Abstract
Hereditary spastic paraparesis type 15 is a recessive complicated form of the disease clinically characterized by slowly progressive spastic paraparesis and mental deterioration with onset between the first and second decade of life. Thinning of corpus callosum is the neuroradiological distinctive sign frequently associated with white matter abnormalities. The causative gene, ZFYVE26, encodes a large protein of 2539 amino acid residues, termed spastizin, containing three recognizable domains: a zinc finger, a leucine zipper and a FYVE domain. Spastizin protein has a diffuse cytoplasmic distribution and co-localizes partially with early endosomes, the endoplasmic reticulum, microtubules and vesicles involved in protein trafficking. In addition, spastizin localizes to the mid-body during the final step of mitosis and contributes to successful cytokinesis. Spastizin interacts with Beclin 1, a protein required for cytokinesis and autophagy, which is the major lysosome-mediated degradation process in the cell. In view of the Beclin 1–spastizin interaction, we investigated the possible role of spastizin in autophagy. We carried out this analysis by using lymphoblast and fibroblast cells derived from four different spastizin mutated patients (p.I508N, p.L243P, p.R1209fsX, p.S1312X) and from control subjects. Of note, the truncating p.R1209fsX and p.S1312X mutations lead to loss of spastizin protein. The results obtained indicate that spastizin interacts with the autophagy related Beclin 1–UVRAG–Rubicon multiprotein complex and is required for autophagosome maturation. In cells lacking spastizin or with mutated forms of the protein, spastizin interaction with Beclin 1 is lost although the formation of the Beclin 1–UVRAG–Rubicon complex can still be observed. However, in these cells we demonstrate an impairment of autophagosome maturation and an accumulation of immature autophagosomes. Autophagy defects with autophagosome accumulation can be observed also in neuronal cells upon spastizin silencing. These results indicate that autophagy is a central process in the pathogenesis of complicated forms of hereditary spastic paraparesis with thin corpus callosum.
Keywords: spastizin, autophagy, Beclin 1, autophagosome maturation, SPG15
Introduction
The hereditary spastic paraplegias constitute a group of clinically and genetically heterogeneous neurodegenerative diseases characterized by progressive spasticity and weakness at the lower limbs due to retrograde axonal degeneration of the corticospinal tracts and posterior columns (Fink et al., 2006; Depienne et al., 2007). Hereditary spastic paraplegias are classified as pure, with isolated progressive spasticity, or complicated, when spasticity is associated with additional neurological features such as ataxia, peripheral neuropathy, cognitive impairment, epilepsy or extraneurological signs. Dominant, recessive and X-linked segregation patterns are observed in addition to sporadic occurrence of the disease (Fink et al., 2006). Among the recessive complicated forms, a distinctive form associated with thin corpus callosum (ARHSP-TCC) and mental impairment (Boukhris et al., 2009; Finsterer et al., 2012) appears to be frequent and it is mostly due to mutations in SPG11 (Stevanin et al., 2007) and ZFYVE26/SPG15 (Hanein et al., 2008), accounting for 20% and 3%, respectively, with almost indistinguishable clinical phenotypes (Goizet et al., 2009; Schüle et al., 2009; Schüle and Schols, 2011). ARHSP-TCC forms are clinically characterized by slowly progressive spastic paraparesis and mental deterioration usually beginning before the second decade of life. In addition, peripheral neuropathy, cerebellar ataxia and distal amyotrophy are frequent. Mutations in the ZFYVE26 gene were initially associated with pigmentary maculopathy, Kjellin syndrome (Webb et al., 1997; Hanein et al., 2008) and were found in breast cancer samples with a frequency of >10% (Sjöblom et al., 2006). Analogously to the ZFYVE26 gene, a single mutation in ZFYVE26 was also found in juvenile parkinsonism associated with spastic paraplegia (Schicks et al., 2011).
The ZFYVE26 gene encodes a large protein of 2539 amino acid residues termed spastizin (or FYVE-CENT), containing three recognizable domains: a zinc finger (1563–1587 aa residues), a FYVE domain (1818–1867 aa) and a leucine zipper domain (2217–2238 aa) (Hanein et al., 2008). FYVE finger proteins bind with much higher affinity to membrane-associated phosphatidylinositol 3-phosphate (PtdIns3P; PI3P) than to its soluble analogues, explaining why most FYVE-finger proteins are associated with endosomal trafficking (Itoh et al., 2002; Stenmark et al., 2002). Consistent with this, spastizin has a diffuse cytoplasmic distribution and partially co-localizes with the early endosomal marker EEA1, the endoplasmic reticulum (Hanein et al., 2008), microtubules and vesicles involved in protein trafficking alongside spatacsin, the protein product of SPG11 (Murmu et al., 2011). In zebrafish spastizin, together with spatacsin, is essential for proper establishment of the motor neuron axonal network; these proteins are indeed necessary for outgrowth and proper targeting of motor neuron axons (Martin et al., 2012).
Spastizin also localizes to the mid-body during the final step of mitosis and it is necessary for successful cytokinesis (Sagona et al., 2010). Spastizin interacts with Beclin 1, a subunit of class III phosphatidylinositol 3-kinase complex (PI3KIII/Vps34), and it is thought to recruit Beclin 1 to the mid-body, thereby promoting cytokinesis (Sagona et al., 2011). Beclin1 is implicated in a variety of biological processes including endocytosis, immunity, tumorigenesis and cell death and plays a central role in autophagy (Wirawan et al., 2012). This is an evolutionary conserved intracellular process that delivers cytoplasmic constituents to lysosomes for degradation of aggregated proteins and recycling of organelles or nutrients through the formation of double-membrane vacuoles, termed autophagosomes (Levine and Kroemer, 2008).
Beclin 1 is key regulator of autophagy by binding PI3KIII/Vps34 to form the Beclin 1–Vps34–Vps15 core complex. By interacting with several cofactors (UVRAG, Atg14L and Rubicon among the others) (Itakura et al., 2008; Sun et al., 2008; Zhong et al., 2009), Beclin 1 mediates the formation of different larger complexes that regulate Vps34 activity and are differentially implicated in autophagosome formation and maturation (Kang et al., 2011). Beclin 1 dysfunction has been implicated in many disorders including cancer and neurodegeneration (Kang et al., 2011). Analogously, dysregulation of autophagy plays a crucial role in the pathogenesis of different types of disorders including metabolic diseases (Rabinowitz and White, 2010), lysosomal storage disorders (Fukuda et al., 2006; Settembre et al., 2008; Fraldi et al., 2010), neurodegenerative diseases (Banerjee et al., 2010; Wong et al., 2011), infectious diseases (Levine et al., 2011) and cancer (Mah and Ryan, 2012; Rubinsztein et al., 2012).
The interaction between spastizin and Beclin 1 prompted us to investigate the possible role of spastizin and its pathogenic mutations in autophagy. We carried out this analysis by using the wild-type and different mutated forms of spastizin identified in patients with spastic paraplegia type 15. The results obtained indicated that spastizin interacts with the autophagy-related Beclin 1-UVRAG-Rubicon multiprotein complex and is involved in autophagosome maturation. Mutations in spastizin disrupt its interaction with Beclin 1 and thus with the complex, impair autophagosome maturation and lead to an accumulation of immature autophagosomes in patient’s fibroblasts. Analogously, an accumulation of autophagosomes was observed in SHSY5Y cells and in primary hippocampal neurons after spastizin silencing, thus indicating that autophagy defects due to spastizin depletion occur both in neuronal and non-neuronal cells.
Materials and methods
Patients and mutation analysis
We studied 65 recessive families with a slowly progressive complicated spastic paraparesis (ARHSP-TCC) with mental impairment. Axonal neuropathy and signs of lower motor neuron degeneration were present in all of them. All affected subjects showed thin corpus callosum and white matter abnormalities, by MRI analysis. Age at symptoms onset was between the first and third decade of life. Clinical data of these patients were obtained by clinical evaluation by the referring neurologist, based on the Harding criteria (Harding, 1983) for the definition of the clinical status and thus excluding any possible alternative cause of spastic paraplegia. IQ, evaluated through the Wechsler Intelligence Scales for Children or for Adult, along with a detailed neuropsychological evaluation, including the Wechsler Memory Scale, were available for all cases. All families had previously been screened for mutations in SPG7, SPG11, SPG21 and SPG35 with negative results. Overall, the clinical picture of these patients overlaps the typical clinical and radiological picture of the SPG11 or ZFYVE26/SPG15 mutated patients. Supplementary Table 1A summarizes the clinical and radiological features of the mutated patients. We obtained blood samples and clinical data from affected and unaffected subjects of the pedigree under a protocol of informed consent approved by the Ethics Committee of the E. Medea Scientific Institute. DNA was purified by using a standard high-salt purification method. Primers used for amplification of all ZFYVE26 exons are available upon request. Sequences were prepared with a BigDye® Terminator sequencing Kit (version 3.1 Applied Biosystem). All variants were checked against a panel of 600 Italian controls and against the SNP and 1000genome databases. Mutation nomenclature is according to the recommendations of the Human Genome Variation Society and refers to the published ZFYVE26 complementary DNA sequences (Accession number: NM_015346.3) with nucleotide +1 corresponding to the A of the ATG translation initiation codon. Missense changes were analysed for possible pathogenic effects on protein function or on splicing by using different prediction software (Supplementary Table 2).
Cell cultures and treatments
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, Life Technologies Corp.) supplemented with 10% foetal bovine serum (Euroclone), 100 U/ml penicillin/streptomycin and 2 mM L-glutamine (Invitrogen, Life Technologies Corp.). SHSY5Y human neuroblastoma cell line was grown in the same medium supplemented with 20% foetal bovine serum. Lymphoblastoid cell lines were established by Epstein-Barr virus infection with a standard protocol and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Euroclone) containing 20% foetal bovine serum, 100 U/ml penicillin/streptomycin and 2 mM L-glutamine. Lymphoblastoid cell lines were established from patients carrying the mutations L243P, I508N and S1312X, from three healthy control individuals and from one healthy parent in the I508N mutated family. Fibroblast cell lines were established from skin biopsies obtained from patients carrying the L243P and S1312X mutations identified in this study and from an affected individual in the previously reported family harbouring the R1209fsX mutation (Hanein et al., 2008, Patient 761/3) and from two healthy control individuals. Fibroblasts were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal bovine serum, 100 U/ml penicillin/streptomycin and 2 mM L-glutamine.
Cells were transiently transfected using Lipofectamine™ 2000 (Invitrogen, Life Technologies Corp.). Autophagy was induced by amino acid and serum starvation in Earle’s Balanced Salt Solution (Euroclone) for the indicated times (Matsunaga et al., 2010). To analyse the autolysosomal degradation of LC3-II, cells were pretreated for 30 min with 200 nM bafilomycin A1 (Sigma-Aldrich) in complete medium, then were incubated in complete or starvation medium (Earle’s Balanced Salt Solution) in the presence of 200 nM bafilomycin A1 for the indicated times (Vergne et al., 2009).
RNA interference
The plasmids for RNA interference studies were purchased from SABiosciences Corporation. Two sets of four different short hairpin RNAs (Cat. N. KH07631G for human gene and KM38929G for mouse gene), all with the green fluorescence protein (GFP) reporter gene, was tested and two different short hairpin RNA vectors (shRNA1 and shRNA2), showing similar silencing efficiency, were chosen for each set. The specificity of human and mouse short hairpin RNA sequences was determined by analysing ZFYVE26 expression levels in HeLa and P19 cells, respectively. Cells were transiently transfected with the short hairpin RNAs or with the scrambled control sequence and the levels of ZFYVE26 were analysed by quantitative real-time PCR. RNA was prepared using TRIzol® (Invitrogen, Life Technologies Corp.) and 1 µg/sample was reverse transcribed into complementary DNA using the SuperScript® First Strand Synthesis System for RT-PCR kit (Invitrogen, Life Technologies Corp.) and random hexamers. The expression levels of ZFYVE26 were analysed by quantitative real-time PCR on an ABI PRISM® 7900HT Fast Real-Time PCR Systems (Applied Biosystems, Life Technologies Corp.) by using specific gene expression assays (ZFYVE26; assays Hs00389635_m1 and Mm01305546_m1, Applied Biosystems, Life Technologies Corp.). β2 microglubulin (B2m, assay Hs99999907_m1) and TATA box binding protein (Tbp, assay Mm00446973_m1) were used for normalization. Untransfected cells were used as endogenous controls. Data were analysed using the delta-delta-Ct method.
Hippocampal cultures
Hippocampal primary cultures were prepared from CD1 mice as described (Martel et al., 2009). Experiments were in accordance with the standard ethical guidelines (National Institute of Health and European Community Guidelines on the Care and Use of Laboratory Animals) and were approved by the local Ethics Committee. Briefly, hippocampi were dissected from embryonic Day 18 embryos in Hank’s balanced salts solution (Invitrogen, Life Technologies Corp.) and mechanically dissociated. Cells were plated onto poly-L-lysine coated coverslips (100 µg/ml) in Dulbecco’s modified Eagle’s medium (50%, Invitrogen Life Science) and Ham’s F12 (50%, Invitrogen Life Science), supplemented with 5 mM HEPES buffer, 0.6% glucose, 20 µg/ml insulin (Sigma-Aldrich) and N2 supplement (Sigma-Aldrich). Neurons were transfected 1 week after plating with a control vector, Spg15 shRNA1 or shRNA2 and processed for immunofluorescence 72 h later as described (Vantaggiato et al., 2011). For autophagy induction neurons were cultured in nutrient-limited media (Dulbecco’s modified Eagle’s medium alone) for 4 h (Young et al., 2009).
Antibodies
Antibodies against Atg7, LC3B, Beclin 1, mTor, phospho-mTor, Atg14L, PI3KIII/Vps34, UVRAG, Rab7 and rabbit IgG isotype were purchased from Cell Signaling. Anti-Atg12, -Atg16, -Rab5 and -β-actin antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-LAMP1, -LAMP2, -ubiquitin and -Rubicon antibodies were purchased from Abcam. Anti-p62 antibody was from Sigma-Aldrich and anti-GFP antibody from Chemicon International Inc. For spastizin detection two different antibodies were used, one directed against the C-terminus (Sigma-Aldrich) and the other against the N-terminus (Santa Cruz Biotechnology).
Confocal immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with PBS containing 0.1% saponin and 1% bovine serum albumin for 30 min. Samples were then incubated for 2 h with primary antibodies and revealed using the secondary antibodies Alexa Fluor® 488, 546 and 647 (Invitrogen, Life Technologies Corp.). For the staining of LC3-positive vesicles, cells were transfected with the pCMVMAP1LC3B-RFP vector (Addgene). Images were acquired using a Leica TCS SP2 AOBS confocal laser scanning microscope with a ×63 oil immersion lens at 1024 × 1024 pixels resolution, at the same laser attenuation. The number of LC3-positive vesicles and Pearson’s correlation coefficients for LC3 and LAMP1 co-localization were determined using Volocity Demo 6.0 (Perkin Elmer Inc.) (Settembre et al., 2008).
Electron microscopy
Cultured skin fibroblast from patients and control cells were fixed in 1% glutaraldehyde in 0.2 M HEPES buffer, post-fixed in uranyl acetate and in OsO4. After dehydration through a graded series of ethanol, the cells were embedded in epoxy resin (Epon 812) and polymerized at 60°C for 72 h. From each sample, thin sections were cut with a Leica EM UC6 ultramicrotome. Electron microscopy images were acquired from thin sections using an FEI Tecnai-12 electron microscope equipped with an ULTRA VIEW CCD digital camera (FEI).
Immunoprecipitation and protein level analysis
HeLa and lymphoblastoid cells were lysed in ice-cold lysis buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 10% glycerol, 5 mM EGTA, 1 mM EDTA, 1% Triton™ X-100, 50 mM NaF, 20 mM Na3VO4, 1 mM PMSF and protease inhibitor cocktail from Sigma-Aldrich) and incubated on ice for 10 min. Cells were sonicated and centrifuged at 13 000 rpm for 10 min at 4°C. Supernatants were assayed and an equal amount of total proteins for each cell line was immunoprecipitated with the indicated primary antibody overnight at 4°C. Protein A-Sepharose (GE Healthcare-Italia) was then added to each sample and rotated at 4°C for 1 h. Total protein extracts from HeLa cells were immunoprecipitated also with an antibody against rabbit IgG isotype conjugated with sepharose beads to determine non-specific immunoprecipitation complexes. Immunoprecipitation analysis in HeLa cells and limphoblastoid cell lines were performed using the same antibodies. Spastizin immunoprecipitation and detection in both cell types was performed using the anti-spastizin antibody directed against the N-terminus of the protein. Spastizin immunoprecipitation in HeLa cells were also confirmed using the antibody directed against the C-terminus of the protein. Immunocomplexes were washed three times with lysis buffer, eluted in 5× sample buffer plus 1 mM DTT at 95°C for 5 min and subjected to SDS-PAGE and western blot as described (Vantaggiato et al., 2009).
Statistical analysis
The results are expressed as means ± standard error of the mean (SEM); n represents the number of individual experiments. Statistical analysis was carried out using the Student’s t-test for unpaired variables (two-tailed), double or triple asterisks refer to statistical probabilities (P < 0.01 and P < 0.001, respectively), measured in the various experimental conditions as detailed in the figure legends. P-values < 0.05 were considered significant.
Results
Spastizin interacts with UVRAG and Rubicon proteins in the autophagy pathway
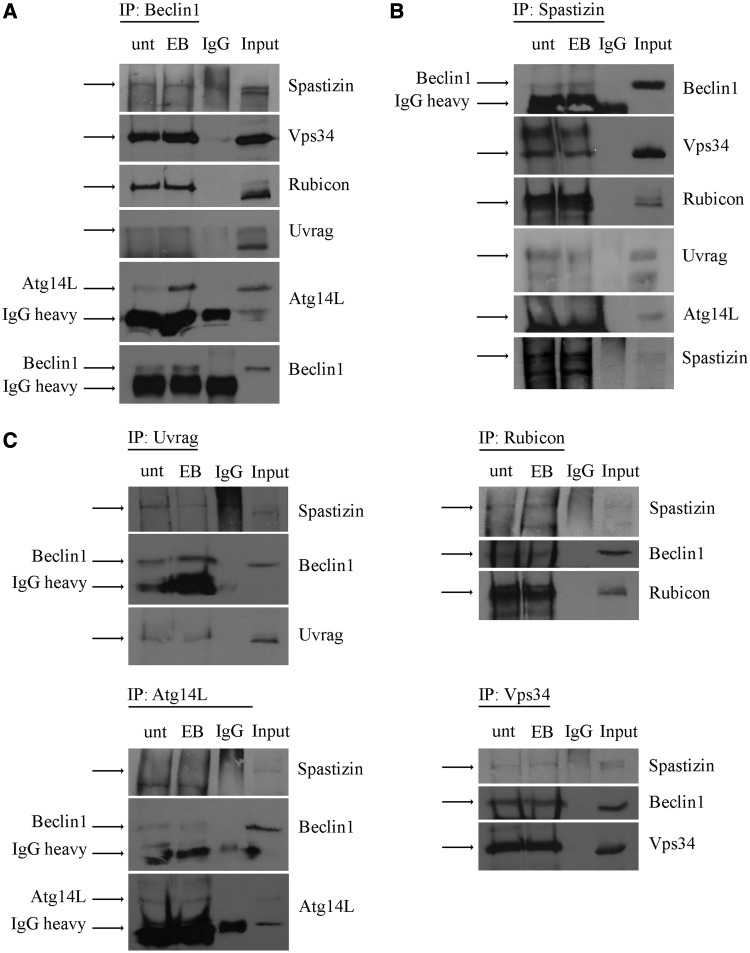
We investigated whether spastizin interacts with key proteins involved in the formation of the core complexes regulating autophagosome formation and maturation. The core complex Beclin 1–Vps15–Vps34 regulates autophagosomes formation and maturation by forming two mutually exclusive larger complexes with Atg14L or UVRAG, respectively (Itakura et al., 2008; Sun et al., 2008). The complex generated with Atg14L regulates PI3P synthesis, the formation of the isolation membranes and the generation of autophagosomes. The complex formed with UVRAG promotes both the formation and the maturation of autophagosomes (Liang et al., 2006, 2008). The binding of Rubicon to this complex negatively regulates autophagosome maturation (Zhong et al., 2009). By immunoprecipitation analysis in HeLa cells using an antibody against Beclin 1, we confirmed that Beclin 1 interacts with Vps34, Rubicon, UVRAG and Atg14L, as previously demonstrated (Matsunaga et al., 2009), both in normal condition and after autophagy induction with starvation in Earle’s Balanced Salt Solution (Fig. 1A). Spastizin was likewise co-immunoprecipitated with Beclin 1 (Fig. 1A).
Figure 1.
Spastizin interacts with Beclin 1, Vps34, Rubicon and UVRAG in HeLa cells. Total protein extracts were prepared from HeLa cells untreated (unt) or starved with Earle’s Balanced Salt Solution (EB) for 2 h and then subjected to immunoprecipitation (IP) with anti-Beclin 1 (A), anti-spastizin (B) and anti-UVRAG,-Rubicon, -Atg14L or -Vps34 antibodies (C). Proteins were loaded on 6% polyacrylamide gels. Total protein extracts used for the immunoprecipitation were loaded as positive control (Input). Total protein extracts were immunoprecipitated also with an antibody against rabbit IgG isotype as negative control (IgG). Specific bands are indicated by the arrows. The 50 kDa rabbit IgG heavy chain is indicated (IgG heavy) where present. Shown is a representative of three reproducible blots.
We then investigated the interaction of spastizin with Vps34, Atg14L, UVRAG and Rubicon using an anti-spastizin antibody. Anti-spastizin antibody co-immunoprecipitated Beclin 1 and Vps34, both in normal and autophagy inducing conditions, thus indicating that spastizin interacts with the Beclin 1–Vps34 core complex (Fig. 1B). Anti-spastizin antibody co-immunoprecipitated also Rubicon and UVRAG, but not Atg14L, suggesting that spastizin interacts with the Beclin 1 complex containing UVRAG and Rubicon, but not with the complex containing Atg14L. Consistently, spastizin was co-immunoprecipitated using antibodies against Rubicon, UVRAG and Vps34, but not with the antibody against Atg14L (Fig. 1C).
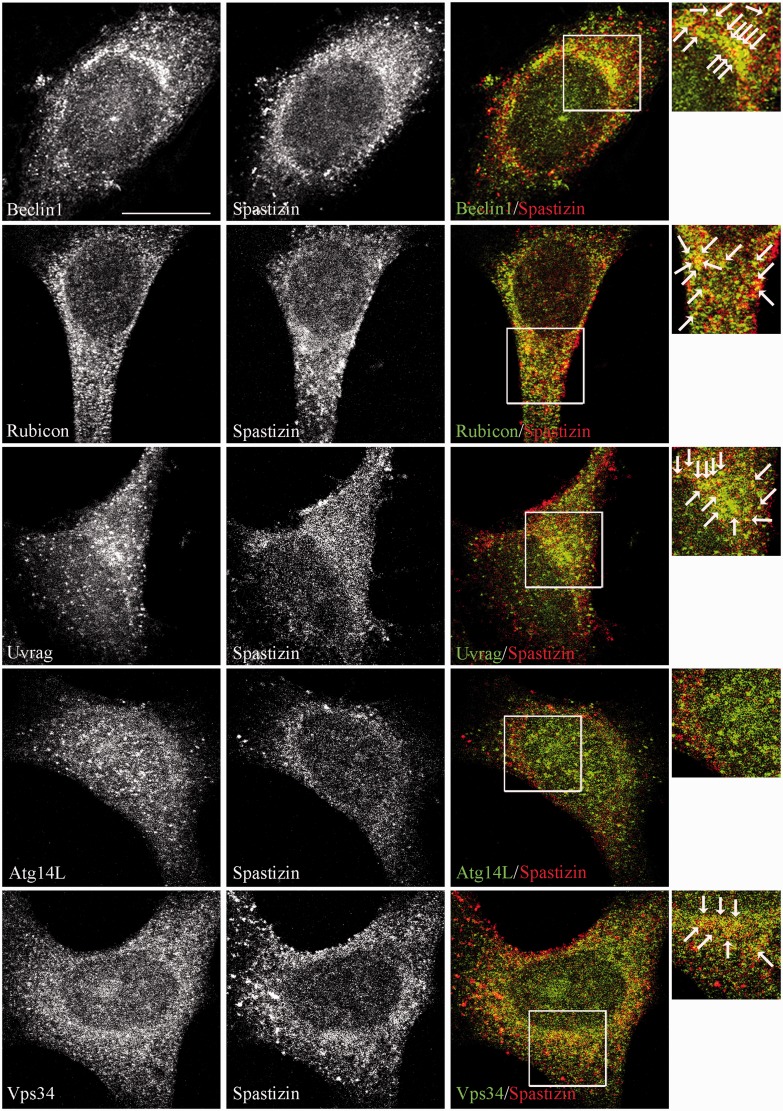
Immunofluorescence analysis in untreated and starved HeLa cells confirmed the interaction of spastizin with Beclin 1, Rubicon, UVRAG and Vps34 and not with Atg14L (Fig. 2 and Supplementary Fig. 1), indicating that spastizin is part of the autophagy complexes, but it is excluded from the Atg14L–Beclin 1 complex.
Figure 2.
Spastizin partially co-localizes with Beclin 1, Rubicon, UVRAG and Vps34 in HeLa cells. Untreated HeLa cells were fixed and immunostained with anti-spastizin (red) and anti-Beclin 1, -Rubicon, -UVRAG, -Atg14 and -Vps34 (green) antibodies. Yellow in the merge images indicates partial co-localization of spastizin with Beclin 1, Rubicon, UVRAG and Vps34. Co-localization is indicated by arrows in the magnified boxed area on the right. Scale bar = 10 µm.
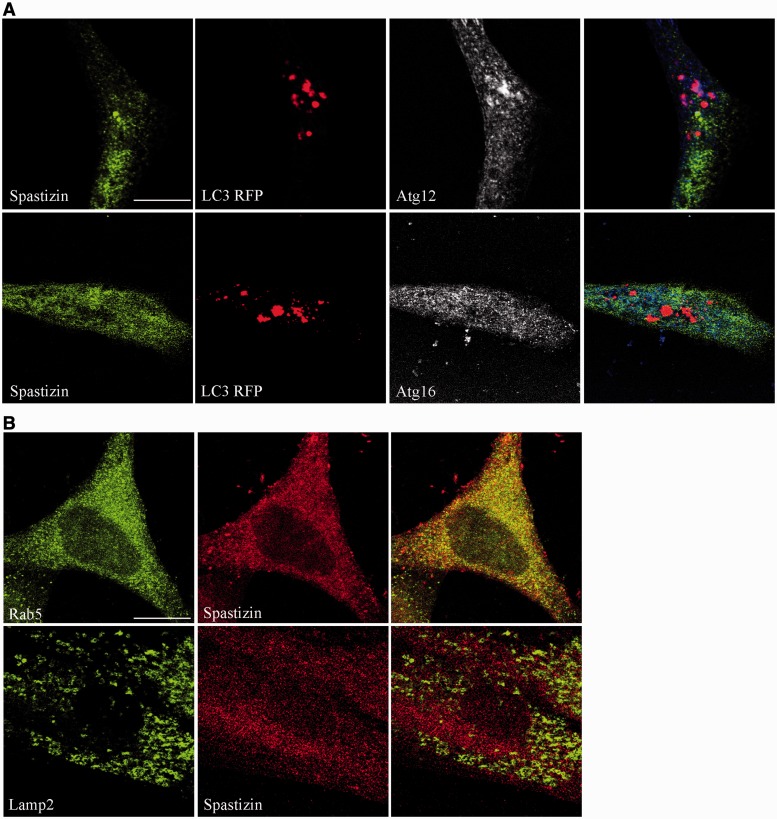
This differential interaction with the Beclin 1 complexes was further supported by spastizin subcellular distribution. Immunofluorescence analysis revealed that spastizin does not co-localize with the autophagosomal marker LC3 or with the isolation membranes proteins Atg12 and Atg16 (Mizushima et al., 2001) in autophagy-inducing conditions (Fig. 3A). This distribution is similar to that of Rubicon and UVRAG (Itakura et al., 2008; Matsunaga et al., 2009), but differs from that of Atg14L, which localizes on early autophagosomes and on the isolation membranes (Itakura et al., 2008). The partial co-localization of spastizin with the early endosomal marker Rab5 (Fig. 3B), while confirming previous data in different cell types (Hanein et al., 2008), extends the overlapping distribution of Rubicon, UVRAG (Itakura et al., 2008; Matsunaga et al., 2009) and spastizin to endosomes. Further, spastizin, unlike Rubicon and UVRAG (Liang et al., 2008), did not co-localize with LAMP2 and was absent from lysosomes (Fig. 3B; Hanein et al., 2008).
Figure 3.
Spastizin does not localize on autophagy-related structures. (A) HeLa cells were transfected with MAP1LC3BRFP for the staining of autophagosomes (red), 24 h later cells were starved with Earle’s Balanced Salt Solution for 30 min, fixed and immunostained with anti-spastizin (green) and anti-Atg12 or Atg16 (blue) antibodies. Spastizin does not co-localize with the autophagosomal marker LC3 or with the isolation membrane markers Atg12 and Atg16. Scale bar = 10 µm. (B) Untreated HeLa cells were fixed and immunostained with anti-spastizin (red) and anti-Rab5 or -LAMP2 (green) antibodies. Yellow in the merge images indicates co-localization of spastizin with early endosomes. Scale bar = 10 µm.
We then decided to investigate the effects of spastizin pathogenic mutations on protein–protein interaction.
Identification of novel spastizin mutations
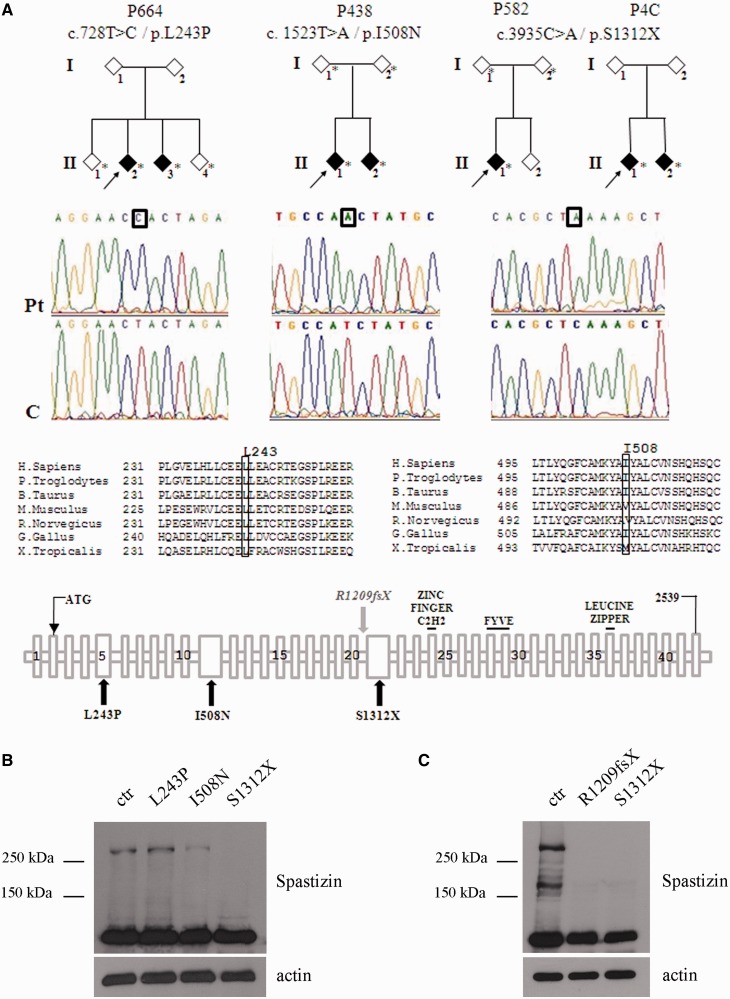
We considered three novel homozygous mutations in the ZFYVE26 gene identified in a screening of 65 families with ARHSP-TCC. Clinical features of the four mutated families (Families P664, P438, P582 and P4C) are summarized in Supplementary Table 1. Overall the clinical features of the new mutated families overlap those of typical patients with spastic paraplegia type 15 (Goizet et al., 2009; Schüle et al., 2009) with the exception of the two affected members of Family P664 who show a later age of onset (34–38 years). Two homozygous missense changes, c.728 T>C (p.L243P) in exon 5 and c.1523T>A (p.I508N) in exon 10 of ZFYVE26 were found in two affected members of Families P664 and P438, respectively (Fig. 4A). A nonsense mutation c.3935C>A, in exon 21 (p.S1312X) was detected in the proband of Family P582 (Fig. 4A). This mutation was also found in two patients (Patients P4C1 and P4C2) from an independent family with undisclosed consanguinity; they were apparently unrelated to Family P582, in spite of the common origin from Sicily (Fig. 4A and Supplementary Table 1). All ZFYVE26 variants were checked against a set of 600 controls and the 1000 Genomes database. The missense changes were predicted to damage protein structure and affect protein function by using different software (Supplementary Table 2). Evolutionary conservation was very high only for the Leu243 residue, whereas for the Ile508 residue only partial conservation was observed (Fig. 4A). We analysed expression of the various mutated spastizin proteins versus the wild-type form. To this end we analysed not only the newly identified mutants, but also a previously described spastizin truncating mutation, p.R1209fsX (Family 761 in Hanein et al., 2008), for which we managed to receive patient-derived fibroblasts. As a control we used cell lines from three healthy controls and a healthy carrier (the parent of Patient I508N mutated proband). No difference in the expression level of spastizin was observed among the four controls used (not shown). The mutations L243P and I508N did not alter the expression level of spastizin, while the truncating mutation S1312X determined the loss of protein expression (Fig. 4B). Likewise no spastizin expression was observed in fibroblast cells carrying the p.R1209fsX truncating mutation (Fig. 4C).
Figure 4.
Patient's families and ZFYVE26 mutations. (A) Pedigrees of the families carrying the ZFYVE26 mutations here identified. Arrows indicate the proband in each family. Asterisks indicate the subjects for whom DNA was available. Elechtropherograms of mutant and control sequences are shown below, with boxes indicating the mutant nucleotide for each mutation. A partial multiple alignment of protein sequences of human spastizin and orthologues from other species with positions of the mutant residues (L243P and I508N) boxed is shown below. A schematic representation of spastizin protein domains is shown with a summary of the spastizin mutations identified in this work (L243P, I508N and S1312X) and analysed (L243P, I508N, S1312X and R1209fsX in Hanein et al., 2008). (B) Spastizin protein levels in spastizin mutated cells. Total extracts prepared from L243P, I508N, and S1312X mutated and control (ctr) lymphoblast cells were run on 6% SDS-polyacrylamide gel and stained with an anti-spastizin antibody directed against the N-terminus of the protein and with anti-actin antibody. Whereas L243P and I508N mutated cells express spastizin at the expected 280 kDa size, no signal is observed in S1312X mutated cells at the size of 154 kDa corresponding to the truncated protein. (C) Spastizin protein levels in R1209fsX mutated fibroblast. Total extracts prepared from R1209fsX, S1312X and control fibroblasts were run on 6% SDS-polyacrylamide gel and stained with an anti-spastizin antibody directed against the N-terminus of the protein and with anti-actin antibody. No signal is observed in R1209fsX mutated cells at the size of 135 kDa corresponding to the truncated protein, analogously to what is observed in S1312X mutated fibroblasts.
Spastizin mutations alter the interaction with Beclin 1, Rubicon, UVRAG and Vps34
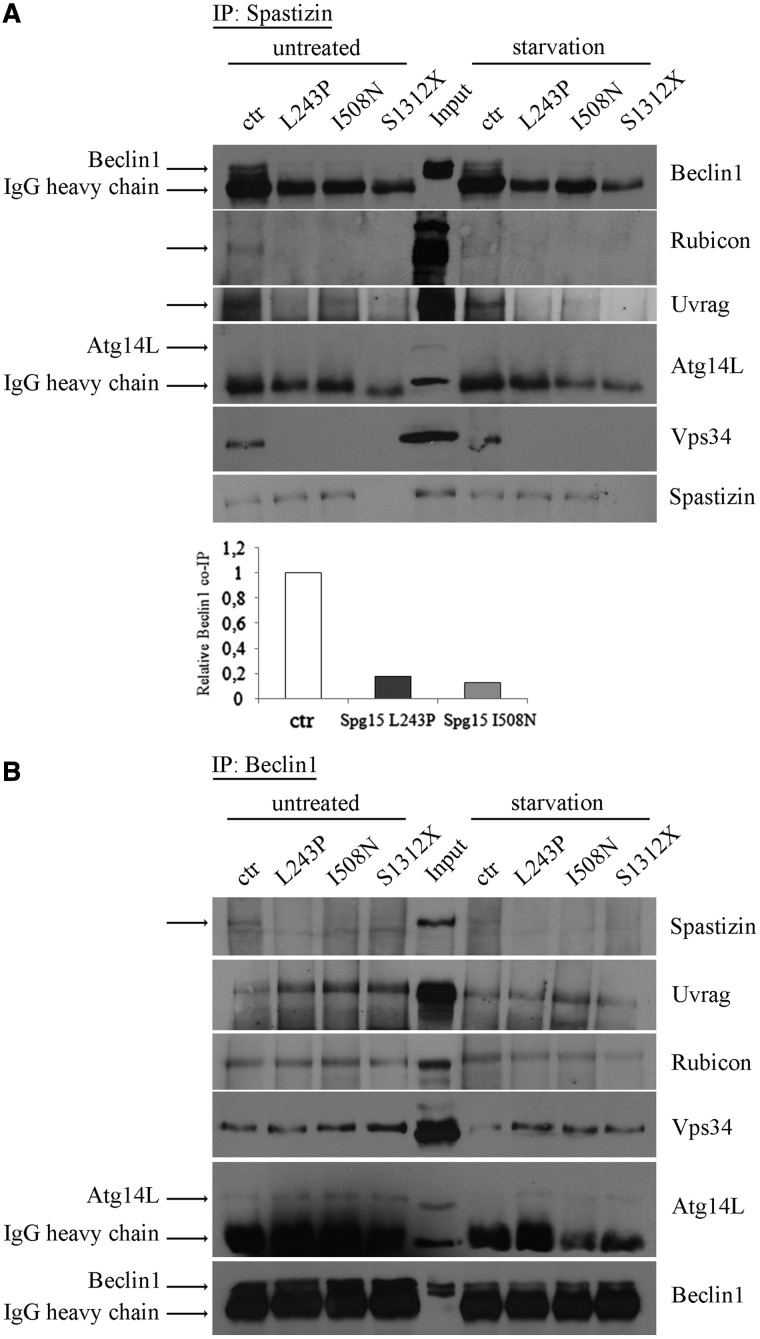
We analysed the effects of patient-derived spastizin mutations on the interaction of spastizin with Beclin 1, Vps34, UVRAG and Rubicon and on the formation of the Beclin 1 complexes. To this purpose we used lymphoblast cell lines established from patients carrying the two point mutations (L243P and I508N) and one of the two cell lines lacking spastizin protein (S1312X).
Cells were analysed both in basal and autophagy-inducing conditions. In control cells, in both conditions, anti-spastizin antibody co-immunoprecipitated Beclin 1 Rubicon, UVRAG and Vps34 (Fig. 5A), and not Atg14L, as observed in HeLa cells (Fig. 1B). By contrast, in L243P and I508N spastizin mutated cells the anti-spastizin antibody co-immunoprecipitated only a minimal amount of Beclin 1 (<20% compared with control cells) and did not co-immunoprecipitate Vps34, Rubicon and UVRAG (Fig. 5A). In S1312X spastizin mutated cells, lacking spastizin protein, the anti-spastizin antibody failed to co-immunoprecipitate Beclin 1, Vps34, Rubicon and UVRAG, as expected, thereby confirming the specificity of spastizin interactions in control cells. No significant differences were found between basal and autophagy-inducing conditions. Anti-Beclin 1 antibody in spastizin mutated and depleted cells still co-immunoprecipitated Rubicon, UVRAG and Vps34 indicating that spastizin mutations do not affect the formation of the Beclin 1–UVRAG–Rubicon multiprotein complex (Fig. 5B). All spastizin interactions in control and mutated cells were confirmed using anti-Rubicon, UVRAG, Atg14L and Vps34 antibodies (Supplementary Fig. 2) and by immunofluorescence in control and L243P spastizin mutated fibroblast cell lines (Supplementary Figs 3 and 4).
Figure 5.
SPG15-related mutations alter spastizin interactions with Beclin 1-complexes. Total protein extracts were prepared from lymphoblastoid cells untreated or starved with Earle’s Balanced Salt Solution for 2 h (starvation) and subjected to immunoprecipitation (IP) with anti-spastizin (A) or anti-Beclin 1 (B) antibodies. Proteins were loaded on 6% SDS-polyacrylamide gels. Total protein extracts used for the IP were tested for the presence of relevant proteins with similar results in all cell lines and control cell extracts were loaded as positive control (Input). Specific bands are indicated by the arrows. Where present, the 50 kDa rabbit IgG heavy chain is also indicated. Shown is a representative of three reproducible blots. Beclin 1 levels in untreated control and L243P, I508N and S1312X mutated cells were quantified from A, normalized on IgG and shown in the graph.
Pathogenic mutations or silencing of spastizin induce accumulation of autophagosomes in fibroblasts and neuronal cells
In view of the role of Vps34, Rubicon, UVRAG and Beclin 1 in autophagy regulation, we analysed the role of spastizin in autophagy. To this end we used the available fibroblast cell lines from healthy controls and patients (L243P, R1209fsX and S1312X) described above (Fig. 4), as well as primary hippocampal neurons and the SHSY5Y human neuroblastoma cell line.
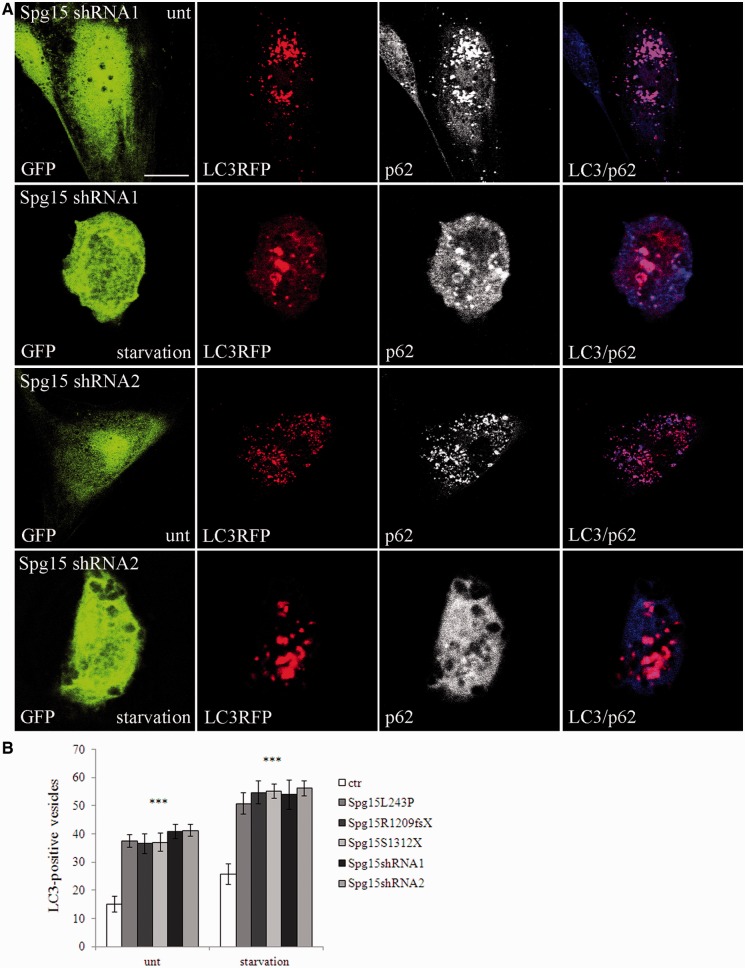
Autophagy was analysed by determining autophagosome numbers both in basal conditions and after autophagy induction with starvation, by using the autophagosomal marker LC3. During autophagosomes formation the cytosolic LC3-I isoform is converted into LC3-II and is incorporated in the autophagosome membrane, thus LC3-II amount correlates with the number of autophagosomes (Kabeya et al., 2000). The number of LC3-positive vesicles detected by immunofluorescence in L243P, R1209fsX and S1312X fibroblast cells was significantly higher than in control cells, both in basal and starved conditions (Figs 6 and 7). Consistently, depletion of spastizin with two specific short hairpin RNAs (shRNA1 and shRNA2), with high silencing efficiency (Supplementary Fig. 5), induced an increased accumulation of autophagosomes (Fig. 7). The autophagosomal nature of the LC3-positive vesicles was confirmed by their co-localization with p62, a protein targeting poly-ubiquitinated protein to autophagosomes for degradation (Figs 6 and 7), and with ubiquitin (Supplementary Fig. 6).
Figure 6.
Spastizin mutations induce accumulation of autophagosomes in fibroblast cells. Control (ctr) and L243P, R1209fsX and S1312X spastizin mutated fibroblast cells were transfected with MAP1LC3B-RFP for the staining of autophagosomal vesicles (red) and 24 h later were starved for 2 h to induce autophagy (starvation), fixed and immunostained with anti-p62 (green) antibody. Yellow in the merge images indicates co-localization of p62 and LC3. LC3-positive vesicles quantification is shown in Fig. 7B. Scale bar = 10 µm. unt = untreated.
Figure 7.
Spastizin depletion induces accumulation of autophagosomes in cells in which spastizin had been silenced. Control fibroblast cells were transfected with two different Spg15 short hairpin RNA vectors (shRNA1 or shRNA2), showing similar silencing efficiency, both with a GFP tag (green) and with MAP1LC3B-RFP for the staining of autophagosomes (red). Forty-eight hours later cells were starved for 2 h (starvation), fixed and immunostained with anti-p62 (blue) antibody. Violet in the merge indicates co-localization of p62 and LC3. Scale bar = 10 µm. (B) Quantification of LC3-positive vesicles from the silenced fibroblasts above and from controls and mutated cells of Fig. 6 is shown in the graph. The graph shows the mean ± SEM of three independent experiments for a total of at least 30 cells for each sample (***P < 0.001).
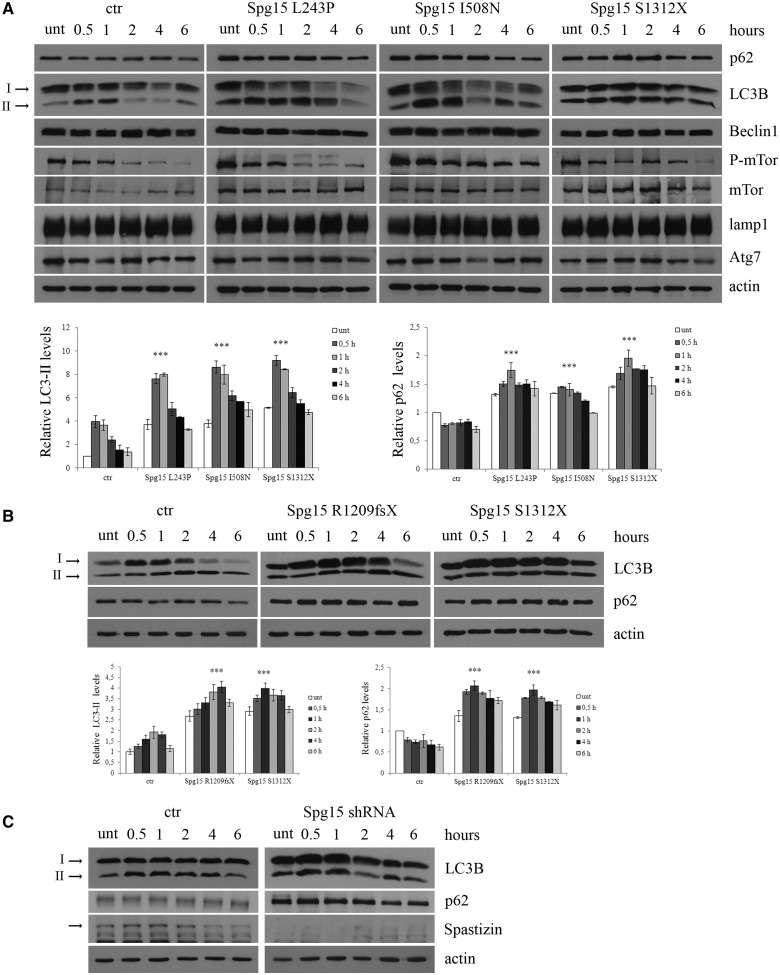
We then quantified LC3-II and p62 expression levels in L243P, I508N and S1312X mutated and control lymphoblast cells. A total of four control cells (three from healthy individuals, Control Subjects 1, 2, 3 and one from the healthy carrier of the I508N family, Control Subject 4) were analysed alongside patient-derived cells. Results from patients-derived cells and Control Subject 1 are shown in Fig. 8A, whereas results from the remaining three controls are reported in Supplementary Fig. 7A. LC3-II expression levels, which were enhanced during starvation in control cells, were enhanced further in patient-derived cells. p62 levels, which were decreased in control cells, as expected because of autophagy induction, were instead increased in patient-derived cells, indicating defective autophagy, despite the increased number of autophagosomes. We assessed whether the increased number of autophagosomes was due to autophagy induction or to inhibition of autophagosome degradation. We did not detect differences in Beclin 1 expression and in mTor phosphorylation, two key players in stimulating or inhibiting autophagy, respectively (Noda and Ohsumi, 1998). Atg7 levels were also normal, thereby excluding alterations in the production of LC3-II (Abounit et al., 2012) and no differences were detected in LAMP1 expression levels, a lysosomal marker.
Figure 8.
Spastizin mutations induce an increase in LC3-II and p62 levels. (A) Lymphoblastoid cells from patients carrying L243P, I508N and S1312X spastizin mutations, three healthy control individuals and one healthy carrier from the I508N mutated family were starved with Earle’s Balanced Salt Solution for different times, from 30 min to 6 h to induce autophagy. Untreated and treated cells were lysed and total protein extracts were run on 6 or 10% SDS-polyacrylamide gels and probed with anti-p62, LC3B, -Beclin 1, -mTor, -phospho-mTor (P-mTor), -LAMP1, -Atg7 and -actin antibodies. In the figure is shown only one control (ctr), whereas results from the remaining three controls are reported in Supplementary Fig. 7A. Arrows indicate the two isoforms LC3-I and LC3-II. LC3-II and p62 levels were quantified and normalized on actin levels. The graphs show the mean ± SEM of three independent experiments (***P < 0.001). The mean of all the four controls is reported in the graphs. (B) Fibroblast cells from patients carrying R1209fsX and S1312X spastizin mutations and from a control were starved as in A to induce autophagy. Total protein extracts were run on 12% SDS-polyacrylamide gels and probed with anti-LC3B, -p62 and -actin antibodies. Shown is a representative of three reproducible blots. LC3-II and p62 levels were quantified and normalized on actin levels. The graphs show the mean ± SEM of three independent experiments (***P < 0.001). (C) Silencing of ZFYVE26 increases LC3-II and p62 levels. HeLa cells were transfected with Spg15 short hairpin RNA or with a control vector (ctr) and 72 h later were starved to induce autophagy. Total extracts were probed with anti-LC3B, -p62, -actin and -spastizin antibodies, the latter to show the residual spastizin levels in ZFYVE26 (Spg15) short hairpin RNA-transfected cells. Shown is a representative of three reproducible blots.
We quantified LC3-II and p62 expression levels also in R1209fsX mutant fibroblasts, in which spastizin is not expressed. In parallel, as internal control, we analysed fibroblast cells carrying the S1312X mutation, also lacking spastizin expression. Both cell lines showed similar increases of LC3-II and p62 levels compared with the control (Fig. 8B). Analogously, HeLa cells transfected with Spg15 short hairpin RNA and starved for the indicated time, showed an increase in LC3-II and p62 levels compared with cells transfected with a control vector (Fig. 8C). This confirms further the link between spastizin depletion and the autophagy defects observed in patients’ cells.
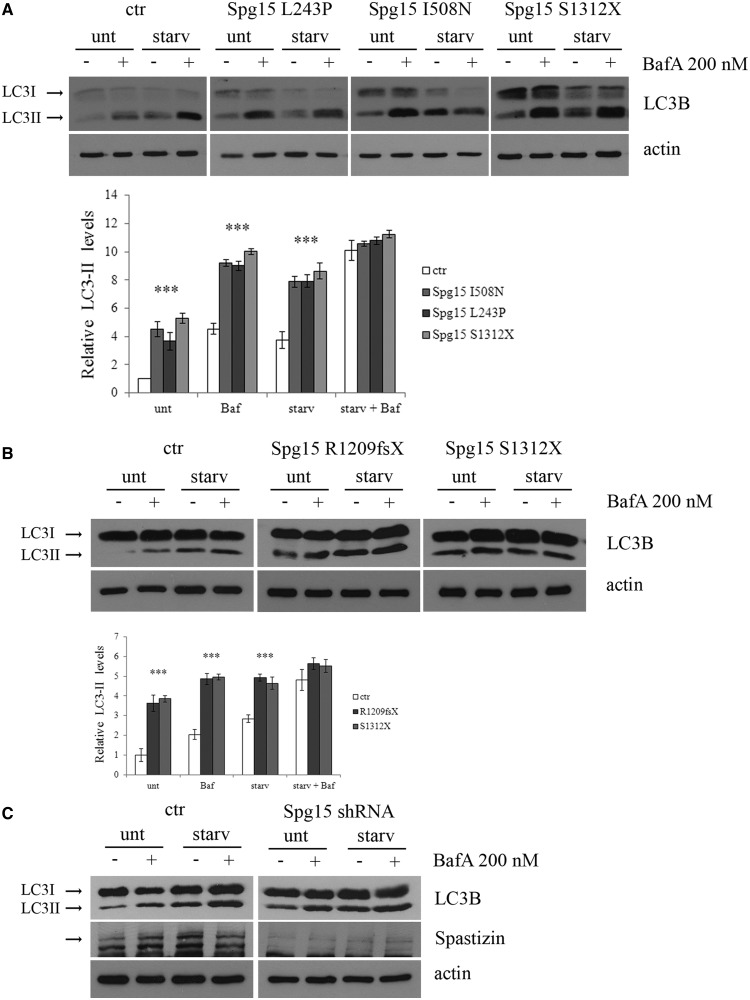
In control and spastizin mutated lymphoblast cells (L243P, I508N and S1312X) treatment with bafilomycin A1, which inhibits autophagosome-lysosome fusion and induces autophagosomes accumulation (Yamamoto et al., 1998), did not result into different LC3-II levels in autophagy-inducing conditions (i.e. starvation) (Fig. 9A and Supplementary Fig. 7B for additional controls). This indicates that LC3-II production and autophagy induction were not increased in spastizin mutated cells. Similar results were obtained in fibroblasts carrying the R1209fsX truncating mutation (Fig. 9B) and in spastizin-silenced HeLa cells (Fig. 9C).
Figure 9.
Analysis of the effect of autophagy inhibition with bafilomycin A1 in control and spastizin (Spg15) mutated cells. (A) L243P, I508N and S1312X spastizin mutated and four different control (one is shown, the others are reported in Supplementary Fig. 7B) lymphoblast cells were incubated with 200 nM bafilomycin A1 in complete medium (unt) or in starvation medium (starv) for 1 h. Total protein extracts were run on 12% SDS-polyacrylamide gels and probed with anti LC3B and actin antibodies. Shown is a representative blot out of three reproducible ones. LC3-II levels were quantified and normalized on actin levels. The mean of the four controls is reported in the graph. The graphs show the mean ± SEM of three independent experiments (***P < 0.001). (B) Fibroblast cells from patients carrying R1209fsX and S1312X spastizin mutations and from a control were incubated with 200 nM bafilomycin A1 and processed as in A. The graphs show the mean ± SEM of three independent experiments (***P < 0.001). (C) HeLa cells were transfected with ZFYVE26 (Spg15) short hairpin RNA or with a control vector (ctr) and 72 h later were incubated with Bafilomycin A1 as described above. Total extracts were probed with anti-LC3B, -actin and -spastizin antibodies, the latter to show the residual spastizin levels in ZFYVE26 (Spg15) short hairpin RNA-transfected cells. Shown is a representative of three reproducible blots.
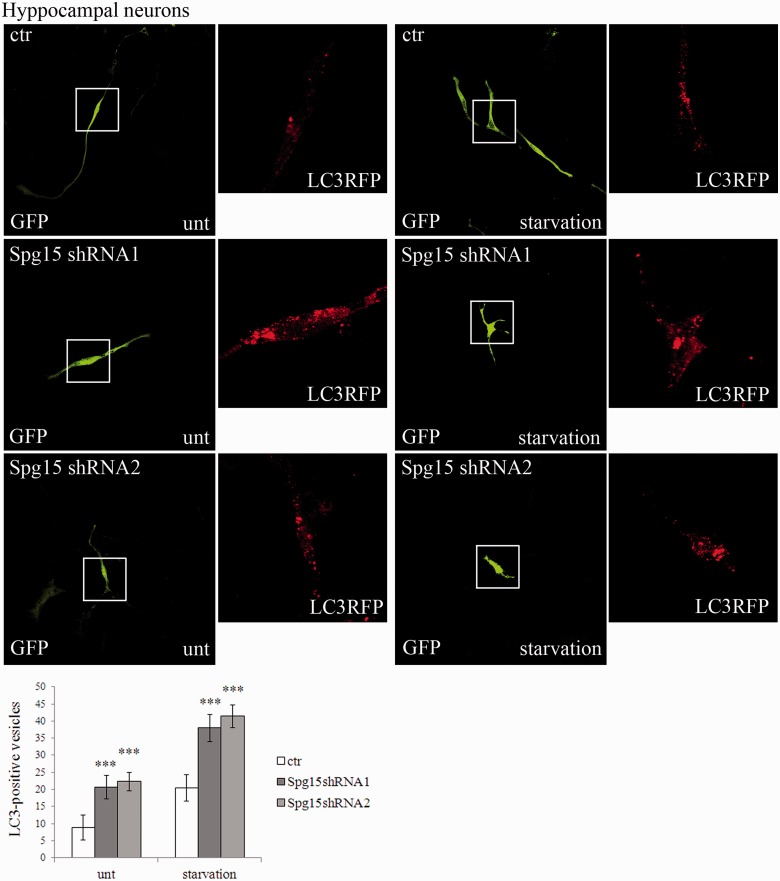
We then investigated whether spastizin depletion induced autophagosome accumulation also in neuronal cells. By using two specific short hairpin RNAs with similar silencing efficiency (Supplementary Fig. 8), silencing of Zfyve26 in hippocampal neurons (Fig. 10) determined an accumulation of LC3-positive vesicles both in basal conditions and after autophagy induction. Accumulation of autophagosomes was observed also in silenced SHSY5Y cells (Supplementary Fig. 9). Thus, spastizin depletion appears to generate the same effects in all cells analysed: lymphoblasts, fibroblasts, HeLa cells, primary neurons and SHSY5Y cells.
Figure 10.
Spastizin depletion induces accumulation of autophagosomes in primary hippocampal neurons. Primary hippocampal neurons were transfected with MAP1LC3B-RFP for the staining of autophagosomal vesicles (red) and with a control vector (ctr) or with two specific Zfyve26 (Spg15) short hairpin RNAs (shRNA1 and shRNA2) to silence the gene, all with a GFP tag. Seventy-two hours later neurons were starved with Dulbecco’s modified Eagle’s medium for 4 h and fixed. LC3-positive vesicles quantification is shown in the graph. The graphs show the mean ± SEM of three independent experiments for a total of at least 30 cells for each sample (***P < 0.001). Scale bar = 10 µm.
Autophagosome maturation is impaired by spastizin mutation or silencing
Clearance of autophagosomes occurs through fusion with lysosomes. We investigated whether accumulation of autophagosomes observed in different cells not expressing spastizin or expressing its mutant forms was due to defective clearance caused by impaired or reduced autophagosome-lysosome fusion.
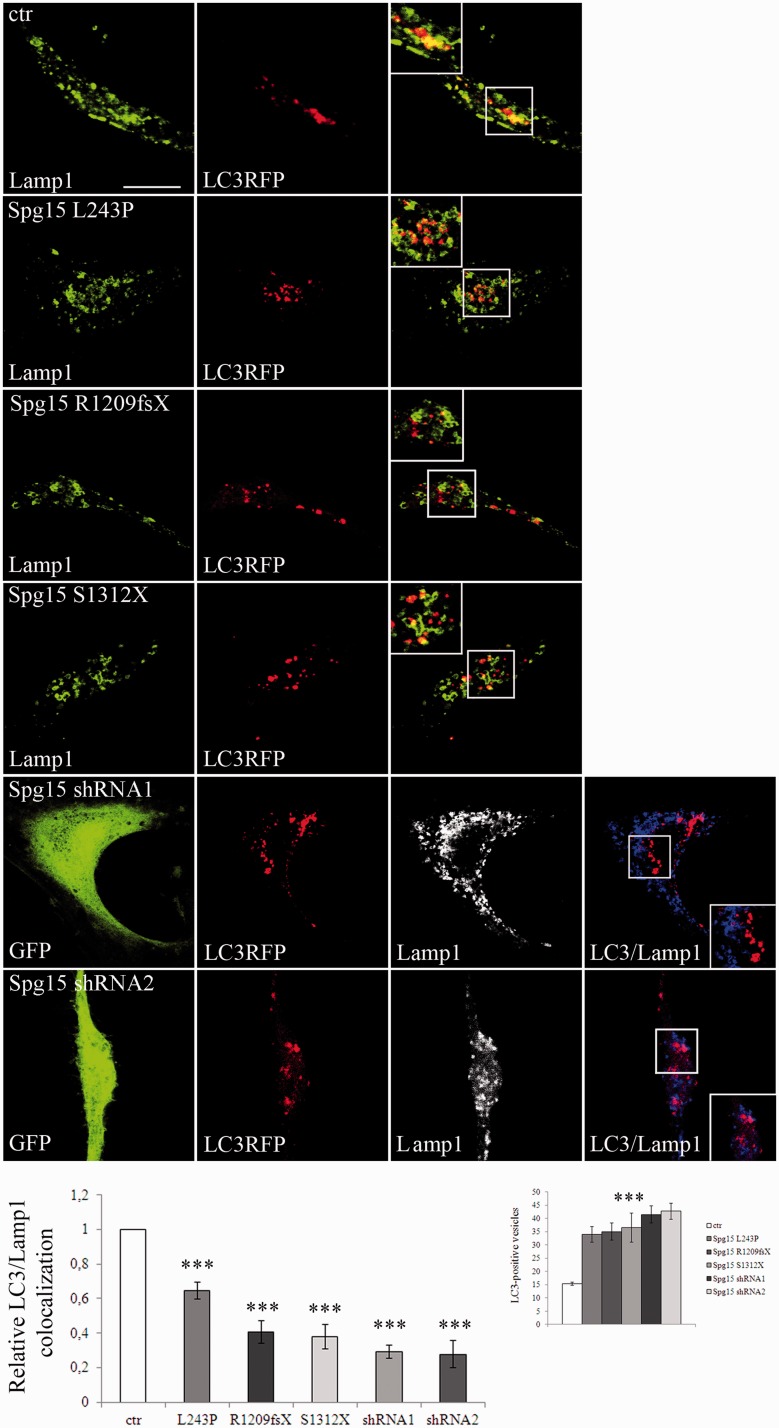
To test this hypothesis, we analysed the subcellular localization of LAMP1 and LC3 by confocal microscopy in fibroblast cell lines (Settembre et al., 2008). The co-localization of LAMP1 and LC3 in basal conditions was significantly reduced in L243P (by 40%), R1209fsX and S1312X cells (both by 60%) compared with control cells (Fig. 11), and decreased further in autophagy-inducing conditions (Supplementary Fig. 10), thus indicating impaired fusion of autophagosomes with lysosomes. The reduction in co-localization observed in R1209fsX and S1312X cells was confirmed in spastizin-silenced cells (Fig. 11 and Supplementary Fig. 10). As shown in Fig. 9A and B, in spastizin mutated cells autophagosome maturation is not completely blocked, as treatment of L243P, I508N, R1209fsX and S1312X cells with bafilomycin A1 induced a further increase in LC3-II levels compared with untreated L243P, I508N, R1209fsX and S1312X cells.
Figure 11.
Spastizin mutations induce a reduction of autophagosome-lysosomes fusion. Control (ctr) and L243P, R1209fsX and S1312X spastizin mutated fibroblast cells were transfected with MAP1LC3B-RFP for the staining of autophagosomes (red) and 24 h later were fixed and immunostained with anti-LAMP1 (green) antibody. Yellow in the merge images indicates co-localization of LC3 and LAMP1. Control fibroblast were transfected with two different ZFYVE26 (Spg15) silencing vectors (shRNA1 or shRNA2), showing similar silencing efficiency, both with a GFP tag (green) and with MAP1LC3B-RFP for the staining of autophagosomes (red). Forty-eight hours later cells were fixed and immunostained with anti-LAMP1 (blue) antibody. Violet in the merge indicates co-localization. Pearson’s correlation coefficient for LC3 and LAMP1 co-localization were determined using Volocity software and normalized on that of control cells. The graphs show the mean ± SEM of three independent experiments for a total of at least 30 cells for each sample (***P < 0.001). LC3-positive vesicles quantification is shown in the small graph on the right. Scale bar = 10 µm.
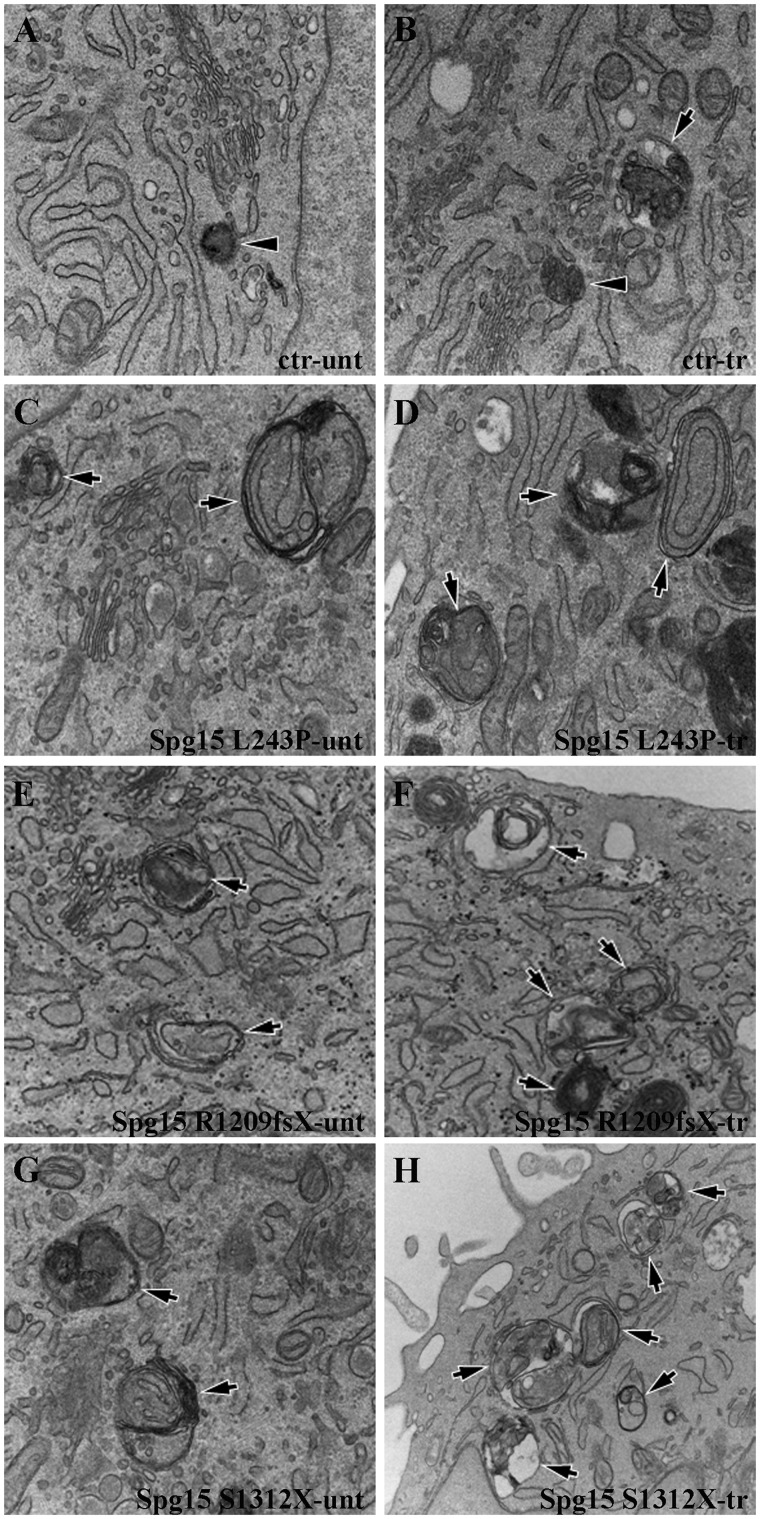
Analysis of fibroblasts at the ultrastructural level revealed that control cells contained mainly electron dense lysosomes surrounded by single membrane (Fig. 12A and B) and exhibited few autophagosomes/autolysosomes (Fig. 12A and B) with heterogeneous luminal content upon starvation. In contrast, numerous autophagosomes were already detected in L243P, R1209fsX and S1312X cells in basal conditions (Fig. 12C, E and G) and increased in number after starvation (Fig. 12D, F and H). Most of such autophagic vacuoles contained double membranes and exhibited remnants of cytosol and membrane organelles in their lumen. This indicates impairment of the fusion of autophagosome with lysosomes and their further maturation into autolysosomes.
Figure 12.
Accumulation of autophagosomes in spastizin mutated fibroblast cells. Control, L243P, R1209fsX and S1312X spastizin mutated fibroblast cells were starved with Earle’s Balanced Salt Solution for 30 min (tr; B, D, F, H) or untreated (unt; A, C, E, G) and processed by electron microscopy. In contrast to control cells, which contain lysosomes (A and B, arrowheads) and few autophagosomes (B, arrow), L243P, R1209fsX and S1312X cells exhibited an increased number of autophagosomes (C–H, arrows).
Discussion
The role of autophagy in different neurodegenerative diseases including Alzheimer’s, Parkinson’s, Huntington’s diseases, spinocerebellar ataxias, (Banerjee et al., 2010; Wong et al., 2011 for review) amyotrophic lateral sclerosis and different forms of motor neuron diseases is well established (Ravikumar et al., 2005; Morimoto et al., 2007; Li et al., 2008; Cox et al., 2010). Each neurodegenerative disease may harbour a different extent of autophagy deregulation, involving different steps in the autophagic pathways. Indeed excessive activation or defects in autophagosome maturation and degradation were described in these diseases and correlated with disease status (Banerjee et al. 2010; Wong et al., 2011). Overall, these studies have shown that constitutive autophagy is essential for neuron and motor neuron survival; defects in any of the steps of this process lead to abnormalities at downstream cellular pathways and thus to cytotoxicity, thereby disrupting neural function (Moreau et al., 2010; Wong et al., 2010).
In hereditary spastic paraplegias, where the neuropathology relates to distal degeneration of the lateral corticospinal tracts, a single gene, TECPR2, was recently identified whose mutations lead to aberrant autophagy in fibroblasts of patients affected by a recessive complicated form of the disease (Oz-Levi et al., 2012, 2013). The TECPR2-associated phenotype is apparently more severe and complicated in terms of clinical presentation, disease progression and age at onset with respect to the spastic paraplegia type 15 subtype. Thinning of corpus callosum at the MRI is shared with the spastic paraplegia type 15 subtype (Oz-Levi et al., 2012).
Here we show that spastizin, the protein mutated in the spastic paraplegia type 15 subtype of hereditary spastic paraplegias, participates to autophagy by interacting with the Beclin 1–Vps34 core complex, both in normal and autophagy induction conditions. In particular, spastizin interacts with the Rubicon–UVRAG–Beclin 1 complex and not with the complex containing Atg14L. The UVRAG–Beclin 1 and Atg14L–Beclin 1 complexes are known to be mutually exclusive (Itakura et al., 2008; Sun et al., 2008): the first promotes both the formation and the maturation of autophagosomes (Liang et al., 2006, 2008), whereas the latter regulates PI3P synthesis, the formation of the isolation membranes and the generation of autophagosomes. Binding of Rubicon to the UVRAG–Beclin 1 complex negatively regulates autophagosome maturation (Zhong et al., 2009). The interaction of spastizin with both UVRAG and Rubicon indicates that the protein is a component of the Rubicon-UVRAG-Beclin 1 complex, although we cannot exclude an interaction of spastizin with the complex in the absence of Rubicon. This represents the first evidence that spastizin participates to large protein complexes actively involved in autophagy.
Further, the immunoprecipation experiments performed with the anti-spastizin antibody in the spastizin missense mutant cells L243P and I508N showed a strong reduction (by >80%) of Beclin 1 protein and the consequent lack of Vps34, Rubicon and UVRAG proteins. The data about the binding of Beclin 1 by the mutant L243P and I508N forms of spastizin indicate also that the N-terminus region of spastizin, containing these mutations, likely cooperates with the C-terminus in binding Beclin 1. This extends the minimal region of spastizin involved in Beclin 1 interaction previously indicated to encompass mainly the C-terminus (Sagona et al., 2011).
An important issue is how spastizin interacts with the specific components of the autophagy machinery, i.e. Rubicon and UVRAG. The complexity of this issue is related to the fact that it is not yet clear how Beclin 1 interacts with the various components of its larger complexes. Indeed, both Rubicon and UVRAG have been identified as Beclin 1 interacting proteins (Liang et al., 2006; Sun et al., 2008; Matsunaga et al., 2009) and Beclin 1 and UVRAG have been demonstrated to form a stable heterodimer through their coiled-coiled domains (Li et al., 2012a, b). However, these data have been questioned by a study showing that only Atg14L, but not Rubicon, Vps34 or UVRAG, interacts directly with Beclin 1 (Sun et al., 2011). Our results indicate that mutated forms of spastizin unable to interact with Beclin 1, cannot immunoprecipitate Rubicon and UVRAG. This suggests that the interaction of spastizin with Rubicon/UVRAG is mediated by Beclin 1.
An intriguing issue is how spastizin recognizes and interacts with Beclin 1 in the Rubicon–UVRAG complex only, whereas the Atg14L-Beclin 1 complex is not recognized. We demonstrated here that spastizin, like UVRAG and Rubicon (Itakura et al., 2008), is present on the endosomes and does not co-localize with autophagosomes or with the isolation membranes, as instead observed for Atg14L (Itakura et al., 2008). This suggests that spastizin distribution is spatially restricted and confined to structures involved in autophagosomes maturation, rather than in the early stage of autophagocytosis, such as autophagosomes formation. This may explain the absence of interaction with the Atg14L–Beclin 1 complex in spite of the ability of spastizin to bind Beclin 1.
Spastizin pathogenic mutations increased the number of autophagosomes in mutated patients’ fibroblasts both in basal and starved conditions. Consistently, the number of autophagosomes was increased also after depletion of spastizin by silencing. The increased number of autophagosomes, however, did not reflect in an enhanced autophagic demise as the cellular expression levels of p62 (a classical cargo of these vesicles, targeting poly-ubiquinated proteins to the autophagosomes) were increased rather than decreased. In addition we did not find differences in Beclin 1 expression levels and in mTor phosphorylation levels, which are known to activate and inhibit autophagy, respectively (Noda and Ohsumi, 1998). Atg7 levels were also normal, thereby excluding alterations in the production of LC3-II (Abounit et al., 2012). Since no differences in LC3-II levels were observed between control and spastizin-mutated cells after treatment with bafilomycin A1 (which inhibits autophagosome-lysosome fusion and induces autophagosomes accumulation) in autophagy-inducing conditions, a role of spastizin in autophagosome formation was excluded. No differences were also detected in LAMP1 expression levels; however, the reduced co-localization between the lysosomal marker LAMP1 and the autophagosomal marker LC3, observed in spastizin mutated or silenced cells, strongly suggests that the role of this protein is to mediate autophagosome maturation. Spastizin depletion or its mutations reduced the fusion rate of autophagosomes with lysosomes, thus impairing autophagosome maturation. Indeed, autophagosome maturation is not completely blocked because inhibition of autophagosome-lysosome fusion with bafilomycin A1 further increased LC3-II levels in spastizin mutated cells.
Analysis of fibroblast cells at ultrastructural level confirmed the abnormal accumulation of autophagosomes in spastizin mutated cells compared to the control. Interestingly, most of such autophagic vacuoles in patients’ cells contained double membrane and exhibited remnants of cytosol and membrane organelles in their lumen suggesting the presence of defective maturation (Boland and Nixon, 2006). Taken together, these data clearly indicate that spastizin is required to generate mature autophagosomes, a finding in line with the subcellular localization and protein–protein interactions data.
To understand how spastizin acts in this process we need to consider the critical steps of autophagosome maturation when the nascent autophagosomes fuse with several types of vesicles from the endosomal/lysosomal pathway including late endosomes and lysososmes to create a fully functional degradative compartment, the autolysosome. In mammalian cells this is not a single-step process, but occurs by sequential fusions with different endosomal populations (Eskelinen, 2005). Although the molecular machinery governing these steps is not fully elucidated yet, several players were already identified including UVRAG, Rubicon, (Liang et al., 2008), VCP (Tresse et al., 2010) and the SNARE (SNAP receptor) complex (Fraldi et al., 2010; Renna et al., 2011) in addition to the late-endosomal marker RAB7 and the lysosomal protein LAMP2 (Jäger et al., 2004; Kimura et al., 2007). Two of the main regulators of these steps are UVRAG and Rubicon that we found to co-immunoprecipitate with spastizin. UVRAG promotes autophagosome maturation by promoting autophagosome fusion with endosomes (Liang et al., 2008). Suppression of UVRAG expression inhibits autophagosome maturation and endocytic trafficking (Liang et al., 2008). Rubicon negatively regulates autophagy, by inhibiting autophagosome maturation and endocytic trafficking.
Spastizin contains a PI3P-binding FYVE domain; therefore a possible function of spastizin in autophagy could be to recruit Beclin 1 and its binding partners UVRAG and Rubicon on structures enriched in PI3P that are involved in autophagosome maturation, at the fusion step between autophagosomes and endosomes. This is reminiscent of what had been already demonstrated in cytokinesis, where spastizin recruits Beclin 1 and the binding partners Vps34 and UVRAG (Thoresen et al., 2010) to the mid-body, to PI3P enriched membranes (Sagona et al., 2010, 2011) thereby allowing cytokinesis.
This putative function of spastizin in autophagosome maturation is supported by the fact that either spastizin mutations or protein silencing did not alter the interaction between Beclin 1 and the components of the complexes. Moreover, Rubicon-associated structures are enriched in PI3P (Zhong et al., 2009). Spastizin could finally collaborate with Rubicon in sequestering UVRAG in the Rubicon–UVRAG–Beclin 1 complex; mutations or depletion of the protein could perturb the equilibrium between the two complexes involved in maturation, thereby affecting this process.
The relevance of the data we present here is linked to two aspects of the spastic paraplegia type 15 subtype of paraparesis. Firstly, the majority of ZFYVE26 mutations are of loss of function type (33 of 38 mutations known are nonsense and frame-shifts, excluding variants of unknown significance), therefore the loss of spastizin activity demonstrated for the two truncating mutations represents the most likely and frequent condition associated with spastic paraplegia type 15. The data about the two missense mutations analysed here, further support this observation. Secondly, the effect of spastizin depletion on autophagy observed in patients’ cells, likely occurs also in neuronal cells, such as hippocampal neurons and SHSY5Y neuroblastoma cell line, thus supporting the hypothesis of defective autophagy in the neurodegenerative process underlying the spastic paraplegia type 15 disease. The correlation of these data with the timing and progression of the degenerative processes of the disease however still needs to be directly investigated in in vivo systems.
Summarizing the data herein reported about spastizin, together with the observations in TECPR2 mutated fibroblast (Oz-Levi et al., 2012), strengthens the role of autophagy in complicated hereditary spastic paraplegias and add complexity to the multifaceted biochemical mechanisms implicated in these diseases, such as axonal transport, mitochondrial processes, endoplasmic reticulum shaping, microtubule organization and stability, endosomal trafficking (Blackstone et al., 2011), sterol (Tsaousidou et al., 2008), ceramid (Hammer et al., 2013; Martin et al., 2013) and lipid metabolism (Schuurs-Hoeijmakers et al., 2012; Tesson et al., 2012).
The relevance of these findings has to be considered also in view of the possible therapeutic implications. Indeed, pharmacological approaches to upregulate or inhibit autophagy are currently receiving considerable attention and therapeutic benefits were proved in certain neurodegenerative diseases such as Huntington’s disease (Rubinsztein et al., 2012) or a human tauopathy (Schaeffer et al., 2012). This issue is of particular interest in hereditary spastic paraplegias because no cure is available besides symptomatic treatment in spite of the multiplicity of pathogenic mechanisms identified so far. The identification of autophagy defects in forms of recessive complicated spastic paraparesis indicates that autophagy might become a potential target for pharmacological intervention also in this group of disease.
Supplementary Material
Acknowledgements
The authors wish to thank the patients and their families for participation to the study, E. Tenderini and A. Tonelli for cell cultures, G. Orso and A. Fraldi for critical reading of the manuscript. Confocal images acquisition was performed in ALEMBIC, an advanced microscopy laboratory at the San Raffaele Scientific Institute.
Glossary
Abbreviations
- ARHSP-TCC
autosomal recessive hereditary spastic paraplegia with thin corpus callosum
- PI3P
phosphatidylinositol 3-phosphate
- Rubicon
RUN domain Beclin 1-interacting and cystein-rich containing protein
Funding
We acknowledge the support of the Italian Ministry of Health [grant no RC2011-2013, RF 2007-75 to M.T.B., N.B.], [grant no. GR-2007-690661 to M.T.B.] and [grant no. 5XMille to M.T.B.], the Italian Telethon Foundation [grant no. TGM11CB6 to A.B.], the European Research Council Advanced Investigator [grant no. 250154 to A.B.], March of Dimes grant no. 6-FY11-306 to A.B.], US National Institutes of Health [grant no. R01-NS078072 to A.B.], Associazione Italiana Ricerca sul Cancro [grant no. AIRC IG11362 to E.C.].
Supplementary material
Supplementary material is available at Brain online.
References
- Abounit K, Scarabelli TM, McCauley RB. Autophagy in mammalian cells. World J Biol Chem. 2012;3:1–6. doi: 10.4331/wjbc.v3.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Beal MF, Thomas B. Autophagy in neurodegenerative disorders: pathogenic roles and therapeutic implications [review] Trends Neurosci. 2010;33:541–9. doi: 10.1016/j.tins.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway [review] Nat Rev Neurosci. 2011;12:31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–19. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Boukhris A, Stevanin G, Feki I, Denora P, Elleuch N, Miladi MI, et al. Tunisian hereditary spastic paraplegias: clinical variability supported by genetic heterogeneity. Clin Genet. 2009;75:527–36. doi: 10.1111/j.1399-0004.2009.01176.x. [DOI] [PubMed] [Google Scholar]
- Cox LE, Ferraiuolo L, Goodall EF, Heath PR, Higginbottom A, Mortiboys H, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) PLoS One. 2010;5:e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Fedirko E, Faucheux JM, Forlani S, Bricka B, Goizet C, et al. A de novo SPAST mutation leading to somatic mosaicism is associated with a later age at onset in HSP. Neurogenetics. 2007;8:231–3. doi: 10.1007/s10048-007-0090-4. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL. Maturation of the autophagic vacuoles in mammalian cells [review] Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- Fink JK. Hereditary spastic paraplegia [review] Curr Neurol Neurosci Rep. 2006;6:65–76. doi: 10.1007/s11910-996-0011-1. [DOI] [PubMed] [Google Scholar]
- Finsterer J, Löscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance [review] J Neurol Sci. 2012;318:1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Fraldi A, Annunziata F, Lombardi A, Kaiser H-J, Medina DL, Spampanato C, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–20. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fukuda T, Ewan L, Bauer M, Mattaliano RJ, Zaal K, Ralston E, et al. Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann Neurol. 2006;59:700–8. doi: 10.1002/ana.20807. [DOI] [PubMed] [Google Scholar]
- Goizet C, Boukhris A, Maltete D, Guyant-Maréchal L, Truchetto J, Mundwiller E, et al. SPG15 is the second most common cause of hereditary spastic paraplegia with thin corpus callosum. Neurology. 2009;73:1111–9. doi: 10.1212/WNL.0b013e3181bacf59. [DOI] [PubMed] [Google Scholar]
- Hammer MB, Eleuch-Fayache G, Schottlaender LV, Nehdi H, Gibbs JR, Arepalli SK, et al. Mutations in GBA2 cause autosomal-recessive cerebellar ataxia with spasticity. Am J Hum Genet. 2013;92:245–51. doi: 10.1016/j.ajhg.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein S, Martin E, Boukhris A, Byrne P, Goizet C, Hamri A, et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am J Hum Genet. 2008;82:992–1002. doi: 10.1016/j.ajhg.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;21:1151–4. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh F, Divecha N, Brocks L, Oomen L, Janssen H, Calafat J, et al. The FYVE domain in Smad anchor for receptor activation (SARA) is sufficient for localization of SARA in early endosomes and regulates TGF-beta/ Smad signalling. Genes Cells. 2002;7:321–31. doi: 10.1046/j.1365-2443.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117(Pt 20):4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis [review] Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease [review] Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation [review] Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–3. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012a;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, He L, Zhang M, Yue Z, Zhao Y. The BECN1 coiled coil domain: an “imperfect” homodimer interface that facilitates ATG14 and UVRAG binding. Autophagy. 2012b;8:1258–60. doi: 10.4161/auto.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah LY, Ryan KM. Autophagy and cancer [review] Cold Spring Harb Perspect Biol. 2012;4:a008821. doi: 10.1101/cshperspect.a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel MA, Wyllie DJ, Hardingham GE. In developing hippocampal neurons, NR2B-containing N-methyl-D-aspartate receptors (NMDARs) can mediate signaling to neuronal survival and synaptic potentiation, as well as neuronal death. Neuroscience. 2009;158:334–43. doi: 10.1016/j.neuroscience.2008.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Yanicostas C, Rastetter A, Naini SM, Maouedj A, Kabashi E, et al. Spatacsin and spastizin act in the same pathway required for proper spinal motor neuron axon outgrowth in zebrafish. Neurobiol Dis. 2012;48:299–308. doi: 10.1016/j.nbd.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Martin E, Schüle R, Smets K, Rastetter A, Boukhris A, Loureiro JL, et al. Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia. Am J Hum Genet. 2013;92:238–44. doi: 10.1016/j.ajhg.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Curr Opin Cell Biol. 2010;22:206–11. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–17. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- Murmu RP, Martin E, Rastetter A, Esteves T, Muriel MP, El Hachimi KH, et al. Cellular distribution and subcellular localization of spatacsin and spastizin, two proteins involved in hereditary spastic paraplegia. Mol Cell Neurosci. 2011;47:191–202. doi: 10.1016/j.mcn.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Oz-Levi D, Ben-Zeev B, Ruzzo EK, Hitomi Y, Gelman A, Pelak K, et al. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am J Hum Genet. 2012;91:1065–72. doi: 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz-Levi D, Gelman A, Elazar Z, Lancet D. TECPR2: a new autophagy link for neurodegeneration. Autophagy. 2013;9:801–2. doi: 10.4161/auto.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism [review] Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–6. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, et al. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J Cell Sci. 2011;124(Pt 3):469–82. doi: 10.1242/jcs.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Cologno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases [review] Nat Rev Drug Discov. 2012;11:3709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestøl K, Poulton J, Rusten TE, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–71. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Bache KG, Haglund K, Bakken AC, Skotheim RI, et al. A tumor-associated mutation of FYVE-CENT prevents its interaction with Beclin 1 and interferes with cytokinesis. PLoS One. 2011;6:e17086. doi: 10.1371/journal.pone.0017086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer V, Lavenir I, Ozcelik S, Tolnay M, Winkler DT, Goedert M. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain. 2012;135(Pt 7):2169–77. doi: 10.1093/brain/aws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicks J, Synofzik M, Pétursson H, Huttenlocher J, Reimold M, Schöls L, et al. Atypical juvenile parkinsonism in a consanguineous SPG15 family. Mov Disord. 2011;26:564–6. doi: 10.1002/mds.23472. [DOI] [PubMed] [Google Scholar]
- Schüle R, Schlipf N, Synofzik M, Klebe S, Klimpe S, Hehr U, et al. Frequency and phenotype of SPG11 and SPG15 in complicated hereditary spastic paraplegia. J Neurol Neurosurg Psychiatry. 2009;80:1402–4. doi: 10.1136/jnnp.2008.167528. [DOI] [PubMed] [Google Scholar]
- Schüle R, Schöls L. Genetics of hereditary spastic paraplegias [review] Semin Neurol. 2011;31:484–93. doi: 10.1055/s-0031-1299787. [DOI] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers JH, Geraghty MT, Kamsteeg EJ, Ben-Salem S, de Bot ST, Nijhof B, et al. Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia. Am J Hum Genet. 2012;91:1073–81. doi: 10.1016/j.ajhg.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–29. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Driscoll PC. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513:77–84. doi: 10.1016/s0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, et al. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet. 2007;39:366–72. doi: 10.1038/ng1980. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–6. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–91. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson C, Nawara M, Salih MA, Rossignol R, Zaki MS, Al Balwi M, et al. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet. 2012;91:1051–64. doi: 10.1016/j.ajhg.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen SB, Pedersen NM, Liestøl K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010;316:3368–78. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–27. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaousidou MK, Ouahchi K, Warner TT, Yang Y, Simpson MA, Laing NG, et al. Sequence alterations within CYP7B1 implicate defective cholesterol homeostasis in motor-neuron degeneration. Am J Hum Genet. 2008;82:510–5. doi: 10.1016/j.ajhg.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantaggiato C, Redaelli F, Falcone S, Perrotta C, Tonelli A, Bondioni S, et al. A novel CLN8 mutation in late-infantile-onset neuronal ceroid lipofuscinosis (LINCL) reveals aspects of CLN8 neurobiological function. Hum Mutat. 2009;30:1104–16. doi: 10.1002/humu.21012. [DOI] [PubMed] [Google Scholar]
- Vantaggiato C, Bondioni S, Airoldi G, Bozzato A, Borsani G, Rugarli EI, et al. Senataxin modulates neurite growth through fibroblast growth factor 8 signalling. Brain. 2011;134(Pt 6):1808–28. doi: 10.1093/brain/awr084. [DOI] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, et al. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–58. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S, Patterson V, Hutchinson M. Two families with autosomal recessive spastic paraplegia, pigmented maculopathy, and dementia. J Neurol Neurosurg Psychiatry. 1997;63:628–32. doi: 10.1136/jnnp.63.5.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M, et al. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8:6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- Wong ASL, Cheung ZH, Ip NY. Molecular Machinery of macroautophagy and its deregulation in diseases [review] Biochim Biophysica Acta. 2011;1812:1490–7. doi: 10.1016/j.bbadis.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases [review] Nat Neurosci. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem. 2009;284:2363–73. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.