Abstract
The central nervous system has a pattern of gene expression that is closely regulated with respect to functional and anatomical regions. DNA methylation is a major regulator of transcriptional activity, and aberrations in the distribution of this epigenetic mark may be involved in many neurological disorders, such as Alzheimer’s disease. Herein, we have analysed 12 distinct mouse brain regions according to their CpG 5’-end gene methylation patterns and observed their unique epigenetic landscapes. The DNA methylomes obtained from the cerebral cortex were used to identify aberrant DNA methylation changes that occurred in two mouse models of Alzheimer’s disease. We were able to translate these findings to patients with Alzheimer’s disease, identifying DNA methylation-associated silencing of three targets genes: thromboxane A2 receptor (TBXA2R), sorbin and SH3 domain containing 3 (SORBS3) and spectrin beta 4 (SPTBN4). These hypermethylation targets indicate that the cyclic AMP response element-binding protein (CREB) activation pathway and the axon initial segment could contribute to the disease.
Keywords: DNA methylation, Alzheimer’s disease, brain regions, epigenetics
Introduction
DNA methylation has a critical role in the regulation of gene expression in tissues and organs (Cedar and Bergman, 2012) and its deregulated patterns are increasingly associated with human diseases (Feinberg, 2007; Heyn and Esteller, 2012), among which feature a range of neurological disorders (Urdinguio et al., 2009; Jakovcevski and Akbarian, 2012). In this latter context, epigenetic marks, such as CpG methylation, may be essential because the CNS is a highly specialized structure that requires fine-tuning for gene expression. This is exemplified by the observations that it expresses more alternatively spliced transcripts (Yeo et al., 2004) and microRNAs (Cao et al., 2006) than any other tissue, and that three out of four genes are active (Johnson et al., 2009). The role of DNA methylation in the function of the CNS has been highlighted by its involvement in memory formation (Miller and Sweatt, 2007; Feng et al., 2010; Guo et al., 2011) and ageing-related cognitive decline (Oliveira et al., 2012). Consequently, increasing interest is now focused on the impact of aberrant DNA methylation patterns in the origin and progression of Alzheimer’s disease. This, the most common type of dementia, is characterized by insidious degeneration of brain networks related to memory and cognition. However, few genes are known to be deregulated by DNA methylation in case-control studies of Alzheimer’s disease (Siegmund et al., 2009; Bakulski et al., 2012; Rao et al., 2012). The explanation for this paucity of results might be associated with the existence of many distinctly functional subregions of the brain that may have different DNA methylation patterns that mask the analyses of the data, a matter that has only recently been addressed, and even then, only for large brain regions (Ladd-Accosta et al., 2007; Xin et al., 2010; Hernandez et al., 2011; Lee et al., 2011; Davies et al., 2012). Most importantly, even if close to 80% of human genes are expressed in the brain, most of them are expressed in a relatively small percentage of cells (i.e. 70.5% of genes are transcribed in <20% of all cells) (Lein et al., 2007), an important handicap to discover epigenetically altered genes in specific cells because their DNA methylation patterns could be diluted amongst the majority of cells with different epigenomic patterns. We have addressed this challenging issue by carefully microdissecting the 12 most relevant brain regions of the mouse and hybridizing the corresponding samples to a DNA methylation microarray. The observed DNA methylomes were used to identify aberrant DNA methylation changes that occurred in two mouse models of Alzheimer’s disease and to translate these findings to patients with Alzheimer’s disease.
Materials and methods
Mice samples
Brain region profiles were derived from C57BL/6J mice obtained from the Charles River laboratories. Twelve P60 samples (three female and three male samples for the array hybridization and three female and three male samples for the pyrosequencing validation) were used. Mice were killed by cervical dislocation, and their brains were dissected out within the first 2 min post-mortem and kept at −80°C. Brain areas were dissected from cryostat brain sections (−20°C) with the help of a ×5 magnifying lens, following their stereotaxic coordinates (Paxinos and Franklin, 2001), as previously described (Roux et al., 2003; Panayotis et al., 2011a, b). For validations of epigenetically deregulated genes in Alzheimer’s disease, frontal cortex samples from two transgenic models were used, namely APP/PSEN1 (Borchelt et al., 1997) and 3xTg-AD (Oddo et al., 2003) mice. Five 12-month-old AβPP/PS1 and five wild-type littermates, and three 18-month-old 3TG and three 3TG control mice, were assayed for DNA methylation using the mouse brain genome-wide promoter DNA methylation array. Additionally, independent sets of 12-month-old AβPP/PS1 and five wild-type littermates, and three 18-month-old 3TG and three 3TG control mice were used for pyrosequencing validations and RNA assays. Four APP/PS1 and four wild-type animals were deeply anaesthetized by intraperitoneal injection (0.2 ml/10 g body weight) with a mixture of ketamine (100 mg/kg) and xylazine (20 mg/kg) before intra-cardiac perfusion with 4% paraformaldehyde in PBS. Brains were removed, post-fixed in the same fixative for 4 h and processed for immunohistochemistry. Another group of four APP/PS1 and four wild-type mice were sacrificed by decapitation and their brains quickly removed, dissected on ice and immediately frozen and stored at −80°C until processing for the immunoblot analysis. All animals were maintained under standard animal house conditions in a 12-h dark-light cycle with free access to food and water. The experimental procedures complied with the European guidelines for the care and use of laboratory animals (EU directive 2010/63/EU) and were approved by the local ethics committee (UB-IDIBELL).
Human samples
Post-mortem tissues were obtained from the IDIBELL Biobank, which is part of the eBrainNet Europe Bank (http://www.brainnet-europe.org/) ‘Network of Excellence’ funded by the European Commission in the 6th Framework Program ‘Life Science’ (LSHM-CT-2004-503039); the CIEN Tissue Bank, CIEN Foundation, Instituto de Salud Carlos III; and the University of California Irvine Alzheimer Disease Research Centre, UCI Institute for Memory Impairments and Neurological Disorders. The collection of all samples conformed to the relevant regulations, ethical considerations and legislation as defined by the European Union and Spain. Samples were dissected and characterized for Braak stage before further examination. DNA and RNA from grey matter samples of frontal cortex were extracted for pyrosequencing and RNA assay validations. Only samples with RIN>6.5 according to the RNA quality test developed using the Agilent 2100 bioanalyzer were included in the study. These filtered samples were DNA and RNA from grey matter of frontal cortex (Brodmann area 9) of 20 controls [65% female; age 71 ± 3 years, post-mortem interval delay (PMI) 9 ± 1 h, mean ± SEM] and 20 Alzheimer’s disease Braak stage V-VI (70% female; age 82 ± 2 years, PMI 8 ± 1 h, mean ± SEM) samples matched for age and gender. Supplementary Table 1 describes in detail the available clinicopathological information for all cases and controls (Braak staging, PMI, age and gender). The PMIs were similar for both groups.
Bisulphite conversion of DNA
The Repli-g whole genome-amplification kit (Qiagen) was used to generate non-methylated DNA following the manufacturer’s protocol. For methylated DNA, part of the resulting DNA was in vitro methylated using SssI enzyme (New England Biolabs). The EZ DNA methylation kit (Zymo Research) was used for bisulphite conversion of all DNA samples used, according to Bibikova et al. (2009).
Mouse brain genome-wide promoter DNA methylation array
The mouse brain genome-wide promoter DNA methylation array is a custom designed Illumina’s VeraCode GoldenGate DNA methylation assay that was specifically enriched in genes related with sensory perception, cognition, neuroplasticity, brain physiology and mental diseases in order to provide a useful tool for subsequent neurological studies. From the starting list of 762 genes we downloaded the transcription start coordinates from http://genome.ucsc.edu using 2007 July mm9 assembly and UCSC Genes track. From genes with more than one transcription starting site we selected the first at the 5' end of the gene. We took 750 bp upstream (5') and 250 bp downstream (3') relative to the marked transcription site to design the specific probe for the array, which was subsequently filtered for single mapping on its specific chromosome position. The 384 genes with the highest scores were included in the final design of the array (Supplementary Table 2). Among the 384 amplicons, the average of interrogated CpGs was 2.7 (Supplementary Table 2).
Data analysis
All data were analysed using the open-source statistical programming language R (version 2.13.1). Quantile normalization of the array was carried out using the lumi package and posterior analysis was done with the genefilter package (Bioconductor). For general analysis, all probes with detection values of P < 0.01 in >15% of the samples (25 probes) and probes located on chromosome X (17 probes) were excluded. Poor-quality samples (>10% failed probes) were also excluded. For unsupervised clustering analysis, probes with a general standard deviation <0.05 were excluded as non-informative. Subsequently, heatmaps were analysed using the gplots package. Spectral map analysis was carried out using the mpm package (Wouters et al., 2003). Statistical analysis was performed using the lumi package (Mann-Whitney and Kruskal-Wallis tests for two- and multiple-group comparisons, respectively, adjusting P-values with the FDR algorithm).
Pyrosequencing
The set of primers for PCR amplification and sequencing (Supplementary Table 3) were designed using the PyroMark assay design program, version 2.0.01.15 (Qiagen); amplification primers hybridize with CpG-free sites to ensure a methylation-independent reaction and one primer (opposite the sequencing primer) is biotinylated to convert the PCR product to single-stranded DNA templates. We used 1 µl of bisulphite-treated DNA for each PCR. In order to prepare single-stranded PCR products, we used the Vacuum Prep Tool (Qiagen), following the manufacturer’s instructions. Pyrosequencing reactions and methylation quantification were performed in a PyroMark Q24 System version 2.0.6 (Qiagen). All pyrosequencing assays were run in triplicate. These technical replicates yielded standard errors of means <1%.
Real-time polymerase chain reaction
Total RNA purification and DNase treatment were performed using TRIzol (Invitrogen) and the Turbo DNA-Free kit (Ambion). RNA was reverse-transcribed using the Thermoscript RT-PCR system (Invitrogen) and each PCR was carried out in triplicate using SYBR Green PCR MasterMaster Mix (Applied Biosystems). Thermocycling conditions were 10 min at 95°C, then 50 cycles of 15 s at 95°C and 1 min at 60°C. Fluorescent signals were acquired by the ABI Prism 7900HT Sequence Detection System (Applied Biosystems), and positive standard deviations were normalized using three housekeeping genes (GUSB, RPL38 and TBP). The primers for real-time PCR are listed in Supplementary Table 4. PCR efficiencies were calculated using standard dilutions and the LinReg software (Ruijter et al., 2009).
Immunofluorescence and confocal microscopy
Tissue samples were embedded in paraffin and 4 -µm coronal sections were cut with a microtome. De-waxed sections were stained with a saturated solution of Sudan Black B (Merck) for 30 min to block the autofluorescence of lipofuscin granules present in cell bodies, then rinsed in 70% ethanol and washed in distilled water. The sections were treated with citrate buffer to enhance antigenicity, and then incubated at 4°C overnight with primary antibodies against Tbxa2r (1:200, rabbit; Acris), F2rl2 (1:50, rabbit; Santa Cruz), Sptbn4 (1:100, rabbit; Sigma-Aldrich). After washing, the sections were incubated with Alexa488 (1:400, Molecular Probes) fluorescence secondary antibody against the corresponding host species. After counterstaining of nuclei with DRAQ5TM (1:2000, Biostatus), the sections were mounted in Immuno-Fluore Mounting medium (ICN Biomedicals), sealed, and dried overnight. Sections were examined with a Leica TCS-SL confocal microscope. The protein levels were evaluated by densitometric quantification in reference to the GFAP immunostained area in five representative pictures taken from the neocortex of each animal using Adobe® Photoshop® CS4.
Immunoblot analysis
Frozen cortical areas were dounce-homogenized in lysis buffer (50 mM Tris/HCl buffer, pH 7.4 containing 2 mM EDTA, 0.2% Nonidet P-40, 1 mM PMSF, protease and phosphatase inhibitor cocktails, Roche Molecular Systems), incubated for 20 min with agitation at 4°C and centrifuged for 30 min at 16 000 g. The supernatant was recovered and stored at −80°C. Protein content was determined by the BCA method (Thermo Scientific). Equal amounts of protein (20 µg per lane) were separated by SDS-PAGE (10%) and transferred onto nitrocellulose membranes (Amersham, GE Healthcare). Non-specific bindings were blocked by incubation in 5% non-fat milk in Tris-buffered saline (100 mM NaCl, 10 mM Tris, pH 7.4) containing 0.2% Tween (TTBS) for 1 h at room temperature. Afterwards, membranes were incubated overnight at 4°C with the polyclonal mouse anti-SORBS3 (1:1000, Abnova) in TTBS with 3% bovine serum albumin. Protein loading was monitored using a mouse monoclonal antibody against β-actin (1:30 000, Sigma-Aldrich). Membranes were then incubated for 1 h in the appropriate horseradish peroxidase-conjugated secondary antibodies (1:2000, Dako) and immunocomplexes were revealed by an enhanced chemiluminescence reagent (ECL AdvanceTM, Amersham Biosciences). Densitometric quantification was carried out with TotalLab v2.01 software (Pharmacia). Protein bands were normalized to β-actin levels and expressed as a percentage of the control group level.
Results
We first carefully microdissected the 12 most relevant brain regions of the mouse (cerebellar granular and Purkinje layers, frontal cortex, hippocampal regions CA1, CA3 and dentate gyrus, basolateral amygdala, caudatus putamen, substantia nigra, hypothalamus, globus pallidus and thalamus) and hybridized the corresponding DNAs obtained to a custom microarray containing the 5’-end regulatory regions of 384 mouse genes (Supplementary Table 2). The list was specifically enriched in genes related to sensory perception, cognition, neuroplasticity, brain physiology and mental diseases. The CpG probes used were mapped to locations between −750 bp and +250 bp of the corresponding transcription start sites. Among the 384 amplicons, the average of interrogated CpGs was 2.7. To test the reliability of the array we used three different technical replicates randomly distributed on the array. After quantile normalization, we obtained excellent coefficients of correlation (r2 = 0.98, 0.99 and 0.99, Supplementary Fig. 1) confirming the high reliability of the method. Two important control samples were also included: DNA fully methylated in vitro using the SssI enzyme (New England Biolabs), as a positive methylated control for all CpG sites, and a whole genome PCR amplified DNA obtained using the Repli-g whole genome-amplification kit that does not retain methylated CpGs (Qiagen), as a negative methylated control for all CpG sites. The VeraCode assay recognizes the SssI-treated DNA as highly methylated and the whole genome amplified DNA as fully unmethylated (Supplementary Fig. 2A). We also interrogated the linear capacity of the array, mixing different proportions of methylated and unmethylated DNA generating a set of five different ratios (1:0, 2:1, 1:1, 1:2 and 0:1). The use of the above described whole-genome amplified DNA as a negative control and in vitro methylated DNA as positive control DNA obtained a high Pearson’s coefficient of correlation (r2 = 0.97, Supplementary Fig. 2B). We further validated the VeraCode assay as a tool to measure DNA methylation by the existence of inactivation of one X-chromosome in females by DNA methylation, when compared with the unmethylated X-chromosome in males. Herein, we observed that those CpG sites located in the X-chromosome in the VeraCode assay were able to discriminate between female and male samples in brain regions (Supplementary Fig. 2C).
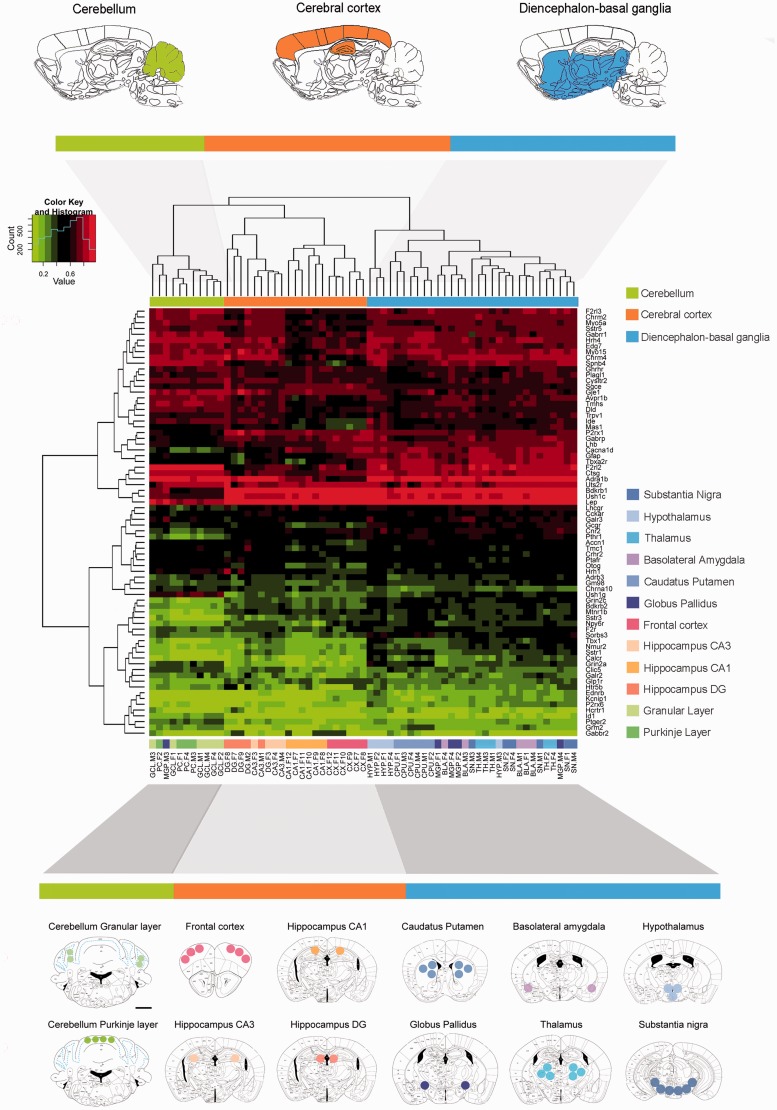
To further validate the array data (obtained using three female and three male samples), we took advantage of the spectral map analysis (Supplementary Fig. 3A) and performed pyrosequencing of the most discordant regions for each of the genes identified using an additional set of biological samples (a distinct group of three female and three male samples). Herein, we obtained similar ratios between both techniques and high overall correlation (r2 = 0.721, Supplementary Fig. 3B and C). In addition, we interrogated the biological relevance of such DNA methylation correlating these data with the RNA expression profiles available in Allen Brain Atlas database (Supplementary Fig. 4). In this regard, we obtained an excellent correlation between both sets of values (Fisher’s exact test P < 0.001) underlying its biological significance. Using this approach, we identified 72 genes (Supplementary Table 5) that were significantly differentially methylated (FDR < 0.05) across the described 12 brain regions of C57BL/6J mice, enabling them to be distinguished by the hierarchical clustering approach (Fig. 1). The observed differences were not associated with cellular heterogeneity at the level of glia versus neuron lineage. When these two cell types were isolated in the mouse brain using A2B5 and PSA-N-CAM antibodies, respectively, followed by magnetic separation (Seidenfaden et al., 2006; Strathmann et al., 2007) no significant differences in DNA methylation were observed for seven studied genes (Spearman’s correlation test, rho = 0.60, P = 0.166; Supplementary Fig. 3D). Interestingly, the cluster analysis identified three main DNA methylation branches corresponding to cerebellum (granular cell layer and Purkinje cells), cerebral cortex (CX, CA1, CA3 and dentate gyrus) and diencephalon-basal ganglia (basolateral amygdala, caudate-putamen, substantia nigra, hypothalamus, globus pallidus and TH). These largely correspond with the major divisions of the brain into hindbrain, forebrain and midbrain (Fig. 1). Thus, consistent with the previously reported RNA expression profiles (Johnson et al., 2009; Lein et al., 2007), our results suggest that major brain divisions are not only structural bisections but also have distinct functional significance. These DNA methylation patterns also reflect the cell type composition of these regions: diencephalon-basal ganglia, which are mainly enriched in GABAergic neurons, and cerebral cortex, which is enriched in glutamate neurons, cluster according to their cell characteristics (Fig. 1). DNA methylation microarray data for 384 genes in the 12 studied mouse brain regions are available to download from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo): GSE47038.
Figure 1.
Unsupervised heatmap clustering of a genome-wide promoter DNA methylation microarray across 12 brain regions in the C57BL/6 mouse. A schematic representation of studied regions was performed on plates adapted from the mouse brain in a stereotaxic coordinates atlas. The circles and areas mark sites from which brain tissue was dissected. The frontal cortex (distance from the bregma, +2.68), caudate-putamen (+0.98), basolateral amygdala (−0.90 to −0.95), hypothalamus (−0.82), globus pallidus (−1.20), thalamus (−1.34), hippocampus (−1.94, hippocampal field CA3, CA1 and dentate gyrus, DG), midbrain area (substantia nigra + ventral tegmental area, −3.08), and cerebellum (−5.80, granular cell layer, GCL; Purkinje cells, PC) were microdissected. Red and green colours indicate high and low levels of DNA methylation, respectively. Scale bars = 1 mm.
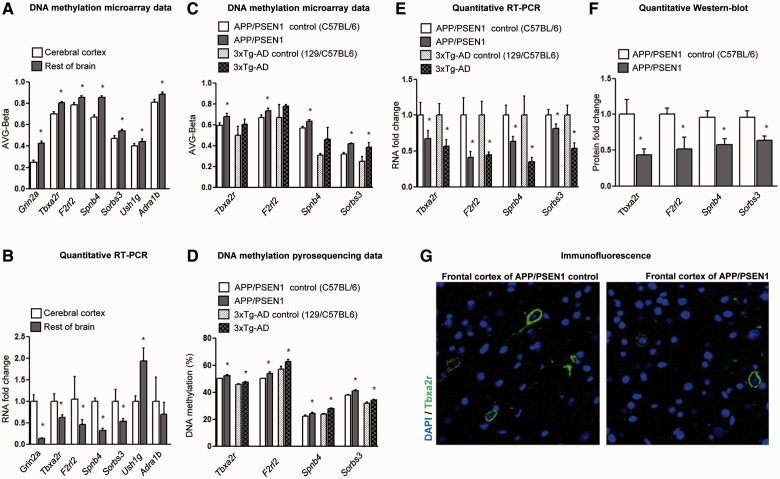
The DNA methylation clustering analyses that recognize cerebral cortex as a distinct branch are relevant to our understanding of Alzheimer’s disease because this is the region most strongly affected in the disorder. Thus, we made a detailed comparison of the DNA methylation patterns in cerebral cortex and the rest of the brain, the latter being regions that are not targeted in Alzheimer’s disease. We identified 52 genes that were differentially methylated between cerebral cortex and the rest of the brain (Supplementary Table 6) (FDR < 0.05) and further investigated the seven genes with the greatest differential methylation (Fig. 2A). For six of these seven (86%) genes, we were able to link the presence of high levels of DNA methylation at their 5’-end regions with downregulation of the corresponding transcripts measured by quantitative real time-PCR (Fig. 2B), highlighting the functional relevance of the observed epigenetic differences. To determine if these genes underwent DNA methylation changes in the disorder, we hybridized the frontal cortex samples from two well-established models of Alzheimer’s disease in mice, APP/PSEN1 and 3xTg-AD (Borchelt et al., 1997; Oddo et al., 2003) to the DNA methylation microarray. Strikingly, we observed that thromboxane A2 receptor (Tbxa2r), coagulation factor II (thrombin) receptor-like 2 (F2rl2), sorbin and SH3 domain containing 3 (Sorbs3) and spectrin beta 4 (Spnb4) were hypermethylated in the frontal cortex of APP/PSEN1 mice observing a similar trend in 3xTg-AD mice (Fig. 2C). DNA methylation microarray data for the 384 genes in the frontal cortex samples from control animals and the two transgenic models APP/PSEN1 and 3xTg-AD are available to download from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo): GSE47036.
Figure 2.
Epigenetic deregulation of target genes in Alzheimer’s disease brain regions in mice. Differential DNA methylation (A) and RNA expression (B) for the top seven candidate genes that discriminate cerebral cortex from the rest of the brain (control C57BL/6 mouse). Differential 5’-end DNA methylation patterns for Tbxa2r, F2rl2, Sorbs3 and Spnb4 according to DNA methylation microarray values (C) and pyrosequencing (D) in the frontal cortex of APP-PSEN1 and 3xTg-AD Alzheimer’s disease mouse models in relation to their corresponding controls. Pyrosequencing values presented correspond to the average CpG methylation across each amplicon. The gain of promoter hypermethylation for Tbxa2r, F2rl2, Sorbs3 and Spnb4 in the frontal cortex of APP-PSEN1 and 3xTg-AD Alzheimer’s disease mouse models is associated with the downregulation of the corresponding RNA transcripts (E) and proteins, measured by western blot (F) and immunofluorescence (G). *P < 0.05 in the Student’s t-test.
To achieve biological replication of the data obtained with the first group of samples, we confirmed the hypermethylation of the four genes identified above using an additional set of samples from the frontal cortex of APP/PSEN1 and 3xTg-AD mice using a different DNA methylation technique, pyrosequencing (Fig. 2D). The presented pyrosequencing values correspond to the average level of CpG methylation across each amplicon and Supplementary Fig. 5 shows methylation values for each CpG within the studied amplicons. In these cases, we also linked the presence of the hypermethylation events in the cortex from the Alzheimer’s mouse models to the downregulation of the corresponding transcripts (Fig. 2E) and proteins in the western blot of F2rl2, Sorbs3 and Spnb4 genes (Fig. 2F and Supplementary Fig. 6) and immunofluorescence of Tbxa2r (Fig. 2F and G).
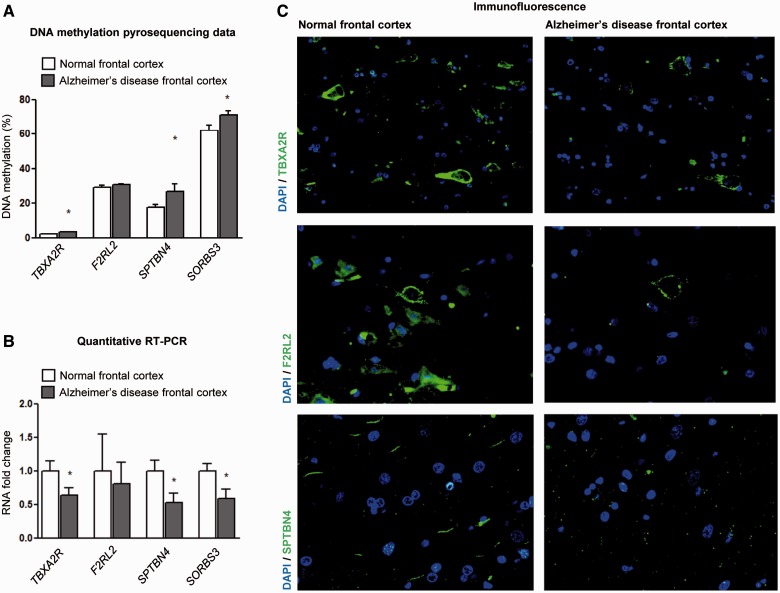
These encouraging results prompted us to investigate human biological samples to address whether the observed epigenetic changes occurred in patients with Alzheimer’s disease. We used pyrosequencing to analyse the DNA methylation status of the four validated genes (from which gain of 5`-end methylation was associated with diminished expression in the Alzheimer’s disease mouse models) in a collection of 20 frontal cortex samples from Alzheimer’s disease Braak stage V-VI cases, comparing them with 20 aged-matched control samples. As in the mouse models, TBXA2R, SORBS3 and SPTBN4 were hypermethylated in the human Alzheimer’s frontal cortex samples (Fig. 3A). The pyrosequencing values presented correspond with the average level of CpG methylation across each amplicon and Supplementary Fig. 7 shows methylation values for each CpG within the studied amplicons. We found an association between the gain in hypermethylation of TBXA2R, SORBS3 and SPTBN4 in the frontal cortex of the patients with Alzheimer’s disease with a reduction of the corresponding RNA transcripts (Fig. 3B) and proteins (Fig. 3C), reinforcing the biological role of the identified epigenetic events. Finally, we found a similar trend for F2RL2 DNA methylation and RNA expression although the great variability of expression among samples precluded a definitive conclusion for this gene.
Figure 3.
Epigenetic deregulation of target genes in the frontal cortex of patients with Alzheimer’s disease. The gain of promoter hypermethylation for the TBXA2R, SORBS3 and SPTBN4 genes in the Alzheimer’s disease frontal cortex determined by pyrosequencing (A) (pyrosequencing values presented correspond to the average CpG methylation across each amplicon) is associated with the downregulation of the corresponding RNA transcripts (B) and proteins, measured by overall immunofluorescence (C). The same trend is observed for F2RL2, but was not statistically significant. *P < 0.05 in the Student’s t-test.
It is worth mentioning that the 5’-ends of the human counterparts (F2RL2, SORBS3, SPTBN4 and TBXA2R) of the mouse genes (F2rl2, Sorbs3, Spnb4 and Tbxa2r) validated as targets of epigenetic disruption in Alzheimer’s disease show structural and functional homologies (Supplementary Fig. 8). In addition to the chromosomic location of the corresponding homologous regions between humans and mice, the 5’-ends of these four genes share, in both species, a high number of identical transcription factor binding sites (Supplementary Fig. 8A). Interestingly, this observation is particularly true for the nucleotide regions that contain the CpG dinucleotides assessed by the DNA methylation microarray and pyrosequencing (Supplementary Fig. 8B). Most importantly, four of the transcription factor binding sites common in both species, and located precisely within the CpGs with differential DNA methylation between cases and controls, show methylation-sensitive binding affinity. These sites were TFAP2A (Kulak et al., 2012), TBP (Bane et al., 2002), GATA-2 (Rui et al., 2013) and GATA-3 (Hutchins et al., 2002) (Supplementary Fig. 8C). In this regard, for promoters without a CpG island, the methylation of particular CpG sites surrounding the transcription start sites can also be linked with transcriptional downregulation (Han et al., 2011; Kelly et al., 2012; Øster et al., 2013). These findings warrant further research in this area.
Discussion
As mentioned previously, ∼80% of total genes are expressed in the brain and most are expressed in a relatively small percentage of cells (Lein et al., 2007). It is an important handicap to elucidate epigenetically deregulated genes in CNS because the characteristic patterns that regulate the expression of concrete genes in specific cells are presumably diluted inside a majority of cells governed by alternative epigenetic patterns. Furthermore, we should consider that neurons show a high variation in DNA methylation profiles according to their unique characteristics (Iwamoto et al., 2011). An additional complication arises from the interindividual variation observed in humans which also contributes to mask such differences. These make the discovery of epigenetic alterations in neurological diseases difficult and, as consequence, few genes have, to date, been reported in widespread non-tumoral disorders such as Alzheimer’s disease (Siegmund et al., 2009; Wang et al., 2008; Bakulski et al., 2012; Rao et al, 2012).
To face this challenge, we focused on genetically homogeneous mouse models because they show a reduced interindividual variation. In spite of the importance of mice models in scientific research, no platforms interrogating mouse DNA methylation have been commercialized and the available alternatives are not sensitive enough to quantify the expected DNA methylation differences in CNS (a comparison of methods is available in Harris et al., 2010). For that, we designed a custom Illumina VeraCode DNA methylation mouse array enriched in genes related to sensory perception, cognition, neuroplasticity, brain physiology and mental diseases. Illumina platforms have been demonstrated to be accurate genome-wide assays, commonly used to study DNA methylation (Bock et al., 2010), and, as consequence, our custom array could be a useful tool to identify DNA methylation differences in mouse brain. However, there is the limitation that only the DNA sequences included in the microarray can be studied and the enrichment of candidate genes selected for their putative role on neurological pathways creates a bias for these types of candidates. Additionally, we have confined our study to the sequences 750 bp upstream and 250 bp downstream of the corresponding transcription start sites and CpGs located in other intragenic or intergenic DNA methylation regions, which could also have an impact on gene activity. Importantly, there are new challenges in the study of DNA methylation in the brain and its disorders that should be considered, such as the presence of hydroxymethylation (Kriaucionis and Heintz, 2009; Münzel et al., 2010; Song et al., 2011; Szulwach et al., 2011; Khare et al., 2012) which, unfortunately, cannot be distinguished by standard bisulphite conversion (Huang et al., 2010) or the recognition that DNA methylation is a dynamic process in brain regions, including those associated with cognition and behaviour (Lubin et al., 2011; Numata et al., 2012). In our study, we accomplished a detailed description of DNA methylation profiles of 12 well-defined mouse brain regions and used this information to highlight the specific characteristics of affected regions in Alzheimer’s disease. Thus, we identified a subset of possible target genes for Alzheimer’s disease that were interrogated in two different Alzheimer’s mouse models and human samples achieving a high correlation between the groups.
The DNA hypermethylation-associated inactivation of the TBXA2R, F2RL2, SORBS3 and SPTBN4 genes discovered here in samples of Alzheimer’s disease could provide additional clues to understand the pathology of the disease. TBXA2R is a member of the family of G protein-coupled receptors that regulates the cAMP response element-binding protein (CREB) (Muja et al., 2001; Obara et al., 2005). In this regard, the CREB activation pathway is critical for neuron activity (Franklin et al., 1992). It has been recently observed that other CREB-related genes are differentially methylated across different cortex regions and cerebellum (Davies et al., 2012) and CREB-signalling alterations occur in Alzheimer’s disease (Saura and Valero, 2011). TBXA2R plays another important role in the pathobiology of Alzheimer’s disease by mediating NMDA excitotoxicity (Mitsumori et al., 2011). NMDA treatment induces apoptosis and the production of thromboxane A2 (TXA2) (Okada et al., 2008). The ligand of TBXA2R establishes a link between NMDA excitotoxicity and TXA2 signalling. Interestingly, amyloid-β treatments also induce TXA2 production and are associated with neurodegeneration, which is, in turn, attenuated by TXA2 antagonists (Yagami et al., 2004). It is also important to mention that F2RL2, identified in the Alzheimer’s mice models but not completely validated in the human samples, is also associated with NMDA excitotoxicity and is necessary for the APC-mediated protection after NMDA treatments (Guo et al., 2004). The second identified target of epigenetic inactivation, SORBS3, is a gene previously reported to be hypermethylated in Alzheimer’s disease (Siegmund et al., 2009). SORBS3, also known as vinexin, is involved in synapsis (Ito et al., 2007) and regulates gene expression (Matsuyama et al., 2005) and further research is necessary to understand its putative role in Alzheimer’s disease.
Finally, the third candidate, SPTBN4, is a prominent member of the axon initial segment and, in combination with the ankyrin G protein, acts as a bridge between the cytoskeleton and voltage-gated channels (Grubb et al., 2010). The correct formation of the axon initial segment is essential for synaptic integration and the firing of the action potential (Palmer and Stuart, 2006; Kress et al., 2010) and its disruption compromises neuronal function, organization and polarity (Hedstrom et al., 2008). Indeed, Spnb4 mutant mice fail to initiate action potentials and show progressive behavioural deficits with altered network excitability (Uemoto et al., 2007; Yang et al., 2007; Winkels et al., 2009). Most importantly for Alzheimer’s disease, the integrity of the axon initial segment is necessary to maintain the axonal localization of tau (Li et al., 2011) and its disruption could result in tau mislocalization, increase of tau presence in the somatodendritic compartment and enhanced exposure to kinases. In this regard, SPTBN4 also anchors CAMKII to the axon initial segment (Hund et al., 2010), and, thus, CAMKII mislocalization upon SPTBN4 epigenetic loss could cause tau hyperphosphorylation, a common finding in Alzheimer’s disease (Xiao et al., 1996). Supporting this hypothesis, RNA interference of the SPTBN4 orthologue in a Drosophila Alzheimer’s disease model also enhances tau toxicity (Shulman et al., 2011). In addition, the mutant mice for ankyrin G (the partner of Spnb4 in axon initial segment) shows neurodegeneration (Zhou et al., 1998), reinforcing the importance of axon initial segment integrity in neuronal viability. Interestingly, the DNA-methylation associated loss of SPTBN4 identified here could also contribute to Alzheimer’s disease by another pathway: the alteration of amyloid precursor protein processing. Alpha and beta-secretases, which are responsible for non-amyloidic and amyloidic processing of amyloid precursor protein, tend to localize in the somatodendritic compartment and plasmatic membrane, respectively (reviewed in Haass et al., 2012). In this context, the non-amyloidic processing of amyloid precursor protein occurs predominantly on the cell surface and could depend on the integrity of the axon initial segment that could be disrupted by SPTBN4 epigenetic silencing. Thus, the observed DNA methylation-associated inactivation of SPTBN4 could relate to both tau hyperphosphorylation and an increase in the amyloidic processing of amyloid precursor protein, the two main hallmarks of Alzheimer’s disease.
Overall, our data suggest that key genes are altered by DNA methylation in Alzheimer’s disease, highlighting the disruption of particular molecular signalling pathways and cellular structures, such as CREB-activation and the axon initial segment.
Funding
This work was supported by the Human Frontiers Science Program (HFSP) Ref.RGP0018/2007-C, the European Community’s Sixth Framework Programme (FP7/2007-2013) under grant agreement PITN-GA-2009-238242—DISCHROM project and grant agreement 278486-DEVELAGE, NIH Grants NIA P50-AG16573 and HD065160-01, the Instituto de Salud Carlos III—Ministerio de Sanidad y Consumo Proyecto FIS (Fondo Investigación Sanitaria, Spain) through the E-RARE EuroRETT network and CureFXS ERare-EU/FIS PS09102673, Grants P50-AG16573, HD065160-01 and SAF2007-6-8, the Fondation Lejeune (France), the Lilly Foundation (Spain), the Lilly Foundation (Spain), Botin Foundation, and the Cellex Foundation (Catalonia). J.V.S-M is affiliated to the Neuroscience PhD program of the Autonomous University of Barcelona, Spain. ME is an ICREA Reseah Professor.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviations
- PMI
post-mortem interval delay
References
- Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP, et al. Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J Alzheimers Dis. 2012;29:571–88. doi: 10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bane TK, LeBlanc JF, Lee TD, Riggs AD. DNA affinity capture and protein profiling by SELDI-TOF mass spectrometry: effect of DNA methylation. Nucleic Acids Res. 2002;30:e69. doi: 10.1093/nar/gnf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, et al. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Müller F, Simmer F, Gu H, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–14. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Ratovitski T, van Lare J, Lee MK, Gonzales V, Jenkins NA, et al. Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron. 1997;19:939–45. doi: 10.1016/s0896-6273(00)80974-5. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–30. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin JL, Johnson EM., Jr Suppression of programmed neuronal death by sustained elevation of cytoplasmic calcium. Trends Neurosci. 1992;15:501–8. doi: 10.1016/0166-2236(92)90103-f. [DOI] [PubMed] [Google Scholar]
- Guo H, Liu D, Gelbard H, Cheng T, Insalaco R, Fernández JA, et al. Activated protein C prevents neuronal apoptosis via protease activated receptors 1 and 3. Neuron. 2004;41:563–72. doi: 10.1016/s0896-6273(04)00019-4. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–51. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb MS, Burrone J. Building and maintaining the axon initial segment. Curr Opin Neurobiol. 2010;20:1–8. doi: 10.1016/j.conb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Cortez CC, Yang X, Nichols PW, Jones PA, Liang G. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum Mol Genet. 2011;20:4299–310. doi: 10.1093/hmg/ddr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom KL, Ogawa Y, Rasband MN. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J Cell Biol. 2008;183:635–40. doi: 10.1083/jcb.200806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–72. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;10:679–92. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 2010. 26 doi: 10.1371/journal.pone.0008888. 5: e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund TJ, Koval OM, Li J, Wright PJ, Qian L, Snyder JS, et al. A β(IV)-spectrin/CaMKII signaling complex is essential for membrane excitability in mice. J Clin Invest. 2010;120:3508–19. doi: 10.1172/JCI43621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- Ito H, Usuda N, Atsuzawa K, Iwamoto I, Sudo K, Katoh-Semba R, et al. Phosphorylation by extracellular signal-regulated kinase of a multidomain adaptor protein, vinexin, at synapses. J Neurochem. 2007;100:545–54. doi: 10.1111/j.1471-4159.2006.04222.x. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–96. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;188:1194–204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress GJ, Dowling MJ, Eisenman LN, Mennerick S. Axonal sodium channel distribution shapes the depolarized action potential threshold of dentate granule neurons. Hippocampus. 2010;20:558–71. doi: 10.1002/hipo.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19:1037–43. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak MV, Cyr AR, Woodfield GW, Bogachek M, Spanheimer PM, Li T, et al. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene. 2012 doi: 10.1038/onc.2012.400. in press. doi: 10.1038/onc.2012.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–15. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Tamashiro KL, Aryee MJ, Murakami P, Seifuddin F, Herb B, et al. Adaptation of the CHARM DNA methylation platform for the rat genome reveals novel brain region-specific differences. Epigenetics. 2011;6:1378–90. doi: 10.4161/epi.6.11.18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li X, Kumar Y, Zempel H, Mandelkow EM, Biernat J, Mandelkow E. Novel diffusion barrier for axonal retention of Tau in neurons and its failure in neurodegeneration. EMBO J. 2011;30:4825–37. doi: 10.1038/emboj.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Gupta S, Parrish RR, Grissom NM, Davis RL. Epigenetic mechanisms: critical contributors to long-term memory formation. Neuroscientist. 2011;17:616–32. doi: 10.1177/1073858411386967. [DOI] [PubMed] [Google Scholar]
- Matsuyama M, Mizusaki H, Shimono A, Mukai T, Okumura K, Abe K, et al. A novel isoform of Vinexin, Vinexin gamma, regulates Sox9 gene expression through activation of MAPK cascade in mouse fetal gonad. Genes Cells. 2005;10:421–34. doi: 10.1111/j.1365-2443.2005.00844.x. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–69. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mitsumori T, Furuyashiki T, Momiyama T, Nishi A, Shuto T, Hayakawa T, et al. Thromboxane receptor activation enhances striatal dopamine release, leading to suppression of GABAergic transmission and enhanced sugar intake. Eur J Neurosci. 2011;34:594–604. doi: 10.1111/j.1460-9568.2011.07774.x. [DOI] [PubMed] [Google Scholar]
- Muja N, Blackman SC, Le Breton GC, DeVries GH. Identification and functional characterization of thromboxane A2 receptors in Schwann cells. J Neurochem. 2001;78:446–56. doi: 10.1046/j.1471-4159.2001.00378.x. [DOI] [PubMed] [Google Scholar]
- Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–7. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–72. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara Y, Kurose H, Nakahata N. Thromboxane A2 promotes interleukin-6 biosynthesis mediated by an activation of cyclic AMP-response element-binding protein in 1321N1 human astrocytoma cells. Mol Pharmacol. 2005;68:670–9. doi: 10.1124/mol.105.012922. [DOI] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, et al. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–21. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- Okada S, Yamaguchi-Shima N, Shimizu T, Arai J, Yorimitsu M, Yokotani K. Centrally administered N-methyl-d-aspartate evokes the adrenal secretion of noradrenaline and adrenaline by brain thromboxane A2-mediated mechanisms in rats. Eur J Pharmacol. 2008;586:145–50. doi: 10.1016/j.ejphar.2008.02.069. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012;15:1111–3. doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- Øster B, Linnet L, Christensen LL, Thorsen K, Ongen H, Dermitzakis ET, et al. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int J Cancer. 2013;132:2303–15. doi: 10.1002/ijc.27921. [DOI] [PubMed] [Google Scholar]
- Palmer LM, Stuart GJ. Site of action potential initiation in layer 5 pyramidal neurons. J Neurosci. 2006;26:1854–63. doi: 10.1523/JNEUROSCI.4812-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotis N, Pratte M, Borges-Correia A, Ghata A, Villard L, Roux JC. Morphological and functional alterations in the substantia nigra pars compacta of the Mecp2-null mouse. Neurobiol Dis. 2011a;41:385–97. doi: 10.1016/j.nbd.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Panayotis N, Ghata A, Villard L, Roux JC. Biogenic amines and their metabolites are differentially affected in the Mecp2-deficient mouse brain. BMC Neurosci. 2011b;12:47. doi: 10.1186/1471-2202-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KB. The mouse brain in stereotaxic coordinates. 2nd edn. San Diego: Academic Press; 2001. [Google Scholar]
- Rao JS, Keleshian VL, Klein S, Rapoport SI. Epigenetic modifications in frontal cortex from Alzheimer's disease and bipolar disorder patients. Transl Psychiatry. 2012;2:e132. doi: 10.1038/tp.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux JC, Mamet J, Perrin D, Peyronnet J, Royer C, Cottet-Emard JM, et al. Neurochemical development of the brainstem catecholaminergic cell groups in rat. J Neural Transm. 2003;110:51–65. doi: 10.1007/s00702-002-0767-7. [DOI] [PubMed] [Google Scholar]
- Rui W, Jin Z, Zhe G, Song H. The methylation of C/EBP β gene promoter and regulated by GATA-2 protein. Mol Biol Rep. 2013;40:797–801. doi: 10.1007/s11033-012-2117-7. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Valero J. The role of CREB signaling in Alzheimer's disease and other cognitive disorders. Rev Neurosci. 2011;22:153–69. doi: 10.1515/RNS.2011.018. [DOI] [PubMed] [Google Scholar]
- Seidenfaden R, Desoeuvre A, Bosio A, Virard I, Cremer H. Glial conversion of SVZ-derived committed neuronal precursors after ectopic grafting into the adult brain. Mol Cell Neurosci. 2006;32:187–98. doi: 10.1016/j.mcn.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Shulman JM, Chipendo P, Chibnik LB, Aubin C, Tran D, Keenan BT, et al. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet. 2011;88:232–8. doi: 10.1016/j.ajhg.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2009;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann FG, Wang X, Mayer-Pröschel M. Identification of two novel glial-restricted cell populations in the embryonic telencephalon arising from unique origins. BMC Dev Biol. 2007;7:33. doi: 10.1186/1471-213X-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–16. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemoto Y, Suzuki S, Terada N, Ohno N, Ohno S, Yamanaka S, et al. Specific role of the truncated betaIV-spectrin Sigma6 in sodium channel clustering at axon initial segments and nodes of ranvier. J Biol Chem. 2007;282:6548–55. doi: 10.1074/jbc.M609223200. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–72. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkels R, Jedlicka P, Weise FK, Schultz C, Deller T, Schwarzacher SW, et al. Reduced excitability in the dentate gyrus network of betaIV-spectrin mutant mice in vivo. Hippocampus. 2009;19:677–86. doi: 10.1002/hipo.20549. [DOI] [PubMed] [Google Scholar]
- Wouters L, Göhlmann HW, Bijnens L, Kass SU, Molenberghs G, Lewi PJ. Graphical exploration of gene expression data: a comparative study of three multivariate methods. Biometrics. 2003;59:1131–9. doi: 10.1111/j.0006-341x.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- Xiao J, Perry G, Troncoso J, Monteiro MJ. alpha-calcium-calmodulin-dependent kinase II is associated with paired helical filaments of Alzheimer's disease. J Neuropathol Exp Neurol. 1996;55:954–63. doi: 10.1097/00005072-199609000-00002. [DOI] [PubMed] [Google Scholar]
- Xin Y, Chanrion B, Liu MM, Galfalvy H, Costa R, Ilievski B, et al. Genome-wide divergence of DNA methylation marks in cerebral and cerebellar cortices. PLoS One. 2010;5:e11357. doi: 10.1371/journal.pone.0011357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagami T, Takahara Y, Ishibashi C, Sakaguchi G, Itoh N, Ueda K, et al. Amyloid beta protein impairs motor function via thromboxane A2 in the rat striatum. Neurobiol Dis. 2004;16:481–9. doi: 10.1016/j.nbd.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ogawa Y, Hedstrom KL, Rasband MN. betaIV spectrin is recruited to axon initial segments and nodes of Ranvier by ankyrinG. J Cell Biol. 2007;176:509–19. doi: 10.1083/jcb.200610128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–304. doi: 10.1083/jcb.143.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.