Abstract
Ewing sarcoma (ES) is the second most common bone tumor in children and young adults, with dismal outcomes for metastatic and relapsed disease. To better understand the molecular pathogenesis of ES and to identify new prognostic markers, we used molecular inversion probes (MIPs) to evaluate copy number alterations (CNAs) and loss of heterozygosity (LOH) in formalin-fixed paraffin-embedded (FFPE) samples which included 40 ES primary tumors and 12 ES metastatic lesions. CNAs were correlated with clinical features and outcome, and validated by immunohistochemistry (IHC). We identified previously reported CNAs, in addition to SMARCB1 (INI1/SNF5) homozygous loss and copy neutral LOH. IHC confirmed SMARCB1 protein loss in 7–10% of clinically-diagnosed ES tumors in three separate cohorts (University of Utah [N=40], Children’s Oncology Group [N=31], and University of Michigan [N=55]). A multifactor copy number (MCN)-index was highly predictive of overall survival (39% vs. 100%, P<0.001). We also identified RELN gene deletions unique to 25% of ES metastatic samples. In summary, we identified both known and novel CNAs using MIP technology for the first time in FFPE samples from patients with ES. CNAs detected by microarray correlate with outcome and may be useful for risk-stratification in future clinical trials.
Keywords: Ewing sarcoma, copy number, outcome, SMARCB1/INI1/SNF5, CEBPB
INTRODUCTION
Ewing sarcoma (ES) is the second most common bone malignancy in childhood and young adults (1). However, less than 300 new cases of ES are diagnosed per year in the United States which makes biology studies difficult to perform (2–4). Even with aggressive therapy, ES survival approaches 65% for localized disease and less than 30% for metastatic disease (1, 5). Survival after relapse remains dismal, and new treatment strategies for metastatic or relapsed disease have been limited and unsuccessful (6–7). The t(11;22)(q24;q12) translocation characterizes over 80% of ES cases and leads to the EWSR1-FLI1 fusion product contributing to sarcomagenesis (8–9). Understanding the additional molecular alterations in ES may lead to new insight into tumorigenesis, prognostics, and targets for developmental therapeutics.
Earlier studies have described the use of traditional array-based comparative genomic hybridization (aCGH) to identify recurrent copy number alterations (CNAs) in ES. These CNAs included gains in chromosome 8, 12, 20 and 1q as well as losses in 16q and 9p21, and were inconsistently associated with poor outcome (10–17). An increased total number of CNAs has been implicated with worse outcome (18–21). However, many of these CNA studies were limited by small sample sizes and brief follow-up periods (9, 22).
Molecular inversion probe (MIP) microarray technology recently has allowed for high-resolution, genome-wide copy number analysis of formalin-fixed paraffin-embedded (FFPE) samples (23–26). This novel genomic technology provides an opportunity to learn about ES molecular genetics because of the wide availability of clinically-archived FFPE samples for analysis. Here we report the first use of MIPs on ES FFPE samples to evaluate CNAs and allelic imbalances. We validate previously reported CNAs and also identify novel CNAs which may be important to ES tumorigenesis and could serve as future therapeutic targets, including unexpected deletions of SMARCB1 (INI1/SNF5). Furthermore, we describe CNA patterns that strongly correlate with event-free survival (EFS) and overall survival (OS) in ES demonstrating the strength of microarray technology as a possible prognostic tool in this disease.
MATERIALS AND METHODS
Cell Lines
Genomic DNA was extracted from 5 ES cell lines (A673, EWS502, SKNMC, TC32, TC71) and from 6 healthy individuals as controls. Cell line DNA was run on the Agilent aCGH 44B array (44K tiled probes) per manufacturer’s directions. 37 ng of cell line and control DNA was submitted for MIP analysis using an initial customized panel of 24,000 cancer gene probes (Affymetrix Research Services Laboratory, Santa Clara, CA). Data from the Agilent aCGH and MIP array platforms were compared using Nexus Copy Number 5.1 (BioDiscovery, Inc., El Segundo, CA).
Clinical sample collection
This study was approved by our Institutional Review Board. De-identified patient material was obtained from the Pathology Department at Primary Children’s Medical Center (PCMC), Salt Lake City, Utah. Clinically-archived FFPE scrolls were retrieved from 51 individual patients diagnosed with ES between November 1997 and June 2010. Twenty (39%) patients were confirmed by split EWSR FISH probe, PCR, or t(11;22) karyogram. Genomic DNA was isolated from the FFPE samples and included 45 primary tumors (PTs), 12 metastatic lesions (MLs, 8 of which were paired to PTs), 5 post-treatment tumor samples (4 paired to PTs), and 1 relapse sample. PT samples were from diagnostic biopsies prior to chemotherapy. FFPE staging bone marrow aspirate clots were used as paired normal references for each patient when available and negative for tumor. See Supplementary Materials for details on FFPE sample preparation. Clinical factors were extracted from the medical records and included: age, sex, primary tumor site (location and soft tissue vs. bony lesion), metastasis, tumor diameter, tumor percentage and tumor viability of diagnostic biopsy (See Supplementary Table 1), percentage necrosis at surgical resection, EFS, and OS.
OncoScan™ FFPE Express Assay
Genomic DNA (3 to 300 ng/sample) from the FFPE clinical samples underwent MIP analysis with the expanded OncoScan™ FFPE Express assay (V1.0) with 330,000 cancer gene and genome-wide probes (Affymetrix, Santa Clara, CA). Each probe in the assay contains 40–60 nucleotides in length that flank the targeted SNP position. Probes were chosen from intragenic sequences of >1,000 genes that have been reported to be specifically involved in cancer development, with the remaining assay probes chosen to fill in the gaps across the genome. This assay provides both copy number and LOH data, and was performed as previously described (24, 26–29). In brief, the entire probe set is introduced to the sample DNA and the following steps are performed as a multiplex reaction: 1) Annealing, 2) Gap fill polymerization, 3) Gap fill ligation, 4) Exonuclease selection of unbound probes, 5) Release of bound probes, 6) Amplification of released probes, 7) Probe hybridization to microarray, and 8) Microarray scanning. Specific details of this assay can be found in previously published technology reviews (30).
Data Visualization and Statistical Analysis
Data visualization and CNA analysis was done with Nexus Copy Number 5.1 (BioDiscovery, Inc., El Segundo, CA). Univariate and multivariate Cox proportional hazards models were fit to recurring CNAs and regions previously reported to correlate with outcome, with reported P-values from log-rank tests. P-values were adjusted using the Benjamini & Hochberg step-up FDR controlling procedure (31). See Supplementary Materials for details on CNA and statistical analyses.
Immunohistochemical (IHC) staining and analysis
IHC was performed for SMARCB1 (INI-1/BAF-47 antibody; BD Transduction Lab, San Jose, CA) and CEBPB (CEBP beta antibody; Abcam, Cambridge, MA). Results were interpreted by a pathologist (HHZ) blinded to patient identity or CNA results.
Tissue Microarrays (TMAs)
A de-identified TMA was obtained from the Children’s Oncology Group (COG) that contained 59 tissues from 31 separate ES patients. The COG TMA (3000-30-P8798) was made from a combination of samples collected from biology studies COG B947 and AEWS02B1. Samples were selected based on availability of blocks at the COG bank, and all samples were diagnosed and clinically treated as ES. A second de-indentified TMA was obtained from the University of Michigan (DL and DT) that contained 44 PTs and 11 recurring MLs, all from individual patients clinically diagnosed and treated as ES. IHC was performed for SMARCB1 protein expression on both TMAs. Results were interpreted by a pathologist (ARP) blinded to patient identity.
Pathology Review
Diagnostic H&E slides from samples staining negative for SMARCB1 protein expression were independently reviewed by an anatomic pathologist with expertise in bone and soft tissue sarcomas (AER). The IHC status of the samples was blinded to the pathologist at the time of the review.
RESULTS
Cell lines
To establish the validity of the MIP array, we compared ES cell lines (n=5) run on the Agilent 44K aCGH to the same cell lines run on the original MIP 24K platform. The same gains and losses were seen across both platforms (Supplementary Materials Fig 1). The cell lines demonstrated similar CNAs as reported in the literature. Trisomy 8 appeared in 3/5 cell lines with an additional focal high gain encompassing the MYC gene. 1q amplification was seen in 2/5 of cell lines. Trisomies 12 and 21 were present in 1/5 of cell lines. Expected losses at 9p21 and 16q appeared in 4/5 and 1/5 of cell lines, respectively. These data demonstrate that the MIP platform reliably identifies CNAs in a similar fashion as aCGH.
FFPE Clinical Samples
Copy number results were derived from 40 FFPE PT samples (5 of the 45 PT samples were excluded due to insufficient DNA quantity) and 32 paired bone marrow aspirate clots that were negative for tumor. See Table 1 for clinical data. Median follow-up length was 3 years (0.7–12.4). Age at diagnosis ≥ 10 years old significantly correlated with EFS (P=.015) and the presence of metastatic disease significantly correlated with both EFS and OS (P=.0016 and P=.00033, respectively). No other clinical factors significantly associated with outcome.
Table 1. Clinical summary.
Clinical data is displayed for the 40 patients in the Utah cohort, including information about their primary tumors.
| Sample ID | Gender | Age at Diagnosis (Years) | Primary site (axial, appendicular, limb girdle) | Primary site (soft tissue, bone) | Metastasis at diagnosis | Tumor diameter >10 cm | Necrosis > 90% | Relapse | Follow-up (Years) | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| EWS-1 PT | female | 14.7 | appendicular | bone | no | yes | > 90% | no | 6.86 | NED |
| EWS-2 PT | male | 16.2 | limb girdle | bone | yes | yes | unknown | yes | 2.92 | DOD |
| EWS-3 PT | male | 13.4 | limb girdle | bone | no | unknown | > 90% | no | 5.13 | NED |
| EWS-4 PT | male | 17.8 | appendicular | soft | no | yes | unknown | no | 4.68 | NED |
| EWS-5 PT | male | 1.4 | appendicular | soft | no | no | > 90% | no | 6.63 | NED |
| EWS-6 PT | male | 9.3 | axial | soft | no | no | > 90% | no | 11.28 | NED |
| EWS-7 PT | male | 5.2 | appendicular | soft | no | yes | < 90% | no | 10.38 | NED |
| EWS-8 PT | female | 15.3 | appendicular | bone | no | no | < 90% | yes | 3.76 | AWD |
| EWS-10 PT | female | 13.4 | limb girdle | bone | no | no | unknown | no | 12.38 | NED |
| EWS-11 PT | male | 18.4 | axial | soft | yes | no | unknown | yes | 1.05 | DOD |
| EWS-13 PT | male | 13.9 | appendicular | bone | no | no | > 90% | no | 9 | NED |
| EWS-14 PT | male | 9.2 | appendicular | bone | no | no | unknown | no | 6.52 | NED |
| EWS-15 PT | male | 7.2 | axial | soft | no | yes | < 90% | no | 4.13 | NED |
| EWS-16 PT | female | 11.7 | limb girdle | bone | yes | yes | unknown | yes | 3.28 | DOD |
| EWS-17 PT | male | 15.6 | limb girdle | bone | yes | yes | > 90% | yes | 4.56 | DOD |
| EWS-18 PT | male | 11.2 | axial | bone | no | yes | > 90% | yes | 8.51 | NED |
| EWS-19 PT | male | 13.8 | appendicular | bone | no | no | > 90% | no | 1.3 | NED |
| EWS-20 PT | male | 16.3 | appendicular | bone | no | unknown | > 90% | no | 1.98 | NED |
| EWS-22 PT | male | 9.7 | axial | soft | no | yes | < 90% | no | 1.26 | NED |
| EWS-25 PT | female | 14.1 | limb girdle | bone | no | unknown | unknown | no | 1 | NED |
| EWS-26 PT | female | 9.3 | appendicular | bone | yes | no | > 90% | no | 1.31 | NED |
| EWS-27 PT | male | 12.7 | appendicular | soft | no | yes | > 90% | no | 3.01 | NED |
| EWS-28 PT | female | 6.2 | appendicular | bone | no | no | > 90% | no | 0.81 | NED |
| EWS-29 PT | female | 12.1 | axial | bone | no | no | unknown | no | 0.65 | NED |
| EWS-30 PT | female | 11.2 | limb girdle | bone | no | no | > 90% | no | 4.53 | NED |
| EWS-31 PT | female | 20.5 | axial | soft | no | no | < 90% | yes | 4.36 | DOD |
| EWS-32 PT | male | 16.9 | axial | bone | yes | no | unknown | yes | 3.59 | DOD |
| EWS-35 PT | female | 15.6 | appendicular | bone | no | yes | > 90% | no | 1.85 | NED |
| EWS-36 PT | male | 0.6 | axial | soft | yes | no | unknown | no | 4.04 | NED |
| EWS-37 PT | male | 15.7 | axial | bone | no | no | unknown | no | 3.37 | NED |
| EWS-40 PT | male | 10.1 | axial | soft | no | no | > 90% | no | 9 | NED |
| EWS-42 PT | female | 4.0 | axial | bone | no | yes | unknown | no | 2.72 | NED |
| EWS-44 PT | male | 17.4 | limb girdle | bone | no | unknown | unknown | yes | 6.13 | AWD |
| EWS-45 PT | male | 17.3 | limb girdle | bone | no | no | < 90% | yes | 2.2 | DOD |
| EWS-46 PT | male | 17.7 | axial | bone | yes | unknown | unknown | no | 4.15 | DOD |
| EWS-47 PT | female | 7.9 | limb girdle | bone | no | unknown | unknown | no | 5.82 | NED |
| EWS-48 PT | male | 5.7 | axial | bone | no | unknown | unknown | no | 3.56 | NED |
| EWS-49 PT | male | 13.9 | limb girdle | bone | yes | unknown | unknown | yes | 5.1 | NED |
| EWS-50 PT | female | 12.2 | axial | soft | no | unknown | unknown | no | 7.03 | NED |
| EWS-51 PT | male | 15.1 | limb girdle | bone | yes | no | < 90% | no | 4.42 | NED |
Overall Copy Number Alterations (CNAs)
We analyzed the 40 pre-treatment PTs for recurring CNAs and LOH (Table 2, Supplementary Materials Fig 2). In a log-rank test, an excess of 3 CNAs correlated with outcome when considered as a continuous variable (EFS P=.017, OS P=.00005).
Table 2.
Recurrent CNAs including high gains and homozygous losses.
| PRIMARY TUMORS (N=40) | REGION | EVENT | LENGTH (Kilobases, KB) | PERCENTAGE |
|---|---|---|---|---|
| CHROMOSOME 1 | 1q21.1-qter | CN Gain/2 High Copy Gains | 104529 KB | 17.5% (7/40) |
| 1q21.2 | High Copy Gain | 956 KB | 2.5% (1/40) | |
| CHROMOSOME 2 | 2p25.3-pcen | CN Gain | 89915 KB | 5% (2/40) |
| 2q11.1-qter | CN Gain | 147991 KB | 5% (2/40) | |
| CHROMOSOME 5 | 5p15.33-pcen | CN Gain | 46368 KB | 10% (4/40) |
| 5q11.1-qter | CN Gain | 131261 KB | 7.5% (3/40) | |
| 5q34 | Homozygous Copy Loss | 127 KB | 2.5% (1/40) | |
| 5q35.1 | High Copy Gain | 537 KB | 2.5% (1/40) | |
| CHROMOSOME 6 | 6p25.3 | High Copy Gain | 501 KB | 2.5% (1/40) |
| 6p22.3 | High Copy Gain | 1343 KB | 2.5% (1/40) | |
| 6p22.2-p22.1 | High Copy Gain | 727 KB | 2.5% (1/40) | |
| CHROMOSOME 7 | 7q11.1-qter | CN Gain | 97752 KB | 5% (2/40) |
| CHROMOSOME 8 | 8p23.3-pcen | CN Gain (includes 1 focal High Gain within Full Gain) | 43868 KB | 35% (14/40) |
| 8q11.1-qter | CN Gain/1 High Copy Gain (includes 1 shorter CN Gain) | 99153 KB | 37.5% (15/40) | |
| CHROMOSOME 9 | 9q31.3-qter | CN Gain | 28340 KB | 5% (2/40) |
| 9p24.1 | Homozygous Copy Loss | 43 KB | 2.5% (1/40) | |
| 9p21.3 | Homozygous Copy Loss | 115 KB | 5% (2/40) | |
| CHROMOSOME 12 | 12p13.33-pcen | CN Gain | 35400 KB | 12.5% (5/40) |
| 12q11.-qter | CN Gain | 95605 KB | 12.5% (5/40) | |
| CHROMSOME 13 | 13q13.1 | CN Gain/1 High Copy Gain | 49 KB | 5% (2/40) |
| CHROMOSOME 14 | 14q11.2-qter | CN Gain | 86854 KB | 7.5% (3/40) |
| CHROMOSOME 16 | 16q11.2-qter | CN Loss | 43685 KB | 10% (4/40) |
| CHROMOSOME 18 | 18q12.1 | High Copy Gain | 871 KB | 2.5% (1/40) |
| CHROMOSOME 20 | 20p13-pcen | CN Gain | 27100 KB | 10% (4/40) |
| 20q11.21-qter | CN Gain (includes 1 focal High Gain within Full Gain) | 33075 KB | 15% (6/40) | |
| 20q13.2 | High Copy Gain | 389 KB | 2.5% (1/40) | |
| CHROMOSOME 21 | 21q11.1-qter | CN Gain | 33279 KB | 10% (4/40) |
| CHROMOSOME 22 | 22q12.2-qter | CN Gain | 21656 KB | 5% (2/40) |
| 22q11.23 | Homozygous Copy Loss | 53 KB | 5% (2/40) | |
| 22q13.31-q13.33 | Homozygous Copy Loss | 3166 KB | 2.5% (1/40) | |
| METASTATIC LESIONS (N=12) | ||||
| CHROMOSOME 1 | 1q21.1-qter | CN Gain | 104529 KB | 5% (2/40) |
| CHROMOSOME 2 | 2p25.3-pcen | CN Gain | 89915 KB | 5% (2/40) |
| CHROMOSOME 5 | 5q11.2 | High Copy Gain | 309 KB | 2.5% (1/40) |
| CHROMOSOME 6 | 6p25.3 | High Copy Gain | 501 KB | 2.5% (1/40) |
| 6p22.3 | High Copy Gain | 890 KB | 2.5% (1/40) | |
| CHROMOSOME 7 | q22.1-q31.1 | CN Gain | 500 KB | 5% (2/40) |
| CHROMOSOME 8 | 8p23.3-pcen | CN Gain/3 High Copy Gains | 43868 KB | 10% (4/40) |
| 8q11.1-qter | CN Gain/3 High Copy Gains | 99153 KB | 10% (4/40) | |
| CHROMOSOME 9 | 9q34.2-q34.3 | High Copy Gain | 923 KB | 5% (2/40) |
| CHROMOSOME 12 | 12p13.33-pcen | CN Gain/1 High Copy Gains | 35400 KB | 10% (4/40) |
| 12q11.-qter | CN Gain/1 High Copy Gains | 95605 KB | 7.5 % (3/40) | |
| CHROMOSOME 16 | 16q11.2-qter | CN Loss | 43685 KB | 5% (2/40) |
| CHROMOSOME 19 | 19p13.13 | Homozygous Copy Loss | 325 KB | 2.5% (1/40) |
SMARCB1 Deletions and LOH
To our surprise, we detected SMARCB1 homozygous deletions in 2 PTs (5%) and focal copy neutral LOH in 1 PT (2.5%) in our Utah cohort. The two homozygous deletions were located within 22q11.23 heterozygous deletions, were quite focal, and spanned 529.1 kilobases (KB) to 1,227.7 KB in length. The common overlapping region of these two focal homozygous deletions was only 53.5 KB in length and included only SMARCB1 and DERL3. The deletion of SMARCB1 was unexpected because SMARCB1 loss is thought to be pathognomonic for rhabdoid tumors (although it can be found in other rare soft tissue sarcomas, as well) (32). Indeed, lack of SMARCB1 staining is often sufficient for pathologists to revise an unknown pediatric solid tumor diagnosis to a rhabdoid tumor. To confirm the consequence of SMARCB1 deletion or LOH in our sample set, we performed IHC protein staining. The 3 PTs with SMARCB1 genetic alterations in the Utah cohort showed loss of nuclear SMARCB1 protein expression, while their paired normal bone marrows showed positive nuclear staining, demonstrating that SMARCB1 expression loss was a somatic event in these three patients. Twelve additional ES samples (6 PTs + 6 MLs) that were diploid for SMARCB1 with normal heterozygosity showed retained nuclear positivity with the INI-1/Baf-47 antibody.
Each of these three SMARCB1-negative tumors was clinically diagnosed and treated as ES based on the histopathology review described below. All of the SMARCB1-negative tumors in our Utah cohort presented in soft tissue structures (buttock, thigh, and calf), showed “small round cell” histology, occurred in young patients from 1.4 to 10.1 years old (median 5.2), and had EFS of 6 to 10 years with NED.
The first SMARCB1-deleted tumor showed sheets of tumor cells with round to oval nuclei, small nucleoli and small to moderate amounts of eosinophilic cytoplasm. Some tumor cells showed eccentric blobs of eosinophilic cytoplasm. A vague nesting pattern was demarcated by irregular bands of fibrous tissue. Rosettes were not identified. Tumor cells were diffusely reactive for vimentin and focally reactive for AE1/AE3 cytokeratin, EMA, S100 and synaptophysin. 013 (CD99) showed cytoplasmic blush staining, but no definitive membranous staining. Tumor cells were negative for smooth muscle actin, muscle specific actin, desmin, myoglobin, and CD45. The karyogram demonstrated a t(11;22). The histologic appearance, IHC profile and t(11;22) lead to the clinical interpretation of this tumor as ES with the atypical feature of absence of 013(CD 99) membranous positivity.
The second SMARCB1-deleted tumor showed sheets of tumor cells with round to oval nuclei, occasional nucleoli and clear to pale cytoplasm. A vague nesting pattern and zone of hemorrhage and necrosis were noted. A PAS and PAS with diastase demonstrated abundant cytoplasmic glycogen. Tumor cells were reactive for vimentin, synaptophysin, S100, CD57, and neuron specific enolase. Focal reactivity was seen with EMA. Tumor cells were negative for HMB45, CD68, muscle specific actin, desmin, smooth muscle actin, AE1/AE3 cytokeratin, cytokeratin 7, TDT, CD1a, CD20 and 013 (CD99). Chromosome analysis demonstrated a derivative chromosome 22 from a translocation between chromosome 22 and another unknown chromosome. This tumor was classified as an atypical ES based on histology, IHC profile and cytogenetic results.
The third specimen, with SMARCB1 LOH, showed small round uniform tumor cells, with small nucleoli and scanty clear to eosinophilic cytoplasm. No fibrous stroma or rosette formation was identified. Tumor cells showed strong positivity for vimentin and FLI-1. Focal membranous staining was seen with 013 (CD99). EMA and muscle specific action also showed focal staining. Tumor cells were negative for CD45, myogenin, desmin and S100. Chromosome analysis demonstrated a normal male karyogram. At the time of diagnosis, this case was seen in consultation by a soft tissue pathologist who concurred with the diagnosis of ES.
SMARCB1 protein loss has not been described in ES, so we sought to validate this finding with two independent sets of patient samples. An ES TMA obtained from COG showed a similar rate of SMARCB1 protein expression loss (N=3/31). The COG tumors originated from the leg, thigh, and scalp/skull, and also occurred in young patients 10 to 11 years old. Outcome data were not available for the COG tumors.
A second TMA was obtained from the University of Michigan and also demonstrated a similar rate of SMARCB1 protein loss (N=3/41 primary tumors, and N=1/10 metastatic/recurrent tumors). The SMARCB1-negative primary tumors were located in the pelvis (extraosseous), nasal cavity, shoulder (osseous), and the recurrent ML was located in the lung. The Michigan TMA contained both pediatric and adult patients clinically diagnosed and treated for ES (PTs from patients 3–49 years old [median 20], and recurrent MLs from patients 12–59 years old [median 20]). The SMARCB1-negative PTs occurred in patients 19 to 39 years old (median 38), and the single SMARCB1-negative recurrent ML occurred in a 20 year old patient. Two of 3 SMARCB1-negative PT patients are still alive without disease, and the patient that died presented with the osseous right shoulder tumor. The SMARCB1-negative metastatic patient with the lung lesion was alive with disease (AWD) at the time of last follow-up (over 1 and half years since relapse diagnosis). All four of these SMARCB1-negative tumors from the University of Michigan were signed out as primitive neuroectodermal tumor (PNET)/Ewing Sarcoma based on morphology.
These SMARCB1-negative tumors (N=10 total, 3 from Utah, 3 from COG, and 4 from Michigan) were from archived clinical samples and only the Michigan samples were available for molecular testing to further confirm the diagnosis of ES. However, H&E slides were obtained from each of these samples along with clinical pathology reports and sent for blind review by an expert bone tumor pathologist. Upon review, it was felt that 4 of 10 of these tumors represented “Conventional ES” (2=COG, 2=Michigan), and the remaining 6 tumors were “Atypical ES” with a mixture of large cells with abundant clear cytoplasm (large cell ES), spindle cells, and intermittent rhabdoid-appearing cells. Of note, none of the atypical cases appeared to look like classic soft-tissue rhabdoid tumors. Both of the conventional-appearing ES tumors from Michigan that were SMARCB1-negative were found to be positive for EWSR rearrangement by FISH using a break-apart probe, which the two atypical cases were FISH negative. See Supplementary Table 2 for individual descriptions of these tumors.
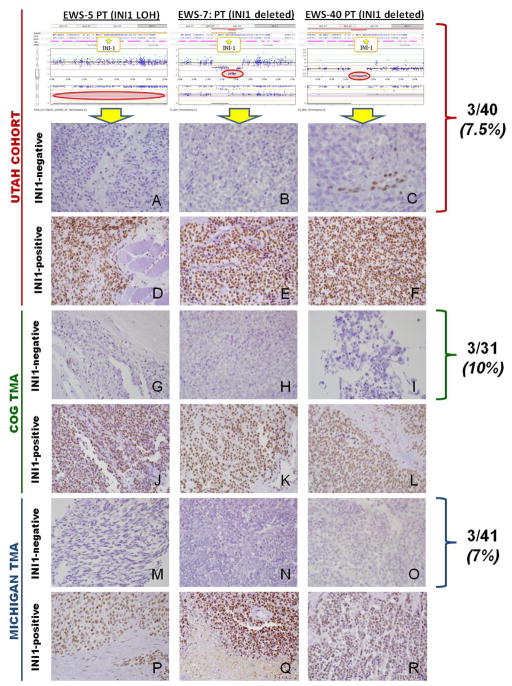
Taken together, these data demonstrate that SMARCB1 protein loss may be found in a very small proportion of conventional ES, and that another small proportion of atypical (or large cell) ES may contain SMARCB1 loss. Due to the presence of rhabdoid-appearing cells mixed with ES large cells, this latter subset may represent a rare but novel type of hybrid rhabdoid-ES tumor that requires further investigation. See Figure 1.
Figure 1. SMARCB1 (INI1/SNF5) LOH, Copy Number, and Immunohistochemistry (IHC) in Ewing Sarcoma.
The top three figures display the probe values and B-allele frequency for OncoScan™ FFPE Express assay, illustrating copy neutral LOH (A:EWS-5) and SMARCB1 homozygous deletions (B:EWS-7, C:EWS-40). IHC staining in the Utah cohort, COG tissue microarray (TMA), and Michigan TMA revealed SMARCB1 protein expression loss in 7–10% of Ewing sarcoma primary tumor samples. Utah cohort: A) Sample A contained SMARCB1 copy neutral LOH and shows loss of SMARCB1 (INI-1/BAF-47 antibody) nuclear expression in tumor cells. B–C) Samples B and C both had SMARCB1 homozygous deletions with loss of SMARCB1 nuclear expression in tumor cells. Note that samples A and C show retained SMARCB1 nuclear positivity in endothelial cells, which serves as an internal, positive control. D–F) All contained normal SMARCB1 copy number and LOH, and each sample shows diffuse SMARCB1 nuclear staining. COG TMA: G–I) Samples show loss of SMARCB1 nuclear expression in tumor cells. Note the retained SMARCB1 nuclear positivity in endothelial cells (Figure G, left). J–L) Samples show diffuse SMARCB1 nuclear staining. MICHIGAN TMA: M–O) Samples show loss of SMARCB1 nuclear expression in tumor cells. P–R) Samples show diffuse SMARCB1 nuclear staining.
Additional CNAs and Outcome
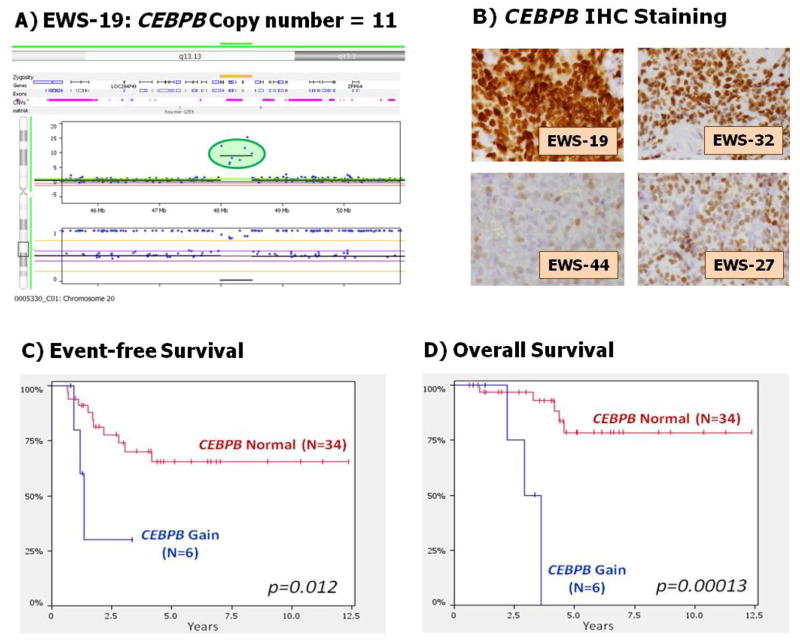
Table 3 summarizes the recurring CNAs that correlated with outcome. Chromosome 20q showed trisomy in 15% (n=6/40) of PTs. Two separate and focal regions in 20q showed additional gains in 2 different patients. The first focal gain on 20q13.2 contains three genes (ZNF217, SUMO1P1, and BCAS1) and was the most significant CNA region to predict poor outcome (EFS P=.00033, OS P=1.9E-7). The second focal region on 20q13.13 contained a very high gain (11 copies) within the trisomy 20q centered on CEBPB (CCAAT/enhancer binding protein beta). IHC showed increased or equal CEBPB nuclear staining in the ES tumors with CEBPB (20q) gain compared to ES tumors without CEBPB gains and a non-tumor control. The ES sample with the high-gain of 11 copies showed the darkest IHC nuclear staining (Fig 2). CEBPB gains correlated with worse outcome (EFS P=.012, OS P=.00013).
Table 3. Copy number and outcome.
Event-free survival (EFS) and overall survival (OS) are displayed for the recurring regions and other commonly reported CNAs in Ewing sarcoma. Both adjusted and unadjusted P values are displayed. Outcome is adjusted for age at diagnosis greater than 10 years and the presence of metastatic disease at diagnosis. The regions on Chromosome 20q remain significant after adjustment and therefore have the strongest independent correlation with clinical outcome.
| EVENT FREE SURVIVAL (N=40) | OVERALL SURVIVAL (N=40) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Alteration | Gain or Loss Present | No gain or Loss | P-value (unadjusted) | P-value (adjusted) | Gain or Loss Present | No gain or Loss | P-value (unadjusted) | P-value (adjusted) | ||||
|
|
|
|||||||||||
| Relapse or Death | 5-year survival
|
Relapse or Death | 5-year survival | Death | 5-year survival
|
Death | 5-year survival
|
|||||
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | |||||||||
| 1q amplification | 1/7 | 0.8 | 11/33 | 0.59 | 0.33 | 0.68 | 0/7 | 1 | 8/33 | 0.64 | 0.14 | 0.33 |
| (0.52–1.00) | (0.43–0.82) | (0.46–0.89) | ||||||||||
|
| ||||||||||||
| Trisomy 5 | 3/3 | 0 | 9/37 | 0.7 | 0.01* | 0.54 | 3/3 | 0 | 5/37 | 0.81 | 0.0027** | 0.22 |
| (0.56–0.89) | (0.67–0.98) | |||||||||||
|
| ||||||||||||
| MYC amplification | 8/16 | 0.35 | 4/24 | 0.8 | 0.0059** | 0.58 | 8/16 | 0.26 | 0/24 | 1 | 0.0000174*** | ---& |
| (0.16–0.80) | (0.64–1.00) | (0.08–0.77) | ||||||||||
|
| ||||||||||||
| Trisomy 8 | 7/15 | 0.38 | 5/25 | 0.77 | 0.032* | 0.95 | 7/15 | 0.28 | 1/25 | 0.96 | 0.00038*** | 0.12 |
| (0.17–0.84) | (0.61–0.97) | (0.09–0.92) | (0.88–1.00) | |||||||||
|
| ||||||||||||
| CDKN2A deletion | 0/2 | 1 | 12/38 | 0.61 | 0.39 | ---& | 0/2 | 1 | 8/38 | 0.7 | 0.66 | ---& |
| (0.46–0.82) | (0.55–0.90) | |||||||||||
|
| ||||||||||||
| Trisomy 12 | 2/5 | 0.5 | 10/35 | 0.66 | 0.85 | 0.35 | 2/5 | 0.38 | 6/35 | 0.76 | 0.43 | 0.65 |
| (0.19–1.00) | (0.51–0.86) | (0.08–1.00) | (0.61–0.95) | |||||||||
|
| ||||||||||||
| 16q24.1 deletion | 4/5 | 0.2 | 8/35 | 0.73 | 0.041* | 0.65 | 3/5 | 0.4 | 5/35 | 0.79 | 0.1 | 0.77 |
| (0.035–1.00) | (0.58–0.91) | (0.14–1.00) | (0.64–0.98) | |||||||||
|
| ||||||||||||
| 16q23.3-24.1 deletion | 4/5 | 0.2 | 8/35 | 0.72 | 0.014* | 0.31 | 3/5 | 0.4 | 5/35 | 0.77 | 0.03* | 0.13 |
| (0.035–1.00) | (0.57–0.91) | (0.14–1.00) | (0.60–0.97) | |||||||||
|
| ||||||||||||
| 16q deletion | 3/4 | 0.25 | 9/36 | 0.7 | 0.11 | 0.68 | 2/4 | 0.5 | 6/36 | 0.74 | 0.26 | 0.47 |
| (0.046–1.00) | (0.55–0.89) | (0.19–1.00) | (0.58–0.95) | |||||||||
|
| ||||||||||||
| 20q13.2 amplification | 4/7 | 0.26 | 8/33 | 0.7 | 0.00033*** | 0.0084** | 4/7 | 0 | 4/33 | 0.81 | 0.00000019*** | 0.0021* |
| (0.053–1.00) | (0.54–0.90) | (0.66–0.99) | ||||||||||
|
| ||||||||||||
| 20q13.13 (CEBPB) amplification | 3/6 | 0.3 | 9/34 | 0.68 | 0.012* | 0.07 | 3/6 | 0 | 5/34 | 0.79 | 0.00013*** | 0.029* |
| (0.063–1.00) | (0.52–0.88) | (0.63–0.98) | ||||||||||
|
| ||||||||||||
| Trisomy 20 (includes partial) | 3/7 | 0.3 | 9/33 | 0.68 | 0.012* | 0.07 | 3/7 | 0 | 5/33 | 0.79 | 0.00013*** | 0.029* |
| (0.063–1.00) | (0.52–0.88) | (0.63–0.98) | ||||||||||
|
| ||||||||||||
| Trisomy 21 | 2/4 | 0.5 | 10/36 | 0.64 | 0.41 | 0.58 | 2/4 | 0.5 | 6/36 | 0.74 | 0.2 | 0.96 |
| (0.19–1.00) | (0.48–0.86) | (0.19–1.00) | (0.58–0.95) | |||||||||
|
| ||||||||||||
| INI1 deletion/loss of heterozygosity | 0/3 | 1 | 12/37 | 0.59 | 0.21 | ---& | 0/3 | 1 | 8/37 | 0.67 | 0.28 | ---& |
| (0.43–0.81) | (0.50–0.90) | |||||||||||
|
| ||||||||||||
| Multifactor Copy Number (MCN)- Index | 9/19 | 0.4 | 3/21 | 0.83 | 0.013* | 0.61 | 8/19 | 0.39 | 0/21 | 1 | 0.00059*** | 0.047* |
| (0.21–0.78) | (0.67–1.00) | (0.19–0.80) | NA | |||||||||
P<0.05,
P<0.01,
P<0.001,
Stratified Cox model did not converge.
Figure 2. CEBPB in Ewing Sarcoma.
Panel A shows OncoScan™ FFPE Express probes with focal 11 copy number gain of CEBPB located within larger trisomy 20q in sample EWS-19. Panel B displays CEBPB IHC for same sample, note the extremely intense and diffuse nuclear staining in EWS-19 (Copy number 11) compared to the relatively strong nuclear staining in EWS-32 (ES tumor with 3 copies), and two ES tumors with less diffuse, lighter staining and normal CEBPB copy number (EWS-44 and EWS-27). Panels C and D display the event-free survival and overall survival for the Ewing sarcoma patients within our Utah cohort with and without the focal 20q13.13 gain that contains the CEBPB gene (P=.012, P=.00013, respectively).
CEBPB regulates cellular growth and differentiation, and is involved in collagen synthesis (33) and chondrocyte terminal differentiation (34). CEBPB also has been implicated in increased bone reabsorption through the RANKL pathway in giant cell tumors (35). Interestingly, the primary sites of cases with CEBPB amplification in our cohort occurred exclusively in bone (femur, fibula, pelvis, rib, L7 vertebra). We examined cases with CEBPB gains for bone destruction and found 4 cases presented with no evidence of bone destruction, 1 case contained notable bone destruction, and 1 case had no available data. These results lead us to believe that while CEBPB gains may be related to occurrence in bone, it does not appear linked to bone destruction. Our data set showed RANKL amplification in only one case which also coincided with CEBPB amplification but no bone destruction.
Previous studies have linked 16q with survival outcomes.(13) Our Utah cohort contained recurring, focal deletions in 16q24.1 and 16q23.3-24.1 that marginally correlated with EFS (P=.014 and .041, respectively) and OS (P=.030 and .10, respectively). Trisomy 8 also has been reported to correlate with worse outcome, with C-MYC as a suspected driver linked to poor prognosis (12, 36). Forty-percent of our cohort contained full or partial trisomy 8, including 1 patient with a focal MYC gain. Full or partial trisomy 8 correlated with outcome (EFS P=.032, OS P=.00038), and this significance increases when including the focal MYC gain (EFS P=.0059, OS P=.0000174). Trisomy 5 is not commonly reported in ES, but our cohort contained 3 patients (7.5%) with a full trisomy. All 3 patients were greater than 10 years old (16.9, 17.8, 20.6 years old), 2 of 3 presented with metastasis, and all of them relapsed and/or died (EFS P=.01, OS P=.0027).
Only 5% of our PTs showed 9p21 (CDKN2A) loss, lower than the reported ranges of 13-56% (10, 37). Eighty-percent of the ES cell lines showed CDKN2A deletions, and this higher rate in cell lines has been observed (17). 9p21 loss has been linked to negative outcome (15), but both patients with CDKN2A deletions in our cohort presented without metastasis and have not relapsed more than 2 and half years after diagnosis. Our limited number of CDKN2A deletions is too small to determine the true correlation with outcome. Current studies validating the prevalence and prognostic significance of CDKN2A deletions in ES are underway through COG.
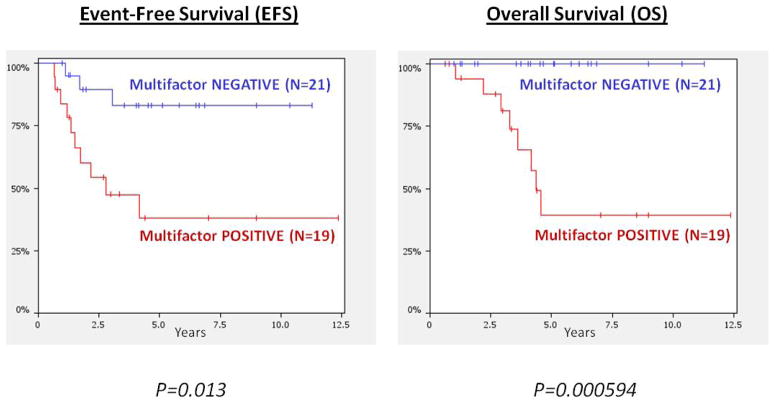
We combined all significant CNA regions correlating with outcome from our unadjusted analyses into a “multifactor copy number (MCN) index” with at least one affected region. The MCN-index significantly correlated with worse EFS (P=.013) and OS (P=.00059) (Fig 3). The MCN-index still predicted OS (P=.03) even when using the extremely stringent bootstrapping analysis technique. See Supplementary Materials for more details on the bootstrapping statistical approach used in the MCN analysis.
Figure 3. Kaplan-Meier curves for Multifactor Copy Number (MCN) Index.
These survival plots display the multifactor copy number (MCN) index comprised of any sample that contains a copy number alteration (CNA) in any of the following regions: 20q13.2 gain, 20q13.13 [CEBPB] gain, MYC gain, 16q24.1 deletion, 16q23.3-24.1 deletion, Trisomy 5, Trisomy 8, Trisomy 20. The lower, red line represents the 19 individuals with a CNA in at least one region (“Multifactor-POSITIVE”) while the blue line represents the 21 individuals with no CNA in any of the regions (“Multifactor-NEGATIVE”). The MCN-index correlated significantly with both EFS (P=.013) and OS (P=.00059).
Metastatic Lesions (MLs)
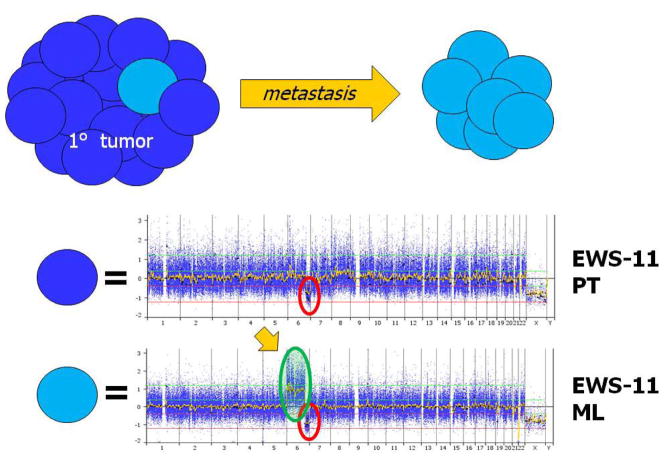
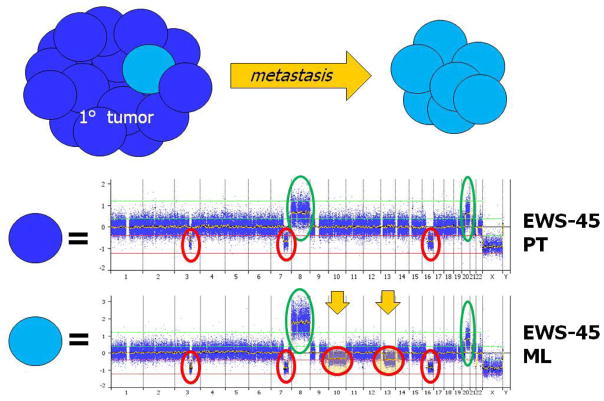
Analysis of paired MLs from archived FFPE samples allowed us to recognize acquired lesions at relapse, see Figures 4a–b. In addition, we compared the 30 PTs without metastases to the 12 MLs in order to identify differences unique to MLs using the two-tailed Fisher’s Exact Test. Two regions were found to be significant including an allelic imbalance in 7q22.1 (0% PTs vs. 25% MLs, P=0.0192) and a gain in 12p12.3 - p12.2 (7% PTs vs. 42% MLs, P=0.0138). No other regions reached statistical significance, see Supplementary Material Figure 3. Further investigation of the 7q22.1 allelic imbalance revealed a nearly 1 MB region deleted in 2 ML samples and copy neutral LOH in 1 ML sample (total n=3/12, 25%). Only one PT contained this same 7q22.1 deletion and it was the PT paired to the ML with the same deletion. The 12p12.3 - p12.2 gain contained only a single predicted gene of unknown function. The focal 7q22.1 region contained 10 unique genes including the RELN (reelin) gene. The reduced expression of RELN has been implicated in other tumor types such as hepatocellular,(38) pancreatic,(39) and gastric carcinomas.(40–41)
Figure 4.
Figures 4a and b. Paired Metastatic Lesions.
Two sets of paired diagnostic and metastatic lesions are displayed. X-axis represents chromosomes labeled and divided by bars, and Y-axis represents linear copy number values re-centered on zero (0=2 copies). Figure 4a) Note the progression of CNAs in the metastatic lesion with the acquisition of trisomy 6p in addition to the 6q deletion already present at diagnosis. Figure 4b) Note the metastatic acquisition of chromosome 10 and 13 deletions in an already genomically unstable diagnostic sample. Also note that the deletions are subclonal in nature with copy number values of 1.5 (ie., only half the metastatic tumor contains subclones with the newly acquired deletions).
DISCUSSION
The introduction of genome-wide, high-resolution SNP microarray technology has led to the discovery of CNAs in a variety of common tumors such as breast cancer (42), ovarian cancer (43), lung cancer (44), glioblastoma (45), neuroblastoma (46–47), pediatric astrocytoma (28, 48), and acute lymphoblastic leukemia (ALL) (29, 49–50). These copy number studies identified specific genes and molecular pathways that have emerged as prognostic markers and therapeutic targets, such as the IKZF1 and the JAK-STAT signaling pathway in childhood ALL (51–52). CNA investigations by new technologies continue to lead to clinical improvement in the common cancers readily available for analysis.
The less common cancers, such as ES, have not been as thoroughly investigated by SNP microarray technologies. The limited number of published CNA studies in ES inconsistently correlate their findings with outcome, most likely due to relatively small sample sizes and differing resolution of aCGH platforms (17). With ES survival ranging between 10–30% for metastatic or relapsed disease (6–7), the identification of new molecular candidates for risk stratification and therapeutic targets remains imperative for this still deadly disease. In this study, we take advantage of novel MIP technology to analyze clinically-archived FFPE diagnostic, metastatic, and post-treatment ES samples that have been accumulating for many years. These FFPE specimens originated from previously treated patients at our institution and so we were able to correlate our molecular findings with clinical factors in the medical record.
The most unexpected finding of our study was the identification and validation of SMARCB1 protein loss in up to 10% of samples initially diagnosed as ES. Inactivation and loss of the tumor suppressor SMARCB1 defines the diagnosis of soft-tissue rhabdoid tumors and atypical teratoid/rhabdoid tumors (ATRTs), but has never been reported in ES despite initial studies that looked for it (53–55). The previous inability to detect SMARCB1 loss in ES may have been due to the quality of the initial antibodies used or the small number of ES cases investigated. In rhabdoid tumors, SMARCB1 inactivation can occur through predisposing germline mutations or deletions (15–20%) as well as somatic changes (65%) (53). Loss of SMARCB1 protein expression has been linked to other tumors such as myxoid (55) and poorly differentiated chordomas (56), epithelioid sarcomas (57–58), epithelioid malignant peripheral nerve sheath tumors (MPNSTs) (32), renal medullary carcinomas (59), pediatric undifferentiated soft tissue sarcomas (60), and hepatoblastoma (61). In one study, a similar gene SMARCA5, also responsible for chromatin remodeling, was found to be a novel fusion product with EWSR1 in t(4;22)(q31;q12) (62), but the gene was not deleted.
All patients in our cohort were clinically diagnosed and treated for ES. Two of the SMARCB1-negative patients from Utah contained what appear to be EWSR1-translocations. Upon blinded review, an anatomic pathologist with expertise in bone tumors and soft tissue sarcomas agreed that 40% (4/10) of the SMARCB1-negative tumors appeared to be classic in their appearance and staining for ES. The remaining 6 tumors (despite being clinically diagnosed and treated as ES) were atypical with a mixture of large cell ES and rhabdoid-like tumor cells. Of note, despite the presence of rhabdoid cells in some of these atypical ES cases, the pathologist did not believe these specimens were classic for the diagnosis of rhabdoid tumors.
All SMARCB1-negative samples in the pediatric cohorts (3 Utah + 3 COG) came from patients 11 years old or less (mean 8 years old), younger than the typical adolescent and young adult commonly diagnosed with ES (2–3). The Michigan TMA included adult patients and was skewed to patients older patients than the Utah or COG pediatric cohorts, so is difficult to evaluate the effect of age in the SMARCB1-negative patients in the Michigan TMA. Eight of the total INI1-negative tumors (80%) in all three cohorts presented in soft tissue structures, and one of the two osseous tumors presented in both the scalp and skull; this extraskeletal predominant presentation in SMARCB1-negative ES is reminiscent of rhabdoid tumor and may represent a common underlying molecular mechanism of tumorigenesis in soft tissue. Outcome data was unavailable from the COG patients, but all 3/3 SMARCB1-negative patients from the Utah pediatric cohort and 2/3 of the SMARCB1-negative PT patients in the Michigan cohort are alive without relapse or disease and the 1 ML patients is currently AWD. This high rate of survival would be unexpected in traditional rhabdoid diagnoses. Perhaps the combination of EWSR1-translocations with SMARCB1 loss confers susceptibility to treatment, or the specific ES therapy itself could have contributed to the favorable outcome in these patients.
Ultimately, the diagnosis of ES, like any other tumor, needs to be based upon careful histopathology, immunohistochemical, and genetic examination. The samples for this study were obtained from clinical archives (Utah) or part of external TMAs (COG, Michigan), so we were unable to perform adequate molecular studies to confirm the diagnosis of ES in all of the SMARCB1-deleted tumors. Nevertheless, careful review by an expert pathologist in bone tumors confirmed that at least 40% of these tumors appeared to be “conventional ES” under the microscope (two of which contained FISH evidence for EWSR1 rearrangement), and most of the remaining 60% contained large cell ES mixed with a variety of spindle and rhabdoid cells. We believe, therefore, that the possibility must be considered that this unique set of tumors represents atypical ES, atypical rhabdoid tumors, or some hybrid combination of both ES-rhabdoid tumors. We are now collecting more cases of atypical ES for further genomic scrutiny of the SMARCB1 region.
Other CNAs detected in our study have been previously described in the ES copy number literature, including gains in chromosome 8, 12, 20 and 1q as well as losses in 16q and 9p21. Using MIP technology, we narrowed 20q gains to smaller, focal regions in 20q13.12 and 20q13.13, and 16q losses to 16q24.1 and 16q23.3-24.1. We demonstrated that increased CEBPB copy number in the 20q13.13 gain correlates with increased nuclear staining and presumably function. Further exploration of the connection between CEBPB gains, biological function, and outcome are now required. We also identified the novel finding of trisomy 5 in older patients with metastatic disease and worse outcome. Many of the genes found within these CNA regions, including SMARCB1, can now be targeted in the laboratory for a better understanding of ES development and progression.
Many of our candidate regions reached only marginal significance when correlated with outcome, especially when adjusting for metastasis and age. We therefore developed a more robust model for outcome prediction by combining these multiple CNAs into a single variable for risk (MCN-index). The MCN-index maintained significance for OS even when using the very strict bootstrapping statistical method. In our cohort, survival was 100% for patients without any CNAs in the MCN-index (20q13.2 gain, 20q13.13 [CEBPB] gain, MYC gain, 16q24.1 deletion, 16q23.3-24.1 deletion, Trisomy 5, Trisomy 8, Trisomy 20). It would be useful to apply the MCN-index analysis to a larger number of clinically annotated, well-characterized cohort of patients to determine its prognostic value in different clinical and genetic groups. If validated in larger cohorts, the use of the MCN-index to accurately identify ES patients at low risk for relapse could be helpful with clinical trial design and the potential to selectively decrease therapeutic intensity in these patients.
We also studied a relatively large number of metastatic samples in our cohort. Twenty-five percent of our ML samples contained genetic alterations in the RELN gene compared to none of the PTs without metastasis. RELN encodes an extracellular protein that regulates cell-cell interactions critical for cell positioning and neuronal migration during brain development (63–64). In breast cancer, promoter methylation controls RELN expression and decreased expression has been linked to increased invasiveness and metastatic potential (65). We believe RELN deletions may be informative about ES metastatic biology, but also can serve as a potential therapeutic candidate for further investigation. Efforts are now underway to collect more of these rare metastatic lesions from ES patients to increase sample size and the likelihood of finding recurrent genomic alterations in MLs. The ability to target metastatic-specific lesions would be very helpful in ES due to limited therapeutic options and poor survival for patients with widespread disease.
In this study, we demonstrate that MIP technology can evaluate molecular alterations in clinically-archived FFPE samples in ES. We identified both novel and known CNAs, including the novel finding of SMARCB1 alterations and protein loss in a unique subset of patients clinically diagnosed as ES. These unusual SMARCB1-deleted tumors may represent atypical cases of large cell ES or perhaps a hybrid of ES-rhabdoid tumors. Finally, we created a MCN-index based on genome-wide microarray findings that identified 100% of surviving patients versus less than 40% who relapsed and died. As these findings are replicated in future studies, genome-wide microarray technology may hold a useful place for patient risk stratification and therapeutic target identification in ES.
Supplementary Material
Acknowledgments
J.D.S. received support for this research from St. Baldrick’s Foundation, Sarcoma Alliance for Research through Collaboration (SARC) Career Development Award, Damon Runyon Clinical Investigator Award, and the Eunice Kennedy Shriver Children’s Health Research Career Development Award NICHD 5K12HD001410. S.L.L. is supported by the NIH (R21 CA138295, R01 CA140394), the Terri Anna Perine Sarcoma Fund, and the University of Utah Department of Pediatrics and Huntsman Cancer Institute/Huntsman Cancer Foundation. We also acknowledge NIH support to the Huntsman Cancer Institute (P30 CA042014) and the Children’s Oncology Group (COG) for the tissue microarray. Finally, we acknowledge Elizabeth Lawlor, MD PhD (University of Michigan) and Grant Rowe MD PhD (Harvard University) who helped to gather the clinical data on the Michigan tissue microarray.
References
- 1.Carvajal R, Meyers P. Ewing’s sarcoma and primitive neuroectodermal family of tumors. Hematol Oncol Clin North Am. 2005 Jun;19(3):501–25. vi–vii. doi: 10.1016/j.hoc.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Esiashvili N, Goodman M, Marcus RB., Jr Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol. 2008 Jun;30(6):425–30. doi: 10.1097/MPH.0b013e31816e22f3. [DOI] [PubMed] [Google Scholar]
- 3.Bleyer A, O’Leary M, Barr R, Ries L, editors. Cancer Epidemiology in Older Adolescents and Young Adults 15 to 29 Years of Age, Including SEER Incidence and Survival: 1975–2000. Bethesda, MD: National Cancer Institute; 2006. NIH Pub. No. 06-5767. [Google Scholar]
- 4.Jawad MU, Cheung MC, Min ES, Schneiderbauer MM, Koniaris LG, Scully SP. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973–2005. Cancer. 2009 Aug 1;115(15):3526–36. doi: 10.1002/cncr.24388. [DOI] [PubMed] [Google Scholar]
- 5.Linabery AM, Ross JA. Childhood and adolescent cancer survival in the US by race and ethnicity for the diagnostic period 1975–1999. Cancer. 2008 Nov 1;113(9):2575–96. doi: 10.1002/cncr.23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grier HE, Krailo MD, Tarbell NJ, Link MP, Fryer CJ, Pritchard DJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003 Feb 20;348(8):694–701. doi: 10.1056/NEJMoa020890. [DOI] [PubMed] [Google Scholar]
- 7.DuBois SG, Krailo MD, Lessnick SL, Smith R, Chen Z, Marina N, et al. Phase II study of intermediate-dose cytarabine in patients with relapsed or refractory Ewing sarcoma: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2009 Mar;52(3):324–7. doi: 10.1002/pbc.21822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turc-Carel C, Aurias A, Mugneret F, Lizard S, Sidaner I, Volk C, et al. Chromosomes in Ewing’s sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12) Cancer Genet Cytogenet. 1988 Jun;32(2):229–38. doi: 10.1016/0165-4608(88)90285-3. [DOI] [PubMed] [Google Scholar]
- 9.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010 Jun 14; doi: 10.1038/onc.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang HY, Illei PB, Zhao Z, Mazumdar M, Huvos AG, Healey JH, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. J Clin Oncol. 2005 Jan 20;23(3):548–58. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 11.Armengol G, Tarkkanen M, Virolainen M, Forus A, Valle J, Bohling T, et al. Recurrent gains of 1q, 8 and 12 in the Ewing family of tumours by comparative genomic hybridization. Br J Cancer. 1997;75(10):1403–9. doi: 10.1038/bjc.1997.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozaki T, Paulussen M, Poremba C, Brinkschmidt C, Rerin J, Ahrens S, et al. Genetic imbalances revealed by comparative genomic hybridization in Ewing tumors. Genes Chromosomes Cancer. 2001 Oct;32(2):164–71. doi: 10.1002/gcc.1178. [DOI] [PubMed] [Google Scholar]
- 13.Hattinger CM, Potschger U, Tarkkanen M, Squire J, Zielenska M, Kiuru-Kuhlefelt S, et al. Prognostic impact of chromosomal aberrations in Ewing tumours. Br J Cancer. 2002 Jun 5;86(11):1763–9. doi: 10.1038/sj.bjc.6600332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurici D, Perez-Atayde A, Grier HE, Baldini N, Serra M, Fletcher JA. Frequency and implications of chromosome 8 and 12 gains in Ewing sarcoma. Cancer Genet Cytogenet. 1998 Jan 15;100(2):106–10. doi: 10.1016/s0165-4608(97)00028-9. [DOI] [PubMed] [Google Scholar]
- 15.Wei G, Antonescu CR, de Alava E, Leung D, Huvos AG, Meyers PA, et al. Prognostic impact of INK4A deletion in Ewing sarcoma. Cancer. 2000 Aug 15;89(4):793–9. doi: 10.1002/1097-0142(20000815)89:4<793::aid-cncr11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Tarkkanen M, Kiuru-Kuhlefelt S, Blomqvist C, Armengol G, Bohling T, Ekfors T, et al. Clinical correlations of genetic changes by comparative genomic hybridization in Ewing sarcoma and related tumors. Cancer Genet Cytogenet. 1999 Oct 1;114(1):35–41. doi: 10.1016/s0165-4608(99)00031-x. [DOI] [PubMed] [Google Scholar]
- 17.Jahromi MS, Jones KB, Schiffman JD. Copy Number Alterations and Methylation in Ewing’s Sarcoma. Sarcoma. 2011;2011:362173. doi: 10.1155/2011/362173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savola S, Klami A, Tripathi A, Niini T, Serra M, Picci P, et al. Combined use of expression and CGH arrays pinpoints novel candidate genes in Ewing sarcoma family of tumors. BMC Cancer. 2009;9:17. doi: 10.1186/1471-2407-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shing DC, Morley-Jacob CA, Roberts I, Nacheva E, Coleman N. Ewing’s tumour: novel recurrent chromosomal abnormalities demonstrated by molecular cytogenetic analysis of seven cell lines and one primary culture. Cytogenet Genome Res. 2002;97(1–2):20–7. doi: 10.1159/000064063. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira BI, Alonso J, Carrillo J, Acquadro F, Largo C, Suela J, et al. Array CGH and gene-expression profiling reveals distinct genomic instability patterns associated with DNA repair and cell-cycle checkpoint pathways in Ewing’s sarcoma. Oncogene. 2008 Mar 27;27(14):2084–90. doi: 10.1038/sj.onc.1210845. [DOI] [PubMed] [Google Scholar]
- 21.Zielenska M, Zhang ZM, Ng K, Marrano P, Bayani J, Ramirez OC, et al. Acquisition of secondary structural chromosomal changes in pediatric ewing sarcoma is a probable prognostic factor for tumor response and clinical outcome. Cancer. 2001 Jun 1;91(11):2156–64. doi: 10.1002/1097-0142(20010601)91:11<2156::aid-cncr1244>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 22.Jahromi MS, Jones KB, Schiffman JS. Copy Number Alterations (CNAs) and Methylation in Ewing’s Sarcoma. Sarcoma. doi: 10.1155/2011/362173. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Carlton VE, Karlin-Neumann G, Sapolsky R, Zhang L, Moorhead M, et al. High quality copy number and genotype data from FFPE samples using Molecular Inversion Probe (MIP) microarrays. BMC Med Genomics. 2009;2:8. doi: 10.1186/1755-8794-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Moorhead M, Karlin-Neumann G, Wang NJ, Ireland J, Lin S, et al. Analysis of molecular inversion probe performance for allele copy number determination. Genome Biol. 2007;8(11):R246. doi: 10.1186/gb-2007-8-11-r246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brewster AM, Thompson P, Sahin AA, Do KA, Edgerton ME, Murray JL, et al. Copy Number Imbalances Between Screen and Symptom-Detected Breast Cancers and Impact on Disease-free Survival. Cancer Prev Res (Phila) 2011 Jul 27; doi: 10.1158/1940-6207.CAPR-10-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiffman JD, Lorimer PD, Rodic V, Jahromi MS, Downie JM, Bayerl MG, et al. Genome wide copy number analysis of paediatric Burkitt lymphoma using formalin-fixed tissues reveals a subset with gain of chromosome 13q and corresponding miRNA over expression. Br J Haematol. 2011 Nov;155(4):477–86. doi: 10.1111/j.1365-2141.2011.08883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Moorhead M, Karlin-Neumann G, Falkowski M, Chen C, Siddiqui F, et al. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33(21):e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiffman JD, Hodgson JG, VandenBerg SR, Flaherty P, Polley MY, Yu M, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010 Jan 15;70(2):512–9. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiffman JD, Wang Y, McPherson LA, Welch K, Zhang N, Davis R, et al. Molecular inversion probes reveal patterns of 9p21 deletion and copy number aberrations in childhood leukemia. Cancer Genet Cytogenet. 2009 Aug;193(1):9–18. doi: 10.1016/j.cancergencyto.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Welch K. Molecular inversion probe assay for allelic quantitation. Methods Mol Biol. 2009;556:67–87. doi: 10.1007/978-1-60327-192-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 32.Hollmann TJ, Hornick JL. INI1-deficient tumors: diagnostic features and molecular genetics. Am J Surg Pathol. 2011 Oct;35(10):e47–63. doi: 10.1097/PAS.0b013e31822b325b. [DOI] [PubMed] [Google Scholar]
- 33.Houglum K, Buck M, Adir V, Chojkier M. LAP (NF-IL6) transactivates the collagen alpha 1(I) gene from a 5′ regulatory region. J Clin Invest. 1994 Aug;94(2):808–14. doi: 10.1172/JCI117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchimochi K, Otero M, Dragomir CL, Plumb DA, Zerbini LF, Libermann TA, et al. GADD45beta enhances Col10a1 transcription via the MTK1/MKK3/6/p38 axis and activation of C/EBPbeta-TAD4 in terminally differentiating chondrocytes. J Biol Chem. 2010 Mar 12;285(11):8395–407. doi: 10.1074/jbc.M109.038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng PK, Tsui SK, Lau CP, Wong CH, Wong WH, Huang L, et al. CCAAT/enhancer binding protein beta is up-regulated in giant cell tumor of bone and regulates RANKL expression. J Cell Biochem. 2010 May;110(2):438–46. doi: 10.1002/jcb.22556. [DOI] [PubMed] [Google Scholar]
- 36.Sollazzo MR, Benassi MS, Magagnoli G, Gamberi G, Molendini L, Ragazzini P, et al. Increased c-myc oncogene expression in Ewing’s sarcoma: correlation with Ki67 proliferation index. Tumori. 1999 May-Jun;85(3):167–73. doi: 10.1177/030089169908500304. [DOI] [PubMed] [Google Scholar]
- 37.Neale G, Su X, Morton CL, Phelps D, Gorlick R, Lock RB, et al. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008 Jul 15;14(14):4572–83. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamura Y, Nomoto S, Kanda M, Hayashi M, Nishikawa Y, Fujii T, et al. Reduced expression of reelin (RELN) gene is associated with high recurrence rate of hepatocellular carcinoma. Ann Surg Oncol. 2011 Feb;18(2):572–9. doi: 10.1245/s10434-010-1273-z. [DOI] [PubMed] [Google Scholar]
- 39.Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006 Feb;130(2):548–65. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Resende C, Ristimäki A, Machado JC. Genetic and epigenetic alteration in gastric carcinogenesis. Helicobacter. 2010 Sep;15(Suppl 1):34–9. doi: 10.1111/j.1523-5378.2010.00782.x. [DOI] [PubMed] [Google Scholar]
- 41.Dohi O, Takada H, Wakabayashi N, Yasui K, Sakakura C, Mitsufuji S, et al. Epigenetic silencing of RELN in gastric cancer. Int J Oncol. 2010 Jan;36(1):85–92. [PubMed] [Google Scholar]
- 42.Karlsson E, Waltersson MA, Bostner J, Perez-Tenorio G, Olsson B, Hallbeck AL, et al. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes Chromosomes Cancer. 2011 Jul 11; doi: 10.1002/gcc.20900. [DOI] [PubMed] [Google Scholar]
- 43.Gorringe KL, George J, Anglesio MS, Ramakrishna M, Etemadmoghadam D, Cowin P, et al. Copy number analysis identifies novel interactions between genomic loci in ovarian cancer. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0011408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007 Dec;450(7171):893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):4323–8. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One. 2007;2(2):e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forshew T, Tatevossian RG, Lawson AR, Ma J, Neale G, Ogunkolade BW, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009 Jun;218(2):172–81. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 49.Paulsson K, Cazier JB, Macdougall F, Stevens J, Stasevich I, Vrcelj N, et al. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: Unexpected similarities with pediatric disease. Proc Natl Acad Sci U S A. 2008 May;105(18):6708–13. doi: 10.1073/pnas.0800408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007 Apr 12;446(7137):758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 51.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. N Engl J Med. 2009 Jan 7; doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009 Jun 9;106(23):9414–8. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eaton KW, Tooke LS, Wainwright LM, Judkins AR, Biegel JA. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatric Blood & Cancer. 2011;56(1):7–15. doi: 10.1002/pbc.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoot AC, Russo P, Judkins AR, Perlman EJ, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am J Surg Pathol. 2004 Nov;28(11):1485–91. doi: 10.1097/01.pas.0000141390.14548.34. [DOI] [PubMed] [Google Scholar]
- 55.Kohashi K, Oda Y, Yamamoto H, Tamiya S, Oshiro Y, Izumi T, et al. SMARCB1/INI1 protein expression in round cell soft tissue sarcomas associated with chromosomal translocations involving EWS: a special reference to SMARCB1/INI1 negative variant extraskeletal myxoid chondrosarcoma. Am J Surg Pathol. 2008 Aug;32(8):1168–74. doi: 10.1097/PAS.0b013e318161781a. [DOI] [PubMed] [Google Scholar]
- 56.Mobley BC, McKenney JK, Bangs CD, Callahan K, Yeom KW, Schneppenheim R, et al. Loss of SMARCB1/INI1 expression in poorly differentiated chordomas. Acta Neuropathol. 2010 Dec;120(6):745–53. doi: 10.1007/s00401-010-0767-x. [DOI] [PubMed] [Google Scholar]
- 57.Hornick JL, Dal Cin P, Fletcher CD. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am J Surg Pathol. 2009 Apr;33(4):542–50. doi: 10.1097/PAS.0b013e3181882c54. [DOI] [PubMed] [Google Scholar]
- 58.Modena P, Lualdi E, Facchinetti F, Galli L, Teixeira MR, Pilotti S, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res. 2005 May;65(10):4012–9. doi: 10.1158/0008-5472.CAN-04-3050. [DOI] [PubMed] [Google Scholar]
- 59.Cheng JX, Tretiakova M, Gong C, Mandal S, Krausz T, Taxy JB. Renal medullary carcinoma: rhabdoid features and the absence of INI1 expression as markers of aggressive behavior. Mod Pathol. 2008 Jun;21(6):647–52. doi: 10.1038/modpathol.2008.44. [DOI] [PubMed] [Google Scholar]
- 60.Kreiger PA, Judkins AR, Russo PA, Biegel JA, Lestini BJ, Assanasen C, et al. Loss of INI1 expression defines a unique subset of pediatric undifferentiated soft tissue sarcomas. Mod Pathol. 2009 Jan;22(1):142–50. doi: 10.1038/modpathol.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trobaugh-Lotrario AD, Tomlinson GE, Finegold MJ, Gore L, Feusner JH. Small cell undifferentiated variant of hepatoblastoma: adverse clinical and molecular features similar to rhabdoid tumors. Pediatr Blood Cancer. 2009 Mar;52(3):328–34. doi: 10.1002/pbc.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sumegi J, Nishio J, Nelson M, Frayer RW, Perry D, Bridge JA. A novel t(4;22)(q31;q12) produces an EWSR1-SMARCA5 fusion in extraskeletal Ewing sarcoma/primitive neuroectodermal tumor. Mod Pathol. 2011 Mar;24(3):333–42. doi: 10.1038/modpathol.2010.201. [DOI] [PubMed] [Google Scholar]
- 63.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011 Feb 10;69(3):482–97. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichols AJ, Olson EC. Reelin promotes neuronal orientation and dendritogenesis during preplate splitting. Cereb Cortex. 2010 Sep;20(9):2213–23. doi: 10.1093/cercor/bhp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stein T, Cosimo E, Yu X, Smith PR, Simon R, Cottrell L, et al. Loss of reelin expression in breast cancer is epigenetically controlled and associated with poor prognosis. Am J Pathol. 2010 Nov;177(5):2323–33. doi: 10.2353/ajpath.2010.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.