Abstract
Sequence-based variation in gene expression is a key driver of disease risk. Common variants regulating expression in cis have been mapped in many eQTL studies typically in single tissues from unrelated individuals. Here, we present a comprehensive analysis of gene expression across multiple tissues conducted in a large set of mono- and dizygotic twins that allows systematic dissection of genetic (cis and trans) and non-genetic effects on gene expression. Using identity-by-descent estimates, we show that at least 40% of the total heritable cis-effect on expression cannot be accounted for by common cis-variants, a finding which exposes the contribution of low frequency and rare regulatory variants with respect to both transcriptional regulation and complex trait susceptibility. We show that a substantial proportion of gene expression heritability is trans to the structural gene and identify several replicating trans-variants which act predominantly in a tissue-restricted manner and may regulate the transcription of many genes.
The risk of developing common complex diseases such as type 2 diabetes and obesity involves multiple genetic and environmental factors. Genome-wide association studies (GWAS) have been successful in identifying common genetic variants associated with these complex human diseases. A suggested approach for finding the additional genetic components is to focus on low frequency and rare variants 1. In parallel, approaches to disentangle the underlying molecular mechanism for identified disease loci are also needed. As the majority of the common genetic variants associated with complex traits map to non-coding regions and may thus alter gene regulation the use of gene expression data integrated with sequence variation, eQTL studies, is a commonly applied approach using various cell 2-5 and tissue samples6-8. More recently, we and others have been able to collect multiple cells and/or tissues from the same individuals9-12 demonstrating the degree of tissue dependency of cis-regulatory effects 11,13.Tissue-dependency seems to be an important feature of disease susceptibility variants regulating gene expression11,14 promoting the use of multiple disease-targeted cell-types in future large-scale eQTL studies. However, despite the success in mapping common cis-regulatory variants in these studies, 8,15,16trans variants have been more difficult to map mainly due to small effect sizes, emphasising the need for well-powered studies and thorough replication efforts in multiple tissues.
To this end, we designed the MuTHER project (Multiple Tissue Human Expression Resource ) to develop a major resource of detailed genomic and transcriptomic data from three disease-relevant tissues (adipose, lymphoblastoid cell lines, and skin) originating from a cohort of 856 deeply phenotyped twins (one-third monozygotic (MZ) and two-thirds dizygotic (DZ)) from the TwinsUK adult registry. The increased sample size compared to our pilot study 11 allow us to utilize the classical twin design for systematic dissection of the genetic (cis and trans) effects on gene expression providing for the first time 1) estimates of additional heritable cis-effects unexplained by common SNPs (MAF > 5%) identified in standard cis-eQTL analysis and 2) in depth characterization of the architecture of trans-regulation of gene expression highlighted by thorough replication efforts in multiple independent data sets. In addition, the large-scale multi-tissue design of our study also allows us to provide the most precise estimates to date of not only gene expression heritability but also of the degree of tissue-dependency of eQTLs. The relevance of these tissue-dependent eQTLs in complex trait susceptibility is further highlighted as for hundreds of the known GWAS SNPs we reveal candidate causal eQTLs with good correlation of the phenotype to the candidate tissue.
RESULTS
Data structure
In total of 856 female twins (154 monozygotic (MZ) twin pairs, 232 dizygotic (DZ) twin pairs and 84 singletons) aged 38.7-84.6 years were recruited from the TwinsUK resource 17and adipose (subcutaneous fat) and skin tissue biopsies as well as peripheral blood samples (for generation of lymphoblastoid cell lines or LCLs) were collected for subsequent genome-wide expression profiling. The TwinsUK cohort has previously shown to be comparable to population singletons in terms of disease-related and lifestyle characteristics 18 . Cohort characteristics are presented in Supplementary Table 1.
Genetic and non-genetic effects on gene expression
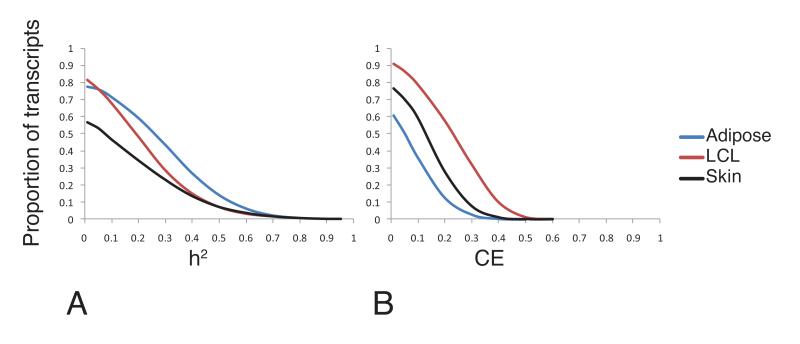
We estimated narrow-sense heritability, h2, for each transcript across the three tissues in all available twin pairs using a variance-component model adjusting for known technical cofactors19. The average h2 estimates of expressed transcripts corresponded to h2adipose=0.26, h2lcl=0.21 andh2skin=0.16 (Fig 1, Supplementary Table 2). The cell-type heterogeneity expected in skin most likely explains the lower estimates in that tissue. We tested how heritability of transcripts compares across tissues and found that around 50% of the top 5000 heritable transcripts in each tissue (corresponding toh2adipose>0.33, h2LCL>0.27 and h2skin>0.22) are in fact heritable across two or more tissues (Supplementary Fig. 1A) with similar results when restricting to transcripts expressed in all three tissues (Supplementary Fig. 1B).
Fig 1. Genetic and non-genetic effect of gene expression across multiple tissues.
Estimation of the proportion of variation in expressed transcripts in adipose (N=11394), LCL (N=10631) and skin tissue (N=11932) attributable to genetic (narrow-sense heritability, h2)(A) and familial non-genetic factors (shared common environment, CE)(B). The y-axis is showing the proportion of transcripts at h2/CE cut-off indicated on the x-axis.
Twin studies also allow calculation of the proportion of phenotypic variation attributable to familial non-genetic factors, i.e. the shared common environment. Notably, we found that as much as 32% of expressed LCL transcripts have a common environment component that explains over30% of the total variance compared to 2% and 8% in adipose and skin tissue (Fig. 1B). This larger shared environmental effect in LCLs most likely reflect the impact of additional correlated sample handling steps not applicable for tissue biopsies such as blood sampling, cell isolation, EBV transformation and cell culture procedures as the study subjects visited the clinic in pairs.
Large-scale cis-eQTL mapping
To map the underlying common, genetic effect of transcript levels, we performed global cis-eQTL mapping associating the 23,596 expression traits with imputed HapMap2 genotypes in a linear mixed (polygenic) model followed by a score test taking the relatedness into account (Supplementary Fig 2). cis-eQTLs were called with a per tissue FDR of 1%, which corresponds to P <5.0×10−5 in adipose, P < 7.8×10−5 in LCL and P < 3.8×10−5 in skin tissue, respectively. Across all transcripts, we detected an abundance of cis-eQTLs per tissue (Nadipose=3529, NLCL=4625, Nskin=2796, Supplementary Table 3) with 14%, 17% and 10% of transcripts with a cis-eQTL in adipose, LCL and skin tissue, respectively having more than one independent cis-eQTLs. For these transcripts associated with at least one cis-variant, the average h2 estimates are 0.31, 0.25 and 0.21 in adipose, LCL and skin tissue, respectively. The probability of detecting large effect sized cis-eQTLs across tissues increases with heritability as shown by average h2 estimates of transcripts associated with a cis-variant at P<5×10-8 where a maximum average h2 seen in adipose tissue is 0.38.
We validated our identified cis-regulatory effects by performing replication studies in independent expression data sets (Supplementary Table 4). Using the list of replication P-values from the different data sets we first estimated π0 which is the overall proportion of true null hypotheses among all tests performed. We could then quantify the proportion of significant replicated cis results in each study, π1, corresponding to π1≡ 1 - π0 20 and noted a high replication rate of cis-eQTLs across studies of similar size (π1= 0.7-0.76) (Supplementary Fig.3).
Most previous efforts to look at tissue dependency of cis-eQTL effects have only used a P-value threshold but this has obvious limitations. Here, we employed several complementary approaches in addition to the threshold-based approach to address this question. First, we assessed tissue dependency by studying shared effects at 1% FDR and found substantial tissue independency of cis-eQTLs (Table 1). For instance, 47% of cis-eQTLs identified at 1% FDR in adipose tissue are shared in at least one other tissue and as many as 22% are seen across all three tissues at a similar FDR threshold. This degree of sharing was further confirmed by estimating the proportion of significant results across tissues (π1=0.5-0.7) (Supplementary Fig. 4). As with previous smaller studies 9-11,13, tissue independent cis-eQTLs had larger effect sizes and were over-represented close to transcription start sites compared to tissue dependent effects (Supplementary Fig. 5). In general, cis-effects located less than 200kb from TSS explain a larger proportion of the variance in expression levels (r2average_adipose=0.07, r2average_LCL=0.08, r2average_skin=0.08) than long-range effects (located >200 kb from TSS) (r2average_adipose=0.04, r2average_LCL=0.04, r2average_skin=0.04) (Supplementary Fig. 6).
Table 1.
Estimated degree of tissue sharing of cis-effects (1% FDR) using (A) a threshold-based approach and (B) a matched co-twin design comparing the p-value distributions across tissueswhereπ1 represents the proportion of true positives
| Reference tissue | Secondary tissue | A | B | |
|---|---|---|---|---|
|
| ||||
| N (%) | Twin 1 π1 | Twin 2 π1 | ||
|
| ||||
| Adipose | LCL | 1221 (34.6) | 0.64 | 0.56 |
| Skin | 1207 (34.3) | 0.79 | 0.77 | |
| LCL and Skin | 767(21.8) | - | - | |
| LCL or Skin | 1661 (47.1) | - | - | |
|
| ||||
| LCL | Adipose | 1118 (24.2) | 0.65 | 0.63 |
| Skin | 978 (21.1) | 0.65 | 0.61 | |
| Adipose and Skin | 728 (15.7) | - | - | |
| Adipose or Skin | 1368 (29.6) | - | - | |
|
| ||||
| Skin | Adipose | 1265 (45.2) | 0.77 | 0.83 |
| LCL | 1104 (39.5) | 0.64 | 0.64 | |
| Adipose and LCL | 790(28.3) | - | - | |
| Adipose or LCL | 1579 (56.5) | - | - | |
We then characterized tissue dependency of regulatory effects in more detail using a matched co-twin design as previously described 11, comparing the p-value distribution of significant SNP-probe pairs within and across tissues (Supplementary Fig. 7).We found that 56-83% of cis-effects were shared across tissues with adipose and skin sharing more with each other than with LCLs (Table 1). eQTLs sharing statistical significance might still have tissue-dependent biological consequences if they have different effect sizes (fold change in expression) across tissues. Thus, we also evaluated tissue-dependency by contrasting expression fold changes between tissues and estimating the predictive value (r2) of each tissue against the other two (Supplementary Fig. 8). After accounting for winner’s curse (subtracted unexplained intra-tissue variance), we estimate that 41-62% of cis-eQTLs are not only tissue-independent but also have a similar magnitude of effect. Taken together, using several complementary approaches we find evidences pointing towards >60% of cis-eQTLs being statistically significant in multiple tissues.
Dissection of the contribution of cis-effects to heritability of gene expression
To estimate the proportion of the heritability of each transcript that is driven in cis by alleles of high frequency (i.e. common SNPs, here defined as SNPs with minor allele frequency (MAF) > 5%) we combined the results from our heritability and cis-eQTL analyses. As the current sample size is not sufficient to obtain reliable h2 estimates of less than 0.1, we focused on transcripts with h2>0.1 which corresponds to 10,027 (43%), 10,219 (44%) and 7,511 (32%) transcripts in adipose, LCL and skin tissue, respectively (Fig. 1, Supplementary Table 2).
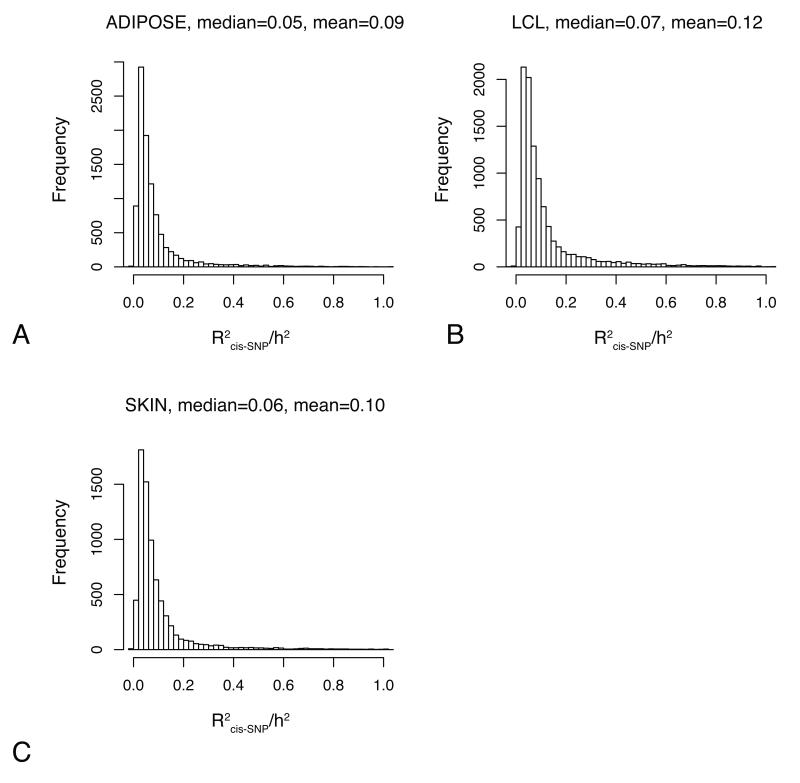
Overall when taking all transcripts into account, we found that common cis-SNPs (MAF >5%) explain on average only 9% (adipose), 12% (LCL), and 10% (skin) of the total genetic variance at each locus (Fig.2). Less than a third (27% (adipose), 33% (LCL) and 21% (skin)) of the transcripts are in fact associated with a cis-variant at 1% FDR so when focusing on these transcripts only and taking independent cis-effects into account, the cis-component accounts for a greater proportion of the genetic variance, namely on average 25% (adipose), 31% (LCL) and 32% (skin). Notably, the effect of common cis-variants increases as heritability increases. If we filter to transcripts that are highly heritable in all tissues (h2>0.6 across all tissues, N=24), ~95% of these transcripts are associated with a cis variant and at 18 of the 24, a single cis-SNP explains over 50% of the genetic variance (Supplementary Table 5).
Fig 2. The contribution of heritable cis-components to gene expression variation.
The contribution of individual cis-SNPs (MAF>5%) to the genetic variance of gene expression (h2>0.1) in adipose (A), LCL (B), and skin tissue (C). The x-axis is showing the proportion of the heritability of each transcript that is explained by independent cis-SNPs.
These results from multiple tissues indicate that a large proportion of the heritability of gene expression remains unexplained by the common SNPs (MAF > 5%) analyzed in standard cis-eQTL analyses. Thus, we asked whether there are other genetic cis-effects which account for the additional genetic variance of gene expression.
We therefore performed quantitative linkage analysis in the cis-regions of those transcripts with h2>0.1 and associated with a common cis-SNP using a global regression approach that analyze the data of all expression traits (Nadipose=2537, NLCL=3157, Nskin=1493) in a single linear regression. We phased all SNPs in each cis-region (~2Mb) and counted the haplotypes shared identity-by-descent (IBD) for all DZ twin pairs. We then estimated the average heritability at each cis region using the global regression approach which is based on the Haseman-Elston algorithm but including all selected transcripts into account. We noted that on average 30% (adipose), 35% (LCL) and 36% (skin) of the total genetic variance is explained by variants in cis which is in fact on average 40% more than if only common cis-SNPs identified from our cis-eQTL analysis are included (Table 2). This added genetic component is likely due to low frequency / rare cis-variants and should be considered as a lower bound as our sample size limited us to conduct the analyses on a subset of the heritable transcripts.
Table 2.
Proportion of the heritability explained by cis-effects
| Tissue | N transcripts | Average h2 | h2cis | h2cis/ h2 | h2SNP | h2SNP/ h2 | h2SNP/ h2cis |
|---|---|---|---|---|---|---|---|
| Adipose | 2537 | 0.40 | 0.12 | 0.30 | 0.072 | 0.18 | 0.60 |
| LCL | 3157 | 0.34 | 0.12 | 0.35 | 0.095 | 0.28 | 0.79 |
| Skin | 1497 | 0.36 | 0.13 | 0.36 | 0.094 | 0.26 | 0.72 |
Integration of cis-eQTL data with disease loci
A major application of eQTL data has been the functional annotation of loci identified in genome-wide association (GWA) studies. We investigated the regulatory impact of GWA variants by integrating cis-eQTLs (1% FDR, Supplementary Table 6) and disease SNPs (NHGRI database, accessed 21.12.10) with the RTC methodology as previously described 14. RTC scores≥0.9 indicate that overlapping eQTL and GWAS signals likely tag the same functional variant. In all three tissues we observed an overrepresentation of high RTC scoring candidates, suggestive of disease effects mediated through gene expression (Supplementary Fig. 9).Of the total interval-disease combinations tested in adipose (N = 765), LCL (N = 887) and skin (N = 639) we detect 181 (23.7%), 225 (25.4%) and 145 (22.7%) signals with RTC ≥0.9 respectively, more than twice the number expected by chance (adipose P = 0.0009, LCL P = 0.008, skin P = 0.0009). The disease eQTL candidates are largely tissue-dependent (~60%), in line with the estimated proportion of tissue-dependent cis-effects (52 of the total non-redundant 358 RTC signals were discovered across all three tissues - Supplementary Table 6).
This suggests that the ability to interpret functionality of GWAS loci is highly dependent on the tissue where gene expression is being interrogated and its relevance to the trait of interest. Indeed, we observe a significant enrichment of immunity-related GWAS signals among high RTC scoring cis-eQTLs in LCLs (P = 6.6×10−5, Fisher’s exact test), much more so than in the other two tissues (adipose P = 0.003, skin P=0.013). Likewise, disease eQTLs detected in adipose and skin samples explain associations with biologically relevant traits (Supplementary Table 7). For example, in adipose we discover regulatory effects potentially explaining associations with triglycerides (rs230413021 - ATP13A1, rs439401 21- APOE)or birth weight (rs900400 22- TIPARP) while in skin, associations with melanoma (rs91087323-ASIP) and skin sensitivity to sun (rs180500724 - DBNDD1) stand out.
Trans-regulation of gene expression across tissues
Although low frequency/rare cis-variants seem to contribute significantly to the total heritable cis-effect , as discussed above, a large proportion of the total heritability (>60%) of gene expression is still unexplained, indicating that the effect of trans-regulatory variants on gene expression are likely to be critical to inter-individual differences in gene expression. We thus proceeded to explore the trans-regulatory landscape across tissues. Given the large number of tests performed and the relatively small effect sizes of trans-eQTLs we chose the P< 5×10−8 GWAS threshold (corresponding to FDR less than 10%) to select possible candidates for further investigation including replication analysis in independent samples (see below). At P< 5×10−8 we found 639, 557, and 609 trans-eQTLs in adipose, LCL and skin respectively (Supplementary Table 8). The relative proportion of trans-eQTLs per tissue is the inverse of that seen in the cis-eQTL analysis, perhaps reflecting the different external environments present for complex tissues vs. cultured cells. In contrast to the cis results, nearly all trans-eQTLs seem to be tissue-dependent, have relatively small effect sizes and are associated with transcripts with lower average h2 (h2adipose=0.19, h2LCL=0.18 and h2skin=0.13).
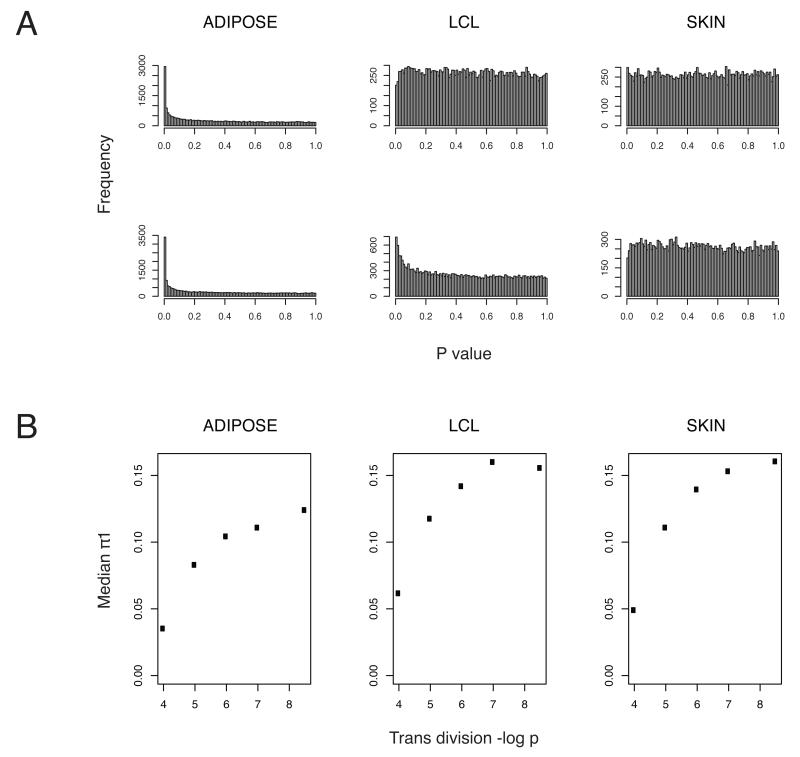
Strikingly, many trans-SNPs at P< 5×10−8 are associated with multiple transcripts, suggesting their role as multi-gene regulators. In adipose tissue, 48 SNPs account for 169 (32%) of the trans-eQTLs and in LCL and skin tissue, 48 SNPs account for 121 (21%) and 44 SNPs for 164(27%) of the trans-eQTLs, respectively. These multi-gene regulators (defined here as trans-SNPs associated with at least two distinct transcripts at P< 5×10−8) consistently show enrichment for additional trans associations with low P-values beneath the 5×10−8 threshold (Fig.3A), indicating that trans-SNPs may regulate additional genes below our P-value threshold. In contrast, the P-value distribution across all measured transcripts in the other two tissues approximates the null (Fig.3). To quantify the genome-wide effect of these trans-SNPs we again used π1 for the estimation of the proportion of true positives in the distribution of P-values from each trans-SNP vs. all transcripts and compared it with similar calculations for trans-SNPs associated with only one transcript at P< 5×10−8 (i.e. single-gene regulators). We find that the multi-gene regulators are enriched for greater number of true positives compared to single-gene regulators (Supplementary Fig. 10, Supplementary Table 9). We further investigated π1 values at trans-SNPs beyond our threshold of P< 5×10−8, and found that the median π1 increases with increasing significance of the top association per trans-SNP (Fig. 3B). As trans-SNPs with more significant association should be enriched for true positives, this confirms that a general property of true trans-SNPs might be regulation of multiple transcripts.
Fig 3. Trans-variants regulating expression of multiple transcripts.
(A)P value distributions (x-axis) of genome-wide associations(N, y-axis) between two potential adipose multi-gene regulators (rs1752223 on chromosome 1, upper panel, and rs7595947 on chromosome 2, lower panel) and transcript levels in adipose (left panel), LCL (middle) and skin tissue (right panel).
(B) Plots showing the median trans-π1 (π1 calculated from p-value distribution of a trans-SNP vs. all probes) at increasing levels of trans-SNP significance in adipose (left panel), LCL (middle) and skin tissue (right panel). The top trans-SNP for each probe was included and trans-SNPs were divided into non-overlapping bins based on the p value of the top trans- association. The median π1(y-axis) is graphed against the -log p (x-axis) of the lower limit of each bin.
We then sought to study the genome-wide regulatory behavior of our LCL trans-SNPs (P<5×10-8) using the calculated π1 values in our replication cohorts (ALSPAC and Oxford-TwinsUK) (Supplementary Table 4). In total 314 trans-SNPs with π1 ≥ 0.10 in the MuTHER discovery cohort were tested of which 61 (19%) and 43 (14%) also have π1 estimates of ≥ 0.10 in the ALSPAC-LCL and/or Oxford-LCL replication cohort, respectively. When comparing not only the π1’s in the replication cohorts but also the top 1000 vs. bottom 1000 associated transcripts for each of the 61 and 43 trans-SNPs, respectively, we find a highly significant enrichment for lower P-values in the top 1000 transcripts (Mann-Whitney, ALSPAC P = 2.6 × 10−6 and Oxford P=3.8 × 10−5). This indicates that not only the genome-wide regulatory behavior but also the ranking of associated genes for a subset of trans-SNPs is consistent across studies but larger sample sizes are needed to confirm the observed effect of gene regulation.
We also performed replication studies of the single trans-loci identified at P< 5×10−8 in independent data sets (Supplementary Table 4) and noted that the replication rate of trans-associations was strikingly lower compared to cis-effects (Supplementary Fig. 3, Table 3) with π1 estimates ranging from 0.0002-0.13 compared to π1 of 0.34-0.76 for cis-replication. However, using a P-value cutoff of <0.05 and taking direction of effect into account we found an up to 3-fold enrichment of replicated trans-eQTLs (Table 3, Supplementary Table 10). Taken together, trans-regulatory effects of gene expression are highly complex with small effect sizes indicating that samples sizes are required to be larger than previously expected.
Table 3.
Replication in independent cohorts of cis – and trans associations
| Tissue | Cohort | N |
trans-hits tested/total hits at P<5×10−8 |
P<0.05* | π 1 |
cis-hits tested/total hits at 1% FDR |
P<0.05* | π 1 |
|---|---|---|---|---|---|---|---|---|
| Adipose (subcutaneous ) |
Decode | 585 | 586/639 | 25/586 (4.3%) |
0.10 | NA | NA | NA |
| Adipose (subcutaneous ) |
MGH | 701 | 514/639 | 27/514 (5.3%) |
0.096 | 2980/3332 | 1751/2980 (59%) |
0.79 |
| LCL | ALSPAC | 931 | 544/557 | 33/544 (6.1%) |
0.13 | 6181/6289 | 4154/6181 (67%) |
0.76 |
| LCL | Oxford (TwinsUK) |
331 | 361/557 | 23/361 (6.4%) |
0.13 | 4608/6289 | 2745/4608 (60%) |
0.75 |
| Skin (fibroblasts) |
GenCORD | 68 | 442/609 | 15/442 (3.4%) |
0.00024 | 2241/3416 | 455/2241 (20%) |
0.34 |
consistent direction
Discussion
We under took a large-scale genetic association study of human gene expression traits in multiple disease-targeted tissue samples (subcutaneous fat, LCL and whole skin) derived from 856 MZ and DZ female twins as part of the MuTHER project. This is the first study performed to date utilizing the twin design for the dissection of genetic and non-genetic components underlying population differences in tissue-independent and dependent expression profiles.
A study using family data sets aiming to partition the heritability of gene expression into cis and trans components recently estimated that 37% of the heritability in blood and 24% in adipose tissue are in fact due to cis-regulation 16. Here we confirm these estimates but decompose the cis component further by utilizing IBD estimates in our DZ subjects. We found that between 30-36% of the heritability is due to cis-components but that up to 40% of the heritable cis-effect or 12% of total heritability is missed when only considering common SNPs from cis-eQTL mapping. However, as our analyses were conducted on heritable transcripts in each tissue for which we observed a significant cis-association from the cis-eQTL mapping approach the estimate of the contribution of undetected regulatory effects to cis genetic variance is most likely an underestimate. Although we acknowledge that common SNPs may in some instances tag low frequency variants 25,26 we expect that a subset of the missing cis-heritability still will be accounted for by low frequency and rare variants supporting large-scale exome and genome re-sequencing initiatives for complex trait mapping. The missing cis-heritability has also intriguing implications for GWA signals in cases where the effect of the lead SNP is mediated via a cis-eQTL, i.e. if a known GWA variant is an eQTL and therefore affects disease risk by modulating expression of a gene, then any additional rare variant modulating expression of the same gene in the same tissue should also affect the same trait. This leads us to predict that on average an additional 40% or more of signal remains to be discovered at cis-eQTL GWAS loci (of which we identify 358 in this study). These estimates are based on calculations within each tissue and thus do not represent tissue-independent cis-heritability. In agreement with a previous study 27 we do not find an enrichment of genetic correlations unequal to zero (data not shown) which could be expected given our high degree of tissue-independency of cis-effects from our cis-eQTL mapping approach. However, as our sample size is limited measurement error in genetic covariance cannot be ruled out.The finding that the majority (>60%) of the genetic effect of expression traits is regulated by components other than those acting in cis, points to the need for studies of the trans-regulatory landscape. Trans-regulatory variants are known to have small effect sizes and thus have previously been difficult to map given limited sample sizes and lack of appropriate replication studies 4. However, the recent findings of disease related trans-variants regulating the expression of multiple genes are promising 28,29. A dilemma with genome-wide eQTL analysis is that only a small proportion of the variants survive multiple testing corrections and by restricting to signals based solely on arbitrary cut-offs, many true hits are likely to be missed. This can be circumvented by analytic methods such as studying the global effect of trans-variants using proportion of true positives (π1) as presented here. By applying this approach and with thorough replication we found evidence of multiple trans-variants acting as multi-gene regulators predominantly in a tissue-dependent manner similar to our previously reported example of the KLF14 locus in adipose tissue 29. For instance, the rs7595947 SNP on chromosome 2 was associated with 27 transcripts in the MuTHER adipose samples and successfully replicated in independent cohorts. In skin, the rs1215608 trans-SNP located within the NUAK1 gene, defined as a multi-gene regulator and associated with three genes (FMO6P, PPM1F, LECT1) in the MuTHER discovery sample, was successfully replicated in the fibroblast cohort. Interestingly, the rs1215608 SNP is also a cis-acting SNP regulating NUAK expression. NUAK1 was recently identified as a key player for cellular senescence and cellular ploidy, mechanisms known to be important in aging30. These examples underscore the potential of utilizing the full transcriptomic architecture to understand biology. However, as demonstrated here by the relative low replication rate the dissection of trans effects and their characteristics such as tissue-dependent are indeed challenging as they are highly complex and require larger sample sizes to be discovered than previously expected.
In conclusion, we present unique twin data using thousands of eQTLs in multiple tissues that extend our understanding of the architecture and regulation of gene expression in multiple ways. We highlight the importance of studying low frequency / rare regulatory variants in complex traits by detecting and mapping missing heritability of gene expression beyond the common cis-variants. We also show that a substantial proportion of gene expression heritability is trans to the structural gene and identify several replicating trans-variants which seem to act predominantly in a tissue-restricted manner and are potentially regulators of many genes.
Online methods
Sample collection
The study included 856 Caucasian female individuals recruited from the TwinsUK Adult twin registry 17 (Supplementary Table 1). Punch biopsies (8mm) were taken from a photo-protected area adjacent and inferior to the umbilicus. Subcutaneous adipose tissue was dissected from each biopsy, weighted and immediately stored in liquid nitrogen. Similarly, the remaining skin tissue was weighted and stored in liquid nitrogen. Peripheral blood samples were collected and lymphoblastoid cell lines (LCLs) were generated by Epstein Barr Virus transformation of the B-lymphocyte component by the European Collection of Cell Cultures agency.
RNA extraction
RNA was extracted from homogenized adipose and skin samples and lysed LCLs using TRIzol Reagent (Invitrogen) according to protocol provided by the manufacturer. RNA quality was assessed with the Agilent 2100 BioAnalyzer (Agilent technologies) and the concentrations were determined using NanoDropND-1000 (NanoDrop Technologies).
Expression profiling
Expression profiling of the samples, each with either two or three technical replicates, were performed using the Illumina Human HT-12 V3 BeadChips (Illumina Inc) including 48,804 probes where 200ng of total RNA was processed according to the protocol supplied by Illumina. All samples were randomized prior to array hybridization and the replicates were hybridized on different beadchips. Raw data was imported to the Illumina Beadstudio software and probes with less than three beads present were excluded. Log2-transformed expression signals were normalized separately per tissue with quantile normalization of the replicates of each individual followed by quantile normalization across all individuals as previously described 11. We acknowledge that quantile normalization does not adjust for shared covariance due to technical factors which may influence subsequent analysis but previous efforts 5 indicate that the impact on the result seems to be minor. Post-QC expression profiles were obtained for 825 (adipose and LCL) and 705 (skin) individuals, respectively. The Illumina probe annotations were cross-checked by mapping the probe sequence to the NCBI Build 36 genome with MAQ 31. Only uniquely mapping probes with no mismatches and either an Ensembl or RefSeq ID were kept for analysis. Probes mapping to genes of uncertain function (LOC symbols) and those encompassing a common SNP (1000G release June 2010) were further excluded leaving 23,596 probes for the analysis.
Genotyping and genotype imputation
Genotyping of the TwinsUK dataset (N = ~6,000) was done with a combination of Illumina HumanHap300, HumanHap610Q, 1M-Duo and 1.2MDuo 1M chips. Intensity data for each of the arrays were pooled separately (with 1M-Duo and 1.2MDuo 1M pooled together) and genotypes called with the Illuminus32 calling algorithm, thresholding on a maximum posterior probability of 0.95 as previously described 29.
Imputation was performed using the IMPUTE software package (v2)26 using two reference panels, P0 (HapMap2, rel 22, combined CEU+YRI+ASN panels) and P1 (610k+, including the combined HumanHap610k and 1M array). Post-imputation, SNPs were filtered at a MAF > 5% and IMPUTE info value of >0.8 resulting in a total of 2,029,988 SNPs available for testing.
Heritability analysis
The classical twin design was applied comparing the similarity of MZ and DZ twins using the ACE model which partitions the variance into: additive genetic (A), common environment (variance due to environmental effects shared within twin pairs)(C) and unique environment (environmental effects not shared within twin pairs)(E). As all twin pairs included in the study visited the clinic in pairs and MZs share 100% of their genes, any differences arising between them in these circumstances are unique (E). The correlation observed between MZ twins provides thus an estimate of A + C. In contrast, DZs have a common shared environment but share on average only 50% of their genes: so the correlation between DZs is a direct estimate of ½A + C . Consequently, twice difference between MZ and DZs gives the genetic additive effect (A) and the common environment (E) is the MZ correlation minus our estimate of the genetic effect (A). A standard linear mixed model was used to estimate these variance components as previously described 19. The following covariates were included in the model: age and experimental batch in adipose and LCL analysis, and age, experimental batch and sample processing in the skin analysis. All available complete twin pairs were included, corresponding to 143 MZ and 214 DZ pairs with adipose profiles, 138 MZ and 221 DZ pairs with LCL and 108 MZ and 162 DZ pairs with skin profiles, respectively.
eQTL analysis
Association of expression levels with probabilities of imputed genotypes were tested in samples of related individuals using a two-step mixed model-based score test developed in the works of Aulchenko et al. and Chen and Abecasis 33,34 and implemented in the GenABEL/ProbABEL packages 35,36. Briefly, the approach is an approximation of a full linear mixed model where the first step includes a mixed model containing all terms but those involving SNPs fitted by maximum-likelihood i.e. fixed effects as well as kinship matrix based on genomic data. Fixed effects included age and experimental batch in the adipose and LCL analysis, while age, batch and sample processing were used in the skin analysis. This step is performed using the GenABEL software 35using the polygenic() function. The resulting object contains the inverse variance-covariance matrix of the estimates and expression trait residuals which are used in the second step together with posterior genotypic probabilities performing the score test in ProbABEL 36, using the “—mmscore” option. In total, 776 adipose, 777 LCL and 667 skin samples had both expression profiles and imputed genotypes and were included in the analysis. Cis analysis was limited to SNPs located within 1MB either side of the transcription start or end site or within the gene body. False discovery rate (FDR) for the cis analysis was calculated from the complete list of p values using the qvalue package20 implemented in R2.11 37. In order to characterize likely independent regulatory effects, the identified cis-eQTLs were mapped to recombination hotspot intervals 38. For each gene, the most significant SNP per hotspot interval were selected followed by additional LD filtering (for each remaining SNP pair with D’ > 0.5 and r2>0.1 the least significant SNP was ignored).
Trans analysis was limited to SNPs located on a different chromosome than the tested transcript. Post-QC analysis of the trans-eQTLs revealed 52 probes with extreme outlier effects which were filtered from further trans analysis. Transcripts associated with a trans-SNP at P<5×10−8 were used for calculations of transcript-wise FDR from the complete list of p values using the qvalue package20 implemented in R2.11 37.
The score test is known to slightly underestimate additive effect sizes 34, so the top association per probe was validated with a linear mixed-effects model in R, using the lmer() function in the lme4 package 39, fitted by maximum-likelihood(Supplementary Fig. 2). The linear mixed-effects model were adjusted for both fixed (age, experimental batch effect and sample processing effect (skin tissue only)) and random effects (family relationship and zygosity). A likelihood ratio test was applied to assess the significance of the SNP effect. The p-value of the SNP effect in each model was calculated from the Chi-square distribution with 1 degree of freedom using -2log(likelihood ratio) as the test statistic.
eQTL analysis using a matched co-twin design
The eQTL analysis was done separately for each tissue, as previously described 11. Within each tissue, twins from the same pair were separated by id in two samples analyzed independently. Related individuals (sister pairs) within a twin set were also removed. This separation resulted in the following sample size for adipose, LCL and skin respectively: Twin 1 (390, 340 and 337) and Twin 2 (384, 338 and 328). For each of the twin–by-tissue sets, associations between genotypes and normalized expression values were conducted using Spearman Rank Correlation (SRC). Age and experimental batch were included as cofactors in the adipose and LCL analysis, and age, batch and sample processing in the skin analysis. We considered a window of <1MB from the TSS for testing SNPs in cis. cis-eQTLs were filtered at a nominal SRC P < 2.5×10−6 corresponding to a 10-3 permutation threshold 11. We contrasted the eQTLs (same SNP-probe combination) and expression fold change (difference in mean expression of homozygous genotypic classes) between twin sets of the same tissue followed by inter-tissue comparison of the same twins (e.g. Twin1 LCL vs. Twin2 LCL, Twin1 LCL vs. Twin1 adipose, Twin1 LCL vs. Twin1 skin).
Global regression
To estimate the proportion of the genetic variance that is due to cis-effects we performed quantitative linkage analysis for the subset of transcripts that have h2>0.1 and associated with a common cis-SNP at 1% FDR. IBD sharing between every pair of DZ twins was calculated by phasing all the SNPs in every cis region (~2Mb) using MERLIN 1.1.2 40 and then counting haplotypes identical in both twins (IBD=0 corresponding to both haplotypes different, IBD=1 one identical or IBD=2 both identical). IBD sharing in cis for all the probes tested for linkage were calculated.
To estimate the average heritability at cis regions a modification of the Haseman-Elston regression method was used that analyzes the data of several traits in a single linear regression 16. Briefly, ygi represents the expression for gene g and individual i, normalized to have mean 0 and variance 1 across all the individuals. Ygij = (ygi * ygj) is a measure of phenotypic similarity between twins i and j in gene g and πgij is the IBD sharing between the DZ pair (i,j) at gene g calculated as described above. Ygij was regressed on the IBD sharing πgij over all genes and all DZ twin pairs. The coefficient of this regression is an estimate of the average variance explained in cis. The quotient of this value with the average of total heritability for the same set of transcripts represents a measure of the proportion of the heritability that is explained by variants in cis. Next, each gene’s expression value was corrected by the cis-eQTL effects and calculated as the residuals of the linear regression of the original gene expression levels on the independent cis-eQTL for each gene. The global regression procedure was then repeated but, in this case, using the gene expression values corrected by the common cis-eQTL effects. The coefficient of this regression represented the estimate of the average gene expression variance explained by variants not discovered in our eQTL analysis, most likely rare variants. Subtracting this value from the total heritability in cis, an estimate of the genetic variance at the cis-regions that is due to common SNPs was obtained. All three tissues were analyzed separately and linear regressions were adjusted using the software R version 2.13.0 37.
Replication cohorts
Characteristics of the replication cohorts are presented in Supplementary Table 4. The deCODE replication sample consists of 585 subcutaneous adipose samples from healthy Icelandic individuals as previously described 8.
The MGH replication sample consists of 701 subcutaneous adipose samples from obese patients undergoing Roux-en Y gastric bypass surgery at Massachusetts General Hospital as previously described 10.
The Oxford replication sample consists of 331 LCLs independently derived from the TwinsUK Adult twin registry and thus do not overlap with MuTHER samples as recently described 41.
The ALSPAC replication sample consists of 931 LCLs derived from The Avon Longitudinal Study of Parents and their Children (ALSPAC) 42. Expression profiling of the samples, each with two technical replicates, were performed using the Illumina Human HT-12 V3 BeadChips (IlluminaInc) and processed as the MuTHER samples described above. ALSPAC individuals were genotyped using the Illumina HumanHap550 genome-wide SNP genotyping platform. Markers with <1% MAF, >5% missing genotypes or which failed an exact test of Hardy-Weinberg equilibrium (P<5×10−7) were excluded from further analysis. Any individuals who did not cluster with the CEU individuals in multidimensional scaling analysis, who had >3% missing data, minimal or excessive heterozygosity (>33% or <31%), evidence of cryptic relatedness (>10% IBD) and any individuals with incorrect gender assignments were also excluded. After data cleaning 315,807 SNPs were left. Imputation was carried out using MACH 1.0.16, Markov Chain Haplotyping43, using CEPH individuals from phase 2 of the HapMap project as a reference set. Associations between SNP genotypes and normalized expression values were conducted using a linear model.
The GenCORD replication sample consist of 68 primary fibroblasts derived from the umbilical cord of newborns of Western European origin born at the maternity ward of the University of Geneva Hospital, for which pregnancies were full term or near full term (38-41 weeks)as previously described 9.
Integration of eQTL data with GWAS hits
The likelihood of a shared functional effect between a GWAS SNP (NHGRI database, accessed 21.12.10) and an eQTL was assessed by quantifying the change in the statistical significance of the eQTL after correcting for the effect of the GWAS SNP as previously described14. The ProbABEL associations of the eQTL genotype with the residuals from the standard linear regression of the “corrected-for” SNP against normalized expression values were re-done. The LD structure in each hotspot interval separately were accounted for by ranking (RankGWAS SNP) the impact on the eQTL (quantified by the adjusted association P-value after correction) of the GWAS SNP correction to that of correcting for all other SNPs in the same interval. By taking into account the total number of SNPs in the interval (NSNPs), this ranking across different genes and intervals can then be compared. For this purpose we define the regulatory trait concordance (RTC) Score ranked below ranging from 0 to 1, with values closer to 1 indicating causal regulatory effects.
Supplementary Material
Web box.
The MuTHER consortium reports an analysis of the genetics of gene expression in three tissues from approximately 850 mono- and dizygotic twins. They systematically dissect cis- and trans- genetic effects and estimate non-genetic effects on gene expression.
Acknowledgements
The MuTHER Study was funded by a program grant from the Wellcome Trust (081917/Z/07/Z) and core funding for the Wellcome Trust Centre for Human Genetics (090532). Additional funding came from European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement, ENGAGE project grant agreement HEALTH-F4-2007-201413, the Swiss National Science Foundation, the Louis-Jeantet Foundation and a National Institutes of Health-NIMH grant (GTEx project). Additional funding for the participating studies and investigators are provided in Supplementary Note.
Footnotes
URLs MuTHER Resource, www.muther.ac.uk; ArrayExpress, www.ebi.ac.uk/arrayexpress/; Genevar data base, www.sanger.ac.uk/resources/software/genevar/.
Supplementary information is linked to the online version of the paper at www.nature.com/ng
Author Information The microarray data has been deposited in the ArrayExpress archive, (accession no. E-TABM-1140) and the full dataset of MuTHER cis-eQTLs is available through the Genevar data base. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–9. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–16. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 4.Grundberg E, et al. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19:1942–52. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 7.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 9.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenawalt DM, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–16. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding J, et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–89. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 16.Price AL, et al. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spector TD, Williams FM. The UK Adult Twin Registry (TwinsUK) Twin Res Hum Genet. 2006;9:899–906. doi: 10.1375/183242706779462462. [DOI] [PubMed] [Google Scholar]
- 18.Andrew T, et al. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–77. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 19.Visscher PM, Benyamin B, White I. The use of linear mixed models to estimate variance components from data on twin pairs by maximum likelihood. Twin Res. 2004;7:670–4. doi: 10.1375/1369052042663742. [DOI] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freathy RM, et al. Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nat Genet. 2010;42:430–5. doi: 10.1038/ng.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown KM, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–40. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 25.Anderson CA, Soranzo N, Zeggini E, Barrett JC. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9:e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickson SP, et al. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell JE, et al. Genetic control of gene expression in whole blood and lymphoblastoid cell lines is largely independent. Genome Res. 2012;22:456–66. doi: 10.1101/gr.126540.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinig M, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–4. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–4. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humbert N, et al. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. Embo J. 2010;29:376–86. doi: 10.1038/emboj.2009.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–6. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–85. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 36.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. [Google Scholar]
- 38.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–4. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 39.Bates DM. lme4: Linear mixed-effects models using S4 classes. R Foundation for Statistical Computing; Vienna, Austria: 2010. M. [Google Scholar]
- 40.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 41.Min JL, et al. The Use of Genome-Wide eQTL Associations in Lymphoblastoid Cell Lines to Identify Novel Genetic Pathways Involved in Complex Traits. PLoS One. 2011;6:e22070. doi: 10.1371/journal.pone.0022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golding J, Pembrey M, Jones R. ALSPAC--the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.