Abstract
p53, a guardian of the genome, exerts its tumor suppression activity by regulating a large number of downstream targets involved in cell cycle arrest, DNA repair, apoptosis, and cellular senescence. Although p53-mediated apoptosis is able to kill cancer cells, a role for cellular senescence in p53-dependent tumor suppression is becoming clear. Mouse studies showed that activation of p53-induced premature senescence promotes tumor regression in vivo. However, p53-mediated cellular senescence also leads to aging-related phenotypes, such as tissue atrophy, stem cell depletion, and impaired wound healing. In addition, several p53 isoforms and two p53 homologs, p63 and p73, have been shown to play a role in cellular senescence and/or aging. Importantly, p53, p63, and p73 are necessary for the maintenance of adult stem cells. Therefore, understanding the dual role the p53 protein family in cancer and aging is critical to solve cancer and longevity in the future. In this chapter, we provide an overview on how p53, p63, p73, and their isoforms regulate cellular senescence and aging.
Keywords: Aging, p53, p63, p73, Senescence
1. Introduction
p53, originally identified as a protein associated with the simian virus 40 large T antigen in 1979 (1–4), guards genomic integrity via regulating numerous cellular processes, including cell cycle arrest, DNA repair, apoptosis, and cellular senescence, in response to various stress signals (5, 6). The p53 pathway is commonly lost in human cancers due to either inactivation of p53 protein or mutations in the TP53 gene, leading to accumulation of damaged cells and cancer progression, (7). Consistent with this, mice deficient in the Tp53 gene are highly prone to spontaneous malignancy at a young age (8, 9). In addition, Li–Fraumeni syndrome patients carrying a TP53 mutation display increased risk of early onset of several types of cancer (10, 11).
Clearance of tumor cells by induction of programmed cell death is well documented as a tumor suppression activity of p53. Recent studies demonstrated that induction of p53-dependent senescence plays a pivotal role in limiting tumor progression in vivo (12–14). Thus, further understanding the mechanism by which p53 is implicated in regulating cellular senescence is of great interest for developing new cancer therapies. Cellular senescence was originally described by Hayflick and Moorhead in the 1960s (15, 16). They found that normal human diploid fibroblasts have a limited lifespan in culture and eventually enter a state of permanent cell cycle arrest called replicative senescence. Senescent cells exhibit enlarged cell size, flattened morphology, inability to synthesize DNA, metabolic active, and expression of the senescence-associated β-galactosidase (SA-β-gal), the latter of which can be detected at pH 6.0 (17, 18). Further studies showed that this life timing is controlled by repeats of telomere, a nucleoprotein structure at chromosome tips, which is undergoing progressive loss during cell divisions (19–21). Due to insufficient telomere after a number of cell doublings exposed or fused chromosome ends trigger DNA damage signals (telomeric stress signals), leading to senescence. As a major mediator of the DNA damage pathway, p53 has been shown to be critical for telomeric stress-induced cellular senescence (22, 23). In addition, multiple stress signals, including aberrant oncogene activation (24–26) and cancer chemotherapeutic drugs (22, 23, 27), are able to induce senescence-like phenotypes (premature senescence) in both primary and tumor cells via activating the p53 and/or p16 pathways (28). In this chapter, we provide an overview of the role of p53, its isoforms, and its family members, p63 and p73, in regulating cellular senescence and aging.
2. The Role of p53 in Cellular Senescence in Response to Different Stress Signals
2.1. The p53 Protein
The wild-type p53 protein is composed of 393 amino acids and contains several functional domains (Fig. 1). These are the N-terminal activation domain 1 (AD1) and 2 (AD2), the proline-rich domain (PRD), the central core DNA-binding domain (DBD), the nuclear localization signal region (NLS), the tetramerization domain (TD), and the C-terminal regulatory basic domain (BD) (29). As a sequence-specific transcription factor, p53 regulates gene expression by directly binding to a p53-responsive element (p53-RE) in the target gene as a tetramer. The consensus p53-RE is composed of two half sites (RRRCA/TA/TGYYY, where R represents purine and Y pyrimidine) separated by up to 13 nucleotides (30). Hundreds of p53 targets have been identified and shown to be involved in a variety of cellular responses, such as cell cycle arrest, DNA repair, apoptosis, and cellular senescence, attributing to the tumor suppressor activity of p53.
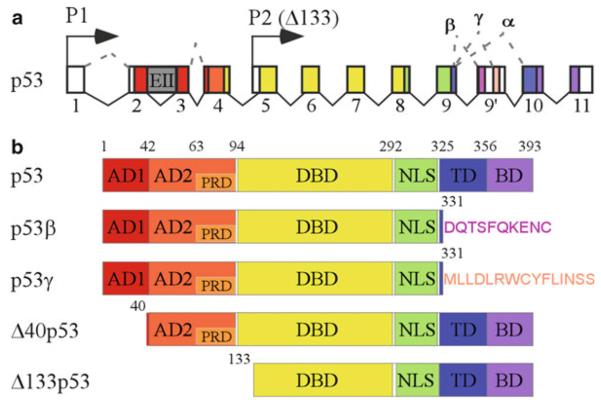
Fig. 1.
The p53 gene and protein structure. (a) TP53 locus, location of the P1 and P2 promoters, and patterns of alternative splicing. p53, also called α isoform, is a full-length p53 generated from the P1 promoter. Transcripts initiated from an internal promoter (P2) in intron 4 leads to production of an N-terminal truncated p53 starting from residue 133, named as Δ133p53. Alternative translation initiation at an AUG codon at position 40 leads to production of a p53 protein with a deletion of the first 39 amino acids (Δ40p53). Although alternative splicing of intron 2 gives rise to a transcript containing intron 2 sequence, the presence of a stop codon in intron 2 also leads to production of Δ40p53. Compared to the α isoforms, the β and γ isoforms have 10 and 15 unique amino acids at C-termini, respectively. (b) p53 isoforms and functional domains. AD activation domain, PRD proline-rich domain, DBD DNA binding domain, NLS nuclear localization domain, TD teramerization domain, BD basic domain.
The level and activity of p53 protein are primarily regulated through posttranslational modifications (Fig. 2). In normal cells, p53 is a short-lived protein which is continuously undergoing ubiquitination and subject to proteasomal degradation. Mouse double minute 2 (MDM2, HDM2 in human), one of the first characterized p53 targets, serves as a major E3 ubiquitin ligase for p53 degradation and thus forms a negative autoregulatory loop to maintain the low level of p53 expression in unstressed conditions (31–34). Upon genotoxic stresses, rapid phosphorylation of p53 at Ser-15 by the Ataxia telangiectasia mutated (ATM), a serine/threonine protein kinase, and at Ser-20 by the checkpoint kinase 2 (Chk2) results in dissociation of p53 from MDM2, leading to p53 stabilization and activation of its downstream processes (35–37). Likewise, phosphorylation of MDM2 at Ser-395 by ATM attenuates the capability of MDM2 in exporting nuclear p53 to cytoplasm for subsequent p53 degradation, thereby enabling p53 accumulation (38, 39). In addition to ubiquitination and phosphorylation, p53 activity is modulated by acetylation (35, 40). Acetylation by histone acetyltransferases (HATs) and deacetylation by histone deacetylases (HDACs) were initially discovered to regulate the extent of histone acetylation and play a critical role in gene transcription (41). p53 was identified as the first non-histone substrate of HATs and HDACs (42, 43). In response to DNA damage, CBP/p300 acetylates p53 on six C-terminal lysine (K) residues (K370, K372, K373, K381, K382, and K386), the same target sites of MDM2-mediated ubiquitination, and hence leads to enhanced stability and DNA binding activity of p53 (44). In addition, acetylation of p53 on K320 by PCAF preferentially directs p53 to activate target genes involved in cell cycle arrest (45), whereas acetylation of p53 on K120 by Tip60/hMOF promotes p53-mediated cell death (46–48). Moreover, acetylation of p53 on K164 by CBP/p300 is required for p53-induced cell cycle arrest and apoptosis (49). Interestingly, emerging evidence showed that TP53 mRNA stability and translation are regulated by multiple RNA binding proteins including HuR (50), ribosomal protein L26 (RPL26) (51, 52), nucleolin (51), and RNPC1 (53), and microRNAs including miR-125b (54), miR-125a (55), and miR-504 (56).
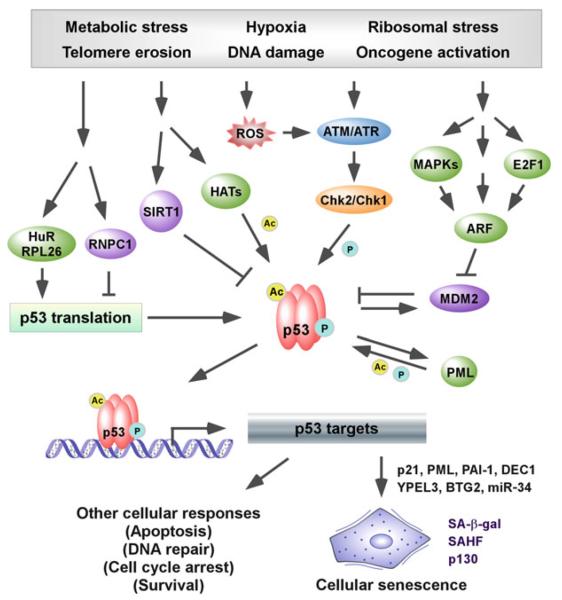
Fig. 2.
The role of p53 in cellular senescence. Upon exposure to numerous stress signals, p53 is regulated by phosphorylation (ATM/ATR-Chk2/Chk1), by acetylation (p300/CBP, PCAF, Tip60/hMOF, and SIRT1), by increased protein stability (ARF-MDM2), and by increased translation rates (RNPC1, HuR, and RPL26). Once activated, p53 regulates a set of downstream targets leading to cellular senescence and other cellular responses. Several senescence markers, including p21, PML, PAI-1, and DEC1, are identi fied as p53 target genes and promote cellular senescence. In addition, PML positively feeds back to promote p53 phosphorylation and acetylation. As a result, senescent cells induced by p53 activation exhibit flattened morphology, enlarged cell size, expression of SA-β-gal, formation of SAHF, and deregulated expression of p53 targets. In some cases, increased hypophosphorylated p130 is detected in cellular senescence induced by p53 and/or DNA damage.
2.2. The Role of p53 in Replicative Senescence
Detection of γ-H2AX foci, a sensitive marker for DNA double-strand break (DSB), on dysfunctional telomeres (shortened telomeres or altered telomere state) in senescent cells indicates that telomere-dependent replicative senescence is stress-dependent (57–59) (Fig. 2). These foci contain multiple DNA damage responsive proteins, such as 53BP1 and MRN complex (MRE11, RAD51, and NBS1), and are indistinguishable from the γ-H2AX foci originated from ionizing radiation-induced DNA DSB (60, 61). Activation of ATM at sites of DNA DSBs elicits phosphorylation of Chk2 and upregulation of p53 (62, 63). ATM is a primary mediator for telomere dysfunction-induced damage signaling. However, in the absence of ATM, ATR (ATM and Rad3-related) substitutes the role of ATM to activate p53 through Chk1-induced p53 phosphorylation at Ser-15 (63). Consistently, enhanced expression of p53 in mice deficient in Terc, the RNA component of telomerase, leads to activation of senescence and reduced tumor formation (64). In contrast, mice deficient in both Terc and p53 bypass senescence and develop tumors (65). These indicate that p53 plays a pivotal role in maintaining telomere dysfunction-initiated senescence.
2.3. The Role of p53 in Oncogene-Induced Senescence
In addition to spontaneous senescence, OIS was discovered as a cancer prevention mechanism in cells exposed to oncogenic Ras. It has been shown that expression of oncogenic Ras promotes acute senescence-like G1 arrest in primary human or rodent cells containing wild-type p53 (24), but initiates cellular transformation in those lacking p53 (66). This indicates that p53 plays a critical role in OIS. There are two underlying mechanisms by which Ras activates the p53-dependent senescence pathway in OIS (67–69) (Fig. 2). First, Ras regulates p53 through the MAPK pathways. It has been shown that Ras utilizes the MAPK signal-transduction pathway, including Raf-1, MEKs (MEK1/2), and MAPKs (ERK1/2), to promote cell cycle arrest in primary cells but malignant transformation in immortal cells (70–72). However, the cell cycle arrest induced by expression of Ras or MEK is abolished in cells lacking p53 (70). In addition, activation of the p38/MAPK pathway, especially its downstream kinase PRAK, leads to p53-dependent senescence by phosphorylating and activating p53 (73, 74). Indeed, ARF can be activated by the Ras/Raf signaling cascade via E2F1 or Dmp1 (75, 76). E2F1 and Dmp1 are capable of directly binding to, and activating, the promoter of the ARF gene (77, 78). p19ARF (p14ARF in human) is encoded from an alternative reading frame within the INK4a/ARF locus which also encodes p16 (79, 80). ARF blocks MDM2-mediated degradation of p53 by repressing MDM2 E3 ligase activity and nuclear export as well as promoting MDM2 degradation (81–84). Interestingly, ARF mediates Ras signaling to p53 in mouse cells but is not required for OIS in human cells (85). Second, Ras regulates p53 by activation of DNA damage response (DDR). It has been shown that ectopic expression of HRasV12 in human diploid fibroblasts induces DNA hyper-replication resulting in prematurely terminated DNA replication and collapse of DNA replication forks, which in turn lead to accumulation of DSBs and subsequent activation of the ATM/ATR-p53 pathway to induce senescence (86–89). In addition, oncogene activation leads to accumulation of reactive oxygen species (ROS), which induces DNA damage (89, 90). Consistent with this, human diploid fibroblasts grown in a restrictive culture condition with low or no ROS production bypass oncogene-induced senescence (91).
2.4. The Role of p53 in Genotoxic Stress-Induced Senescence
Given the importance of the DDR-p53 pathway in both replicative senescence and OIS, it is not surprising that numerous stress signals, such as DNA damage, oxidative stress, insufficient nutrients/growth factors, and improper cell contacts, induce premature senescence in normal human and murine cells (27, 92). Since induction of permanent cell cycle arrest is a potential cancer prevention mechanism by eliminating damaged pre-cancer cells at the initial stage of transformation, it was postulated that evasion of senescence might be a prerequisite for pre-cancer cells to complete transformation into cancer cells (93–95). Surprisingly, senescence-like growth arrest is induced by ectopic expression of a tumor suppressor gene or treatment with chemotherapeutic drugs in a broad range of human cancer cells, including those derived from colon, ovarian, lung, prostate, and cervical carcinomas (25). Importantly, reports showed that restoration of p53-mediated premature senescence is able to promote cancer regression in mice with hepatocarcinoma and sarcomas (12, 96). Indeed, ectopic expression of wild-type p53 triggers premature senescence in tumor cells lacking endogenous p53 (97, 98). In addition, p53-dependent premature senescence is induced by a variety of chemotherapeutic drugs, including camptothecin, doxorubicin, etoposide, cisplatin, and resveratrol (99). However, treatment with therapeutic agents also induces p53-independent premature senescence. For instance, doxorubicin induces senescence in a number of cancer cell lines deficient in p53 (25, 100, 101).
3. Characteristics of p53-Mediated Cellular Senescence
3.1. Cell Cycle Arrest
Since senescent cells must initially undergo cell cycle arrest, DNA histogram analysis is expected to show decreased number of cells in S phase (cell uptake of BrdU) and increased number of cells in G1 or sometimes also G2/M phase, but no change in the number of cells in sub-G1 phase (dead cells) (101, 102). However, the cell cycle profile for senescent cells is indistinguishable from that for quiescent cells and terminally differentiated cells.
3.2. SA-β-Gal
SA-β-gal is a commonly used senescence biomarker, which can be detected at pH 6.0 (18). This phenomenon is due to the fact that senescent cells express a high level of lysosomal β-gal (103–105). It is important to note that overexpression of lysosomal β-gal itself is not able to initiate senescence (103). Nevertheless, SA-β-gal staining is a convenient and accepted method to detect cellular senescence in vitro and even in vivo (18, 26). However, β-gal positive alone is not sufficient to judge whether a cell is in the senescent state. For example, TGF-β stimulation induces SA-β-gal independent of senescence in both cultured human prostate basal cells and epithelial cells in benign prostatic hyperplasia (106). In addition, expression of lysosomal galactosidase beta-1 (GLB1), which accounts for the SA-β-gal activity in some cancer cells, is not correlated with senescence (103). Moreover, in order to measure cellular senescence in vivo, fresh or frozen tissues are required for this staining assay.
3.3. Senescence-Associated Heterochromatic Foci
Cellular senescence is accompanied by formation of facultative heterochromatin, also called senescence-associated heterochromatic foci (SAHF), wherein DAPI staining would show spot-staining patterns (107). In contrast, DAPI staining would show homogenous staining patterns in non-senescent cells (107). SAHF contains histone H3K9 methylation whereas euchromatin contains histone H3K9 acetylation and K4 methylation (107). Meanwhile, the increased incorporation of heterochromatin protein 1γ (HP1γ) into SAHF is distinct from the pericentric heterochromatin (107). Furthermore, γ-H2AX colocalizes with SAHF (108). Importantly, a combination of positive SAHF and γ-H2AX with negative Ki67 (a proliferation index) is a reliable indicator of senescence for cells in culture and in tissues (109, 110). However, SAHF formation is often detected but not a prerequisite for p53-dependent senescence (107, 111).
3.4. p53 Targets Genes
As mentioned above, activation of the p53 pathway is a common cellular response leading to cellular senescence upon exposure to stress signals (Fig. 2). As a result, senescent cells are characterized by enhanced expression of some p53 targets, such as p21 (30), PML (112), plasminogen activator inhibitor (PAI-1) (113), and DEC1 (101), all of which are recognized as senescence markers, and in turn are able to induce senescence themselves (69, 95, 114–117). p21, a cyclin-dependent kinase inhibitor, plays a critical role in inducing G1 cell cycle arrest by inhibiting the activity of cyclin-CDK2/4 complexes, E2F, and PCNA (118–121). Promyelocytic leukemia protein PML, an essential component of PML nuclear bodies (PML-NBs) that accumulates in senescent cells, is induced by several factors, including oncogenic stress and p53 (112, 116). In turn, PML recruits p16, p53, and pRb/E2F complex to the PML-NBs and hence modulates the expression of downstream target genes of these factors, leading to senescence (116, 122). Importantly, PML forms a positive feedback loop with p53 to trigger cellular senescence by promoting p53 acetylation and phosphorylation or inhibiting p53 degradation by MDM2 (115, 116, 123–125). By contrast, inhibition of p53 acetylation by SIRT1 (a NAD+-dependent class III histone deacetylase) blocks PML-mediated premature senescence (126). PAI-1 induces cellular senescence by inhibiting the activity of uPA, a secreted protein which is capable of activating the MAPK pathway, and subsequently promoting G1/S transition (127). DEC1, a basic helix-loop-helix transcription factor, is capable of inducing DNA damage-induced premature senescence via inhibiting ID1, an oncogene which is found to be downregulated in arrested or senescent cells (128, 129). A recent report showed that Yippee-like-3 (YPEL3), a member of the putative zinc finger motif coding gene family, is regulated by p53 and triggers premature senescence in normal and tumor cells (130). In addition, BTG2, a member of the antiproliferative BTG gene family, is a p53-responsive gene and plays an essential role in replicative senescence (131). Moreover, the miR-34 family is a p53 target and elicits cellular senescence in both primary and tumor cells in response to DNA damage and oncogenic stress (132, 133). It is important to note that p130 is the major pocket protein associated with induction of premature senescence via the DNA damage-p53 signaling cascade in both normal and tumor cells (134–136). However, the mechanism by which p53 increases p130 activity during senescence is still not clear.
Detection of p53 and its downstream effectors can be used to measure the state of senescence for cells both in vitro and in vivo. However, these genes are not always induced in senescent cells by p53 in response to a specific stress signal. Therefore, a combination of multiple senescence markers discussed above together with one or more p53 targets is required to implicate the state of p53-mediated cellular senescence.
4. The Role of p53 Isoforms in Cellular Senescence
The TP53 gene encodes at least nine different isoforms due to the use of two promoters, two translation initiation sites, and alternative splicing (137) (Fig. 1). The TP53 gene contains 11 exons. p53, also called α isoform, is a full-length p53 generated from the P1 promoter. An internal promoter in intron 4 leads to production of an N-terminal truncated p53 starting from residue 133, named as Δ133p53 (138). In addition, internal initiation of translation at an AUG codon at position 40 or alternative splice of intron 2 leads to expression of a p53 protein with deletion of the first 39 amino acids, named as Δ40p53 (also called ΔNp53, p47, p53/47, and p44) (138–142). Moreover, alternative splicing of intron 9 produces β and γ isoforms (138) , which lack C-terminal oligomerization domain but contain 10 and 15 unique amino acids at C-termini, respectively (Fig. 1). As a result, nine p53 isoforms, p53α (full-length p53, referred to as p53 in this chapter), p53β (also called p53I9 (143)), p53γ, Δ40p53α, Δ40p53β, Δ40p53γ, Δ133p53α, Δ133p53β, and Δ133p53γ, are expressed. These p53 isoforms are found to be expressed in normal human tissues in a tissue-dependent manner as well as in some tumor tissues (137). This suggests that the expression pattern of p53 isoforms may play a role in tumor formation and affect the outcomes of cancer therapy.
Under an unstressed condition, p53β itself preferentially binds to the p21 and Bax (a pro-apoptotic gene) promoters rather than to the MDM2 promoter, but has no effect on p53 activity (138). However, in response to a stress signal, p53β specifically enhances the Bax gene transcription through physical interaction with p53 and slightly increases p53-mediated apoptosis (138). In contrast, Δ133p53, which lacks the entire N-terminal AD, is dominant-negative over p53 and diminishes p53-mediated apoptosis (138). Interestingly, it has been shown that induction of p53β and suppression of Δ133p53 lead to induction of p21 and miR-34, resulting in senescence in cultured cells and colon adenomas (144). This indicates that the biological activity of full-length p53 is likely modulated by its isoforms in normal and tumor tissues. However, other groups have reported that p53β, which lacks the C-terminal BD and most of the TD, is deficient in DNA binding activity and unable to modulate p53-dependent stress responses (143, 145). Furthermore, ΔNp53 is implicated in G2 cell cycle arrest (146). Therefore, further studies are needed to clarify the mechanism by which p53 isoforms affect each other's functions.
5. The Role of p53 Protein Isoforms in Aging
It has been proposed that aging is characterized by decreased capability to maintain and repair somatic cells (147). One of the causes for aging is associated with senescence-mediated decline in the number and/or proliferative capacity of adult stem cells (148, 149). Indeed, elevated expression of senescence-associated markers, SA-β-gal and p16, is detected in aging tissues (18, 150). As a major mediator of cellular senescence, activation of p53 not only inhibits neoplasia but also promotes organismal aging. Because Tp53 homozygous knockout mice die from tumors at a young age (8), several mouse models with altered p53 activity have been generated to analyze the role of p53 in aging. These are Tp53+/m, Δ40p53-knockin, pL53, and super p53 mouse models (9, 151).
Tp53+/m mice contain a mutant p53 m allele with a deletion mutation in the first six exons of the Tp53 gene and express an N-terminal truncated p53 protein, called M protein (152). Tp53+/m mice display reduced tumor formation and shortened longevity compared with wild-type littermates (152). However, Tp53m/m mice die before birth and Tp53−/m mice develop tumors similarly to Tp53−/− mice. This suggests that M protein-induced aging phenotypes in Tp53+/m are p53-dependent. Indeed, it has been shown that M protein interacts with and stabilizes p53 by facilitating p53 nuclear localization in the absence of a stress (153). Therefore, it is possible that the resulting phenotypes observed in Tp53+/m mice are due to increased p53 activity. However, this mutant allele also contains a deletion of 24 genes, including Aloxe3, Alox12b, Alox15b, Chd3, Aurkb, and Per1, adjacent to the Tp53 gene (154). Thus, it is necessary to further analyze whether haploinsufficiency of these 24 genes contributes to altered p53 activity and decreased tumor formation but accelerated aging phenotypes.
Similar to Tp53+/m mice, Δ40p53-knockin mice, that express a naturally occurring N-terminal truncated p53 isoform, exhibit decreased lifespan and several premature aging phenotypes, including shortened reproductive span, lordokyphosis, and a reduced tumor incidence (155). These phenotypes are likely due to enhanced wild-type p53 activity and hyperactivation of insulin-like growth factor 1 (IGF-1) signaling cascade (155), the latter of which is associated with lifespan in many species (156, 157). The elevated IGF-1 signaling in Δ40p53 mice leads to sustained induction of p21 through the MAPK pathway and consequently causes cellular senescence and premature aging phenotypes (155). Similar premature aging phenomena were observed in another line of transgenic mice containing multiple copies of a temperature-sensitive mutant allele p53V135A derived from BALB/c liver DNA, named as pL53 (158). Together, these results indicate that enhanced p53 activity contributes to aging. This hypothesis is supported by several other accelerated aging mouse models, such as mice deficient in Terc (64), Ku80 (also called Ku86, a regulatory unit of DNA-PK) (159), BRCA1 (a tumor suppressor) (160), and Zmpste24 (also called FACE-1, a metalloproteinase involved in the maturation of Lamin A) (161). Conversely, mice deficient in p66shc, a cytoplasmic signal transducer from activated receptors to Ras, live longer along with decreased p53 activity than wild-type controls (162). However, this hypothesis is challenged by another mouse model, called the “super p53” mice.
The super p53 mice carry extra copies of a complete p53 gene, including the p53 promoter and upstream regulatory region (163). As expected, these mice display augmented p53 activity in response to DNA damage and are tumor-resistant. Interestingly, the super p53 mice have normal lifespan without showing any sign of accelerated aging. Similarly, a “super Ink4a/ARF” mouse strain, which carries a transgenic copy of the entire Ink4a/ARF locus (164), and a mouse strain, which carries a hypomorphic allele of MDM2 (165), were generated, both of which have decreased MDM2 activity and hence enhanced p53 activity. Like the super p53 mice, MDM2 hypomorphic and super Ink4a/ARF mice are resistant to spontaneous and carcinogene-induced tumor formation without premature aging. Moreover, the super p53/p19ARF mice, carrying additional copies of both Tp53 and ARF alleles, were generated by cross-breeding the super p53 mice with the super Ink4a/ARF mice and hence p53 can be further activated (166). Surprisingly, the super p53/p19ARF mice are not only cancer free but also have a significantly extended lifespan. These findings suggest that there is no causal relationship between increased p53-mediated tumor suppression and decreased longevity. Instead, p53 activation in certain circumstances exerts benefits on longevity. There are several potential explanations. First, aging is a consequence of accumulation of DNA damage in tissues, and therefore, by modulating the DDR, p53 is able to reduce age-associated DNA damage and accumulation of damaged cells. Second, p53 activation plays a role in maintaining the stem-cell pool in adult tissues. Third, p53 regulates aging process by modulating other signaling pathways affecting longevity, such as the insulin/IGF-1 and mTOR signaling (167–169). Fourth, p53 may have pro-longevity effects by restraining secretion of the senescence-associated pro-inflammatory cytokines (170, 171), which are known to contribute to chronic age-related inflammation. However, the precise mechanism by which p53 controls the balance between anti-aging and pro-aging needs to be further explored.
6. The Role of p63 and p73 in Cellular Senescence and Aging
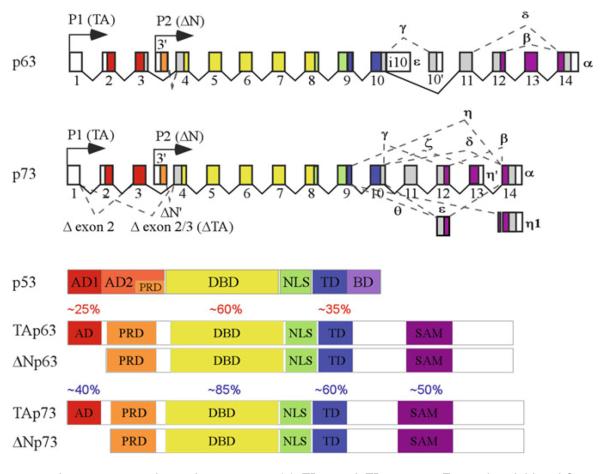
p63 and p73, two p53-ancestral genes (172, 173), share highly amino-acid identity with p53 within the AD, DBD, and TD (174, 175) (Fig. 3). The highest degree of homology among the three members is observed within the DBD (>60% amino-acid identity between p53 and p63/p73, and ~85% amino-acid identity between p63 and p73). Unlike p53, the alpha isoforms of p63 and p73 contain a C-terminal sterile alpha motif (SAM), a protein interaction domain involved in developmental regulation. In addition, multiple protein products are generated from both p63 and p73 genes due to the usage of two promoters and alternative RNA splicing (Fig. 3). Therefore, p63 and p73 isoforms possess one of two N termini, transcriptional active (TA) and N-terminally deleted (ΔN), each with five different C termini for p63 (α – ε) (173, 176) and at least ten different C termini for p73 (α – η, η1, η′, and θ) (172, 177–180). Furthermore, alternative splicing of p73 exon 2 and 3 gives rise to two additional p73 isoforms, ΔN′ p73 and ΔTAp73 (ΔEx2 and ΔEx2/3) (172, 179, 181).
Fig. 3.
The p63 and p73 genes and protein structures. (a) TP63 and TP73 genes. Transcripts initiated from P1 and P2 promoters give rise to TA and ΔN protein isoforms, respectively. Alternative splicing at C-termini of TP63 transcripts leads to production of p63 α, β (Δ exon 13), γ (Δ exons 11–14 and incorporated with additional sequence from exon 10′), δ (Δ exons 12 and 13), and ε (generated by alternative termination in exon 10). Alternative splicing at C-termini of TP73 transcripts leads to production of p73 α, β (Δ exon 13), g (Δ exon 11), δ (Δ exons 11–13), ζ (Δ exons 11 and 12), ε (Δ exons 11 and 13), θ (Δ exons 10, 11, and 13), η (Δ exons 10–13), η 1 (Δ 2 nucleotides at 3′ -end of exon 9 and contains 18 nucleotides from exon 13, compared with h), and η′ (generated by alternative termination in exon 13). Alternative splicing at N-termini of TP73 transcripts leads to production of ΔN′p73 and ΔTAp73 (ΔEx2 and ΔEx2/3). Δ, deleted. (b) Homology among the p53 family members. Compared to p53, the a isoforms of p63 and p73 have a unique C-terminal SAM (sterile alpha motif) domain. A high degree of sequence identity is seen between p53 and p63/p73 (on top) and between p63 and p73 (at bottom).
Unlike Tp53 knockout mice that are prone to tumor formation, Tp63 knockout mice show severe defect in limb formation and epidermal development (182, 183) and p73-null mice exhibit developmental defects in central nervous system and inflammatory response (184). These suggest that p63 and p73 play a key role in development. However, due to the high sequence similarity among the p53 family, p63 and p73 act synergistically or antagonistically with p53 in regulating p53-mediated biological activities. Indeed, p63 and p73, especially their TA isoforms, are able to bind to p53-responsive elements and transactivate a set of p53 targets, such as p21, MDM2, Bax, and PUMA, and regulate cell cycle arrest and cell death (174, 185, 186). Recent reports showed that p63 and p73 are also involved in regulating cellular senescence (Fig. 4).
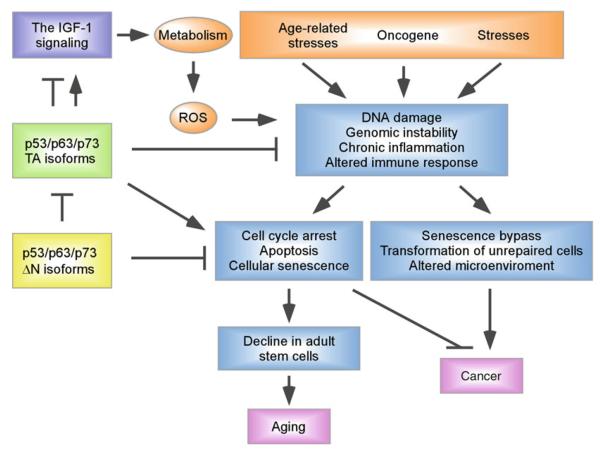
Fig. 4.
The role of p53 family proteins in cancer and aging. Accumulation of abnormal cells caused by aging-associated damage, aberrant oncogenic activity, and other stress signals, contributes to both aging and cancer. Activation of the p53 pathway in response to these stress signals is able to protect adult stem cells to promote longevity and prevent malignant transformation by repairing or eliminating damaged cells. However, accumulation of senescent cells in tissues due to activation of TA isoforms of p53 family members, leads to decline in adult stem cells and accelerated aging. In addition, p53 family members are capable of targeting the IGF-1 signaling cascade, a pathway known to be involved in organismal aging, to regulate longevity. In contrast, ΔN isoforms of p53 family are found to be required for maintenance of stem cell proliferation and act as dominate negative inhibitors of TA counterparts.
Results obtained from Tp63 knockout mice suggest that p63 is an essential regulator for epidermal homeostasis (182, 183). Indeed, ΔNp63 plays a predominant role in epidermal commitment during embryogenesis (187, 188). In addition, ΔNp63 is highly expressed in the basal layer of stratified epithelia to maintain proliferative potential and direct proper differentiation of progenitor cells (189, 190). Moreover, TAp63-specific knockout mice do not display the same impaired skin development as p63-null mice (191), whereas ΔNp63-null mice develop lesions that resemble those seen in AEC (ankyloblepharon ectodermal dysplasia and clefting) patients, an ectodermal dysplasia characterized by skin fragility (192). This further confirms the critical role of ΔNp63 in epidermal morphogenesis. However, as a p53 homolog, mice deficient in TAp63 develop metastatic tumors (193). In addition, loss of p63 or TAp63 increases the number of metastatic tumors in Tp53+/− and Tp53−/− mice (193, 194). Surprisingly, although Tp53 heterozygous mice develop tumors, Tp63 heterozygous mice are tumor-resistant or develop a few tumors along with a decreased lifespan and accelerated aging phenotypes, similar to Tp53+/m mice (195). Consistently, ablation of p63 induces cellular senescence in primary kerotinocytes (195). In addition, germline- or tissue-specific p63 deficiency also leads to enhanced expression of senescence markers, including SA-β-gal and PML, concomitant with premature aging features (195). Importantly, a report showed that four females from a family with Rapp–Hodgkin syndrome (RHS), a disease associated with TP63 mutations, present not only typical RHS (anhidrotic ectodermal dysplasia with cleft lip and palate) but also ophthalmic anomalies such as corneal dystrophy and premature menopause (around 30 years) (196). These data suggest that p63 is implicated in aging progression. It has been shown that overexpression of ΔNp63α inhibits OIS and promotes tumorigenesis in vivo (197), whereas ectopic expression of TAp63 induces senescence and inhibits progression of established tumors in vivo (198). However, the TAp63−/− mice exhibit signs of premature aging, including hair loss, impaired wound healing, kyphosis, and a shortened lifespan (191). It is postulated that TAp63 is necessary for the survival of adult skin stem cells and thus prevents premature aging (191). In addition, TAp63 was shown to regulate senescence and aging via modulating the p53, p16, and IGF pathways (199, 200). These together indicate that TA and ΔN isoforms of p63 play a dual role in cellular senescence and tissue aging in cell-type and stress-specific manners.
Like p53, p73 is activated in response to various stress signals, including DNA damage and oncogenic activation, and regulates a set of p53-dependent and -independent genes involved in cell cycle arrest and apoptosis (185, 201, 202). In the absence of p53, p73 can compensate for p53 function to induce apoptosis in cancer cells and the status of the TP73 gene is correlated with the survival rate for cancer patients (201, 203–207). Likewise, mice heterozygous of both Tp53 and Tp73 exhibit increased tumor burden and metastasis compared to mice heterozygous of Tp53 alone (194). Mice specifically deficient in TAp73 isoform develop spontaneous and carcinogen-induced tumors and cells from TAp73-null mice display genomic instability associated with enhanced aneuploidy (208). By contrast, cells from ΔNp73-null mice are more sensitive to DNA damage-induced apoptosis in a p53-dependent manner and high levels of ΔNp73 expression associates with poor prognosis in human cancers (209). These results demonstrate that TAp73 is a tumor suppressor whereas ΔNp73 acts as an oncogene. However, to date, knowledge on the role of p73 in senescence is limited. Similar to p53 and p63, p73 plays a function in the maintenance of stem cells (Fig. 4). Specifically, it has been shown that ΔNp73 is a potent prosurvival factor for neurons (210–212) and TAp73 is required for the long-term maintenance of neural stem cells to elicit neurogenesis in adults (213). As a result, mice heterozygous of TA or ΔNp73 isoforms display age-related neurodegeneration (214). Similar to p53 and p63, p73 has been shown to inhibit expression of IGF-1 receptor (200, 215, 216). This indicates that the IGF-1 signaling is a common target of the p53 family to modulate aging (Fig. 4). Importantly, a recent report provided evidence for the role of TAp73 in aging. It has been shown that tumor-free TAp73-null mice develop premature aging phenotypes with impaired ROS scavenging compared to wild-type littermates (217). Indeed, TAp73 is crucial to maintain proper mitochondrial functions by directly regulating the expression of mitochondrial complex IV subunit Cox4i1 (cytochrome C oxidase subunit 4 isoform 1) (217). However, the molecular basis for p73 in aging-associated disorders needs to be further uncovered.
7. Summary and Remarks
Proliferating cells can exit from the cell cycle and enter a state of permanent cell cycle arrest when they encounter stress signals originated from telomere shortening, aberrant oncogenic activities, and other DNA damage signals. Senescent cells are distinct from quiescent cells by expression of senescence-associated markers, including SA-β-gal, SAHF, and activation of the p53 or p16 pathway followed by altered gene expression profile. Emerging evidence showed that cellular senescence associates with multiple pathological disorders, including cancer and aging. Importantly, it has been demonstrated in vivo that induction of cellular senescence is a tumor suppression mechanism by blocking malignant transformation at the initial step of tumorigenesis and by clearance of cancer cells at the late stage of tumor progression. However, the benefit of the cellular senescence-mediated tumor suppression is countered by the cost of accelerated aging. Therefore, a future challenge to develop senescence-based cancer therapy is to reveal the target and pathways that control the balance between cancer and longevity.
As we discussed in this chapter, the p53 family plays a pivotal role in cellular senescence and thus affect cancer progression and aging (Fig. 4). Although induction of cellular senescence by activation of the p53 family contributes to tumor suppression and accelerated aging, reports also showed that the p53 family promotes longevity and maintains tissue homeostasis through protecting renewable stem cells. In addition, studies with the “super p53” mouse models indicate that cancer resistance could be achieved by modulation of p53 activity without causing undesirable effects. Importantly, a combination of increased p53 and ARF activities not only inhibits tumor formation but also increases longevity. Therefore, the p53 pathway is a promising target for senescence-based cancer treatment. However, there are still unsolved questions. For instance, how are p53 isoforms, p63 and p73, involved in making a balance between cancer and aging? What is the molecular basis for p63/p73-mediated cellular senescence? How does p53 family integrate with other signaling pathways related to aging? Therefore, further exploration of these puzzles in vitro and in vivo will enhance our knowledge about the p53 family in senescence-associated diseases and may provide applicable clues for clinical use.
References
- 1.DeLeo AB, Jay G, Appella E, Dubois GC, Law LW, Old LJ. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc Natl Acad Sci U S A. 1979;76:2420–2424. doi: 10.1073/pnas.76.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 3.Linzer DI, Levine AJ. Characterization of a 54 K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 4.Melero JA, Stitt DT, Mangel WF, Carroll RB. Identification of new polypeptide species (48–55 K) immunoprecipitable by antiserum to purified large T antigen and present in SV40-infected and -transformed cells. Virology. 1979;93:466–480. doi: 10.1016/0042-6822(79)90250-2. [DOI] [PubMed] [Google Scholar]
- 5.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 8.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 9.Donehower LA. Using mice to examine p53 functions in cancer, aging, and longevity. Cold Spring Harb Perspect Biol. 2009;1:a001081. doi: 10.1101/cshperspect.a001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li–Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 11.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 12.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 17.Matsumura T, Zerrudo Z, Hayflick L. Senescent human diploid cells in culture: survival, DNA synthesis and morphology. J Gerontol. 1979;34:328–334. doi: 10.1093/geronj/34.3.328. [DOI] [PubMed] [Google Scholar]
- 18.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.d'Adda di Fagagna F, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 20.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 21.Hayflick L. The illusion of cell immortality. Br J Cancer. 2000;83:841–846. doi: 10.1054/bjoc.2000.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimri GP. What has senescence got to do with cancer? Cancer Cell. 2005;7:505–512. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Artandi SE, Attardi LD. Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem Biophys Res Commun. 2005;331:881–890. doi: 10.1016/j.bbrc.2005.03.211. [DOI] [PubMed] [Google Scholar]
- 24.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 25.Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, Kandel ES, Lausch E, Christov K, Roninson IB. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- 26.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–1883. [PubMed] [Google Scholar]
- 27.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 29.Harms KL, Chen X. The functional domains in p53 family proteins exhibit both common and distinct properties. Cell Death Differ. 2006;13(6):890–897. doi: 10.1038/sj.cdd.4401904. [DOI] [PubMed] [Google Scholar]
- 30.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 31.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 33.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 34.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 35.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 36.Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 37.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 38.Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 43.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 44.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol. 2006;173:533–544. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 48.Charvet C, Wissler M, Brauns-Schubert P, Wang SJ, Tang Y, Sigloch FC, Mellert H, Brandenburg M, Lindner SE, Breit B, Green DR, McMahon SB, Borner C, Gu W, Maurer U. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol Cell. 2011;42:584–596. doi: 10.1016/j.molcel.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 52.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, Skorupski K, Chen H, Chen X. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 2011;25:1528–1543. doi: 10.1101/gad.2069311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Gao JS, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, Ramratnam B. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583:3725–3730. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, Tang LH, Levine AJ, Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38:689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 58.von Zglinicki T, Saretzki G, Ladhoff J, d'Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126:111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 59.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 60.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 61.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 62.Pandita TK. ATM function and telomere stability. Oncogene. 2002;21:611–618. doi: 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- 63.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 64.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 65.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 66.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 67.McDuff FK, Turner SD. Jailbreak: oncogene-induced senescence and its evasion. Cell Signal. 2011;23:6–13. doi: 10.1016/j.cellsig.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 68.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian Y, Chen X. Tumor suppression by p53: making cells senescent. Histol Histopathol. 2010;25:515–526. doi: 10.14670/hh-25.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67:2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 72.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 73.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, Dong MQ, Frangou CG, Yates JR, 3rd, Wright PE, Han J. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 74.Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, Sun P. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–3403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 77.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 78.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci U S A. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, 3rd, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 80.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 81.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 82.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 84.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei W, Hemmer RM, Sedivy JM. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol Cell Biol. 2001;21:6748–6757. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 87.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 88.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 89.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 91.Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 92.Lloyd AC. Limits to lifespan. Nat Cell Biol. 2002;4:E25–E27. doi: 10.1038/ncb0202-e25. [DOI] [PubMed] [Google Scholar]
- 93.Wynford-Thomas D. Cellular senescence and cancer. J Pathol. 1999;187:100–111. doi: 10.1002/(SICI)1096-9896(199901)187:1<100::AID-PATH236>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 94.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 95.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 96.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 97.Wang Y, Blandino G, Oren M, Givol D. Induced p53 expression in lung cancer cell line promotes cell senescence and differentially modifies the cytotoxicity of anti-cancer drugs. Oncogene. 1998;17:1923–1930. doi: 10.1038/sj.onc.1202113. [DOI] [PubMed] [Google Scholar]
- 98.Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci U S A. 1997;94:9648–9653. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ewald JA, Desotelle JA, Wilding G, Jarrard DF. Therapy-induced senescence in cancer. J Natl Cancer Inst. 2010;102:1536–1546. doi: 10.1093/jnci/djq364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, Roninson IB. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 101.Qian Y, Zhang J, Yan B, Chen X. DEC1, a basic helix-loop-helix transcription factor and a novel target gene of the p53 family, mediates p53-dependent premature senescence. J Biol Chem. 2008;283:2896–2905. doi: 10.1074/jbc.M708624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fang L, Igarashi M, Leung J, Sugrue MM, Lee SW, Aaronson SA. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene. 1999;18:2789–2797. doi: 10.1038/sj.onc.1202615. [DOI] [PubMed] [Google Scholar]
- 103.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 104.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113(Pt 20):3613–3622. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 105.Le Gall JY, Khoi TD, Glaise D, Le treut A, Brissot P, Guillouzo A. Lysosomal enzyme activities during ageing of adult human liver cell lines. Mech Ageing Dev. 1979;11:287–293. doi: 10.1016/0047-6374(79)90008-3. [DOI] [PubMed] [Google Scholar]
- 106.Untergasser G, Gander R, Rumpold H, Heinrich E, Plas E, Berger P. TGF-beta cytokines increase senescence-associated beta-galactosidase activity in human prostate basal cells by supporting differentiation processes, but not cellular senescence. Exp Gerontol. 2003;38:1179–1188. doi: 10.1016/j.exger.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 107.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 108.Zhang R, Chen W, Adams PD. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27:2343–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rastogi S, Joshi B, Dasgupta P, Morris M, Wright K, Chellappan S. Prohibitin facilitates cellular senescence by recruiting specific corepressors to inhibit E2F target genes. Mol Cell Biol. 2006;26:4161–4171. doi: 10.1128/MCB.02142-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lawless C, Wang C, Jurk D, Merz A, Zglinicki T, Passos JF. Quantitative assessment of markers for cell senescence. Exp Gerontol. 2010;45:772–778. doi: 10.1016/j.exger.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 111.Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53 – > p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 113.Kunz C, Pebler S, Otte J, von der Ahe D. Differential regulation of plasminogen activator and inhibitor gene transcription by the tumor suppressor p53. Nucleic Acids Res. 1995;23:3710–3717. doi: 10.1093/nar/23.18.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 115.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 116.Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 117.Mu XC, Higgins PJ. Differential growth state-dependent regulation of plasminogen activator inhibitor type-1 expression in senescent IMR-90 human diploid fibroblasts. J Cell Physiol. 1995;165:647–657. doi: 10.1002/jcp.1041650324. [DOI] [PubMed] [Google Scholar]
- 118.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 119.Afshari CA, Nichols MA, Xiong Y, Mudryj M. A role for a p21-E2F interaction during senescence arrest of normal human fibroblasts. Cell Growth Differ. 1996;7:979–988. [PubMed] [Google Scholar]
- 120.Li R, Waga S, Hannon GJ, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 121.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 122.Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011;25:41–50. doi: 10.1101/gad.1975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bischof O, Kirsh O, Pearson M, Itahana K, Pelicci PG, Dejean A. Deconstructing PML-induced premature senescence. EMBO J. 2002;21:3358–3369. doi: 10.1093/emboj/cdf341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 125.Louria-Hayon I, Grossman T, Sionov RV, Alsheich O, Pandolfi PP, Haupt Y. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J Biol Chem. 2003;278:33134–33141. doi: 10.1074/jbc.M301264200. [DOI] [PubMed] [Google Scholar]
- 126.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57:25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qian Y, Chen X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem. 2008;283:22410–22416. doi: 10.1074/jbc.M800643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 130.Kelley KD, Miller KR, Todd A, Kelley AR, Tuttle R, Berberich SJ. YPEL3, a p53-regulated gene that induces cellular senescence. Cancer Res. 2010;70:3566–3575. doi: 10.1158/0008-5472.CAN-09-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wheaton K, Muir J, Ma W, Benchimol S. BTG2 antagonizes Pin1 in response to mitogens and telomere disruption during replicative senescence. Aging Cell. 2010;9:747–760. doi: 10.1111/j.1474-9726.2010.00601.x. [DOI] [PubMed] [Google Scholar]
- 132.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kapic A, Helmbold H, Reimer R, Klotzsche O, Deppert W, Bohn W. Cooperation between p53 and p130(Rb2) in induction of cellular senescence. Cell Death Differ. 2006;13:324–334. doi: 10.1038/sj.cdd.4401756. [DOI] [PubMed] [Google Scholar]
- 135.Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol Cell Biol. 2006;26:2501–2510. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Helmbold H, Komm N, Deppert W, Bohn W. Rb2/p130 is the dominating pocket protein in the p53–p21 DNA damage response pathway leading to senescence. Oncogene. 2009;28:3456–3467. doi: 10.1038/onc.2009.222. [DOI] [PubMed] [Google Scholar]
- 137.Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Courtois S, Verhaegh G, North S, Luciani MG, Lassus P, Hibner U, Oren M, Hainaut P. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21:6722–6728. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- 140.Ghosh A, Stewart D, Matlashewski G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol. 2004;24:7987–7997. doi: 10.1128/MCB.24.18.7987-7997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 142.Rovinski B, Munroe D, Peacock J, Mowat M, Bernstein A, Benchimol S. Deletion of 5′ -coding sequences of the cellular p53 gene in mouse erythroleukemia: a novel mechanism of oncogene regulation. Mol Cell Biol. 1987;7:847–853. doi: 10.1128/mcb.7.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Flaman JM, Waridel F, Estreicher A, Vannier A, Limacher JM, Gilbert D, Iggo R, Frebourg T. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene. 1996;12:813–818. [PubMed] [Google Scholar]
- 144.Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ, Bowman ED, Mathe EA, Schetter AJ, Pine SR, Ji H, Vojtesek B, Bourdon JC, Lane DP, Harris CC. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11:1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Graupner V, Schulze-Osthoff K, Essmann F, Janicke RU. Functional characterization of p53beta and p53gamma, two isoforms of the tumor suppressor p53. Cell Cycle. 2009;8:1238–1248. doi: 10.4161/cc.8.8.8251. [DOI] [PubMed] [Google Scholar]
- 146.Olivares-Illana V, Fahraeus R. p53 isoforms gain functions. Oncogene. 2010;29:5113–5119. doi: 10.1038/onc.2010.266. [DOI] [PubMed] [Google Scholar]
- 147.Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 148.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 149.Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113:160–168. doi: 10.1172/JCI20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 151.Papazoglu C, Mills AA. p53: at the crossroad between cancer and ageing. J Pathol. 2007;211:124–133. doi: 10.1002/path.2086. [DOI] [PubMed] [Google Scholar]
- 152.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee Park S, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 153.Moore L, Lu X, Ghebranious N, Tyner S, Donehower LA. Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity. Mech Ageing Dev. 2007;128:717–730. doi: 10.1016/j.mad.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gentry A, Venkatachalam S. Complicating the role of p53 in aging. Aging Cell. 2005;4:157–160. doi: 10.1111/j.1474-9726.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- 155.Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 157.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol Cell Biol. 1989;9:3982–3991. doi: 10.1128/mcb.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol. 2000;20:3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 162.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 163.Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Matheu A, Pantoja C, Efeyan A, Criado LM, Martin-Caballero J, Flores JM, Klatt P, Serrano M. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mendrysa SM, O'Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 167.Feng Z, Lin M, Wu R. The regulation of aging and longevity: a new and complex role of p53. Genes Cancer. 2011;2:443–452. doi: 10.1177/1947601911410223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Korotchkina LG, Leontieva OV, Bukreeva EI, Demidenko ZN, Gudkov AV, Blagosklonny MV. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging (Albany NY) 2010;2:344–352. doi: 10.18632/aging.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.de Keizer PL, Laberge RM, Campisi J. p53: pro-aging or pro-longevity? Aging (Albany NY) 2010;2:377–379. doi: 10.18632/aging.100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 173.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 174.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 176.Mangiulli M, Valletti A, Caratozzolo MF, Tullo A, Sbisa E, Pesole G, D'Erchia AM. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 2009;37:6092–6104. doi: 10.1093/nar/gkp674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.De Laurenzi VD, Catani MV, Terrinoni A, Corazzari M, Melino G, Costanzo A, Levrero M, Knight RA. Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta. Cell Death Differ. 1999;6:389–390. doi: 10.1038/sj.cdd.4400521. [DOI] [PubMed] [Google Scholar]
- 179.Ishimoto O, Kawahara C, Enjo K, Obinata M, Nukiwa T, Ikawa S. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 2002;62:636–641. [PubMed] [Google Scholar]
- 180.Scaruffi P, Casciano I, Masiero L, Basso G, Romani M, Tonini GP. Lack of p73 expression in mature B-ALL and identification of three new splicing variants restricted to pre B and C-ALL indicate a role of p73 in B cell ALL differentiation. Leukemia. 2000;14:518–519. doi: 10.1038/sj.leu.2401698. [DOI] [PubMed] [Google Scholar]
- 181.Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem. 2002;277:14177–14185. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 182.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 183.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 184.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 185.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]
- 186.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–3205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 187.Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, Melino G, Aberdam D. DeltaNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, Aberdam D. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3:e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- 190.Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, Miller FD, Flores ER. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Koster MI, Marinari B, Payne AS, Kantaputra PN, Costanzo A, Roop DR. DeltaNp63 knockdown mice: a mouse model for AEC syndrome. Am J Med Genet A. 2009;149A:1942–1947. doi: 10.1002/ajmg.a.32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, Flores ER. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 195.Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Holder-Espinasse M, Martin-Coignard D, Escande F, Manouvrier-Hanu S. A new mutation in TP63 is associated with age-related pathology. Eur J Hum Genet. 2007;15:1115–1120. doi: 10.1038/sj.ejhg.5201888. [DOI] [PubMed] [Google Scholar]
- 197.Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H, Guo X, Garcia EL, Michurina TV, Enikolopov G, Muthuswamy SK, Mills AA. DeltaNp63alpha is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Keyes WM, Mills AA. p63: a new link between senescence and aging. Cell Cycle. 2006;5:260–265. doi: 10.4161/cc.5.3.2415. [DOI] [PubMed] [Google Scholar]
- 200.Nahor I, Abramovitch S, Engeland K, Werner H. The p53-family members p63 and p73 inhibit insulin-like growth factor-I receptor gene expression in colon cancer cells. Growth Horm IGF Res. 2005;15:388–396. doi: 10.1016/j.ghir.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 201.Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 202.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 203.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG., Jr Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 204.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–645. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- 205.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 206.Zhu J, Nozell S, Wang J, Jiang J, Zhou W, Chen X. p73 cooperates with DNA damage agents to induce apoptosis in MCF7 cells in a p53-dependent manner. Oncogene. 2001;20:4050–4057. doi: 10.1038/sj.onc.1204516. [DOI] [PubMed] [Google Scholar]
- 207.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero M, Wang JY. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 208.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, Kaplan DR, Miller FD, Mak TW. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 211.Pozniak CD, Barnabe-Heider F, Rymar VV, Lee AF, Sadikot AF, Miller FD. p73 is required for survival and maintenance of CNS neurons. J Neurosci. 2002;22:9800–9809. doi: 10.1523/JNEUROSCI.22-22-09800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet AM. DeltaNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci U S A. 2009;106:16871–16876. doi: 10.1073/pnas.0903191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, Mak TW, Wojtowicz MJ, Miller FD, Kaplan DR. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20:2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 214.Wetzel MK, Naska S, Laliberte CL, Rymar VV, Fujitani M, Biernaskie JA, Cole CJ, Lerch JP, Spring S, Wang SH, Frankland PW, Henkelman RM, Josselyn SA, Sadikot AF, Miller FD, Kaplan DR. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer's disease. Neuron. 2008;59:708–721. doi: 10.1016/j.neuron.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 215.Werner H, Karnieli E, Rauscher FJ, LeRoith D. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci U S A. 1996;93:8318–8323. doi: 10.1073/pnas.93.16.8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 217.Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, Mak TW. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26:2009–2014. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]