SUMMARY
The inhibition of transcriptional elongation plays an important role in gene regulation in metazoans, including C. elegans. Here, we combine genomic and biochemical approaches to dissect a role of ZFP-1, the C. elegans AF10 homolog, in transcriptional control. We show that ZFP-1 and its interacting partner DOT-1.1 have a global role in negatively modulating the level of polymerase II (Pol II) transcription on essential widely expressed genes. Moreover, the ZFP-1/DOT-1.1 complex contributes to progressive Pol II pausing on essential genes during development and to rapid Pol II pausing during stress response. The slowing down of Pol II transcription by ZFP-1/DOT-1.1 is associated with an increase in H3K79 methylation and a decrease in H2B monoubiquitination, which promotes transcription. We propose a model wherein the recruitment of ZFP-1/DOT-1.1 and deposition of H3K79 methylation at highly expressed genes initiates a negative feedback mechanism for the modulation of their expression.
INTRODUCTION
Transcription is a highly regulated step in gene expression. The most detailed mechanistic understanding of the regulatory mechanisms governing transcription comes from studies in bacteria, yeast, and cultured cells, but much less is known about global transcriptional regulation in developing multicellular organisms.
Specific histone modifications are known to associate with actively transcribed genes. However, their effect on transcription is far from understood. The histone methyltransferase Dot1 was identified as a factor modulating telomere silencing in yeast and later recognized to possess H3K79 methylation activity (reviewed in Wood et al., 2005, and Nguyen and Zhang, 2011). In mammals, DOT1-like (DOT1L) enzyme was found to be associated with a number of nuclear proteins that are often fused to mixed-lineage leukemia (MLL) protein in oncogenes causing leukemia (reviewed in Mohan et al., 2010b) and that do not exist in yeast. Two of these MLL fusion partners, acute lymphoblastic leukemia 1-fused gene from chromosome 10 (AF10) and AF9, have clear homologs in the nematode Caenorhabditis elegans, zinc finger protein 1 (ZFP-1) and GAS41-like 1 (GFL-1), respectively. Here, we present insight on the regulation of transcriptional elongation by C. elegans DOT-1.1, which is most similar to mammalian DOT1L, and its interacting partner ZFP-1.
Methylation of H3K79 is tightly linked to and promoted by H2B ubiquitination, and H2B monoubiquitination (H2Bub1) by a conserved Rad6-Bre1 E2-E3 complex is facilitated by polymerase II (Pol II)-associated factor (PAF) (reviewed in Wood et al., 2005, Weake and Workman, 2008, and Nguyen and Zhang, 2011). Therefore, both H3K79 methylation and H2B ubiquitination correlate with transcription. The important question is how do they affect transcription? Although no clear cause and effect relationship has been shown for H3K79me, the causal link between high levels of H2Bub1 at coding regions and efficiency of transcriptional elongation has been established (reviewed in Weake and Workman, 2008, and Braun and Madhani, 2012). This effect is most likely due to the disruption of chromatin compaction facilitated by H2Bub1 (Fierz et al., 2011) and a role of H2Bub1 in promoting the function of the facilitates chromatin transcription (FACT) complex (Pavri et al., 2006; Fleming et al., 2008).
Here, we report that the recruitment of C. elegans DOT-1.1 to highly transcribed genes enriched in H2Bub1 leads to a reduction in the levels of this modification and to an inhibitory effect on transcriptional elongation, although this effect is modulatory and not silencing. We propose that H2B ubiquitination serves two purposes: (1) it promotes transcription through chromatin, and (2) it initiates a negative feedback loop that modulates transcriptional elongation. We show that the ZFP-1/DOT-1.1 complex contributes to Pol II pausing that occurs on essential widely expressed genes during development and also participates in the regulation of transcription during stress response. We propose that ZFP-1 and DOT-1.1 ensure the precise level of transcription of highly expressed genes by opposing H2B ubiquitination and transcriptional elongation.
RESULTS
DOT-1.1 Is a Major Interacting Partner of ZFP-1(AF10) in C. elegans
We became interested in ZFP-1, the C. elegans homolog of the human AF10 protein, because it was shown to promote the efficiency of RNAi (Dudley et al., 2002; Kim et al., 2005), including RNAi-induced transcriptional gene silencing (Grishok et al., 2005). ZFP-1 is a ubiquitous nuclear protein expressed at all developmental stages (Avgousti et al., 2013). To elucidate the function of ZFP-1, we used an unbiased proteomic approach to identify its interacting partners and the chromatin immunoprecipitation (ChIP)-chip method to map its genome-wide chromatin localization. Extracts from mixed embryos of ZFP-1::FLAG transgenic (Mansisidor et al., 2011; Avgousti et al., 2013) and control nontransgenic strains were used to perform immunoprecipitations with an antibody against FLAG followed by mass spectrometry (MS) analysis of copurified proteins. We identified DOT-1.1 (Y39G10AR.18), a putative H3K79 methyltransferase, as a major interacting partner of both ZFP-1 isoforms (Figure S1A and Table S1 available online).
The Dot1 methyltransferase in S. cerevisiae catalyzes the methylation of H3K79 (reviewed in Khan and Hampsey, 2002), and there is only one Dot1 enzyme in yeast, Drosophila, and humans (Feng et al., 2002). However, the C. elegans genome encodes five putative methyltransferases of the Dot1 family (Feng et al., 2002), which we named DOT-1.1–DOT-1.5 (Figure S2). Like the human DOT1L protein, Y39G10AR.18, which we named DOT-1.1, has an extended C-terminal region several hundred amino acids (aa) long that is bare of known protein domains and marked by low compositional complexity. No such extended C-terminal region exists in the yeast enzyme Dot1p or in C. elegans DOT-1.2–DOT-1.5. Using iterative sequence searches with position-specific scoring (PSI-BLAST) (Altschul et al., 1997), we were able to demonstrate the conservation of a 190 aa core region within this C-terminal portion of C. elegans Y39G10AR.18 across major metazoan groups, including nematodes, vertebrates, insects, and sponges (Figure S3).
The identification of DOT-1.1 as a ZFP-1 interacting partner is consistent with the recent characterization of mammalian DOT1L and AF10 complexes (Mohan et al., 2010a) and a reported DOT1L interaction with MLL-AF10 (Okada et al., 2005). We confirmed the interaction between C. elegans ZFP-1 and DOT-1.1 by reciprocal coimmunoprecipitation experiments (Figures 1A and S1B). The leucine zipper and octapeptide motifs present in the C-terminal region of AF10 were previously implicated in the interaction with DOT1L (Okada et al., 2005). To test whether the interaction with DOT-1.1 occurs through the C-terminal region of ZFP-1, we used transgenic worms expressing the 200 aa N-terminal fragment of ZFP-1 fused to FLAG (Avgousti et al., 2013) (Figure 1A, top) in coimmunoprecipitation experiments. Consistent with the results published for AF10, only full-length ZFP-1::FLAG was found to interact with the two DOT-1.1 isoforms (Figure 1A, bottom). We conclude that, as in mammals, ZFP-1/AF10 and DOT-1.1 specifically interact in nematodes through the C-terminal domain of ZFP-1, which is conserved in metazoans.
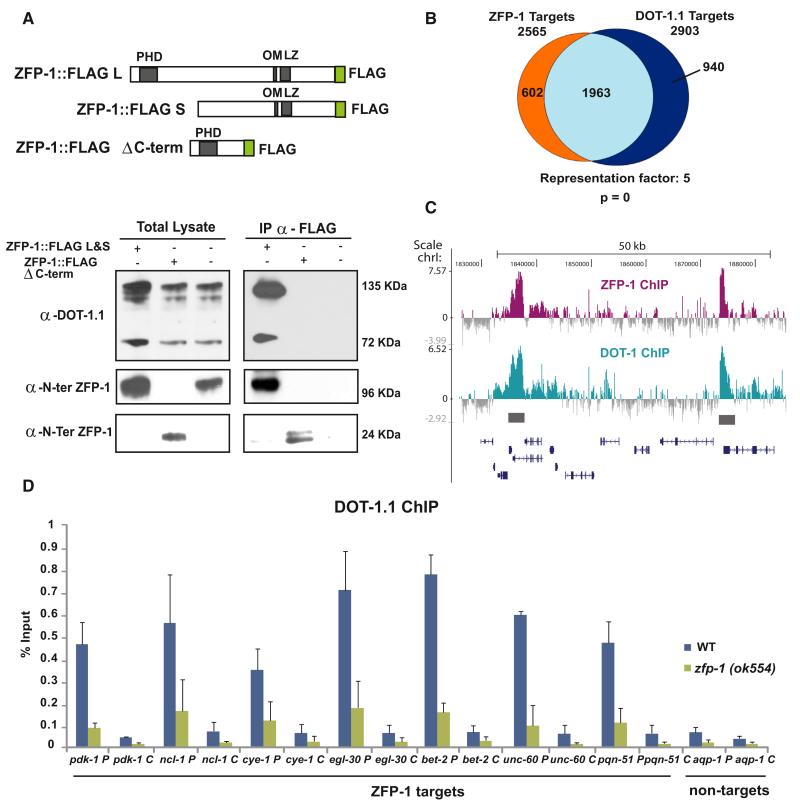
Figure 1. ZFP-1(AF10) and DOT-1.1 Interact and Colocalize to Promoters of Target Genes.
(A) The truncated ZFP-1 protein similar to that present in the zfp-1(ok554) mutant strain does not interact with DOT-1.1. Top, a schematic of the two FLAG-tagged WT ZFP-1 isoforms and a truncated ZFP-1 protein missing the C-terminal portion, which is identical to that present in the zfp-1(ok554) mutant. Bottom, western blots with an antibody against DOT-1.1 and an antibody against ZFP-1 of total lysates (left) and proteins immunoprecipitating with an antibody against FLAG (right) in indicated transgenic strains or WT nontransgenic worms, rightmost lanes.
(B) Venn diagram showing an overlap between ZFP-1 and DOT-1.1 target genes identified by ChIP-chip. A Fisher’s exact test was used for calculating p values for overlaps.
(C) Example of a genomic region with ZFP-1 (top) and DOT-1.1 (bottom) ChIP-chip binding enrichment at the promoters of target genes (grey bars). Gene models are based on the UCSC Genome Browser (WS220/ce10).
(D) ChIP-qPCR results with antibodies against DOT-1.1 in WT and zfp-1(ok554) demonstrating the role of ZFP-1 in recruiting DOT-1.1 to the promoters of the target genes.
Here, and in the following figures, “P” after the gene name indicates promoter, and “C” after the gene name indicates coding region. The results of three independent experiments are shown. Error bars represent SD.
See also Figures S1–S4.
ZFP-1 and DOT-1.1 Colocalize to Promoters of Highly Expressed Genes
To find the endogenous target genes regulated by ZFP-1 and DOT-1.1, we performed a genome-wide binding analysis of the endogenous ZFP-1 and DOT-1.1 by ChIP-chip. We used antibodies recognizing C-terminal portions of DOT-1.1 and ZFP-1 (Figure 1A) (Mansisidor et al., 2011; Avgousti et al., 2013) and identified DNA sequences enriched in ChIP samples. Both ZFP-1 and DOT-1.1 bind to the same genomic regions significantly enriched at the promoters of target genes (Figures 1B, 1C, and S4A and Table S2). The target genes recognized by both ZFP-1 and DOT-1.1 belong to the category of highly expressed genes (Figure S4B).
We confirmed that both DOT-1.1 and ZFP-1 localize to the promoters of select target genes by ChIP quantitative PCR (ChIP-qPCR; Figures 1D and S4C). Then, we determined whether the zfp-1(ok554) loss-of-function mutation, which leads to a truncated version of the protein lacking the C-terminal domain essential for DOT-1.1 interaction (Figure 1A) (Cui et al., 2006; Avgousti et al., 2013), affects DOT-1.1 localization to chromatin. We found a strong decrease in DOT-1.1 binding to the promoters of ZFP-1/DOT-1.1 target genes at the third larval stage (L3) in zfp-1(ok554) (Figure 1D). These results indicate that ZFP-1 enhances the binding of DOT-1.1 to chromatin.
The ZFP-1/DOT-1.1 Complex Negatively Regulates the Transcription of Its Targets
Our previous analysis of gene expression in the zfp-1(ok554) mutant larva revealed that equal numbers of genes were upregulated and downregulated in zfp-1(ok554) (Grishok et al., 2008). Therefore, we asked how many up- and down-regulated genes are direct ZFP-1 targets defined by ChIP-chip. We found that genes expressed at higher levels in the mutant were very significantly enriched for ZFP-1 binding, whereas genes expressed at lower levels in the mutant ware depleted of ZFP-1 binding (Figure 2A and Table S2). This analysis indicated that ZFP-1 is preferentially involved in the negative regulation of highly expressed genes during the larval stages of nematode development.
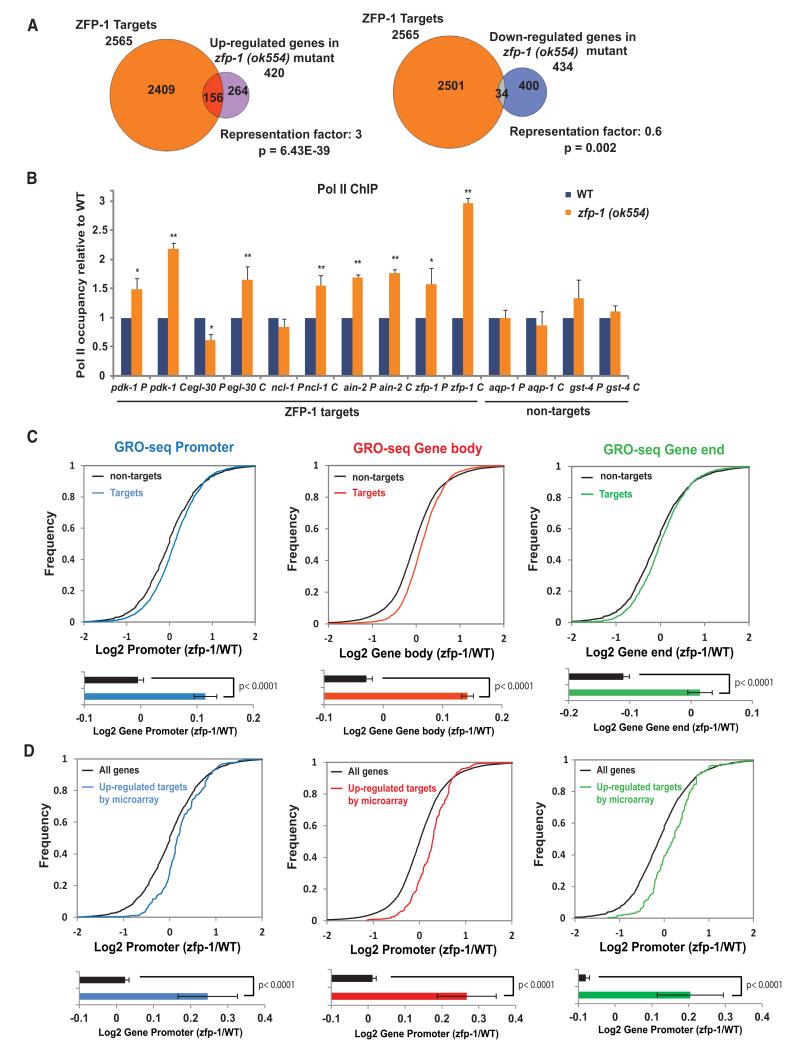
Figure 2. ZFP-1 and DOT-1.1 Inhibit Transcription.
(A) Venn diagrams demonstrating an enrichment of ZFP-1 ChIP-chip targets among genes expressed at a higher level in the zfp-1(ok554) mutant larvae, but not among genes expressed at a lower level (Grishok et al., 2008). A Fisher’s exact test was used for calculating p values for overlaps.
(B) Pol II occupancy analysis on ZFP-1 target genes and control nontarget genes by ChIP-qPCR in WT and zfp-1(ok554) mutant L3 larvae. The results of three independent experiments are shown. Error bars represent SD. * indicates a significance of p < 0.05, and ** indicates p < 0.01 in comparison WT larvae according to a Student’s t test.
(C) Top, cumulative distribution plots of GRO-seq data presented as in Core et al., 2012, and showing the overall effect of zfp-1(ok554) mutation on polymerase density at ZFP-1 target (colored lines) and nontarget genes (black lines). The transcriptome was divided into promoters (−600 bp TSSs, +200 bp TSSs), gene bodies (+200 bp TSSs, −200 bp TTSs), or gene ends (−200 bp TTSs, +400 bp TTSs). Bottom, the average of the log2 ratio between zfp-1(ok554) and WT normalized reads for promoter, gene body, and gene end. Error bars represent a 95% confidence interval for the mean. Two-tailed p values show the statistical significance of difference between ZFP-1 target (colored bars) and nontarget (black bars) means.
(D) GRO-seq analysis performed as in (C) considering the 156 ZFP-1 target genes found upregulated in zfp-1(ok554) by microarray versus all genes. See also Figure S4 and S5.
To further investigate the molecular mechanism of gene regulation by ZFP-1/DOT-1.1, we selected a set of direct target genes of ZFP-1/DOT-1.1 that are upregulated in zfp-1(ok554). We confirmed the increase in the messenger RNA (mRNA) levels in zfp-1(ok554) mutant larvae by quantitative RT-PCR (qRT-PCR;Figure S5A, left). To assess whether the upregulation of the target genes was at the transcriptional level, we measured the pre-mRNA levels. We found a significant increase in the pre-mRNA levels of the target genes in zfp-1(ok554) mutant larva and did not find an increase in pre-mRNA of the control genes (Figure S5A, right). Next, we performed Pol II ChIP-qPCR experiments with the 8WG16 antibody, which recognizes all Pol II isoforms. We found a significant increase in Pol II occupancy on the ZFP-1/DOT-1.1 target genes in zfp-1(ok554) mutant larvae in comparison to wild-type (WT) larvae (Figure 2B). The increase was especially pronounced at the coding sequences of the ZFP-1/DOT-1.1 target genes, which was in accordance with the expression data.
To confirm that the increase in Pol II occupancy reflected active transcription and to extend our analyses genome wide, we adapted the global run-on sequencing (GRO-seq) method, which detects nascent transcripts (Core et al., 2008), to C. elegans (see the Supplemental Experimental Procedures for details). This genome-wide approach allowed us to precisely map the transcriptionally engaged Pol II in WT and zfp-1(ok554) mutant larvae. We detected 14,079 active genes in L3 larvae in our GRO-seq experiments (more than 68% of all C. elegans genes). On the basis of annotated transcription start sites (TSSs) and transcription termination sites (TTSs), we divided C. elegans genes into three regions: promoter (from −600 TSSs to +200 TSSs), gene body (from +200 TSSs to −200 TTSs), and gene end (from −200 TTSs to +400 TTSs). Then, we quantified the relative GRO-seq signals. Although a recent CapSeq analysis mapped the transcription start sites more precisely (Gu et al., 2012), these TSSs still largely fall within the −600 to +200 promoter bin, as defined above (Figures S4D–S4F). In zfp-1(ok554), we detected a global increase in transcriptionally engaged Pol II on ZFP-1 target genes in comparison to nontarget genes along the entire portion of the gene (Figures 2C and S5B). Also, we analyzed GRO-seq signals at the 156 upregulated ZFP-1 target genes identified by microarray (Figure 2A) and found a significant increase in transcription in zfp-1(ok554) (Figure 2D), which confirmed our gene expression analysis at the mRNA level. Altogether, these experiments suggest that the recruitment of DOT-1.1 to promoters of highly expressed genes by ZFP-1 negatively regulates transcription.
The ZFP-1/DOT-1.1 Complex Contributes to an Increase in Pol II Pausing on Essential Widely Expressed Genes during Development
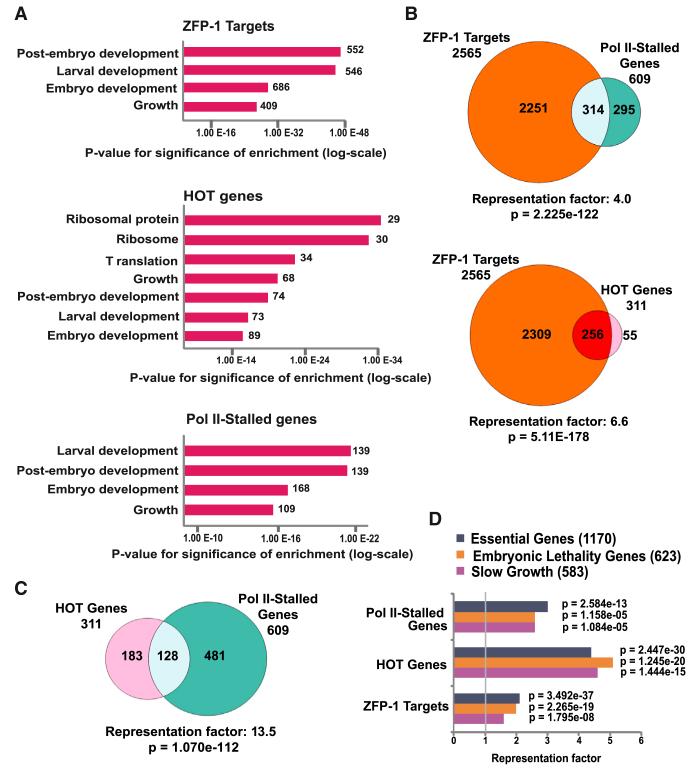
The functional annotation of ZFP-1 targets using the Gene Ontology (GO) database revealed that these genes are associated with embryonic and larval development and the regulation of growth rate (Figure 3A and Table S3). These same GO annotations have been described for genes with paused RNA Pol II (Muse et al., 2007; Zeitlinger et al., 2007; Gilchrist et al., 2010) and high-occupancy target (HOT) genes defined by ChIP-chip as the sites of increased occupancy by many transcriptional factors (Gerstein et al., 2010; Niu et al., 2011) (Figure 3A and Table S3). Notably, HOT genes are broadly expressed (Gerstein et al., 2010). Therefore, we compared a list of ZFP-1/DOT-1.1 target genes with the sets of (1) genes with confirmed Pol II stalling (Zhong et al., 2010) and (2) annotated HOT genes (Gerstein et al., 2010) (Table S2). We found a very significant overlap between these datasets (Figure 3B), suggesting that ZFP-1 may contribute to gene expression regulation by affecting Pol II stalling on broadly expressed genes. Genes classified as transcriptionally stalled and HOT genes also mutually overlapped (Figure 3C).
Figure 3. ZFP-1/DOT-1.1 Targets Represent Essential Developmental Genes that Overlap with Previously Described Genes with Stalled Pol II and High-Occupancy Target Genes.
(A) Gene Ontology analysis of ZFP-1 targets, HOT genes (Gerstein et al., 2010), and genes with stalled Pol II (Zhong et al., 2010). Data sets listing Pol II stalling in the embryo and in the L1 (see Table S2) were combined for statistical analyses. The top enriched functional categories are shown. See Table S3 for complete functional enrichment analysis.
(B) Venn diagrams showing overlaps between ZFP-1 targets and (1) genes with Pol II stalling (top) or (2) HOT genes (bottom).
(C) Venn diagram demonstrating an overlap between HOT genes and genes with stalled Pol II.
(D) Statistical analysis showing the enrichment of Pol II-stalled genes, HOT genes, and ZFP-1 targets among essential gene sets identified by genome-wide RNAi (Kamath et al., 2003). Fisher’s exact test was used for calculating p values.
We also compared the above gene sets with the list of C. elegans genes that are essential for embryonic and larval development and growth, as shown by genome-wide RNAi experiments (Kamath et al., 2003) (Table S2). We found a very significant overlap with ZFP-1/DOT-1.1 targets and Pol II-stalled and HOT genes (Figure 3D), suggesting that genes essential for developmental processes may be transcriptionally regulated by ZFP-1/DOT-1 through the effect on Pol II elongation.
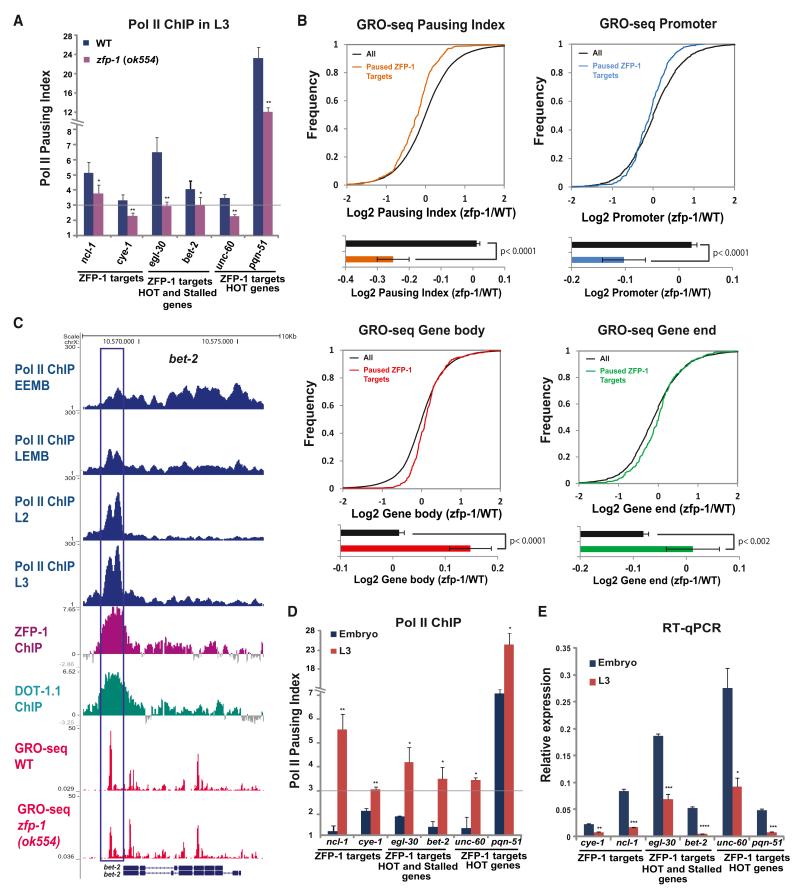
Next, we performed experiments aimed at testing these predictions. We analyzed the enrichment of Pol II at the promoters relative to the coding regions of ZFP-1 target genes by ChIP-qPCR (Figure 4A, left); i.e. we determined the Pol II pausing index. Then, we selected genes to be analyzed from the list of ZFP-1/DOT-1.1 targets, some of which also belonged to the lists of Pol II-stalled genes (Zhong et al., 2010) (Figure 4A, center) or HOT genes (Gerstein et al., 2010) (Figure 4A, center and right). We found a significant level of Pol II pausing on the genes tested, and, most importantly, it was affected by the zfp-1(ok554) mutation (Figure 4A), indicating that DOT-1.1 recruitment by ZFP-1 to the promoters of these genes can modulate their transcription.
Figure 4. Increased Pausing of Pol II Is Accompanied by a Decrease in Gene Expression of ZFP-1/DOT-1.1 Targets during Development.
(A) Pol II pausing index, as measured by the ratio of Pol II ChIP-qPCR signals at the promoter and coding region, at ZFP-1 target genes in WT larvae and zfp-1(ok554). A 3-fold increase in the Pol II promoter occupancy in comparison to the coding region was considered to be the lower threshold of Pol II pausing. Some genes shown were previously annotated as Pol II stalled (Zhong et al., 2010) and/or HOT (Gerstein et al., 2010). The results of three independent experiments are shown. Error bars represent SD. * indicates a significance of p < 0.05, and ** indicates p < 0.01 in comparison to WT larvae.
(B) Top, cumulative distribution plots of GRO-seq data (as in Figures 2C and 2D) showing the effect of the zfp-1(ok554) mutation on 331 ZFP-1 target genes with paused Pol II. Bottom, the average of the log2 ratio between zfp-1 (ok554) and WT normalized reads for pausing index, promoter, gene body, and gene end. Error bars represent a 95% confidence interval for the mean. The two-tailed p value indicates the statistical significance of the difference in means between paused ZFP-1 target genes (colored bars) and all genes (black bars).
(C) An example of gene with progressive pausing of Pol II during development. Top four tracks: Pol II occupancy in early embryos (EEMB), late embryos (LEMB), and L2 larva and L3 larva (based on the Pol II ChIP-seq data available from modENCODE). Other tacks show DOT-1.1 and ZFP-1 binding identified by ChIP-chip (modENCODE) and GRO-seq normalized reads in WT and zfp-1(ok554). Gene models are based on the University of California, Santa Cruz, Genome Browser (ce06): bet-2 (F57C7.1b (top) and F57C7.1a (bottom).
(D) Pol II pausing index at indicated genes in the mixed embryo (blue) and in the L3 larvae (red). The results of two independent experiments are shown. Error bars represent SD. * indicates a significance of p < 0.05, and ** indicates p < 0.01 in comparison to the embryo stage.
(E) Expression of the indicated genes in the mixed embryo (blue) and L3 larvae (orange) relative to act-3 mRNA expression as measured by qRT-PCR. The results of two independent experiments are shown. Error bars represent SD. * indicates a significance of p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.005 in comparison to the embryo stage.
See also Figure S6.
Given that ChIP-qPCR cannot indicate whether Pol II is engaged in transcription, we analyzed the regulation of the ZFP-1 target genes with stalled polymerase by GRO-seq to determine whether there is pausing in active transcription. The pausing indexes for GRO-seq data were calculated as the log2 of the ratio between promoter and gene-body-normalized reads. Pol II transcription at the ZFP-1 target gene was considered paused if the pausing index was 1 or higher. This defined 331 genes (Table S2), including 14 genes from the group of 156 ZFP-1 targets found upregulated by the microarray. Next, we determined the change in Pol II pausing index in zfp-1(ok554) in comparison to WT worms and found a significant decrease in Pol II pausing on ZFP-1 targets (Figure 4B, top left). Importantly, the decrease in Pol II pausing was due to a significant reduction of the transcriptionally engaged Pol II at promoters (Figure 4B, top right) and a concomitant increase on gene bodies and gene ends (Figure 4B, bottom). These results suggest that the ZFP-1/DOT-1 complex negatively modulates the escape of paused Pol II into productive elongation.
Because the genes negatively regulated by ZFP-1/DOT-1.1 are not inactive but, instead, are highly transcribed and essential during embryonic and larval development, we considered the possibility that the level of transcription of these genes needs to be developmentally regulated, perhaps by modulating the level of Pol II pausing. Therefore, we used modENCODE Pol II ChIP-seq data to analyze Pol II-stalled genes (Zhong et al., 2010) at different developmental stages from early embryo to L3 larva and found a global increase in Pol II ChIP signal around the TSSs of these genes over the course of C. elegans development (Figures 4C and S6A). We also detected a peak of GRO-seq signals corresponding to Pol II enrichment (Figures 4C andS6B), which indicates that Pol II is engaged in transcription. We confirmed these results by Pol II ChIP-qPCR comparing the mixed embryo stage and L3 worms (Figure 4D). Importantly, the increase in Pol II pausing in L3 stage worms (Figure 4D) was correlated with a decrease in the mRNA levels (Figure 4E) and with an increase in DOT-1.1 occupancy at the promoters (Figure S6C). These data suggest that the escape of Pol II into elongation is regulated during development, most likely to ensure the precise transcriptional level of essential active genes, and that the ZFP-1/DOT-1.1 complex contributes to this developmental regulation of transcription.
The ZFP-1/DOT-1.1 Complex Promotes Pol II Pausing during Heat Shock
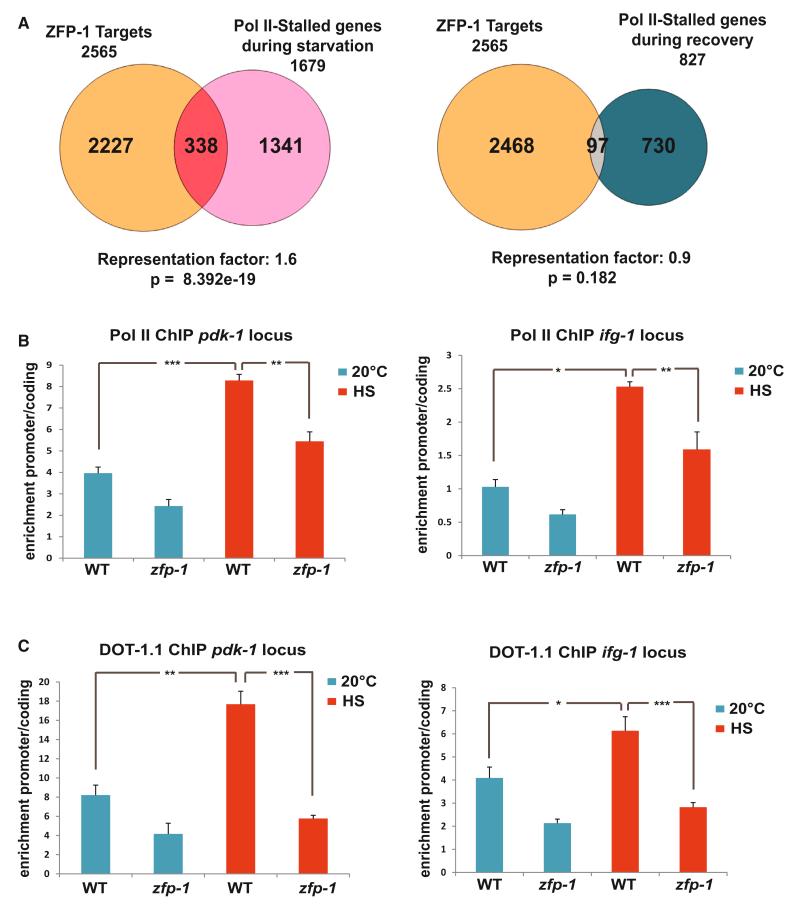
Regulation of transcription by pause release of Pol II can be faster than regulation through the assembly of preinitiation complexes at the promoter (reviewed in Levine, 2011). Indeed, inducible heat shock genes were among the first in which this type of regulation has been demonstrated (reviewed in Guertin et al., 2010). Given that the inhibition of Pol II elongation has been observed in C. elegans during starvation and no mechanisms have been found to account for it (Baugh et al., 2009), we considered the possibility that ZFP-1/DOT-1.1 could regulate Pol II dynamics during stress. We analyzed the lists of genes where (1) Pol II accumulated at the promoters upon starvation of C. elegans larva and (2) where Pol II enrichment at the promoters occurred after feeding and recovery (Baugh et al., 2009). We found that only the first set of genes was enriched in ZFP-1/DOT-1.1 targets (Figure 5A and Table S2). This analysis suggests that ZFP-1/DOT-1.1 may promote transcriptional inhibition upon stress. Consistent with this suggestion, we have recently described the stress sensitivity of the zfp-1(ok554) mutant and demonstrated that increased transcription of the ZFP-1 target gene pdk-1, a component of a conserved insulin-signaling pathway, significantly contributed to the stress sensitivity phenotype (Mansisidor et al., 2011). We have also shown that genes encoding translation factors, such as ifg-1, the C. elegans ortholog of eIF4G, were enriched among genes expressed at a higher level in the zfp-1(ok554) mutant (Grishok et al., 2008). Translation factors belong to the category of genes that are inhibited upon starvation (Baugh et al., 2009). Therefore, we used pdk-1 and ifg-1 genes as models to investigate the possible role of ZFP-1/DOT-1.1 in promoting transcriptional inhibition during stress. We confirmed that both pdk-1 and ifg-1 are down-regulated upon heat shock treatment (Figures S7A–S7C) and performed Pol II and DOT-1.1 ChIP-qPCR experiments in worms grown at either 20°C or after heat shock. We found a significant increase in Pol II pausing on both pdk-1 and ifg-1 genes after heat shock treatment (Figure 5B). This increase was accompanied by a significant enrichment of DOT-1.1 at the promoters of these genes (Figure 5C). We conducted the same experiments in the zfp-1(ok554) mutant animals and observed a reduced recruitment of DOT-1.1 upon heat shock and a reduction in Pol II pausing that did not reach the same levels as in WT worms upon stress (Figures 5B and 5C). Altogether, these experiments demonstrate that ZFP-1 and DOT-1.1 contribute to transcriptional inhibition of genes by promoting Pol II pausing upon stress.
Figure 5. ZFP-1 and DOT-1.1 Stimulate Pol II Pausing on Environmentally Regulated Genes during Stress.
(A) Venn diagrams illustrating overlaps between ZFP-1 targets and genes regulated by Pol II stalling during starvation (left) or after recovery from starvation (right) according to Baugh et al., 2009. A Fisher’s exact test was used for calculating the p values for overlaps.
(B) Increase in Pol II pausing upon heat shock on pdk-1 and ifg-1 (eIF4G) genes in WT larvae and zfp-1(ok554). The results of three independent ChIP-qPCR experiments are shown. Error bars represent SEM. * indicates a significance of p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.005 in comparison to the WT larvae grown at 20°C or upon heat shock (HS).
(C) Increase in DOT-1.1 occupancy upon HS at the promoters of pdk-1 and ifg-1 (eIF4G) genes in WT larvae and zfp-1(ok554). The results of three independent ChIP-qPCR experiments are shown. Error bars represent SEM. * indicates a significance of p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.005 in comparison to the WT larvae grown at 20°C or upon HS.
See also Figure S7.
ZFP-1/DOT-1.1 Inhibit Pol II Elongation by Antagonizing H2B Ubiquitination
From studies in yeast and mammalian cells, it is known that the Paf1 complex association with Pol II during an early elongation step facilitates histone H2B monoubiquitination through the action of Rad6 ubiquitin-conjugating enzyme and Bre1 ubiquitin ligase (reviewed in Osley et al., 2006, Shilatifard, 2006, Weake and Workman, 2008, and Braun and Madhani, 2012). H2Bub1 was preferentially found at transcribed regions of highly expressed genes in human cells (Minsky et al., 2008) and was shown to have a positive role in enhancing transcriptional elongation in vitro (Pavri et al., 2006) and in vivo (Fleming et al., 2008; Batta et al., 2011).
Interestingly, a decrease in DOT1L interaction with endogenous AF10 in the leukemia cell line that expresses dominant CALM-AF10 fusion was correlated with a global increase in H2Bub1 (Darwanto et al., 2010). This suggests that DOT1L recruitment to chromatin by AF10 can antagonize global deposition of H2Bub1. These observations prompted us to investigate the possible connection between ZFP-1/DOT-1.1 and the H2B ubiquitination pathway.
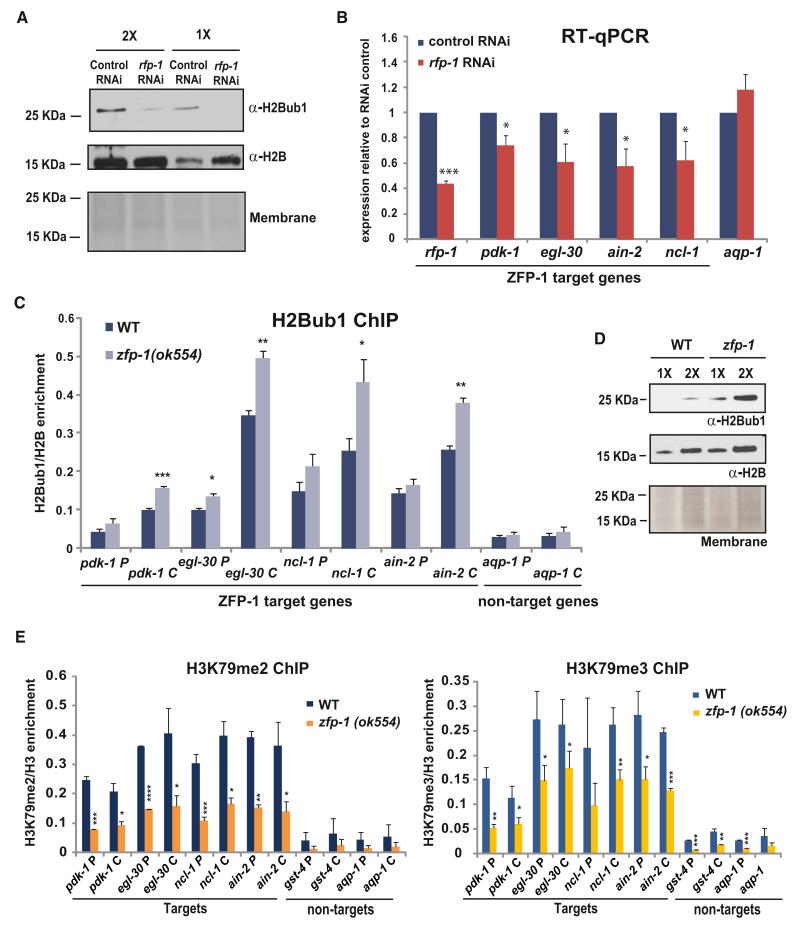
The regulation of H2B monoubiquitination has not been explored in C. elegans, although the interaction between the C. elegans Bre1 homolog RFP-1 and Rad6 homolog UBC-1 has been demonstrated (Crowe and Candido, 2004). We detected monoubiquitinated H2B in C. elegans extracts (Figure 6A), and, importantly, the level of H2Bub1 was reduced in the worms treated with RNAi against rfp-1 (Figure 6A). This confirms that H2B ubiquitination by Bre1 is conserved in C. elegans. Then, we tested whether the expression of ZFP-1/DOT-1.1 target genes was influenced by H2B ubiquitination. We performed qRT-PCR in worms treated with rfp-1 RNAi and control RNAi and found a significant decrease in the mRNA levels of the ZFP-1/DOT-1.1 target genes upon rfp-1 downregulation, whereas the expression of genes not bound by ZFP-1/DOT-1.1, such as aquaporin (aqp-1), did not depend on rfp-1 (Figure 6B). These results are consistent with an active role of H2B ubiquitination in promoting transcription and also highlight the point that not all genes are equally dependent on this chromatin modification for their expression. In order to confirm that the ZFP-1/DOT-1.1 target genes are the direct targets of H2B ubiquitination, we performed H2Bub1 ChIP-qPCR. We observed an enrichment of this chromatin mark on the ZFP-1/DOT-1.1 target genes in comparison to the nontargets that were expressed at similar high levels (Figure 6C). The H2Bub1 was enriched on the body of the genes and also present at the promoters, which was in agreement with previous studies in other species (Pavri et al., 2006; Fleming et al., 2008).
Figure 6. H2B Monoubiqutination Promotes the Transcription of ZFP-1 Target Genes and Is Suppressed by ZFP-1.
(A) Western blot demonstrating that inhibition of C. elegans Bre1 homolog rfp-1 by RNAi leads to a decrease in H2B ubiqutination. Hereafter, H2B western and Ponceau S staining of the membrane are shown as loading controls. (B,C, and E) * indicates a significance of p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.005 in comparison to corresponding controls.
(B) Expression of ZFP-1 target genes is decreased in rfp-1(RNAi) animals as measured by qRT-PCR. act-3 mRNA expression is used as an internal control. The results of two biological replicas are shown. Error bars represent SD.
(C) The level of H2Bub1 is increased on ZFP-1 target genes in the zfp-1(ok554) mutant. The results of two independent ChIP-qPCR experiments are shown. Error bars represent SD.
(D) Western blot demonstrating a global increase in H2Bub1 in zfp-1(ok554).
(E) The level of H3K79me2 and H3K79me3 is decreased on ZFP-1 target genes in the zfp-1(ok554) mutant. The results of two independent ChIP-qPCR experiments are shown. Error bars represent SD.
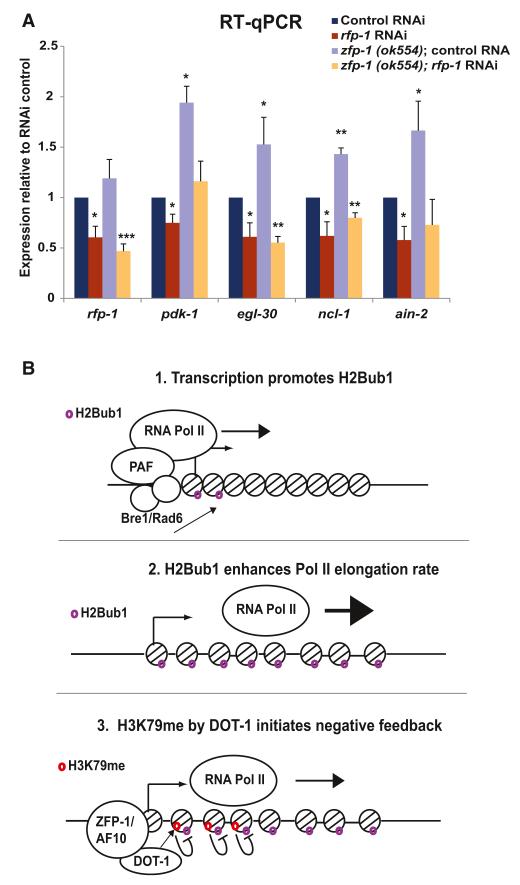
Next, we tested whether the recruitment of DOT-1.1 by ZFP-1 could antagonize the accumulation of H2Bub1 on their target genes while promoting H3K79 methylation. First, we performed ChIP-qPCR to detect H3K79 methylation. We found enrichment in H3K79 methylation at the promoters and coding regions of ZFP-1/DOT-1 target genes, and, most importantly, we found a reduction in H3K79 di- and tri-methylation in the zfp-1(ok554) mutant, suggesting that DOT-1.1 has the predicted H3K79 methyltransferase activity (Figure 6E). Next, we compared the global levels of H2Bub1 by western blotting with protein extracts from zfp-1(ok554) mutant and WT larvae and found a global increase in H2Bub1 in zfp-1(ok554) mutant (Figure 6D). Then, we performed H2Bub1 ChIP-qPCR on ZFP-1/DOT-1.1 target genes and detected a significant increase in H2Bub1 in zfp-1(ok554) mutants in comparison to WT worms, especially on the coding region of the genes (Figure 6C). These results are consistent with the observed increase in Pol II transcriptional elongation at the target genes in zfp-1(ok554) mutants described earlier. Finally, in order to determine whether the increased transcription of ZFP-1 target genes in the zfp-1(ok554) mutant was dependent on the increase in H2Bub1, we knocked down rfp-1 by RNAi in WT and zfp-1(ok554) mutant worms and found that reducing RFP-1 activity indeed suppressed the elevated levels of transcription seen in the zfp-1(ok554) mutant (Figure 7A). We propose that the ZFP-1/DOT-1.1 complex is a part of a negative feedback mechanism modulating the transcription of highly expressed genes enriched in H2B ubiqutination (Figure 7B).
Figure 7. H2Bub1 Is Required for Increased Expression of ZFP-1/DOT-1.1 Target Genes in zfp-1(ok554), Consistent with a Negative Feedback Model between H2Bub1 and H3K79me.
(A) Increased expression of ZFP-1 target genes in zfp-1(ok554) larvae is suppressed by rfp-1(RNAi). act-3 mRNA expression is used as an internal control. The results of three independent experiments are shown. Error bars represent SD. * indicates a significance of p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001 in comparison to corresponding controls.
(B) A model illustrating the proposed relationship between H2Bub1, ZFP-1/DOT-1.1, and transcription.
DISCUSSION
Relevance of the C. elegans System for Studies of Transcriptional Regulation by Mammalian AF10 and DOT1L
Our discovery of the overwhelming negative effect of ZFP-1 on gene expression was unexpected, given the known connection between H3K79 methylation and transcriptional activation of MLL-AF10 target genes (Okada et al., 2005).
Because H3K79 methylation is often associated with active transcription and many proteins coimmunoprecipitating with DOT1L were also found in the super elongation complex (SEC), it was commonly believed that DOT1L promotes transcriptional elongation, but this notion is changing in light of new results (reviewed in Mohan et al., 2010b). The fact that the DOT1L complex (DotCom) is distinct from SEC is becoming evident (Lin et al., 2010; Mohan et al., 2010a; Biswas et al., 2011; He et al., 2011). Moreover, DOT1L was found to inhibit the transcription of the HIV-1 LTR-luciferase reporter in HeLa cells (He et al., 2011). Also, there are several examples where H3K79 methylation at the promoter was correlated with a repressed state of the gene in mammals (Zhang et al., 2006; Buttner et al., 2010; Yu et al., 2010).
Here, we identify natural biological targets of the ZFP-1/DOT-1.1 complex during C. elegans development genome wide. Our experimental system uses whole animals and reveals the role of ZFP-1/DOT-1.1 in a global negative modulation of highly and widely expressed genes, such as cyclin E, translation factor genes, which are involved in actin metabolism and microRNA function, and other essential genes. The precise control of global regulators, including ZFP-1 itself, is critical for the proper development and fitness of animals. Previously, we have shown that a 2-fold increase in cyclin E mRNA levels leads to a large effect on nuclear divisions in the intestine of C. elegans (Grishok and Sharp, 2005), whereas a 2- to 4-fold increase in the abundance of 3-phosphoinositide-dependent kinase 1 (pdk-1) mRNA leads to reduced lifespan and impaired resistance to oxidative stress (Mansisidor et al., 2011).
Our results suggest the possibility that the leukemic fusions of AF10, MLL-AF10 and CALM-AF10, titrate DOT1L away from the endogenous targets that it normally inhibits, such as cyclin E, whose increased expression may contribute to cancer progression.
Regulation of Pol II Pausing by Chromatin Factors in Metazoans
Early studies of gene regulation have focused primarily on RNA Pol II recruitment to promoters followed by preinitiation complex assembly. However, transcription is also regulated at subsequent steps, such as transcriptional elongation and termination (reviewed in Guertin et al., 2010, Levine, 2011, and Nechaev and Adelman, 2011). Highly expressed genes that are critical for development and cell growth and stimulus-responsive regulatory pathways are most affected by RNA polymerase pausing (Gerstein et al., 2010; Gilchrist et al., 2010). The regulation of Pol II pausing requires the coordinated activity of positive and negative elongation factors, such as the DRB sensitivity-inducing factor and negative elongation factor (NELF) (Muse et al., 2007; Gilchrist et al., 2010; Rahl et al., 2010). In addition, a possible role of chromatin factors in Pol II pausing has been recently proposed (Gilchrist and Adelman, 2012). Because high levels of promoter accumulation of Pol II have been observed in the C. elegans embryo (Zhong et al., 2010) and in L1 larva (Baugh et al., 2009; Zhong et al., 2010) and because nematodes lack NELF homologs in their genome, they represent an excellent model for finding additional conserved mechanisms that regulate Pol II pausing. Here, we provide evidence that the ZFP-1(AF10)/DOT-1.1 complex serves this role.
Given that both ZFP-1 and DOT-1.1 are conserved from nematodes to humans, we propose that their homologs may have a similar effect on transcription in other metazoans.
Dynamic Regulation of H2B Ubiquitination and H3K79 Methylation
The requirement of H2Bub1 for H3K79me was first demonstrated in yeast in studies using strains deficient for H2Bub1, which also lacked H3K79 and H3K4 methylation (Briggs et al., 2002; Ng et al., 2002; Wood et al., 2003). This histone “crosstalk” is conserved in higher eukaryotes (reviewed in Osley et al., 2006, Shilatifard, 2006, and Weake and Workman, 2008). However, the reverse is not true—i.e., H2Bub1 is not dependent on the methylation of H3K79 (Ng et al., 2002). Moreover, H2Bub1 is much less stable than H3 methylation marks and is dynamically regulated during transcription through the action of the deubiquitinating enzymes Ubp8 and Ubp10 (Schulze et al., 2011).
We have demonstrated the antagonistic relationship between H2Bub1 and H3K79me in connection to transcription of ZFP-1 target genes (Figure 7). The precise mechanism of the inverse correlation between H2Bub1 and H3K79me on ZFP-1/AF10 target genes requires additional investigation. We believe that our findings in C. elegans provide unexpected directions for mechanistic studies of the modulation of transcriptional elongation, which is highly relevant to animal development and human disease.
EXPERIMENTAL PROCEDURES
Strains
Strains were maintained at 20°C with standard methods (Brenner, 1974) unless otherwise noted. Bristol N2 was the WT strain used. All other alleles are listed in the Supplemental Experimental Procedures. For the heat shock treatments, synchronous populations of worms were grown for 36 hr posthatching at 20°C on OP-50 E. coli at a density of approximately 100,000 animals per 15 cm Petri dish and incubated at 32°C for 4 hr.
Immunoprecipitation and Mass Spectrometry
Details of the immunoprecipitation (IP) and MS experiments are described in the Supplemental Experimental Procedures. In brief, mixed embryos were suspended in the extraction buffer and sonicated. Protein extracts were spanned and incubated with an antibody, and immune complexes were precipitated with IgG Dynabeads (Invitrogen). For MS analyses, five IP samples were pulled together and resolved on SDS gels. Gels were stained with SilverSNAP (Thermo Scientific), and bands were excised and sent to Midwest Bio Services for MS analysis. The antibodies used were anti-FLAG M2 (Sigma-Aldrich) and anti-DOT-1.1 (modENCODE, SDQ4129_Y39G10AR18).
Western Blotting
C. elegans proteins were extracted as described for IP except that, for histone proteins, the extraction buffer contained 1 M NaCl, and the extract was resolved on precast NuPAGE Novex 12% Bis-Tris gels (Invitrogen). The proteins were transferred to a nylon membrane with a semidry transfer (BioRad) at a constant current of 0.12 A for 30 min. The antibodies used were anti-H2B (Abcam, ab-1790), anti-H2Bub1 (Millipore, 05-1312), anti-ZFP-1 C-terminal antibody (modENCODE, JL00006_ZFP1), anti-N-terminal ZFP-1 (modENCODE, SDQ3517_ZFP-1), anti-DOT-1.1 (SDQ4129_Y39G10AR18), anti-IFG-1 (E6 23, generously provided by B. Keiper,), antiactin (Millipore, MAB1501R), anti-mouse IgG HRP labeled (PerkinElmer), and anti-rabbit IgG HRP labeled (PerkinElmer).
Chromatin Immunoprecipitation
ChIP was performed as described in Mansisidor et al., 2011. Details of the experiments are described in the Supplemental Experimental Procedures.
modENCODE Protocols
The protocols used for generating ZFP-1 and DOT-1.1 (Y39G10AR.18) ChIP-chip data and data sets can be found at http://www.modencode.org/lieb.
GRO-Seq
GRO-seq experiments were performed as described in Core et al., 2008, with some modifications. Details are described in the Supplemental Experimental Procedures.
RNA Extraction and qRT-PCR
RNA extraction and qRT-PCR was performed as in Mansisidor et al., 2011, with the exception that synchronous populations of worms were grown for 36 hr posthatching at 20°C on OP-50 E. coli at a density of approximately 100,000 animals per 15 cm Petri dish.
RNAi
Synchronous populations of L1 animals were grown at 20°C on plates seeded with bacteria that produced double-stranded RNA of interest and collected at L3 stage 36 hr posthatching.
Data Analysis
Gene list overlaps were analyzed as described previously (Grishok et al., 2008; Mansisidor et al., 2011). Additional information about the data analysis is provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Mann and T. Maniatis for critical reading of the manuscript; A. Leshinsky, R.F. Cook, and C.A. Whittaker from Swanson Biotechnology Center at the MIT Koch Institute for Integrative Cancer Research for the sequencing of nascent transcript libraries; S. O’Keeffe for bioinformatic advice; A. Mansisidor for technical assistance; J. Lieb and B. Keiper for providing reagents; and members of the Grishok lab for discussions. The zfp-1(ok554) strain was provided by the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation, which is part of the International C. elegans Gene Knockout Consortium. Some strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by 3260-07 Special Fellow Award from the Leukemia and Lymphoma Society, an Arnold and Mabel Beckman Foundation Young Investigator Award, and an NIH Director’s New Innovator Award (1 DP2 OD006412-01) to A.G.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession number for the GRO-seq data generated with WT L3 and zfp-1(ok554) mutant larvae is GSE47132.
Supplemental Information contains Supplemental Experimental Procedures, seven figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2013.06.002.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgousti DC, Cecere G, Grishok A. The conserved PHD1-PHD2 domain of ZFP-1/AF10 is a discrete functional module essential for viability in Caenorhabditis elegans. Mol. Cell. Biol. 2013;33:999–1015. doi: 10.1128/MCB.01462-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc. Natl. Acad. Sci. USA. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Madhani HD. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012;13:619–630. doi: 10.1038/embor.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Buttner N, Johnsen SA, Kugler S, Vogel T. Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2010;107:7042–7047. doi: 10.1073/pnas.0912041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cel Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe E, Candido EP. Characterization of C. elegans RING finger protein 1, a binding partner of ubiquitin-conjugating enzyme 1. Dev. Biol. 2004;265:446–459. doi: 10.1016/j.ydbio.2003.09.037. [DOI] [PubMed] [Google Scholar]
- Cui M, Kim EB, Han M. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2006;2:e74. doi: 10.1371/journal.pgen.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwanto A, Curtis MP, Schrag M, Kirsch W, Liu P, Xu G, Neidigh JW, Zhang K. A modified “cross-talk” between histone H2B Lys-120 ubiquitination and H3 Lys-79 methylation. J. Biol. Chem. 2010;285:21868–21876. doi: 10.1074/jbc.M110.126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley NR, Labbe JC, Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Adelman K. Coupling polymerase pausing and chromatin landscapes for precise regulation of transcription. Biochim. Biophys. Acta. 2012;1819:700–706. doi: 10.1016/j.bbagrm.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sharp PA. Negative regulation of nuclear divisions in Caenorhabditis elegans by retinoblastoma and RNA interference-related genes. Proc. Natl. Acad. Sci. USA. 2005;102:17360–17365. doi: 10.1073/pnas.0508989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Hoersch S, Sharp PA. RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2008;105:20386–20391. doi: 10.1073/pnas.0810589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr., Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Petesch SJ, Zobeck KL, Min IM, Lis JT. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 2010;75:1–9. doi: 10.1101/sqb.2010.75.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. USA. 2011;108:E636–E645. doi: 10.1073/pnas.1107107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Khan AU, Hampsey M. Connecting the DOTs: covalent histone modifications and the formation of silent chromatin. Trends Genet. 2002;18:387–389. doi: 10.1016/s0168-9525(02)02746-4. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansisidor AR, Cecere G, Hoersch S, Jensen MB, Kawli T, Kennedy LM, Chavez V, Tan MW, Lieb JD, Grishok A. A Conserved PHD Finger Protein and Endogenous RNAi Modulate Insulin Signaling in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002299. doi: 10.1371/journal.pgen.1002299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010a;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer. 2010b;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Osley MA, Fleming AB, Kao CF. Histone ubiquitylation and the regulation of transcription. Results Probl. Cell Differ. 2006;41:47–75. doi: 10.1007/400_006. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Shilatifard A. Cross-talking histones: implications for the regulation of gene expression and DNA repair. Biochem. Cell Biol. 2005;83:460–467. doi: 10.1139/o05-116. [DOI] [PubMed] [Google Scholar]
- Yu Z, Kong Q, Kone BC. CREB trans-activation of disruptor of telomeric silencing-1 mediates forskolin inhibition of CTGF transcription in mesangial cells. Am. J. Physiol. Renal Physiol. 2010;298:F617–F624. doi: 10.1152/ajprenal.00636.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J. Biol. Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.