, Hillebrand A, Jensen O, Jerbi K, Litvak V, Maess B, Oostenveld R. Good practice for conducting and reporting MEG research. Neuroimage. 2013;65:349–363. doi: 10.1016/j.neuroimage.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Kujala J, Hamalainen M, Timmermann L, Schnitzler A, Salmelin R. Dynamic imaging of coherent sources: studying neural interactions in the human brain. Proceedings of the National Academy of Sciences. 2001;98:694–699. doi: 10.1073/pnas.98.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggisberg AG, Honma SM, Findlay AM, Dalal SS, Kirsch HE, Berger MS, Nagarajan SS. Mapping functional connectivity in patients with brain lesions. Annals of neurology. 2008;63:193–203. doi: 10.1002/ana.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari R, Salmelin R. Magnetoencephalography: from SQUIDs to neuroscience: Neuroimage 20th anniversary special edition. Neuroimage. 2012;61:386–396. doi: 10.1016/j.neuroimage.2011.11.074. [DOI] [PubMed] [Google Scholar]
- Hathout GM, Gopi RK, Bandettini P, Gambhir SS. The lag of cerebral hemodynamics with rapidly alternating periodic stimulation: modeling for functional MRI. Magn Reson Imaging. 1999;17:9–20. doi: 10.1016/s0730-725x(98)00150-7. [DOI] [PubMed] [Google Scholar]
- Hauk O, Wakeman DG, Henson R. Comparison of noise-normalized minimum norm estimates for MEG analysis using multiple resolution metrics. Neuroimage. 2011;54:1966–1974. doi: 10.1016/j.neuroimage.2010.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmholtz H.v. Ueber einige Gesetze der Vertheilung elektrischer Strome in koerperlichen Leitern mit Anwendung auf die thierisch-elektrischen Versuche. Annalen der Physik. 1853;165:211–233. [Google Scholar]
- Henson RN, Mouchlianitis E, Friston KJ. MEG and EEG data fusion: simultaneous localisation of face-evoked responses. Neuroimage. 2009;47:581–589. doi: 10.1016/j.neuroimage.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillebrand A, Barnes GR, Bosboom JL, Berendse HW, Stam CJ. Frequency-dependent functional connectivity within resting-state networks: An atlas-based MEG beamformer solution. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nature neuroscience. 2012;15:884–890. doi: 10.1038/nn.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey C, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Thivierge JP, Sporns O. Can structure predict function in the human brain? Neuroimage. 2010;52:766. doi: 10.1016/j.neuroimage.2010.01.071. [DOI] [PubMed] [Google Scholar]
- Howard MW, Rizzuto DS, Caplan JB, Madsen JR, Lisman J, Aschenbrenner-Scheibe R, Schulze-Bonhage A, Kahana MJ. Gamma oscillations correlate with working memory load in humans. Cerebral Cortex. 2003;13:1369–1374. doi: 10.1093/cercor/bhg084. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A, Oja E. Independent component analysis: algorithms and applications. Neural Networks. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical image analysis. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jensen O, Gelfand J, Kounios J, Lisman JE. Oscillations in the alpha band (9-12 Hz) increase with memory load during retention in a short-term memory task. Cerebral Cortex. 2002;12:877–882. doi: 10.1093/cercor/12.8.877. [DOI] [PubMed] [Google Scholar]
- Jurkiewicz MT, Gaetz WC, Bostan AC, Cheyne D. Post-movement beta rebound is generated in motor cortex: evidence from neuromagnetic recordings. Neuroimage. 2006;32:1281. doi: 10.1016/j.neuroimage.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Kaminski M, Ding M, Truccolo WA, Bressler SL. Evaluating causal relations in neural systems: Granger causality, directed transfer function and statistical assessment of significance. Biological cybernetics. 2001;85:145–157. doi: 10.1007/s004220000235. [DOI] [PubMed] [Google Scholar]
- Khader PH, Jost K, Ranganath C, Rosler F. Theta and alpha oscillations during working-memory maintenance predict successful long-term memory encoding. Neuroscience letters. 2010;468:339–343. doi: 10.1016/j.neulet.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebel SJ, Garrido MI, Moran RJ, Friston KJ. Dynamic causal modelling for EEG and MEG. Cognitive neurodynamics. 2008;2:121–136. doi: 10.1007/s11571-008-9038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimesch W. Binding principles in the theta frequency range. In: Zimmer HD, Mecklinger A, Lindenberger U, editors. Handbook of binding and memory. Oxford University Press; Oxford: 2006. pp. 115–144. [Google Scholar]
- Kobayashi E, Bagshaw AP, Gotman J, Dubeau F. Metabolic correlates of epileptic spikes in cerebral cavernous angiomas. Epilepsy research. 2007;73:98–103. doi: 10.1016/j.eplepsyres.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Lachaux JP, Rodriguez E, Martinerie J, Varela FJ. Measuring phase synchrony in brain signals. Human brain mapping. 1999;8:194–208. doi: 10.1002/(SICI)1097-0193(1999)8:4<194::AID-HBM4>3.0.CO;2-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated Talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior LJ, Power JD, Vincent JL, Nolan TS, Coalson RS, Zempel J, Snyder AZ, Schlaggar BL, Raichle ME, Petersen SE. Progress in Brain Research. Elsevier; 2011. Chapter 18 - Modulation of the brain’s functional network architecture in the transition from wake to sleep; pp. 277–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F-H, Witzel T, Ahlfors SP, Stufflebeam SM, Belliveau JW, Hamalainen MS. Assessing and improving the spatial accuracy in MEG source localization by depth-weighted minimum-norm estimates. Neuroimage. 2006;31:160–171. doi: 10.1016/j.neuroimage.2005.11.054. [DOI] [PubMed] [Google Scholar]
- Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- Luckhoo H, Hale J, Stokes M, Nobre A, Morris P, Brookes M, Woolrich M. Inferring task-related networks using independent component analysis in magnetoencephalography. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri C, Schridde U, Murayama Y, Panzeri S, Logothetis NK. The amplitude and timing of the BOLD signal reflects the relationship between local field potential power at different frequencies. The Journal of Neuroscience. 2012;32:1395–1407. doi: 10.1523/JNEUROSCI.3985-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Della Penna S, Marzetti L, de Pasquale F, Pizzella V, Corbetta M, Romani GL. A signal-processing pipeline for magnetoencephalography resting-state networks. Brain Connect. 2011;1:49–59. doi: 10.1089/brain.2011.0001. [DOI] [PubMed] [Google Scholar]
- Marcus D, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, Hodge M, Horton W, Herrick R, Olsen T, McKay M, House M, Hileman M, Reid E, Harwell J, Coalson TS, Schindler J, Elam J, Curtis C, Van Essen D, for the WU-Minn HCP Consortium Human Connectome Project Informatics: quality control, database services, and data visualization. NeuroImage Special Issue “Mapping the Connectome”. 2013 doi: 10.1016/j.neuroimage.2013.05.077. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus D, Harwell J, Olsen T, Hodge M, Glasser M, Prior F, Jenkinson M, Laumann T, Curtiss S, Van Essen D. Informatics and Data Mining Tools and Strategies for the Human Connectome Project. Frontiers in neuroinformatics. 2011:5. doi: 10.3389/fninf.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino J, Honma SM, Findlay AM, Guggisberg AG, Owen JP, Kirsch HE, Berger MS, Nagarajan SS. Resting functional connectivity in patients with brain tumors in eloquent areas. Annals of neurology. 2011;69:521–532. doi: 10.1002/ana.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti L, Della Penna S, Snyder A, Pizzella V, Nolte G, de Pasquale F, Romani G, Corbetta M. Frequency specific interactions of MEG resting state activity within and across brain networks as revealed by the Multivariate Interaction Measure. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalareas G, Schoffelen JM, Paterson G, Gross J. Investigating causality between interacting brain areas with multivariate autoregressive models of MEG sensor data. Human brain mapping. 2012 doi: 10.1002/hbm.21482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milchenko M, Marcus D. Obscuring Surface Anatomy in Volumetric Imaging Data. Neuroinformatics. 2012:1–11. doi: 10.1007/s12021-012-9160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K, Zanos S, Fetz E, Den Nijs M, Ojemann J. Decoupling the cortical power spectrum reveals real-time representation of individual finger movements in humans. The Journal of Neuroscience. 2009;29:3132–3137. doi: 10.1523/JNEUROSCI.5506-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra PP, Pesaran B. Analysis of dynamic brain imaging data. Biophysical journal. 1999;76:691–708. doi: 10.1016/S0006-3495(99)77236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann M, Ritter P, Krastel I, Brink A, Thees S, Blankenburg F, Taskin B, Obrig H, Villringer A. Correlates of alpha rhythm in functional magnetic resonance imaging and near infrared spectroscopy. Neuroimage. 2003;20:145–158. doi: 10.1016/s1053-8119(03)00344-6. [DOI] [PubMed] [Google Scholar]
- Muthukumaraswamy SD, Singh KD. A cautionary note on the interpretation of phase-locking estimates with concurrent changes in power. Clinical neurophysiology. 2011;122:2324–2325. doi: 10.1016/j.clinph.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Nolte G. The magnetic lead field theorem in the quasi-static approximation and its use for magnetoencephalography forward calculation in realistic volume conductors. Physics in medicine and biology. 2003;48:3637. doi: 10.1088/0031-9155/48/22/002. [DOI] [PubMed] [Google Scholar]
- Nolte G, Bai O, Wheaton L, Mari Z, Vorbach S, Hallett M. Identifying true brain interaction from EEG data using the imaginary part of coherency. Clinical neurophysiology. 2004;115:2292–2307. doi: 10.1016/j.clinph.2004.04.029. [DOI] [PubMed] [Google Scholar]
- Nolte G, Ziehe A, Nikulin VV, Schlogl A, Kramer N, Brismar T, Muller KR. Robustly estimating the flow direction of information in complex physical systems. Physical review letters. 2008;100:234101. doi: 10.1103/PhysRevLett.100.234101. [DOI] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, Schoffelen JM. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci. 2011;2011:156869. doi: 10.1155/2011/156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelle JE, Davis MH. Neural oscillations carry speech rhythm through to comprehension. Frontiers in Psychology. 2012:3. doi: 10.3389/fpsyg.2012.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfurtscheller G, Lopes da Silva F. Event-related EEG/MEG synchronization and desynchronization: basic principles. Clinical neurophysiology. 1999;110:1842–1857. doi: 10.1016/s1388-2457(99)00141-8. [DOI] [PubMed] [Google Scholar]
- Pulvermueller F. Brain embodiment of syntax and grammar: Discrete combinatorial mechanisms spelt out in neuronal circuits. Brain and language. 2010;112:167–179. doi: 10.1016/j.bandl.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Renvall H, Formisano E, Parviainen T, Bonte M, Vihla M, Salmelin R. Parametric merging of MEG and fMRI reveals spatiotemporal differences in cortical processing of spoken words and environmental sounds in background noise. Cerebral Cortex. 2012;22:132–143. doi: 10.1093/cercor/bhr095. [DOI] [PubMed] [Google Scholar]
- Riera JJ, Wan X, Jimenez JC, Kawashima R. Nonlinear local electrovascular coupling. I: A theoretical model. Human brain mapping. 2006;27:896–914. doi: 10.1002/hbm.20230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Daunizeau J, Friston K. EEG-fMRI integration: a critical review of biophysical modeling and data analysis approaches. Journal of integrative neuroscience. 2010;9:453–476. doi: 10.1142/s0219635210002512. [DOI] [PubMed] [Google Scholar]
- Roux F, Wibral M, Mohr HM, Singer W, Uhlhaas PJ. Gamma-Band Activity in Human Prefrontal Cortex Codes for the Number of Relevant Items Maintained in Working Memory. The Journal of Neuroscience. 2012;32:12411–12420. doi: 10.1523/JNEUROSCI.0421-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, Van Leeuwen C, Breakspear M. Symbiotic relationship between brain structure and dynamics. BMC neuroscience. 2009;10:55. doi: 10.1186/1471-2202-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeringa R, Fries P, Petersson K-M, Oostenveld R, Grothe I, Norris DG, Hagoort P, Bastiaansen MCM. Neuronal Dynamics Underlying High- and Low-Frequency EEG Oscillations Contribute Independently to the Human BOLD Signal. Neuron. 2011;69:572–583. doi: 10.1016/j.neuron.2010.11.044. [DOI] [PubMed] [Google Scholar]
- Schoffelen JM, Gross J. Source connectivity analysis with MEG and EEG. Human brain mapping. 2009;30:1857–1865. doi: 10.1002/hbm.20745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffelen JM, Oostenveld R, Fries P. Neuronal coherence as a mechanism of effective corticospinal interaction. Science Signalling. 2005;308:111. doi: 10.1126/science.1107027. [DOI] [PubMed] [Google Scholar]
- Schummers J, Yu H, Sur M. Tuned responses of astrocytes and their influence on hemodynamic signals in the visual cortex. Science Signalling. 2008;320:1638. doi: 10.1126/science.1156120. [DOI] [PubMed] [Google Scholar]
- Schwarz AJ, Gass N, Sartorius A, Zheng L, Spedding M, Schenker E, Risterucci C, Meyer-Lindenberg A, Weber-Fahr W. The low-frequency BOLD functional connectivity signature of the hippocampal-prefrontal network in the rat brain. Neuroscience. 2013;228:243–258. doi: 10.1016/j.neuroscience.2012.10.032. [DOI] [PubMed] [Google Scholar]
- Sekihara K, Owen JP, Trisno S, Nagarajan SS. Removal of spurious coherence in MEG source-space coherence analysis. Biomedical Engineering, IEEE Transactions on. 2011;58:3121–3129. doi: 10.1109/TBME.2011.2162514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer W. Synchronization of cortical activity and its putative role in information processing and learning. Annual review of physiology. 1993;55:349–374. doi: 10.1146/annurev.ph.55.030193.002025. [DOI] [PubMed] [Google Scholar]
- Skudlarski P, Jagannathan K, Calhoun VD, Hampson M, Skudlarska BA, Pearlson G. Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. Neuroimage. 2008;43:554–561. doi: 10.1016/j.neuroimage.2008.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR. Correspondence of the brain’s functional architecture during activation and rest. Proceedings of the National Academy of Sciences. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AZ, Raichle ME. Studies of the human brain combining functional neuroimaging and electrophysiological methods. Simultaneous EEG and fMRI: Recording, Analysis, and Application: Recording, Analysis, and Application. 2010:47. [Google Scholar]
- Sotero RC, Trujillo-Barreto NJ. Biophysical model for integrating neuronal activity, EEG, fMRI and metabolism. Neuroimage. 2008;39:290–309. doi: 10.1016/j.neuroimage.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Sporns O. The human connectome: a complex network. Annals of the New York Academy of Sciences. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The Human Connectome: A Structural Description of the Human Brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley GB. Reading and writing the neural code. Nature neuroscience. 2013;16:259–263. doi: 10.1038/nn.3330. [DOI] [PubMed] [Google Scholar]
- Ugerbil K, Xu J, Auerback EJ, Mueller S, Vu A, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, Strupp J, Sapiro G, De Martino F, Wang D, Harel N, Garwood M, Chen L, Feinberg D, Smith S, Miller KL, Sotiropoulos SN, Jbabdi S, Andersson JL, Behrens TEJ, Glasser MF, Van Essen D, Yacoub E, Consortium, -.f.t.W.M.H. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. 2013 doi: 10.1016/j.neuroimage.2013.05.012. In press, NeuroImage Special Issue “Mapping the Connectome”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D, Smith S, Barch D, Behrens T, Yacoub E, Ugerbil K, for the WU-Minn HCP Consortium The WU-Minn Human Connectome Project: An Overview. 2013 doi: 10.1016/j.neuroimage.2013.05.041. In press, NeuroImage Special Issue “Mapping the Connectome”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E. The Human Connectome Project: A data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veen BD, van Drongelen W, Yuchtman M, Suzuki A. Localization of brain electrical activity via linearly constrained minimum variance spatial filtering. IEEE Trans Biomed Eng. 1997;44:867–880. doi: 10.1109/10.623056. [DOI] [PubMed] [Google Scholar]
- Van’t Ent D, Van Soelen IL, Stam KJ, De Geus EJ, Boomsma DI. Genetic influence demonstrated for MEG-recorded somatosensory evoked responses. Psychophysiology. 2010;47:1040–1046. doi: 10.1111/j.1469-8986.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- Varela F, Lachaux JP, Rodriguez E, Martinerie J. The brainweb: phase synchronization and large-scale integration. Nature reviews neuroscience. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wibral M, Bledowski C, Kohler A, Singer W, Muckli L. The timing of feedback to early visual cortex in the perception of long-range apparent motion. Cerebral Cortex. 2009;19:1567–1582. doi: 10.1093/cercor/bhn192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter WR, Nunez PL, Ding J, Srinivasan R. Comparison of the effect of volume conduction on EEG coherence with the effect of field spread on MEG coherence. Statistics in Medicine. 2007;26:3946–3957. doi: 10.1002/sim.2978. [DOI] [PubMed] [Google Scholar]
- Xiang J, Liu Y, Wang Y, Kotecha R, Kirtman EG, Chen Y, Huo X, Fujiwara H, Hemasilpin N, deGrauw T. Neuromagnetic correlates of developmental changes in endogenous high-frequency brain oscillations in children: A wavelet-based beamformer study. Brain research. 2009;1274:28–39. doi: 10.1016/j.brainres.2009.03.068. [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu T, Szarkowski R, Rios C, Ashe J, He B. Negative covariation between task-related responses in alpha/beta-band activity and BOLD in human sensorimotor cortex: An EEG and fMRI study of motor imagery and movements. Neuroimage. 2010;49:2596. doi: 10.1016/j.neuroimage.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]