Abstract
We analyzed a new hypomorphic mouse model containing a targeted intronic insertion of a neomycin cassette within the mechanistic target of rapamycin (mTOR) locus. Mice with two hypomorphic (mTORΔ/Δ) alleles are viable but express mTOR at approximately 25% of wild type levels. These animals demonstrate reduced mTORC1 and mTORC2 activity and exhibit an approximate 20% increase in median survival. While mTORΔ/Δ mice are smaller than wild type mice, these animals do not demonstrate any alterations in normalized food intake, glucose homeostasis or metabolic rate. Consistent with their increased lifespan, mTORΔ/Δ mice exhibited a reduction in a number of aging tissue biomarkers. Functional assessment suggested that as mTORΔ/Δ mice age, they exhibit a marked functional preservation in many but not all organ systems. Thus, in a mammalian model, while reducing mTOR expression markedly increases overall lifespan, it affects the age-dependent decline in tissue and organ function in a segmental fashion.
Inhibiting TOR activity appears to extend lifespan in various model systems including yeast, worms and flies (Bjedov et al., 2010; Kaeberlein et al., 2005; Kapahi et al., 2004; Medvedik et al., 2007; Vellai et al., 2003). Moreover, deletion of the TOR1 gene in yeast results in an increase in replicative lifespan that cannot be further extended by nutrient restriction (Kaeberlein et al., 2005). Evidence also suggests that mTOR plays a role in regulating mammalian lifespan. Treatment of mice beginning at 20 months of age with rapamycin, a pharmacological inhibitor of mTOR, results in an extension of lifespan that averages 9% for males and 13% for females (Harrison et al., 2009). When rapamycin was initiated at 9 months of age, median survival was increased to 10% for males and 18% for females (Miller et al., 2011). Similarly, deletion of ribosomal S6 protein kinase 1 (S6K1), a downstream effector of mTOR, extends the median lifespan of female S6K1−/− mice by approximately 19% (Selman et al., 2009). Very recently, an additional genetic model consisting of mice heterozygous for deletion of both mTOR and mLST8 (mammalian lethal with Sec13 protein 8) also demonstrated lifespan extension, again only evident in female mice (Lamming et al., 2012).
In mammals, mTOR exists in two distinct complexes termed mTORC1 and mTORC2. Each of these mTOR complexes has distinct protein components, although both share the catalytic mTOR subunit as well as mLST8 (Dazert and Hall, 2011; Laplante and Sabatini, 2012; Zoncu et al., 2011). Agents such as rapamycin are known to acutely inhibit mTORC1, although chronic treatment can also affect the activity of mTORC2 (Lamming et al., 2012; Sarbassov et al., 2006). How reducing mTOR activity extends lifespan remains incompletely understood. In addition, whether manipulations of pathways that regulate mammalian lifespan will slow aging and age-related pathologies in a uniform or segmental fashion remains largely unexplored. Here, using a new genetic model of reduced mTOR expression, we provide evidence that reducing mTOR activity produces a marked increase in overall lifespan while also regulating an important, but not universal, subset of tissue-specific, age-dependent parameters.
Results
Reduced mTOR expression increases survival
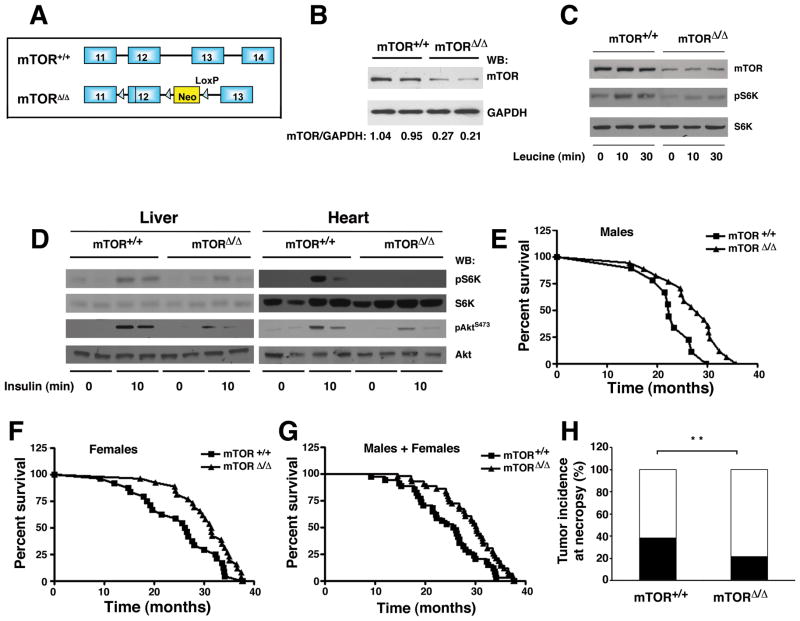
To assess the role of mTOR in mammalian aging we employed a model of hypomorphic mTOR expression that has been recently described (Zhang et al., 2011). This model results from a floxed neomycin cassette inserted between exon 12 and 13 of the mTOR locus that results in the partial disruption of mTOR transcription (Figure 1A). While complete disruption of Raptor, Rictor, mLST8 or mTOR is embryonically lethal (Gangloff et al., 2004; Guertin et al., 2006; Murakami et al., 2004), mTORΔ/Δ mice were viable in a mixed 129/C57BL/6 background. Analysis of tissues derived from mTORΔ/Δ mice revealed that the level of mTOR protein was reduced to approximately 25% of wild type levels (Figure 1B and Figure S1A). Mouse embryonic fibroblasts (MEFs) derived from mTORΔ/Δ mice also exhibited reduced mTOR expression with no apparent alteration in the expression of associated proteins such as Raptor and Rictor (Figure 1C and Figure S1B). When MEFs derived from mTORΔ/Δ mice were analyzed, levels of TORC1 and TORC2 complexes appeared to be similarly reduced (Figure S1B). As expected, mTORΔ/Δ MEFs had reduced activation of S6 kinase following leucine addition (Figure 1C), although the overall level of protein translation was not altered (Figure S1C). We noted that mTORΔ/Δ mice also exhibited a decrease in mTOR signaling in vivo. In particular, the activation of S6 kinase following insulin administration was markedly attenuated in mTORΔ/Δ mice (Figure 1D). Similarly, the mTORC2 dependent serine 473 phosphorylation of Akt was also reduced in these mice.
Figure 1.
A mouse model of reduced mTOR expression extends lifespan. A) Genomic organization of the wild type allele (+) and the hypomorphic mTOR allele (Δ). B) Representative mTOR protein expression in the liver of two wild type (mTOR+/+) and two mTORΔ/Δ mice. GAPDH is used as a loading control and the normalized expression (WT=1) of mTOR to GAPDH is shown for each animal. C) Leucine-stimulated S6 Kinase phosphorylation (pS6K) in primary mouse embryonic fibroblasts isolated from wild type or mTORΔ/Δ mice. D) Insulin-stimulated mTOR activity in pairs of wild type or mTORΔ/Δ mice. E) Survival of a cohort of male WT and mTORΔ/Δ mice. F) Survival of female members of the cohort. G) Survival of the overall cohort. H) Incidence of malignant tumors found at necropsy denoted by shaded portion of each bar. While the overall incidence of cancer was different between the two genotypes, the spectrum of tumors observed was similar. **p<0.001 Fisher’s Exact test.
We next asked whether this reduction in mTOR activity was sufficient to provide an extension in lifespan. Median survival of the mTORΔ/Δ male mice was significantly higher than observed in mTOR+/+ (WT) male mice (Figure 1E; WT median survival 22.9 months (n=10), mTORΔ/Δ 28.0 months (n=17); 22% extension, p=0.02 by Log-rank (Mantel-Cox) test). Similarly, the observed median survival for WT female mice was 26.5 months (n=24), while for female mTORΔ/Δ mice (n=26) median survival was 31.5 months (Figure 1F; 19% extension, p=0.047 by Log-rank test). For the overall combined cohort, median survival was 26.2 months for WT mice and 30.3 months for mTORΔ/Δ mice (n=34 for mTOR+/+ mice and n=43 for mTORΔ/Δ mice, p = 0.0057 by Cox regression using sex and genotype as predictors, Figure 1G). We also assessed whether mTORΔ/Δ mice had an increase in maximal lifespan by using the number of mice in each group still alive after 90% of the pooled distribution of WT and the mTORΔ/Δ mice were dead (Wang et al., 2004). Of the 77 animals in the total cohort, eight animals met this criteria of which 1 was WT and 7 were mTORΔ/Δ mice (p=0.071 by Fisher Exact test and p=0.061 when both genotype and sex were used as predictors). A similar analysis using an 80th percentile cutoff, demonstrated that using this less restrictive threshold, mTORΔ/Δ mice exhibited an increase in maximal lifespan (p=0.005 using genotype and sex as predictors).
Mice involved in the lifespan analysis were not subject to any physiological testing and received no treatment except if they developed a visible superficial infection. In such cases, the facility staff provided a short course of oral, subcutaneous or topical antibiotics with or without ibuprofen (5 out of 34 WT and 10 out of 43 mTORΔ/Δ mice received some treatment). If the infection persisted or worsened, to the point the animal was felt to be in significant pain or functionally impaired, then the animal was euthanized. In some older mice, the initial superficial infection was not discovered until it had progressed to such a degree that the staff believed the condition was too severe to respond to standard treatment, and as such, the animal was euthanized without any prior treatment. We noted that the percentage of mice euthanized because of severe or progressive superficial infections was much higher in the mTORΔ/Δ mice cohort (WT mice = 17 % and mTORΔ/Δ mice = 37 %, p<0.01 Fisher Exact test). In contrast, mTORΔ/Δ mice demonstrated an apparent reduction in the incidence of malignant tumors found at necropsy (Figure 1H and S1D&E; 10 of 26 wild type mice (38.5%) sent to necropsy, versus 8 of 36 (21%) mTORΔ/Δ mice sent to necropsy, **p<0.01 Fisher Exact test). Besides the observed change in rates of malignancies and infections, there were no other marked differences in post-mortem pathologies observed between wild type and mTORΔ/Δ mice.
No alterations in glucose homeostasis or metabolism in the mTORΔ/Δ mice
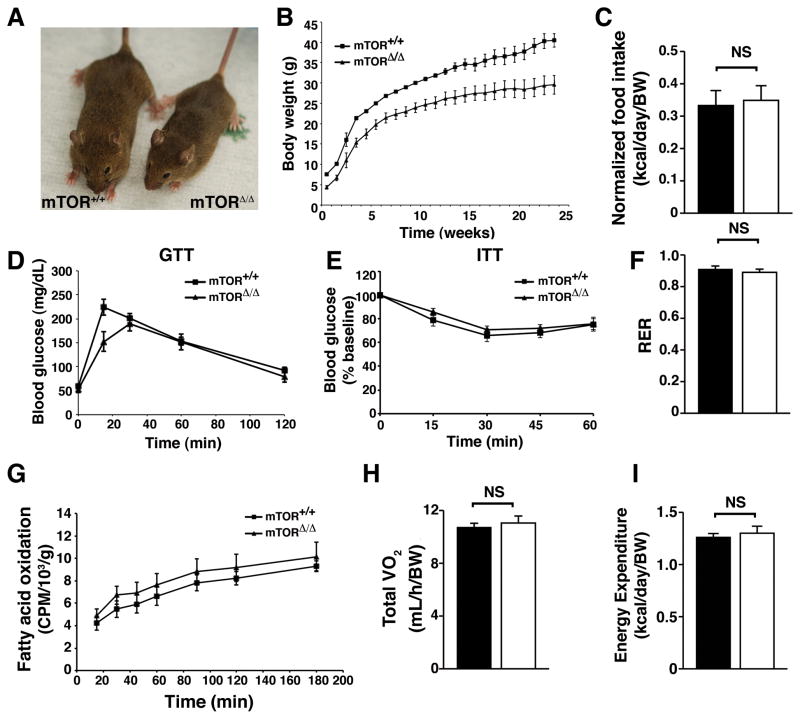
Analysis of body size (Figure 2A) and body weight (Figure 2B) revealed that mTORΔ/Δ mice were consistently smaller than their wild type littermates, although normalized body composition was unchanged (Figure S2A). When food intake was normalized to body weight, mTORΔ/Δ mice and wild type mice consumed equivalent amount of calories (Figure 2C). While rapamycin treatment in mice results in alteration in glucose homeostasis (Cunningham et al., 2007; Lamming et al., 2012), analysis of young mTORΔ/Δ mice revealed no significant alterations in glucose tolerance (Figure 2D) or insulin sensitivity (Figure 2E). Indeed, while a reduction in insulin signaling is associated with increased lifespan (Kenyon, 2011), fasting levels of insulin were slightly higher in the mTORΔ/Δ mice (Figure S2B). This appeared to relate in part to a cell autonomous increase in insulin secretion from pancreatic islets isolated from mTORΔ/Δ mice (Figure S2C&D). Analysis of older wild type and mTORΔ/Δ mice revealed that there was also no marked difference in glucose tolerance as these mice aged (Figure S2E). Serum analysis revealed no significant differences in various lipid parameters (Figure S2F–H). Finally, the respiratory exchange ratio (Figure 2F), in vivo fatty acid oxidation rates (Figure 2G), the overall metabolic rate (Figure 2H) and normalized energy expenditure (Figure 2I) were all unaltered in mTORΔ/Δ mice. Thus, as previously observed, the lifespan extension observed by reducing mTOR expression does not appear to result from a significant alteration in energetic or metabolic parameters (Lamming et al., 2012).
Figure 2.
The mTORΔ/Δ mice are smaller but have no significant alterations in glucose homeostasis and metabolism. All measurements were performed using male mice. A) Representative size of a wild type and mTORΔ/Δ adult mouse. B) Body weight of wild type and mTORΔ/Δ mice (n=7 WT and n=7 mTORΔ/Δ, curves are statistically different using a one-way Anova followed by two tailed t-test, p<0.01). C) Daily food intake is indistinguishable between wild type (shaded bar) and mTORΔ/Δ mice (open bar) (n=7 WT and n=6 mTORΔ/Δ, food intake is normalized to body weight). D) Glucose tolerance of 8–12 week old wild type and mTORΔ/Δ mice (n=7 WT and n=6 mTORΔ/Δ). E) Insulin tolerance test of 8–12 week old wild type and mTORΔ/Δ mice (n=12 WT and n=7 mTORΔ/Δ). F) Respiratory exchange ratio (RER) of WT (n=7) and mTORΔ/Δ (n=5). G) Measurement of rates of total body fatty acid oxidation normalized to body weight in WT (n=7) and mTORΔ/Δ mice (n=5). H) Total oxygen consumption normalized to body weight (n=7 WT and n=5 mTORΔ/Δ). I) Total daily energy expenditure is not altered in mTORΔ/Δ mice (n=7 WT and n=5 mTORΔ/Δ). For all panels, shaded bars represent the wild type mice and the open bars represent the mTORΔ/Δ mice. Where indicated, metabolic parameters are adjusted to body weight raised to the 0.75 power as indicated by the symbol (BW). All pooled data is presented as mean ± SEM.
Biomarkers of aging are reduced in the mTORΔ/Δ mice
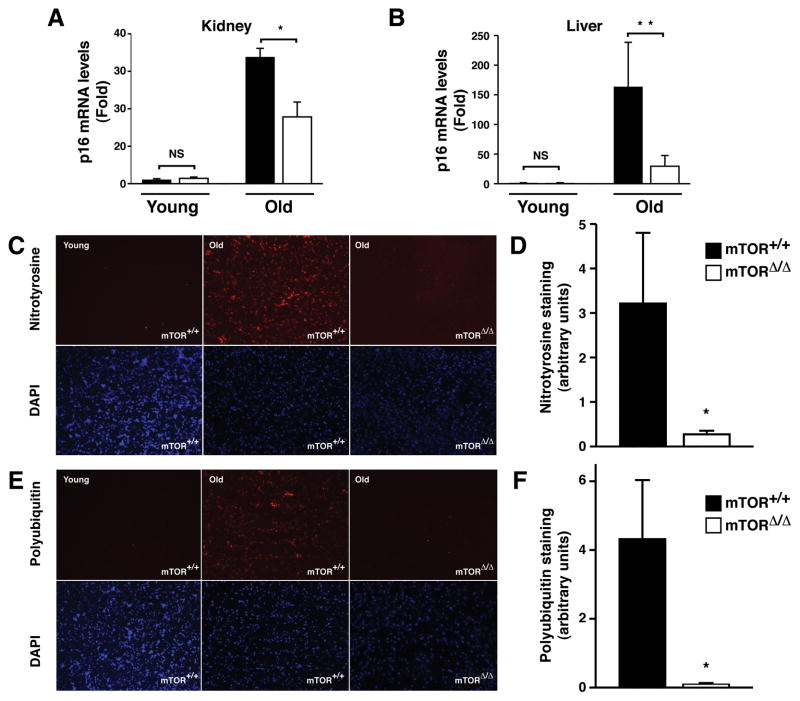
When compared to a cohort of young mice, we observed a significant increase in p16Ink4A mRNA in the tissues of old wild type mice (Krishnamurthy et al., 2004). This age-dependent increase in p16Ink4a mRNA was significantly reduced when kidney and livers of aged matched mTORΔ/Δ mice were assessed (Figure 3A&B). Aging tissues also exhibit evidence of increased oxidative stress and accumulation of protein aggregates (Kastle and Grune, 2011; Schoneich, 2006; Shang and Taylor, 2011). As previously described (Schoneich, 2006), when compared to young wild type mice, tissues from old wild type mice exhibited a marked increase in nitrotyrosine staining (Figure 3C and Figure S3A&B). When compared to age-matched wild type mice, old mTORΔ/Δ mice had significantly reduced levels of tissue nitrotyrosine staining (Figure 3C&D). Older tissues also accumulate aggregates of polyubiquitinated proteins (Kastle and Grune, 2011). These proteins are cleared in part by the mTOR regulated process of autophagy. Older tissues of wild type mice demonstrated a clear increase in the accumulation of polyubiquitin proteins, and this accumulation was less evident in mTORΔ/Δ tissues (Figure 3E&F and Figure S3C&D).
Figure 3.
Molecular and biochemical biomarkers of aging are reduced in old mTORΔ/Δ mice. A) Assessment of the age-dependent increase in kidney mRNA levels for the cell cycle inhibitor p16INK4a normalized to GAPDH expression (n=3 mice per genotype and age with each animal performed in triplicate). B) A similar assessment in old and young liver samples (n=6 young WT, n=5 young mTORΔ/Δ, n=4 old WT and n=4 old mTORΔ/Δ with each sample performed in triplicate). C) Representative brain sections stained for nitrotyrosine (red, upper panels) obtained from young WT mice, old WT mice and old mTORΔ/Δ mice. Cell nuclei with stained concurrently with DAPI (blue, lower panels). D) Intensity of nitrotyrosine staining in the brains of old WT (n=3 animals with three to five determinations per animal) and mTORΔ/Δ (n=4 animals with three to five determinations per animal) mice. E) Staining for polyubquitinated proteins in brain tissue sections obtained from young WT mice, old WT mice or old mTORΔ/Δ mice. Upper panels (red) are stained with an antibody that recognizes proteins that are polyubiquitinated, lower panels are analyzed by nuclear DAPI staining. F) Quantification of polyubiquitinated protein levels in brain sections of WT (n=3 animals with three to five determinations per animal) and mTORΔ/Δ (n=4 animals with three to five determinations per animal) mice. All pooled data is presented as mean ± SEM. *p<0.05, **p<0.01.
mTORΔ/Δ mice have selective improvement in tissue and organ aging
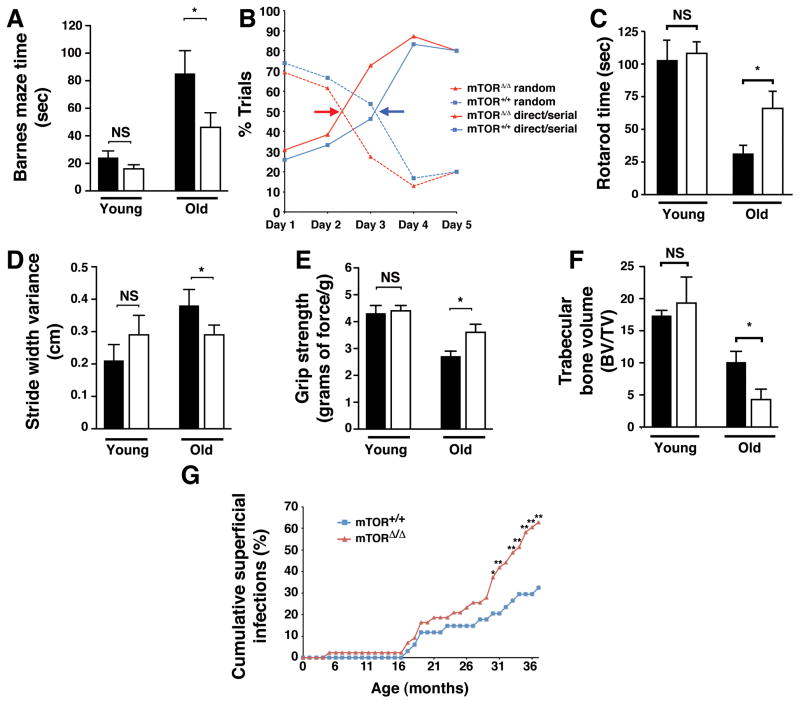
We next sought to evaluate a variety of age-dependent parameters that might be important determinants of improved quality of life, independent of median lifespan. We first evaluated spatial learning and memory using the Barnes maze test, a non-invasive assessment of hippocampal function (Kennard and Woodruff-Pak, 2011). We noted no differences in the latency time to find the escape hole between young wild type and mTORΔ/Δ mice (Figure 4A). Latency times significantly increased in old WT mice, consistent with the well known age-dependent decline in spatial learning and memory. While latency times also increased in old mTORΔ/Δ mice, this age-dependent impairment was significantly less than what was observed in wild type mice. By manually tracking the mice, we could also assess the learning strategies employed during the training period. In both young and old mice, initially animals seek the escape hole using a random strategy. Over time, as spatial memory is encoded, the approach becomes more serial and directed. The amount of training required to have the latter strategy predominate (indicated by the colored arrows) was approximately one day longer in old wild type mice compared to old mTORΔ/Δ animals (Figure 4B). This is consistent with old mTORΔ/Δ mice having a better preserved capacity for acquiring new spatial memory.
Figure 4.
The effects of reduced mTOR expression on a range of tissue specific age-related parameters. A) Escape latency times on day 3 of training for the Barnes maze test for both young female (n=6 mice per genotype) and old female (n=9 WT and n=13 mTORΔ/Δ) mice. Wild type mice are shown in the shaded bars, while the open bars are mTORΔ/Δ mice. *p <0.05. B) Learning strategy of old mice in the acquisition phase for training in the Barnes maze. Arrows indicate transition point between random to directed searching, an indicator of the speed in which new spatial learning is obtained (n=9 WT female mice and n=13 mTORΔ/Δ female mice). C) Duration on the Rotarod, a measure of coordination and balance (n=6 male mice per genotype for young animals and n=4 old male WT, n=7 old male mTORΔ/Δ mice, *p <0.05). D) Stride width variance in young and old mice (n= 6 young female mice per genotype and n=6 old WT male and female mice and n=13 old male and female mTORΔ/Δ mice, *p <0.05). E) Grip strength, normalized to gram of body weight, in young female mice (n=6 per genotype) and old female mice (n=4 WT and n=11 for mTORΔ/Δ mice, *p <0.05). F) Assessment of the age-dependent decline in bone volume (BV) to tissue volume (TV) (n= 4 young mice per genotype and n=6 old mice per genotype, *p <0.05). G) Age-dependent incidence of visibly apparent superficial infections of the skin, eyes or mouth of the total cohort of wild type and mTORΔ/Δ mice (n=34 WT and n=43 mTORΔ/Δ mice, statistical analysis by Fisher’s Exact test, *p<0.05, **p<0.01). All bar graph data is presented as mean ± SEM.
We next assessed the balance and coordination using a Rotarod apparatus. We noted no difference in this functional parameter between young wild type and mTORΔ/Δ mice (Figure 4C). When compared to young wild type mice, older wild type mice were unable to remain as long on the spinning Rotarod apparatus. This is again consistent with the known decrement in balance and coordination as animals age (Barreto et al., 2010). Again, this decline in performance was significantly less marked in the mTORΔ/Δ mice (Figure 4C and S4A). We observed a similar pattern when we assessed gait parameters of the animals. Stride width variability has been closely associated with falls in the elderly population (Hausdorff et al., 2001; Maki, 1997). Again, this parameter was similar in our cohorts of young wild type and mTORΔ/Δ mice (Figure 4D). As wild type mice aged, stride width variability, an integrative measure of neurological, muscular and postural control, increased. Again, this change was less evident in mTORΔ/Δ mice (Figure 4D). Similar, assessment of grip strength, a measure of muscle strength, demonstrated that once again, mTORΔ/Δ mice were protected from an age-dependent decline in function (Figure 4E).
While the mTORΔ/Δ mice appeared to have slower decline in various age-dependent parameters, this was not universally true. Measurement of bone volume revealed that the age-dependent decrease in trabecular bone volume was actually more pronounced in the mTORΔ/Δ mice (Figure 4F). Similarly, we noted that mTOR hypomorphic mice suffered a significant increase in the age-dependent increase in infections that predominantly affected the mouth, eye and skin (Figure 4G). Thus, it would appear that in contrast to the other functional parameters measured, the age-dependent decline in bone volume and immune function were seemingly exacerbated in the mTORΔ/Δ mice.
Discussion
In summary, we describe a new genetic model for reduced mTOR expression and activity that results in a robust increase in lifespan. The magnitude of lifespan extension in our model was larger than previously observed when mice were given rapamycin (Harrison et al., 2009). There are numerous possibilities that might explain these differences. First, in the initial report employing rapamycin, the drug was initiated at 20 months of age. This contrasts with our model in which our genetic reduction of mTOR activity begins in the embryo. Indeed, a subsequent study in which rapamycin was initiated at 9 months of age, saw slightly larger effects on lifespan (Miller et al., 2011). Another possibility is that the reduction in mTOR activity was greater in our model than can be achieved in animals treated pharmacologically. It should be noted that while mTORΔ/Δ mice were viable, when we bred mTOR+/Δ mice, we consistently generated less than 25% of pups that were mTORΔ/Δ. This suggests that the reduction in mTOR expression we observed is close to what may be the lower limit needed for embryonic viability. Another possibility is that rapamycin is primarily an inhibitor of mTORC1 activity although as noted, with chronic administration it can have effects on mTORC2 activity as well (Lamming et al., 2012; Sarbassov et al., 2006). In contrast, our model leads to a balanced reduction in both mTORC1 and mTORC2 activity. Finally, it is important to note that we were dealing with a relatively small cohort of mice and the observed median survival of our WT mice cohort (26.2 months) is on the short end of the published spectrum. As such, the precise magnitude of lifespan extension seen with the mTORΔ/Δ mice must be viewed with caution until it is replicated in other facilities.
Our data suggests that a number of molecular, biochemical and functional parameters of aging were reduced or slowed in the mTORΔ/Δ mice. This is consistent with the notion that reducing mTOR expression does indeed slow the entire aging process in mammals (Wilkinson et al., 2012). Interestingly, our data clearly indicates that not all age-related parameters are regulated in an mTOR-dependent fashion. While the age-dependent increase in infection observed in mTORΔ/Δ mice are likely related to the known role of mTOR in immune function (Chi, 2012; Zhang et al., 2011), these immune effects are less likely to explain the observed accelerated decline in bone volume in mTORΔ/Δ mice. Perhaps more relevant are a set of recent observations suggesting a role for mTOR in modulating the age-dependent decline in bone mass (Xian et al., 2012). Finally, recent data suggesting that long term rapamycin treatment may accelerate cataract formation and augment testicular degeneration (Wilkinson et al., 2012). While these parameters were not part of our standard necropsy analysis, we did a separate analysis for a limited number of WT and mTORΔ/Δ mice. While we saw no evidence for alterations in the testes (Figure S4B&C), our preliminary data is consistent with a potential role for mTOR activity in delaying cataract formation (Figure S4D).
Taken together, our observations suggest that single individual genetic pathways that extend lifespan will likely have non-uniform effects on the rate that individual tissues manifest their age-dependent decline in function. One interpretation of these observations would be that tissue aging and organismal aging are governed by interconnected but separable regulatory control mechanisms. One potential analogy might be circadian rhythms, whereby multiple distinct and independent peripheral clocks coexist in a confederation with a stronger central clock. In a similar fashion, our data suggests the interesting possibility that the rate of tissue aging may be viewed as influenced but not completely subservient to the rate of organismal aging. Alternatively, our results could suggest the possibility that some interventions that slow aging may also have unintended, negative tissue-specific side effects. A very relevant precedent perhaps already exists for this phenomenon as people who undergo voluntary caloric restriction appear to have a corresponding reduction in their bone mineral density (Villareal et al., 2011). Further analysis of this and related genetic models should help distinguish between these possibilities and will hopefully help guide potential therapies aimed at extending lifespan and healthspan in people.
Experimental Procedures
Mice
The generation of mTORΔ/Δ mice has been previously described (Zhang et al., 2011). mTORΔ/+ mice were crossed to generate mTOR+/+ and mTORΔ/Δ littermate mice used for this study. Mice were a mix background consisting of 129S1 and C57BL/6Ncr strains. The proportion of mTOR mice generated from parents bearing the mTOR/+ genotype was less than the predicted 25% (97 mTORΔ/Δ mice pups out of 697 live births, 13.9%), suggestive of some degree of embryonic lethality. All animal experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee, National Heart Lung and Blood Institute, NIH. For genotype analysis, tissues were analyzed by PCR primers and conditions as previously described (Zhang et al., 2011). Unless stated otherwise, young mice represent animals that are between 3–6 months in age and old mice represent animals between 17–27 months in age. Specific ages, number and sex of mice used varied and are listed under the relevant specific test.
Lifespan Analysis
For the determination of lifespan, male and female mTOR+/+ and mTORΔ/Δ mice were housed in a specific pathogen free (SPF) facility. This means that mice entering the facility must be pathogen free with the exception of the specific pathogens Helicobacter, mouse parvovirus (MPV) or mouse norovirus (MNV). Mice were maintained in microisolator cages in ventilated racks with HEPA filtered supply and exhaust air. All cages were opened and changed inside biosafety cabinets or clean air hoods using microisolator technique. Mice were fed a regular chow diet consisting of 24% of calories derived from protein, 14% from fat and 62% from carbohydrates (NIH-31/Harlan Teklad diet). Mice of different genotypes (mTOR+/+ and mTORΔ/Δ mice) were housed together but males and female mice were maintained in separate cages. The maximum density was five mice per cage. The only animals withdrawn from the study occurred within the first six months when a limited number of WT and mTORΔ/Δ male mice were excluded due to excessive fighting. Time of death is calculated using the date mice were found dead, or the time when the mice were determined to be moribund and/or displaying such severe discomfort that veterinary technicians recommended euthanasia. We sought to perform full autopsies on every mouse at the time of their withdrawal from the study however for various logistical reasons, a small fraction of mice of each genotype were not expeditiously forwarded to the pathologist (7 of 43 mTORΔ/Δ mice and 8 of 34 WT mice). Pathological findings including malignant tumors were identified by a team of trained animal pathologists at a central core facility on the NIH Intramural campus. Mice involved in the lifespan analysis did not participate in any metabolic or physiological testing. Statistical analysis for the entire cohort used a Cox regression analysis using genotype and sex as parameters. The statistics for male and female survival was determined by the Log-Rank Test calculated by PRISM.
Supplementary Material
Highlights.
In mammals, decreased mTOR expression produces a profound increase in lifespan.
Reduced mTOR expression results in lower rates of spontaneous tumor formation.
Age-related benefits of reduced mTOR expression are tissue specific.
Acknowledgments
We are grateful to Kumiko Torisu for helpful advice, Zu-Xi Yu for help with tissue sectioning, Lauren Brinster for animal pathology advice, William Jou and Tatyana Chanturiya for help with the metabolic testing and David Allison, Timothy Mark Beasley and John Dawson for statistical assistance. This work was supported by the NIH Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barreto G, Huang TT, Giffard RG. Age-related defects in sensorimotor activity, spatial learning, and memory in C57BL/6 mice. J Neurosurg Anesthesiol. 2010;22:214–219. doi: 10.1097/ANA.0b013e3181d56c98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23:744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausdorff JM, Rios DA, Edelberg HK. Gait variability and fall risk in community-living older adults: a 1-year prospective study. Arch Phys Med Rehabil. 2001;82:1050–1056. doi: 10.1053/apmr.2001.24893. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastle M, Grune T. Protein oxidative modification in the aging organism and the role of the ubiquitin proteasomal system. Curr Pharm Des. 2011;17:4007–4022. doi: 10.2174/138161211798764898. [DOI] [PubMed] [Google Scholar]
- Kennard JA, Woodruff-Pak DS. Age sensitivity of behavioral tests and brain substrates of normal aging in mice. Front Aging Neurosci. 2011;3:9. doi: 10.3389/fnagi.2011.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki BE. Gait changes in older adults: predictors of falls or indicators of fear. J Am Geriatr Soc. 1997;45:313–320. doi: 10.1111/j.1532-5415.1997.tb00946.x. [DOI] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Schoneich C. Protein modification in aging: an update. Exp Gerontol. 2006;41:807–812. doi: 10.1016/j.exger.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Kotyk JJ, Armamento-Villareal RC, Kenguva V, Seaman P, Shahar A, Wald MJ, Kleerekoper M, Fontana L. Reduced bone mineral density is not associated with significantly reduced bone quality in men and women practicing long-term calorie restriction with adequate nutrition. Aging Cell. 2011;10:96–102. doi: 10.1111/j.1474-9726.2010.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on “maximum lifespan”. Mech Ageing Dev. 2004;125:629–632. doi: 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, et al. Rapamycin slows aging in mice. Aging Cell. 2012;11:675–682. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian L, Wu X, Pang L, Lou M, Rosen CJ, Qiu T, Crane J, Frassica F, Zhang L, Rodriguez JP, et al. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat Med. 2012;18:1095–1101. doi: 10.1038/nm.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Readinger JA, DuBois W, Janka-Junttila M, Robinson R, Pruitt M, Bliskovsky V, Wu JZ, Sakakibara K, Patel J, et al. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood. 2011;117:1228–1238. doi: 10.1182/blood-2010-05-287821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.