Abstract
We describe a previously unreported syndrome characterized by secondary (post-natal) microcephaly with fronto-temporal lobe hypoplasia, multiple pituitary hormone deficiency, seizures, severe visual impairment and abnormalities of the kidneys and urinary tract in a highly consanguineous family with six affected children. Homozygosity mapping and exome sequencing revealed a novel homozygous frameshift mutation in the basic helix-loop-helix transcription factor gene ARNT2 (c.1373_1374dupTC) in affected individuals. This mutation results in absence of detectable levels of ARNT2 transcript and protein from patient fibroblasts compared with controls, consistent with nonsense-mediated decay of the mutant transcript and loss of ARNT2 function. We also show expression of ARNT2 within the central nervous system, including the hypothalamus, as well as the renal tract during human embryonic development. The progressive neurological abnormalities, congenital hypopituitarism and post-retinal visual pathway dysfunction in affected individuals demonstrates for the first time the essential role of ARNT2 in the development of the hypothalamo-pituitary axis, post-natal brain growth, and visual and renal function in humans.
Keywords: hypothalamus, congenital blindness, brain development, molecular genetics, malformations of cortical development
Introduction
Development of the CNS involves an extremely complex series of events, requiring intricate spatial and temporal coordination to control cellular signalling, migration, proliferation and differentiation. Post-natally, these processes affect brain volume, weight and surface area, gyration, cell migration, germinal matrix involution and myelination (Barkovich et al., 2001). A consistent feature central to the differentiation of neuroepithelial cells into neurons, regardless of where and when they form, is the involvement of proneural basic helix-loop-helix (HLH) transcription factors (Kintner, 2002). Basic HLH factors are an extensive protein family characterized by a basic DNA binding region adjacent to a helix-loop-helix dimerization region, both of which are required for the formation of functional DNA binding complexes (Powell and Jarman, 2008). In the embryonic CNS, basic HLH factors have a central role in mechanisms controlling cell fate specification, differentiation and cell-cycle maintenance (Bertrand et al., 2002).
One of the most evolutionarily conserved and fundamental regions of the brain is the hypothalamus. Our current knowledge of the molecular cascades involved in hypothalamic development and their relevance to the pathophysiology of human disease is extremely limited (Liu et al., 2003). Congenital hypopituitarism is frequently associated with other abnormalities, notably affecting the forebrain or visual function, suggesting that many cases are the result of disruption of normal brain development affecting several structures or other tissues (Kelberman et al., 2009). Several genes associated with phenotypes involving abnormal pituitary function identified in the mouse have also been implicated in human development by the identification of mutations in their human orthologues in patients with various hypopituitary phenotypes (HESX1, BMP4, OTX2 or SOX2) (Dattani et al., 1998; Ragge et al., 2005; Kelberman et al., 2006, 2008; Bakrania et al., 2008).
Here, we describe a highly consanguineous family with six affected children exhibiting a previously unreported syndrome comprising secondary (post-natal) microcephaly with frontal and temporal lobe hypoplasia, variable combined pituitary hormone deficiency, central diabetes insipidus, seizures, global developmental delay, severe visual impairment and congenital abnormalities of the kidneys and urinary tract. Using a combination of homozygosity mapping and exome sequencing we have identified a homozygous frameshift mutation in the basic HLH transcription factor ARNT2 (c.1373_1374dupTC) that results in reduced levels of mutant transcript and protein, most likely effected by nonsense-mediated decay. The severity of the clinical phenotype, combined with the demonstration of ARNT2 expression in the developing CNS, highlights the essential function of ARNT2 in the developing human brain and renal tract and its importance in the maintenance of normal post-natal brain growth.
Materials and methods
Patients
Studies had ethics committee approval from the Joint Great Ormond Street Hospital/UCL Institute of Child Health Research Ethics Committee and were undertaken following written informed parental consent.
All patients underwent assessment for pituitary function by measurement of free thyroxine and thyroid stimulating hormone, prolactin, 8 a.m. cortisol, insulin-like growth factor 1, insulin-like growth factor binding protein 3 and overnight growth hormone profiles (blood samples for growth hormone taken every 20 min overnight for 12 h). All hormone concentrations were measured using standard radio-immunoassays (thyroid stimulating hormone, thyroxine, prolactin, cortisol: chemiluminescent microparticle immunoassay; insulin-like growth factor 1, insulin-like growth factor binding protein 3 and growth hormone: solid-phase, enzyme labelled chemiluminescent immunometric assay). Visual function was assessed by unilateral flash electroretinogram combined with visual-evoked potential using LED goggles placed directly over the eyes. Standard electroencephalograms, skeletal surveys, bilateral hip X-rays, MRI of the spine and brain (including 3 mm slices through the hypothalamo-pituitary axis), and renal imaging studies were performed. Microcephaly was defined as an occipital-frontal head circumference <3 standard deviation scores below normal for age and sex matched control subjects (Woods, 2004).
Homozygosity mapping and exome sequencing
Available DNA samples from affected individuals Patients VI:3, V:9, V:13 and V:17 underwent whole genome single nucleotide polymorphism genotyping using the Illumina HumanCytoSNP-12 BeadChip following the manufacturer’s protocols. Data were analysed using Beadstudio v3.2 (Illumina) to identify regions of extended (>50) consecutive homozygous single nucleotide polymorphisms common to all affected individuals and sufficient to identify all regions containing potential candidate genes.
Whole exome sequencing was performed to identify candidate genetic variants. DNA (3 µg) from Patient V:17 was used to prepare a sequencing library using SureSelect Human All Exon Kit version 1 following the manufacturer’s instructions (Agilent). Paired-end adaptors were ligated for sequencing, which was performed as a 76 bp paired-end run on an Illumina Genome Analyzer IIx platform. Reads were aligned to the human reference genome (Hg19 NCBI build 37.1) using Novoalign software version 2.07.04 (http://www.novocraft.com/main/index.php), a gapped alignment tool proven to be optimal for reducing false-positive variant calls. Novoalign was launched with the additional hard clipping option based on read base quality (-H) and the default adaptor removal option (-a). Variant calling and bam file manipulation were performed with version 0.1.7-6 (r530) of SAMtools, and together with Dindel version 1.01 (http://www.sanger.ac.uk/resources/software/dindel/) were used to call insertions and deletions (Indels) from the Novoalign output. Dindel was launched with the default values. All variants were called using a depth threshold of 15× (each variant base achieving a minimum of 15-fold coverage). Due to the large number of potential candidate genes within mapped regions (n = 113), we undertook whole exome sequencing of Patient V:17 to identify candidate variants, with a minimum single nucleotide polymorphism and Indel quality of 30 and a minimum single nucleotide polymorphism and gap mapping quality of 50.
Analysis of ARNT2 expression
Real-time polymerase chain reaction
Endogenous ARNT2 gene expression was measured in total RNA extracted from primary fibroblast cultures from two patients (Patients V:13 and V:17) and three control subjects using a quantitative reverse transcription PCR assay with gene-specific primers normalized to levels of endogenous GAPDH. Total RNA was extracted from patient and control fibroblasts using the RNeasy® Mini kit (Qiagen) following the manufacturers’ protocol with DNase digestion (Ambion). RNA (1 µg) was reverse transcribed into complementary DNA using avian myeloblastosis virus-reverse transcriptase (Roche) with random hexamer primers (Promega). Real-time PCR reactions to quantify levels of ARNT2 expression were run on an ABI 7500 Fast cycler using SYBR-based MESA blue reagent (Eurogentec) and repeated for fibroblast complementary DNA from two patients (Patients V:13 and V:17) and three independent age-matched controls. Triplicate reactions used 10–50 ng complementary DNA per reaction as template. Results were normalized to endogenous levels of GAPDH and results were analysed using the deltaCt method. Primer sequences for ARNT2 spanning intron 2 were as follows: forward 5’-GAAATGCTCCTTTGGACCAC-3’ and reverse 5’-ACCACAGCATATTGGGCTTC-3’.
Western blot analysis
Whole cell lysates were prepared from primary fibroblast cultures using RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholdate, 1% Triton™ X-100) containing protease inhibitor cocktail. Total protein (10 µg) underwent electrophoresis through a 10% SDS polyacrylamide gel and transferred to a Protran BA83 nitrocellulose membrane (GE Healthcare). Membranes were blocked in 2% milk in PBS containing 0.5% tween (PBST) then hybridized with anti-ARNT2 (Abcam) and anti-ACTB (Sigma) antibodies at 1:100 and 1:10 000 dilutions, respectively. Membranes were then washed in PBST and hybridized with horseradish peroxidase-conjugated secondary antibodies (Dako) and visualized using ECL Plus western blotting detection reagents (GE Healthcare).
RNA in situ hybridization
Expression of ARNT2 during human embryonic development was studied by RNA in situ hybridization on 7-µm sections of paraffin-embedded human embryos at 8 weeks of development using ARNT2-specific digoxigenin-labelled antisense riboprobes following methods previously described using IMAGE clone 52275941 (Genbank accession number BC036099) as a template (Gaston-Massuet et al., 2011).
Results
Patient clinical phenotypes
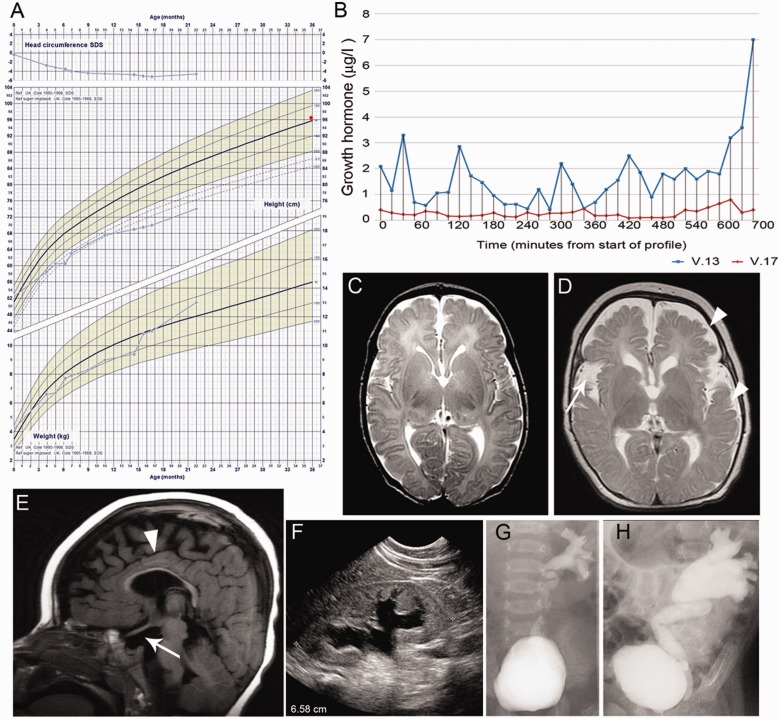
Six children born within a consanguineous family of Saudi Arabian origin presented in the first month of life with hypernatreamia secondary to central diabetes insipidus and cortisol insufficiency (Table 1). Diagnosis of central diabetes insipidus was made due to high serum sodium concentrations and plasma osmolalities in association with low urine osmolalities, as well as response to desmopressin treatment with an increase in urine osmolality and an associated reduction in urine output (Supplementary Table 1). Hypoglycaemia was not recorded in any child. Four children also presented with or developed central hypothyroidism. External genitalia appeared normal in affected females, whereas the single affected male had a normal sized phallus with bilaterally undescended testes in association with low luteinizing hormone and follicle stimulating hormone. Four children demonstrated an abnormal growth curve, with either growth failure or maintenance of linear growth in conjunction with obesity (Fig. 1A and Supplementary Fig. 1). Overnight growth hormone profiles were performed in two children, Patient V:13 (4.5-years-old at time of test) and Patient V.17 (1.5-years-old). Both were abnormal and one child (Patient V:17) was diagnosed with growth hormone deficiency on the basis of an inadequate peak growth hormone (mean overnight growth hormone was 0.27 µg/l, peak overnight growth hormone was 0.8 µg/l). The growth hormone profile for Patient V:13 was also abnormal, with a mean overnight growth hormone of 1.69 µg/l and one single overnight peak of 7 µg/l (Fig. 1B).
Table 1.
Clinical phenotype of affected individuals
| Patient | V:3 | V:9 | V:12 | V:13 | V:17 | VI:3 |
|---|---|---|---|---|---|---|
| Sex | Female | Female | Female | Female | Male | Female |
| Birth weight (kg) | 3.6 | 3.1 | 2.9 | 3.4 | 3.5 | 2.7 |
| Age at presentation (days) | 8 | 10 | 10 | 10 | 2 | 7 |
| Current age (years) | Died (5 years) | 5.4 | Died (4.2 years) | 5.2 | 2 | Died (1.2 years) |
| Investigations at presentation | ||||||
| Sodium (NR: 135–145 mmol/l) | 155 | 145–158 | 148–155 | 155–172 | 147–170 | 149–159 |
| Plasma osmolality (NR: 275–295 mosmol/kg) | ND | 315 | 334 | 323 | 335 | 320 |
| Urine osmolality (NR: 300–900 mosmol/kg) | ND | 180 | 195 | 151 | 150 | 200 |
| Thyroxine (NR: 9–19 pmol/l) | ND | 12.8 | 6.2 | 5.2 | 8.9 | 9 |
| TSH (NR: 0.35–4.94 mIU/l) | ND | 4.7 | 7.9 | 0.85 | 3.8 | 2.1 |
| 0800 cortisol (nmol/l) | ND | 9.1 | <6 | 47 | <27 | 49 |
| ACTH (NR:10–50 ng/l) | ND | <10 | <10 | <10 | 15 | <10 |
| Prolactin (NR: 73.0–557.0 mIU/l) | ND | ND | ND | 241.3 | 145.5 | ND |
| Medications | ||||||
| Hydrocortisone, thyroxine | DDAVP, HC, T4 | DDAVP, HC | DDAVP, HC, T4 | DDAVP, HC, T4 | DDAVP, HC, T4 | DDAVP, HC, T4 |
| Dynamic endocrine function tests, IGF1 and IGFBP3 (not on medication) | ||||||
| Synacthen: baseline cortisol (nmol/l) | ND | 60 | ND | ND | 225 | ND |
| Synacthen: 60 min cortisol (nmol/l) | ND | 76 | ND | ND | 252 | ND |
| Overnight profile GH peak (μg/l) | ND | ND | ND | 7 | 0.8 | ND |
| Insulin-like growth factor 1 ng/ml (normal range) | ND | 110 (51–303) | 100 (51–303) | 95 (50–286) | 96 (51–303) | 85 (49–283) |
| Insulin-like growth factor binding protein 3 μg/ml (normal range) | ND | 3.5 (0.8–5.2) | 2.3 (1.1–5.2) | 2.4 (1.1–5.2) | 2.2 (0.8–5.2) | 1.9 (0.8–3.9) |
| Skeletal, growth and renal phenotype | ||||||
| Hip dysplasia | Bilateral | Bilateral | Bilateral | Left | Normal | Normal |
| Height (cm) SDS | ND | −3.6 | −1.16 | −3.24 | −3.69 | −0.2 |
| Weight (kg) SDS | ND | 0.23 | −1.36 | −0.6 | 0.71 | 2.54 |
| Head circumference (cm) SDS | ND | −6.4 | −4.7 | −6 | −6.1 | −3.4 |
| Grade of vesicouretericreflux (Report of the International Reflux Study Committee) | Bilateral | Left-III Right-V | Left-V Right-IV | Left-III | Right-IV | Left-I Right-V |
| Hydronephrosis | Bilateral | Bilateral | Bilateral | Bilateral | Bilateral | Right |
ND = not done; NR = normal range; GH = growth hormone; ACTH = adrenocorticotropic hormone; TSH = thyroid stimulating hormone; HC = hydrocortisone; T4 = thyroxine; SDS = standard deviation score; DDAVP = desmopressin.
Figure 1.
Clinical phenotype, growth hormone concentrations, kidney and urinary tract dysplasia and MRI brain abnormalities in affected patients. (A) Growth chart of Patient V:17 demonstrating initial cerebral sparing (normal initial head circumference) with subsequent progressive microcephaly indicated by head circumference <3 standard deviation scores below normal for age and sex matched controls, and the abnormal growth curve with early growth failure and increasing obesity. Mid-parental height on 50th centile is denoted by a red dot. (B) Growth hormone profiles of affected Patients V:13 and V:17 with sampling at 20 min intervals for 12 h overnight. (C and D) Sequential MRI brain of Patient V:17 performed shortly after birth (C) and at 18 months (D). Arrowhead indicates hypoplasia of the frontal and temporal lobes. Arrow indicates large Sylvian fissure. Some mature myelin (represented by the dark line on these axial T2-weighted images) is seen in the posterior limb of the internal capsule and is demonstrated to have progressed since birth. However, the rest of the white matter is abnormal and immature for age, both in terms of signal return (interpreted as immature myelin) as well as volume (interpreted as under-developed), demonstrating a pattern of myelination and white matter development, on the combination of T2 and T1-weighted images, that is approximately equivalent to ∼8–10 months of age. (E) Midline sagittal T1-weighted MRI brain of Patient V:17 (18 months of age), arrowhead indicates thin corpus callosum, arrow shows absent posterior pituitary bright spot and hypoplastic anterior pituitary gland. (E) Renal ultrasound of Patient V:17 at 12 months of age. Note poor corticomedullary differentiation and dilatation of collection system. (F and G) Left micturating cystourethrogram of Patient V:17 showing renal reflux on left side, which is increased at 16 months of age (G) compared with 4 weeks (F).
MRI of the brain revealed a strikingly similar pattern of abnormalities in all children. The hypothalamo-pituitary axis was abnormal with an absent posterior pituitary bright spot, thin pituitary stalk, and a hypoplastic anterior pituitary gland (Fig. 1E). Frontal and temporal lobes were hypoplastic, with a thin corpus callosum and a global delay in brain myelination, particularly in the motor and occipital cortices. The optic nerves and chiasm and lateral geniculate body had a normal appearance. One child (Patient V:17) underwent MRI brain in the neonatal period and again at 18 months. At birth, brain MRI appearance was normal (excluding the hypothalamo-pituitary axis); subsequent scanning revealed frontal and temporal lobe hypoplasia and global delay in brain myelination (Fig. 1C and D). Despite initial cerebral sparing (normal initial head circumference), all six children developed secondary microcephaly (Fig. 1A and Supplementary Fig. 1), severe global developmental delay and generalized tonic-clonic/partial seizures (onset 8 days to 1 year, Supplementary Table 2). There was no evidence to support mitochondrial disease from metabolic screening (normal plasma and urine amino acids, acylcarnitine, blood lactate and ammonia, and muscle biopsy). Neurological examination revealed total body spastic cerebral palsy (Gross Motor Function Classification System Grade V) in all children, with abnormal hip posture and hip dislocation confirmed by X-ray before the age of 1 year in four.
All children demonstrated no clinical response to light with minimal pupil responses. Ophthalmic examination of the globe and fundi were normal. Electrodiagnostic testing was performed in three children (Patient V:13 at 2 years of age, Patient V:17 at 1 year, and Patient VI:3 at 4 months). Single flash electroretinograms showed well-defined waveforms providing evidence of generalized retinal activation (summating both rod and cone activity), indicating photoreceptor function in response to stimulation. In two children (Patients V:13 and V:17) there was evidence of post-retinal pathway dysfunction indicated by attenuated and poorly defined flash visual-evoked potentials. Flash visual-evoked potentials in Patient VI:3 showed a clear crossed asymmetry indicative of chiasmal disproportion or dysfunction.
All affected individuals were dysmorphic with a prominent forehead, deep-set eyes, a well-grooved philtrum, and retrognathia in addition to severe gastro-oesophageal reflux that required maximal medical management, with three needing surgical intervention. Hydronephrosis, vesicoureteric reflux and a neurogenic bladder were present in all affected individuals (Fig. 1F–H). Renal glomerular function and spinal MRI were normal. Three affected children have subsequently died due to sepsis. Immune work-up including flow cytometry and tetanus toxoid IgG were performed in Patients V:13 and V:17; these were normal. The oldest surviving affected child is 5.3 years at the time of writing.
Homozygosity mapping and exome sequencing identifies a mutation in ARNT2
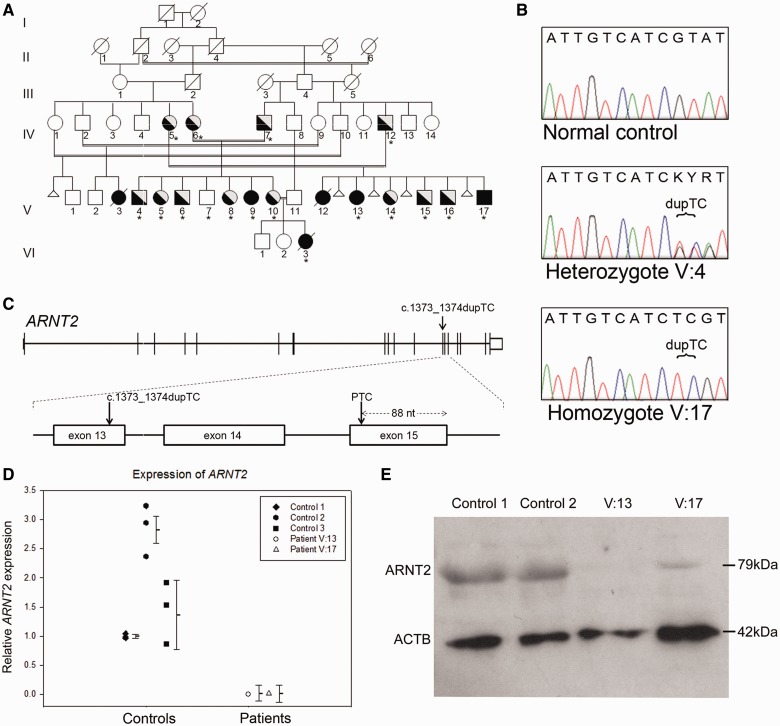
Assuming an autosomal recessive mode of inheritance and given the multiple consanguineous unions within the family, we applied a homozygosity mapping strategy to identify the causative mutation. Analysis of whole genome single nucleotide polymorphism genotype data from four affected individuals for which DNA was available identified nine common candidate regions containing extended consecutive homozygous single nucleotide polymorphisms (>50) ranging in size from 201 Kb to 1.8 Mb of DNA (Supplementary Table 3).
Due to the large number of potential candidate genes within mapped regions (n = 113), we undertook whole exome sequencing of Patient V:17 to identify candidate variants. A total of 5.15 Gb of mappable sequence data was generated, providing a mean read depth of 62-fold for the total 38 Mb of target captured sequence, and sufficient for at least 5-fold coverage at >95% of bases, with average depth of coverage in all regions of shared homozygosity sufficient for comprehensive variant detection (>15-fold) of all exons and for all annotated genes in these regions. From the aligned sequence reads, we identified a total of 21 144 coding single nucleotide variants, 8485 of which were homozygous, in addition to 997 coding insertion/deletion variants (indel) across the entire captured exome. Of these, a total of 253 candidate homozygous single nucleotide variants and a single coding indel mapped within candidate regions identified by homozygosity mapping. Further filtering revealed only a single novel (defined as not present in NCBI dbSNPbuild 131, or in our local database containing variant information from 172 in-house exomes with a minor allele frequency >0.1) duplication of two base pairs situated on chromosome 15 at position g.80,866,545 –80,866,546 (Hg 19 NCBI build 37.1). This insertion lies within exon 13 of the ARNT2 gene (c.1373_1374dupTC; NM_014862.3) (Fig. 2C). No other novel coding nucleotide variants or indels were identified in candidate homozygous regions of interest in Patient V:17 by exome sequencing. This variant is also not present in the latest release of dbSNP (version 137) or the National Heart Lung and Blood Institute Exome Variant Server database containing information on genetic variation from 6500 exomes (ESP6500, 2012).
Figure 2.
Mutation analysis of ARNT2. (A) Pedigree of family: affected individuals indicated by black symbol are all homozygous for the ARNT2 c.1373_1374dupTC mutation, heterozygous unaffected individuals are indicated by half-shaded symbols. Asterisks denote individuals for which samples were available for genotyping. Known spontaneous miscarriages are denoted by a small triangle. (B) Example of Sanger sequence traces showing the ARNT2 c.1373_1374dupTC mutation in a heterozygous and homozygous individual as compared with a normal control subject. (C) Schematic illustration of the ARNT2 gene showing the location of the mutation in exon 13 and introduction of a premature termination codon in exon 15. (D) Quantitative PCR showing expression levels of ARNT2 in primary skin fibroblasts from two patients (Patients V:13 and V:17) relative to three control primary fibroblast cultures. Error bars denote the standard error of the mean for triplicate experiments. (E) Western blot analysis showing greatly reduced levels of ARNT2 protein (79 kDa) in fibroblast cultures from two patients (Patients V:13 and V:17) compared with control subjects. Protein loading is indicated by anti-β-actin control (ACTB, 42 kDa).
The ARNT2 c.1373_1374dupTC allele was confirmed by Sanger sequencing and shown to be homozygous in all four affected individuals for whom DNA was available within the pedigree. All parents of affected patients were heterozygous with respect to the mutation and analysis of available unaffected siblings showed that they were either heterozygous (n = 8) or homozygous for the normal allele (n = 1, Fig. 2A and B). Furthermore, this variant was not identified in 200 ethnically matched control subjects of Saudi Arabian origin.
Functional effect of the ARNT2 c.1374_1375dupTC mutation
The mutation identified in ARNT2 introduces a frameshift at residue p.Tyr459 and introduction of a premature termination codon 52 amino acids downstream of the mutation site (p.Tyr459Argfs*52). We hypothesized that this mutation would evoke nonsense-mediated decay mechanisms, which exist to eliminate messenger RNA species containing premature termination codons (Maquat, 2005; Brogna and Wen, 2009; Schoenberg and Maquat, 2012). To test this, we performed quantitative reverse transcription PCR using total RNA extracted from fibroblast cells derived from Patients V:13 and V:17 as well as from three normal control individuals. As shown in Fig. 2D, the patient samples had a 50 to 150-fold reduction in ARNT2 transcript levels compared with controls. Consistently, western blot analysis showed a reduction in detectable levels of ARNT2 protein in patient fibroblasts (Fig. 2E).
Expression of ARNT2 in the developing human embryo
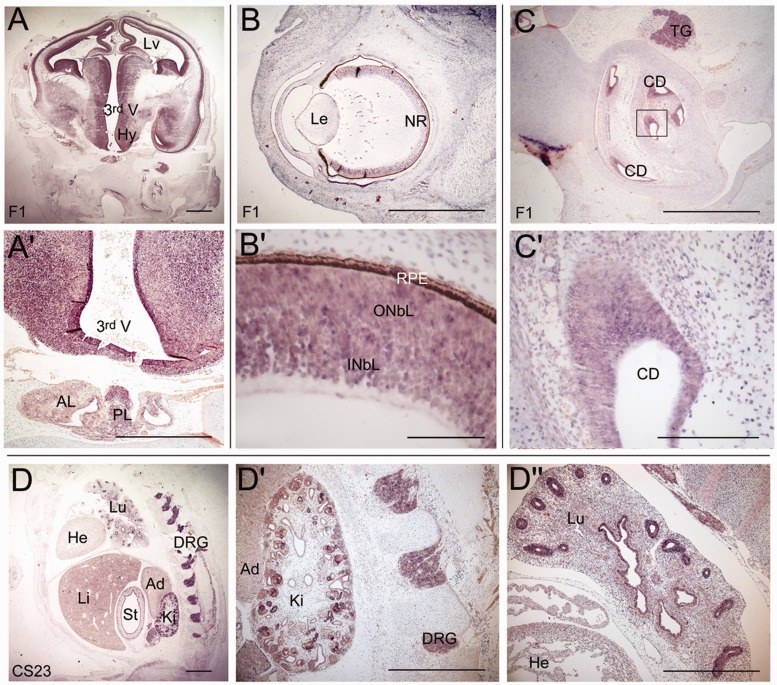
The expression profile of Arnt2 has been extensively studied in mouse embryos (Aitola and Pelto-Huikko, 2003) where high levels of expression were detected throughout the developing CNS, including the cortex, hypothalamus, inner layer of the retina and spinal cord. Other sites of expression include the dorsal root ganglia and developing tubules in the kidneys. To determine the degree of conservation of the expression pattern of ARNT2, we performed RNA in situ hybridization in human embryos. These analyses revealed the presence of ARNT2 transcripts in the CNS, notably the telencephalic cortex, hypothalamus, anterior and posterior pituitary, thalamus and neural retina (Fig. 3). Expression was also detected in neural crest-derived dorsal root ganglia, epithelia of bronchiolar buds of the lung, inner layer of the stomach and in the kidney, with particularly strong expression in the tubules.
Figure 3.
Expression analysis of ARNT2 during human embryonic development. In situ hybridization using riboprobes against ARNT2 reveals expression in the developing brain and pituitary gland at 8 weeks of development (A). Strong expression can be seen in the hypothalamus and in the posterior lobe of the pituitary and weak expression is detected in the anterior lobe of the pituitary (A’). (B) Expression in the eye is detected in the developing neural retina (B’). (C) ARNT2 expression is detected in the inner ear, specifically in the cochlear ducts (C’). Expression is also observed in the trigeminal ganglion (C). (D) ARNT2 is expressed in the dorsal root ganglia, the developing kidney (D’) and in the lung, with strong expression in the bronchioles (D’’). Expression is also detected in the inner lining of the stomach. Scale bars: A, A’, B, D, D’ and D’’ = 100 µm; B’ = 10 µm; C = 50 µm; C’ = 20 µm. Lv = lateral ventricle; 3rd V = third ventricle; Hy = hypothalamus; PL = posterior lobe; AL = anterior lobe; Le = lens; NR = neural retina; INbL = inner neuroblastic layer; ONbL = outer neuroblastic layer; RPE = retinal pigmented epithelium; CD = cochlear duct; TG = trigeminal ganglion; Lu = lungs; He = heart; Li = liver; St = stomach; Ki = kidney; Ad = adrenal; DRG = dorsal root ganglia.
Discussion
ARNT2 mutation causes a congenital brain malformation syndrome
We have identified a homozygous mutation in the ARNT2 gene segregating between affected individuals as the cause of a congenital brain malformation syndrome with a highly unusual and distinct combination of clinical manifestations. The severity of the phenotype is the likely cause of premature death in three affected individuals. The observation of multiple miscarriages in one branch of the family is also suggestive of a degree of prenatal mortality associated with the condition. This mutation (c.1373_1374dupTC) resides within exon 13 (of 19) and is predicted to result in a frame-shift introducing a premature termination codon (p.Y459Rfs*52) at the beginning of exon 15, 88 nucleotides upstream of the exon 15–16 boundary (Fig. 2C). Nonsense-mediated decay in mammalian cells generally degrades messenger RNAs that contain a premature termination codon more than 50–55 nucleotides upstream of an exon–exon junction. Consistent with this, loss of ARNT2 messenger RNA and protein was demonstrated in patient fibroblasts compared with controls, highly suggestive of nonsense-mediated decay activation and loss of ARNT2 function. Furthermore, this is the only coding genetic variant segregating between the affected individuals and situated within mapped candidate regions that could account for the observed combination of phenotypes. These data, together with expression of ARNT2 in all tissues affected in these patients during human embryonic development, demonstrate that this mutation is the most likely cause of the condition.
Role of ARNT2 in brain development
ARNT2 is a member of the basic HLH-PAS (Per-ARNT-Sim homology) subfamily of transcription factors, containing a basic HLH DNA-binding domain in addition to a PAS domain that mediates heterodimerization with other basic HLH/PAS proteins in order to form functional DNA binding complexes (Drutel et al., 1996; Hirose et al., 1996; Kewley et al., 2004). One of the known dimerization partners of ARNT2 is SIM1, a homologue of the Drosophila single-minded transcription factor, which is a critical regulator of neuronal differentiation during CNS development (Nambu et al., 1991; Michaud et al., 2000). In the mouse, Arnt2 is expressed in the developing CNS, including the hypothalamus and neural retina, as well as the kidney and urinary tract in a pattern similar to that we observe in human development, suggesting a conserved function between species (Hirose et al., 1996; Jain et al., 1998; Aitola and Pelto-Huikko, 2003). However, Arnt2 null mice do not display gross morphological abnormalities of their CNS at birth (Hosoya et al., 2001) and the impact of loss of function on post-natal brain development has not been studied due to early lethality. Affected members of the family we describe had normal brain volume at birth, but all subsequently developed secondary microcephaly within the first few months of life (independent of when their seizures started), with a specific pattern of cerebral hypoplasia affecting the frontal and temporal lobes. Secondary (post-natal) microcephaly indicates a progressive neurodegenerative condition potentially caused by decreased dendritic connection or activity of a (near) normal number of neurons (Woods, 2004). The progressive nature of the neurological phenotype could be explained by a post-natal requirement for ARNT2 expression, as demonstrated in the post-natal and adult rodent brain, where it has a suggested role in neuronal maintenance, regulating cell-cycle progression and inhibiting apoptosis (Drutel et al., 1999). Whether specification of neuronal and glial progenitors in the infant brain relies on the same transcriptional mechanisms as in the embryo remains unknown. The limited differentiation potential of post-natal progenitors could reflect differences in the regulation of proneural factors operating in both embryonic and adult brain, or alternatively the recruitment of a different set of cell fate determinants to control neurogenesis and gliogenesis at post-natal stages (Kintner, 2002).
Requirement for ARNT2 in the hypothalamic-pituitary axis
Murine phenotypes
Homozygous Arnt2 null mice show normal Mendelian ratios at late embryonic stages (embryonic Day 18.5) but die within the first 24 h after birth, most likely due to disrupted hypothalamic development and loss of pituitary hormone secretion (Hosoya et al., 2001; Keith et al., 2001). The hypothalamus is the principal neural structure regulating homeostasis, mediated by the function of specific neuroendocrine neurons residing within discrete hypothalamic nuclei (Kelberman et al., 2009). Magnocellular neurons within the paraventricular and supraoptic nuclei produce oxytocin, required during parturition and lactation, and arginine vasopressin, which is involved in the regulation of osmotic balance. These neurons project directly into the posterior pituitary where their peptide hormones are transported to the axonal terminals and released as required. Parvocellular neurons resident in the paraventricular nuclei secrete thyrotropin-releasing hormone, corticotropin-releasing hormone and somatostatin (a negative regulator of growth hormone secretion) into the hypophyseal portal blood system to the anterior pituitary where they regulate endocrine cell proliferation and hormone synthesis and release. The supraoptic and paraventricular nuclei in the hypothalamus of Arnt2 null mice are hypocellular, with failure of terminal differentiation of magnocellular and dopaminergic parvocellular neurons and loss of detectable levels of their neurosecretory hormones (Hosoya et al., 2001; Keith et al., 2001). Homozygous Sim1 null mice also show a similar phenotype with failed terminal differentiation of these neuroendocrine neurons and deficiencies in oxytocin, arginine-vasopressin, thyrotropin-releasing hormone, corticotropin-releasing hormone and somatostatin (Michaud et al., 1998, 2000). Further evidence for the conserved role of Sim1a and Arnt2 in hypothalamic development comes from recent studies in zebrafish where they contribute to differentiation of a defined population of dopaminergic and neuroendocrine neurons (Schweitzer et al., 2013).
Human phenotypes
The affected children in the family we describe display several features of hypothalamic insufficiency, including obesity, diabetes insipidus, adrenocorticotropic hormone and thyroid stimulating hormone deficiency, consistent with abnormal development of the paraventricular and supraoptic nuclei. Although hypothalamic nuclei cannot easily be identified using conventional neuroimaging (Jones, 2011), the pattern of pituitary hormone deficiencies observed in the patients parallels the neuroendocrine phenotype of homozygous Arnt2 null mice (Hosoya et al., 2001; Keith et al., 2001). We would therefore predict that oxytocin and somatostatin may also be deficient in these patients. This report may represent the first human cases where somatostatin deficiency would be predicted to occur in the absence of the deficiency of growth hormone releasing hormone (extrapolated from the normal appearance of the arcuate nucleus in Arnt2-null mice) (Hosoya et al., 2001). Whilst the role of somatostatin in the regulation of growth hormone secretion is well established (Hindmarsh et al., 1991), the effect of somatostatin deficiency on growth hormone release and long-term growth has not been studied in humans. It is therefore interesting that the growth pattern, either growth failure or maintenance of linear growth in conjunction with obesity (four individuals), and growth hormone secretion (two individuals) are significantly abnormal (Fig. 1A and B and Supplementary Fig. 1). Although there is no linear growth phenotype in somatostatin null mice, they do have reduced pituitary growth hormone content (50%) with significantly increased circulating growth hormone (hypothesized to reflect an altered balance between growth hormone synthesis and release), and by 18 weeks, males develop mild obesity (Low et al., 2001). Low concentrations of oxytocin have also been associated with obesity in humans (Onaka et al., 2012). The underlying mechanism for the growth phenotype we observe remains unclear but could possibly reflect a combination of oxytocin deficiency and dysregulation of growth hormone production.
Pleiotropic role of ARNT2
The effect of loss of Arnt2 on visual pathway or renal function in mice has not previously been assessed as perinatal lethality precludes later analysis, which would require the generation of conditional or inducible loss-of-function alleles. Importantly, we can now also document a critical role for ARNT2 in human renal tract development with all affected children presenting with congenital abnormalities of the kidneys and urinary tract. Additionally, congenital severe visual impairment likely due to post-retinal visual pathway and chiasmal dysfunction is a component of this multisystem disorder. The presence of a mixed rod cone ERG confirmed generalized retinal function. Single flash parameters used in this electrophysiological study generated well-defined ERG waveforms in stark contrast to the simultaneously acquired flash visual-evoked potential waveforms that were disproportionately ill-defined and attenuated. This is highly indicative of post-retinal pathway dysfunction. Notably, in the rat and mouse, Arnt2 is expressed in the developing visual pathway, with high levels of expression in the developing lateral geniculate nucleus and superior colliculus, as well as the occipital cortex (the visual processing centre) where expression is maintained in the adult (Drutel et al., 1999; Aitola and Pelto-Huikko, 2003). Here, we also observed expression of ARNT2 in the developing human retina. From the data available we cannot rule out the presence of a subtle dysfunction affecting either rods or cones. Further investigations, not feasible in the current study, would be necessary to clarify whether the abnormalities in the visual pathway are the result of both retinal and post-retinal dysfunction.
Conclusion
Loss of ARNT2 function has a profound impact on normal CNS development, particularly affecting the hypothalamo-pituitary axis and visual pathway, in addition to the maintenance of normal post-natal growth of the brain in humans. The combination of multiple pituitary hormone deficiencies observed is consistent with a key role for ARNT2 in the development of specific neurosecretory neurons in the human hypothalamus. Further analysis of conditional mouse models and identification of other genes involved in pathways regulated by ARNT2 (and SIM1) will improve our understanding of CNS development and/or neuronal maintenance.
Supplementary Material
Acknowledgements
We thank the patients and their families for their help with this study. We thank Miss Nouh Doaa, research coordinator at King Abdulaziz Medical City, King Abdullah Medical Research Centre. We thank the additional members of the GOSgene Scientific Advisory Board (G.E. Moore, M. Bitner-Glindzicz, B.G. Gaspar, M. Hubank, R.H. Scott). We are grateful to Kerra Pearce for technical assistance. The human embryonic and foetal material was provided by the Human Developmental Biology Resource (http://www.hdbr.org).
Glossary
Abbreviations
- HLH
helix-loop-helix
- PAS
Per-ARNT-Sim homology
Funding
This work was supported by the Child Growth Foundation, an unrestricted educational grant from Novo Nordisk Ltd, the Ulverscroft Foundation, the NIHR Specialist Paediatric Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and UCL Institute of Child Health the Medical Research Council UK (grant number G0700089) and the Wellcome Trust (grant number GR082557). J.C.S. and M.T.D. are funded by the Great Ormond St Hospital Children’s Charity. GOSgene* (* Gudrun E. Moore, Maria Bitner-Glindzicz, Robert Kleta, Bobby G. Gaspar, Mike Hubank, Richard H Scott) at the UCL Institute of Child Health is supported by the Great Ormond Street Hospital (GOSH) Biomedical Research Centre (BRC) of the National Institute for Health Research (NIHR). The human embryonic and fetal material was provided by the Joint MRC (grant number G0700089)/Wellcome Trust (grant number GR082557) Human Developmental Biology Resource (www.hdbr.org).
Supplementary material
Supplementary material is available at Brain online.
References
- Aitola MH, Pelto-Huikko MT. Expression of Arnt and Arnt2 mRNA in developing murine tissues. J Histochem Cytochem. 2003;51:41–54. doi: 10.1177/002215540305100106. [DOI] [PubMed] [Google Scholar]
- Bakrania P, Efthymiou M, Klein JC, Salt A, Bunyan DJ, Wyatt A, et al. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am J Hum Genet. 2008;82:304–19. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. Classification system for malformations of cortical development: update 2001. Neurology. 2001;57:2168–78. doi: 10.1212/wnl.57.12.2168. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–30. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16:107–13. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- Dattani MT, Martinez-Barbera JP, Thomas PQ, Brickman JM, Gupta R, Mårtensson IL, et al. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet. 1998;19:125–33. doi: 10.1038/477. [DOI] [PubMed] [Google Scholar]
- Drutel G, Heron A, Kathmann M, Gros C, Macé S, Plotkine M, et al. ARNT2, a transcription factor for brain neuron survival? Eur J Neurosci. 1999;11:1545–53. doi: 10.1046/j.1460-9568.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- Drutel G, Kathmann M, Heron A, Schwartz JC, Arrang JM. Cloning and selective expression in brain and kidney of ARNT2 homologous to the Ah receptor nuclear translocator (ARNT) Biochem Biophys Res Commun. 1996;225:333–9. doi: 10.1006/bbrc.1996.1176. [DOI] [PubMed] [Google Scholar]
- Gaston-Massuet C, Andoniadou CL, Signore M, Jayakody SA, Charolidi N, Kyeyune R, et al. Increased Wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. Proc Natl Acad Sci USA. 2011;108:11482–7. doi: 10.1073/pnas.1101553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindmarsh PC, Brain CE, Robinson IC, Matthews DR, Brook CG. The interaction of growth hormone releasing hormone and somatostatin in the generation of a GH pulse in man. Clin Endocrinol (Oxf) 1991;35:353–60. doi: 10.1111/j.1365-2265.1991.tb03548.x. [DOI] [PubMed] [Google Scholar]
- Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, et al. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt) Mol Cell Biol. 1996;16:1706–13. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Oda Y, Takahashi S, Morita M, Kawauchi S, Ema M, et al. Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes Cells. 2001;6:361–74. doi: 10.1046/j.1365-2443.2001.00421.x. [DOI] [PubMed] [Google Scholar]
- Jain S, Maltepe E, Lu MM, Simon C, Bradfield CA. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech Dev. 1998;73:117–23. doi: 10.1016/s0925-4773(98)00038-0. [DOI] [PubMed] [Google Scholar]
- Jones SE. Imaging for autonomic dysfunction. Cleve Clin J Med. 2011;78(Suppl 1):S69–74. doi: 10.3949/ccjm.78.s1.12. [DOI] [PubMed] [Google Scholar]
- Keith B, Adelman DM, Simon MC. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc Natl Acad Sci USA. 2001;98:6692–97. doi: 10.1073/pnas.121494298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D, de Castro SC, Huang S, Crolla JA, Palmer R, Gregory JW, et al. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93:1865–73. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D, Rizzoti K, Avilion A, Bitner-Glindzicz M, Cianfarani S, Collins J, et al. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest. 2006;116:2442–55. doi: 10.1172/JCI28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30:790–829. doi: 10.1210/er.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Kintner C. Neurogenesis in embryos and in adult neural stem cells. J Neurosci. 2002;22:639–43. doi: 10.1523/JNEUROSCI.22-03-00639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Goshu E, Wells A, Fan CM. Identification of the downstream targets of SIM1 and ARNT2, a pair of transcription factors essential for neuroendocrine cell differentiation. J Biol Chem. 2003;278:44857–67. doi: 10.1074/jbc.M304489200. [DOI] [PubMed] [Google Scholar]
- Low MJ, Otero-Corchon V, Parlow AF, Ramirez JL, Kumar U, Patel YC, et al. Somatostatin is required for masculinization of growth hormone-regulated hepatic gene expression but not of somatic growth. J Clin Invest. 2001;107:1571–80. doi: 10.1172/JCI11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118:1773–6. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech Dev. 2000;90:253–61. doi: 10.1016/s0925-4773(99)00328-7. [DOI] [PubMed] [Google Scholar]
- Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–75. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–67. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Onaka T, Takayanagi Y, Yoshida M. Roles of oxytocin neurones in the control of stress, energy metabolism, and social behaviour. J Neuroendocrinol. 2012;24:587–98. doi: 10.1111/j.1365-2826.2012.02300.x. [DOI] [PubMed] [Google Scholar]
- Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–7. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragge NK, Brown AG, Poloschek CM, Lorenz B, Henderson RA, Clarke MP, et al. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76:1008–22. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Report of the International Reflex Study Committee: Medical versus surgical treatment of primary vesicoureteral reflux. Pediatrics 1981; 67: 392–400. [PubMed]
- Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer J, Lohr H, Bonkowsky JL, Hubscher K, Driever W. Sim1a and Arnt2 contribute to hypothalamo-spinal axon guidance by regulating Robo2 activity via a Robo3-dependent mechanism. Development. 2013;140:93–106. doi: 10.1242/dev.087825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG. Human microcephaly. Curr Opin Neurobiol. 2004;14:112–7. doi: 10.1016/j.conb.2004.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.