Abstract
Background
TRPS-1 is a new GATA transcription factor that is differentially expressed in breast cancer (BC) where it been found recently to regulate epithelial-to-mesenchymal transition (EMT).
Patients and methods
We carried out a quantitative immunohistochemistry (qIHC) analysis of TRPS-1 expression in 341 primary-stage I–III BC samples in relation to patient clinical characteristics as well as its prognostic value, especially in an estrogen receptor-positive (ER+) subgroup.
Results
Higher TRPS-1 expression was significantly associated with a number of clinical and pathological characteristics as well as with improved overall survival (OS) and disease-free survival (DFS). Among stage I/II ER+ BC patients who received endocrine therapy alone, those with high TRPS-1 expression had significantly longer OS and DFS. There was also a strong association between TRPS-1 levels and the EMT marker E-cadherin in the ER+ invasive ductal carcinoma cases. Analysis of gene expression data on a panel of BC lines found that TRPS-1 expression was low or absent in BC lines having enriched mesenchymal features.
Conclusions
Our data indicated that TRPS-1 is an independent prognostic marker in early-stage BC and a new EMT marker that can distinguish patients with ER+ BC who will respond longer to adjuvant endocrine therapy.
Keywords: breast cancer, immunohistochemistry, prognostic marker, TRPS-1
introduction
TRPS-1 is one of the most prevalent genes expressed in breast cancer (BC) based on microarray and immunohistochemistry (IHC) screening [1–3]. It was initially discovered as the gene associated with three rare autosomal genetic disorders, called trichorhinophalangeal (TRP) syndromes, caused by a loss of heterozygosity at the q24 locus on chromosome 8 [4, 5]. TRPS-1 is a member of the GATA transcription factor family [4, 6], powerful transcription factors regulating cellular differentiation in multiple-cell lineages [7]. It has a single GATA zinc finger domain and may have a unique transcriptional regulatory role [8]. Although originally characterized as a transcriptional repressor of GATA factor-induced genes [8, 9], TRPS-1 is also emerging as a positive transcriptional regulator by its interaction with other transcription elements, such as Gli3, during cellular differentiation [10]. A recent bioinformatics analysis showed that TRPS-1 was one of the 22 transcription factors identified to be enriched in terminal end buds and mature ducts of mammary glands [11], suggesting that TRPS-1 may play a role in luminal progenitor cell differentiation in the mammary gland.
Molecularly, TRPS-1 is emerging to be a critical regulator of mesenchymal-to-epithelial transition (MET) during embryonic development in a number of tissues, including kidney, cartilage, and bone, where it drives the down-regulation of mesenchymal cell genes, such as ZEB-1 and Sox9, and increases the expression of epithelial markers such as E-cadherin [10, 12–15 ]. This role for TRPS-1 is attributed to its regulation of other transcription factors, such as Sox9, Gli3, Runx2, and RNF4 [9, 10, 16, 17]. A similar function in regulating MET during ductal tubule differentiation in the breast is also likely due to its specific expression in ductal epithelial cells of the breast and not in other structures [2, 18, 19]. The reverse process of MET, epithelial-to-mesenchymal transition (EMT), is associated with cancer progression and metastasis [20]. Recent data have found that TRPS-1 may play a critical role in regulating BC initiation and progression through regulating these EMT process. For example, Stinson et al. [18] recently found that TRPS-1 is an EMT inhibitor targeted by miR221/222 in BC. TRPS-1 expression was repressed by miR221/222, leading to a loss of E-cadherin and a gain of vimentin. TRPS-1 was found to repress ZEB2, a key regulator of EMT that inhibits E-cadherin and other epithelial genes [18]. These results suggest that measuring TRPS-1 levels in BC, especially ER+ BC, may be a new and useful prognostic marker. Understanding the function of TRPS-1 in human BC and its clinical relevance may supply a useful and new tool for BC diagnosis and therapy.
We recently developed a robust and reproducible quantitative IHC (qIHC) technique to measure TRPS-1, as well as estrogen receptor-α (ERα), expression [2]. Using this qIHC assay, we analyzed the prognostic value of TRPS-1 in 341 stage I–III BC patients treated at MD Anderson Cancer Center (MDACC) and the Veterans Administration Hospital, Baylor College of Medicine (VA) between 1985 and 2006. Our results demonstrate validity of TRPS-1 as a prognostic marker in early-stage BC and its possible role as a new marker for prognosis in stage I–II ER+ BC patients receiving anti-hormone therapy.
materials and methods
patients
Patients were eligible for the study if they had not received neoadjuvant chemotherapy, as described in the consort diagram in supplementary Figure S1, available at Annals of Oncology online. The retrospective review of the medical records and identification and analysis of tumor blocks were approved by the Institutional Review Board of MDACC and the VA Hospital. The histological type of the tumor specimens, clinical staging, tumor staging, and regional lymph node staging were defined according to the World Health Organization's Classification System [21]. The nuclear grade was defined according to the modified Black's nuclear grading system [22]. The follow-up time was updated in April 2011.
immunohistochemistry and quantitative IHC analysis
IHC staining for TRPS-1, ER, HER2/neu, and E-cadherin was carried out as previously described [2] and as described in supplementary Methods, available at Annals of Oncology online. The quantitation of TRPS-1 and ER IHC staining using a Quick Score (QS) after digital image analysis has been reported previously [2] and as described in supplementary methods, available at Annals of Oncology online. The quantitation of TRPS-1 protein expression was established both by using a threshold of QS ≥ 4 after comparing different cutoff points and by its expression as a continuous variable.
analysis of ER, progesterone receptor (PR), HER2, Ki-67, and E-cadherin expression
All tumors were evaluated for HER2 expression by IHC using the HercepTest scoring system according to the ASCO/CAP guidelines [23, 24]. Nuclear staining ≥10% was considered positive for ER, PR, and Ki-67 according to pathological diagnosis after biopsy [2, 25, 26]. E-cadherin membrane staining was scored as described in supplementary methods, available at Annals of Oncology online [27, 28].
in vitro assay on breast cancer cell lines
The culture of BC cell lines, real-time PCR, western blot, and immunofluorescence methods and data analysis are described in supplementary methods, available at Annals of Oncology online.
gene expression profiling
EMT-related gene expression analysis is described in supplementary methods, available at Annals of Oncology online, using published databases [29, 30].
statistical methods
Since TRPS-1 QS does not meet the normality assumption, the association between TRPS-1 QS and clinical characteristics was tested using the Kruskal–Wallis test (nonparametric test). The log-rank test and Kaplan–Meier curve analysis were used to compare overall survival (OS, time from the date of surgery to death or last follow-up) and disease-free survival (DFS, time from the date of surgery to the first local or distance recurrence, death, or last follow-up) between TRPS-1 groups. Univariate and multivariate Cox proportional hazards models were carried out. The forward (Wald) method was used to select significant clinical factors into the final multivariate model. All P values were two-tailed and considered significant at α = 0.05. Analyses were conducted using SPSS® (version 17.0, SPS Inc., Chicago, IL). Power calculation was done for the log-rank test of OS and DFS among a subgroup of 50 ER+ samples from patients who only received endocrine therapy. It was carried out by Stata/IC 12.0 for Windows (64-bit x86-64) (StataCorp LP, College Station, TX).
results
patients' information
Of the 341 patient samples, 226 (66.3%) were ER+, 68 (19.9%) were HER2-amplified (defined as 3+ by IHC), and 57 (16.7%) were triple negative (ER-negative, PR-negative, and HER2-non-amplified). The patients' median age was 55 (26–99) years. The median follow-up time was 9.7 (0.19–23.68) years. OS from the date of surgery ranged from 1.12 years to 23.68 years (median = 11.53 years). The time to death ranged from 2.28 months to 19.63 years (median = 5.76 years). The time to recurrence ranged from 2.04 months to 16.04 years (median = 5.85 years). DFS from the date of surgery ranged from 0.72 months to 23.20 years (median = 8.30 years). Clinical and pathological characteristics are detailed in Table 1.
Table 1.
Association between TRPS-1 QS and other characteristics among 341 patients with stage I–III breast cancer (BC)
| Characteristic | N (%) | TRPS-1 quick score (QS) |
P* | ||
|---|---|---|---|---|---|
| Maximum | Minimum | Median | |||
| Age | |||||
| ≥50 years | 225 (66) | 38.27 | 0.02 | 14.92 | 0.319 |
| <50 years | 116 (34) | 33.07 | 0.01 | 16.56 | |
| Histology | |||||
| Ductal | 285 (83.6) | 38.27 | 0.01 | 16.02 | 0.694 |
| Lobular | 29 (8.5) | 35.23 | 0.33 | 12.35 | |
| Ductal and lobular | 18 (5.3) | 34.31 | 0.33 | 11.98 | |
| Other | 9 (2.6) | 19.98 | .014 | 13.12 | |
| N stage | |||||
| N0 | 158 (46.3) | 36.42 | 0.02 | 18.00 | 0.001 |
| N1, N2, N3 | 183 (53.7) | 38.27 | 0.01 | 13.38 | |
| T stage | |||||
| T1, T2 | 306 (89.7) | 38.27 | 0.01 | 16.23 | 0.002 |
| T3, T4 | 35 (10.3) | 34.31 | 0.22 | 6.86 | |
| Clinical stage | |||||
| I | 81 (23.8) | 36.42 | 0.24 | 19.69 | 0.000 |
| II | 217 (63.6) | 38.27 | 0.01 | 13.85 | |
| III | 43 (12.6) | 34.31 | 0.22 | 10.45 | |
| Black nuclear grade | |||||
| 1 or 2 | 218 (63.9) | 36.42 | 0.02 | 16.18 | 0.036 |
| 3 | 110 (32.3) | 38.27 | 0.01 | 12.98 | |
| Missing | 13 (3.8) | ||||
| ER status | |||||
| ≥10% (positive) | 226 (66.3) | 38.27 | 0.10 | 15.52 | 0.681 |
| <10% (negative) | 95 (27.9) | 35.95 | 0.01 | 18.60 | |
| Missing | 20 (5.9) | ||||
| ER QS | |||||
| ≥4 | 235 (68.9) | 38.27 | 0.10 | 16.13 | 0.035 |
| <4 | 101 (29.6) | 34.83 | 0.01 | 13.12 | |
| Missing | 5 (1.5) | ||||
| PR status | |||||
| ≥10% (positive) | 186(54.5) | 36.42 | 0.14 | 16.18 | 0.275 |
| <10% (negative) | 132 (38.7) | 38.27 | 0.01 | 15.32 | |
| Missing | 23 (6.7) | ||||
| HER2 status (IHC) | |||||
| Positive (3+) | 68 (19.9) | 35.95 | 0.58 | 16.56 | 0.017 |
| Negative (0–2+) | 273 (80.1) | 38.27 | 0.01 | 14.54 | |
| Subtype | |||||
| HER2+ | 68 (19.9) | 35.95 | 0.58 | 16.75 | 0.031 |
| ER/PR+, HER2- | 199 (58.4) | 38.27 | 0.10 | 15.43 | |
| Triple negative | 57 (16.7) | 34.83 | 0.01 | 13.12 | |
| Missing | 17 (5.0) | ||||
| Adjuvant chemotherapy | |||||
| No | 153 (44.9) | 34.83 | 0.14 | 15.22 | 0.509 |
| Yes | 183 (53.7) | 38.27 | 0.01 | 15.89 | |
| Missing | 5 (1.5) | ||||
| Adjuvant radiation therapy | |||||
| No | 191 (56) | 38.27 | 0.10 | 15.87 | 0.568 |
| Yes | 144 (42.2) | 34.31 | 0.01 | 15.27 | |
| Missing | 6 (1.8) | ||||
| Adjuvant anti-hormonal therapy | |||||
| No | 157 (46) | 34.83 | 0.01 | 15.61 | 0.713 |
| Yes | 177 (51.9) | 38.27 | 0.02 | 15.43 | |
| Missing | 7 (2.1) | ||||
*P values were determined by the Kruskal–Wallis test.
correlation of TRPS-1 expression with clinical and pathological characteristics in stage I–III BC cohort
TRPS-1 protein showed heterogeneous expression across the 341 stage I–III BC samples. It was also expressed in normal ductal epithelial cells at a low level compared with the adjacent tumor area. By expressing ER staining and TRPS-1 staining as continuous variables, we found a positive correlation between TRPS-1 QS and ER QS (P < 0.05, r2 = 0.160; Spearman's correlation coefficient). The mean rank of TRPS-1 QS was significantly greater in tumors from node-negative patients (P < 0.001), in T1–T2 tumors (P < 0.05), in clinical stage I disease (P < 0.001), in tumors with Black's nuclear grade 1 or 2 (P < 0.05), in tumors with ER QS ≥ 4 (P < 0.05), in HER2-positive tumors (P < 0.05), and in the HER2-positive subtype (P < 0.05) (Table 1).
survival analysis in relation to TRPS-1 expression
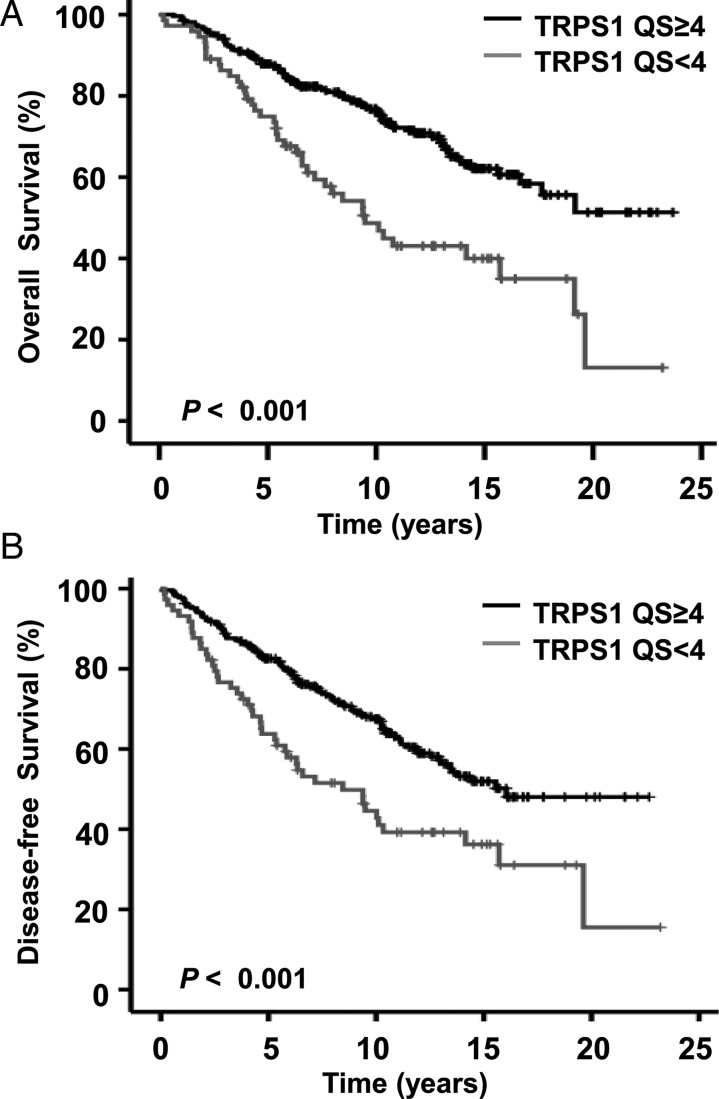
OS and DFS among patients were analyzed according to TRPS-1 expression by qIHC. When analyzed using univariate Cox regressive analysis, patients who had a TRPS-1 QS ≥ 4 had significantly better OS (HR = 0.44, P < 0.001) and DFS (HR = 0.53, P < 0.001) durations (Table 2). The log-rank test and Kaplan–Meier survival analysis revealed significantly longer OS (Figure 1A) and DFS (Figure 1B) durations in patients with a TRPS-1 QS ≥ 4 versus < 4 (P < 0.001 for both OS and DFS). ER status did not achieve statistical significance in predicting OS and DFS when measured either using the 10% cutoff or QS.
Table 2.
Univariate and multivariate Cox proportional hazards regression analyses predicting hazards of death or recurrence based on TRPS-1 QS and other clinical parameters
| Variable | Level | Overall survival (OS) |
Disease-free survival (DFS) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate |
Multivariate |
Univariate |
Multivariate |
||||||||||
| HR | 95% CI | P | HR | 95% CI | P* | HR | 95% CI | P | HR | 95% CI | P* | ||
| TRPS-1 QS | QS ≥ 4 | 0.44 | 0.30–0.64 | <0.001 | 0.59 | 0.38–0.90 | 0.016 | 0.53 | 0.37–0.75 | <0.001 | 0.61 | 0.41–0.90 | 0.014 |
| QS < 4 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||
| Patient age at | ≥50 years | 1.93 | 1.27–2.93 | 0.002 | 1.89 | 1.12–3.19 | 0.017 | 1.40 | 0.98–2.01 | 0.068 | |||
| diagnosis | <50 years | 1.00 | 1.00 | 1.00 | |||||||||
| Histologic type | Ductal | 1.74 | 0.43–7.07 | 0.612 | 2.21 | 0.55–8.92 | 0.651 | ||||||
| Lobular | 1.63 | 0.36–7.45 | 1.90 | 0.42–8.60 | |||||||||
| Ductal and lobular | 2.71 | 0.55–13.52 | 2.63 | 0.53–13.08 | |||||||||
| Other | 1.00 | 1.00 | |||||||||||
| Black nuclear | I or II | 0.58 | 0.39–0.85 | 0.005 | 0.75 | 0.52–1.06 | 0.106 | ||||||
| grade | III | 1.00 | 1.00 | ||||||||||
| T classification | T1 or T2 | 0.31 | 0.19–0.53 | <0.001 | 0.41 | 0.25–0.67 | <0.001 | ||||||
| T3 or T4 | 1.00 | 1.00 | |||||||||||
| N classification | N0 | 0.65 | 0.45–0.93 | 0.020 | 0.75 | 0.54–1.05 | 0.092 | ||||||
| N1-N3 | 1.00 | 1.00 | |||||||||||
| Stage | I | 0.16 | 0.08–0.29 | <0.001 | 0.07 | 0.03–0.15 | <0.001 | 0.23 | 0.13–0.41 | <0.001 | 0.13 | 0.07–0.26 | <0.001 |
| II | 0.29 | 0.18–0.47 | 0.14 | 0.08–0.26 | 0.39 | 0.25–0.62 | 0.25 | 0.15–0.42 | |||||
| III | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||
| Adjuvant | Yes | 0.76 | 0.53–1.08 | 0.129 | 0.48 | 0.30–0.77 | 0.003 | 0.78 | 0.56–1.09 | 0.142 | 0.51 | 0.35–0.75 | 0.001 |
| chemotherapy | No | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| Adjuvant anti- | Yes | 1.10 | 0.77–1.58 | 0.608 | 0.97 | 0.70–1.34 | 0.838 | ||||||
| Hormone therapy | No | 1.00 | 1.00 | ||||||||||
| Adjuvant | Yes | 0.98 | 0.68–1.42 | 0.933 | 1.01 | 0.72–1.40 | 0.981 | ||||||
| radiation therapy | No | 1.00 | 1.00 | ||||||||||
| Subgroup | HER2+ | 0.68 | 0.38–1.21 | 0.155 | 0.67 | 0.36–1.26 | 0.038 | 0.89 | 0.52–1.50 | 0.488 | |||
| ER/PR+, HER2- | 0.66 | 0.42–1.01 | 0.53 | 0.32–0.86 | 0.78 | 0.51–1.19 | |||||||
| TNBC | 1.00 | 1.00 | 1.00 | ||||||||||
| HER2 status | Positive (3+) | 0.99 | 0.62–1.61 | 0.986 | 1.15 | 0.75–1.74 | 0.529 | ||||||
| (IHC) | Negative (0–2+) | 1.00 | 1.00 | ||||||||||
| PR status | ≥10% (positive) | 0.82 | 0.56–1.18 | 0.283 | 0.97 | 0.69–1.37 | 0.882 | ||||||
| <10% (negative) | 1.00 | 1.00 | |||||||||||
| ER QS | ≥4 | 0.74 | 0.51–1.09 | 0.133 | 0.80 | 0.56–1.14 | 0.220 | ||||||
| <4 | 1.00 | 1.00 | |||||||||||
| ER status | ≥10% (positive) | 0.81 | 0.55–1.20 | 0.289 | 0.87 | 0.61–1.24 | 0.435 | ||||||
| <10%(negative) | 1.00 | 1.00 | |||||||||||
*P values are obtained from the multivariate Cox proportional model after using the forward (Wald) selection method at the significance level = 0.05.
Figure 1.
Kaplan–Meier analysis of OS (A) and DFS (B) durations in 341 patients with stage I–III BC. Survival probability (in years from primary surgery) was evaluated for TRPS-1 QS at ≥4 versus < 4. The P values were calculated using log-rank analysis. Patients with a TRPS-1 QS of ≥4 had significantly longer OS (A) and DFS (B) durations versus <4.
Multivariate Cox regressive analysis was carried out using the clinical and pathological characteristics that were used in the univariate model. As shown in Table 2, patients with tumor TRPS-1 QS ≥ 4 had significantly longer OS (HR = 0.59, P < 0.05) and DFS (HR = 0.61, P < 0.05).
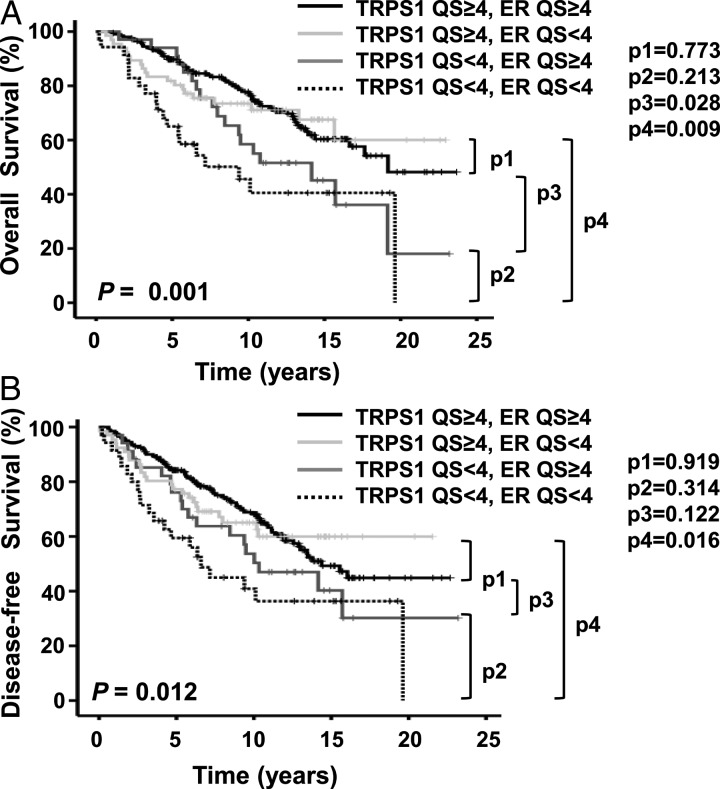
Since patients with ER+ BC have a better prognosis than those with HER2-positive and triple-negative BC [31–34 ], and since there appeared to be an association between median ER QS and median TRPS-1 QS (Table 1), it is possible that the survival differences seen could have been related to hormonal status alone, irrespective of TRPS-1 status. We therefore examined OS and DFS in patient subgroups based on TRPS-1 and ER expression. As shown in Figure 2, patients with TRPS-1 QS ≥ 4 tumors had prolonged OS and DFS durations irrespective of ER status (ER QS ≥ 4 or <4).
Figure 2.
Kaplan–Meier analysis of OS (A) and DFS (B) durations in 336 patients with stage I–III BC. Survival probability (in years from primary surgery) was evaluated for TRPS-1 QS and ER QS. The P values were calculated using log-rank analysis. Patients with TRPS-1 QS ≥ 4 tumors had significantly longer OS and DFS durations and patients with TRPS-1 QS < 4 tumors had a poor prognosis regardless of ER expression (QS ≥ 4 or <4).
prognostic value of TRPS-1 expression in patients with ER+ BC treated with endocrine therapy alone
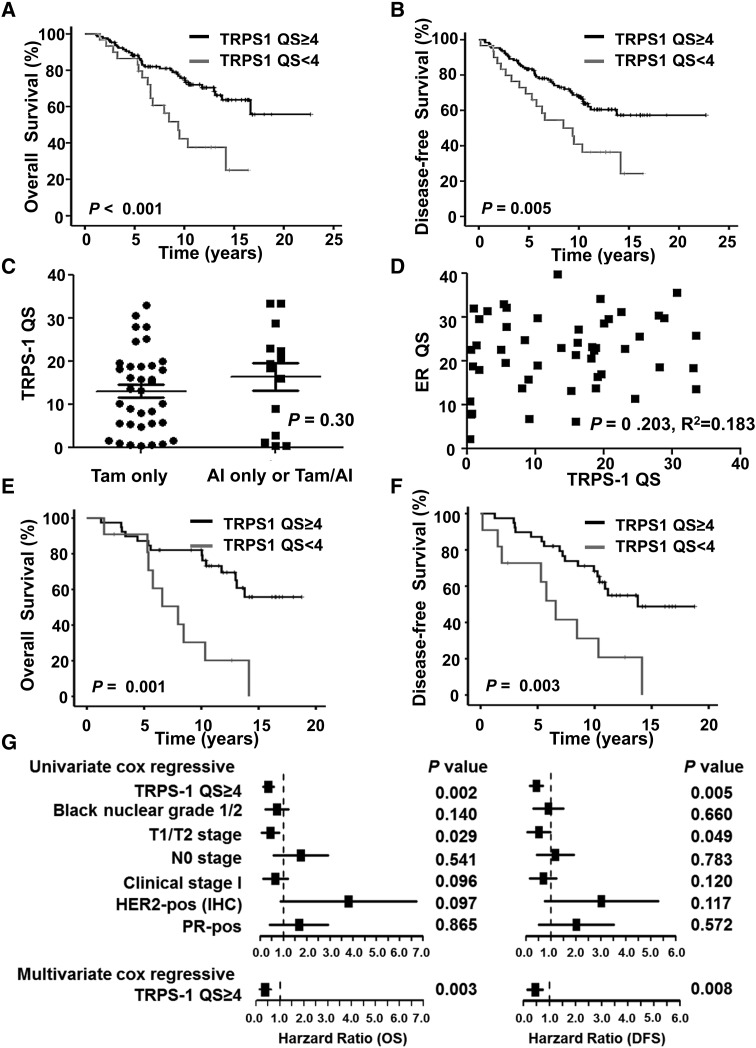
We were also interested in examining whether patients treated with endocrine therapy could be divided into good and poor prognostic groups based on TRPS-1 expression. We carried out Kaplan–Meier survival analysis for 159 patients with ER+ BC treated with endocrine therapy with or without chemotherapy and/or radiation therapy. As shown in Figure 3, patients with a TRPS-1 QS ≥ 4 still had significantly longer OS (P = 0.001, Figure 3A) and DFS (P = 0.005, Figure 3B) durations than did patients with a QS of <4. Among 114 early-stage ER+ BC treated with endocrine therapy, with or without chemotherapy and/or radiation therapy, there was no significant correlation between TRPS-1 QS and % of Ki-67 expression when plotted as continuous variables (P = 0.858, r2 = 0.017, Spearman's correlation coefficient). Although lower Ki-67 (<10%) had a trend toward better OS (P = 0.060), multivariate cox regressive analysis demonstrated that only TRPS-1 QS ≥ 4 and black nuclear grade 1 or 2 remained in the OS model and TRPS-1 QS ≥ 4 in the DFS model (supplementary Figure S2, available at Annals of Oncology online).
Figure 3.
Kaplan–Meier analysis of TRPS-1 QS and OS (A) and DFS (B) durations for 159 patients with ER+ BC treated with endocrine therapy, with or without chemotherapy and radiation therapy. Survival probability (in years from primary surgery) was evaluated for TRPS-1 QS at ≥ 4 versus <4. The P values were calculated using log-rank analysis. (C–G) The prognostic relevance of TRPS-1 QS in the subset of 50 patients with stage I and II disease who received endocrine therapy alone. (C) Scatter plot demonstrating no significant difference of mean ± SEM TRPS-1 expression between the group that received tamoxifen alone and the group that received AI or tamoxifen followed by an AI (P = 0.30). (D) Scatter plot showing no significant correlation between TRPS-1 QS and ER QS as continuous variables (P = 0.203, r2 = 0.183; Spearman's correlation coefficient). Kaplan–Meier survival analysis of OS (E) and DFS (F) in patients with a TRPS-1 QS ≥ 4 versus < 4 (log-rank, P = 0.001 and 0.003, respectively). (G) Forest plot demonstrating the hazard ratio for OS and DFS for TRPS-1 as well as other parameters, as assessed by both univariate and multivariate models.
We were further interested in TRPS-1 expression related to the outcome of patients treated with endocrine therapy alone. TRPS-1 QS was determined in tumors from 50 stage I/II patients, 36 of whom received tamoxifen alone and 14 who received an aromatase inhibitor (AI) alone or tamoxifen followed by an AI. Eight patients had tumors that were both ER+ and HER2-amplified. There was no difference in TRPS-1 expression between patients treated with tamoxifen alone versus those treated with an AI alone or tamoxifen followed by an AI (Figure 3C). As shown in Figure 3D, there was no significant correlation between TRPS-1 QS and ER QS when plotted as continuous variables (P > 0.05, r2 = 0.183; Spearman's correlation coefficient) in these ER+ BC subset receiving endocrine therapy alone, indicating that this was simply due to ER regulation of TRPS-1. In addition, treatment of two ER+ BC cell lines (T47D and MCF-7) with charcoal-stripped serum to remove estradiol did not induce significant changes in TRPS-1 expression, while ER-sensitive genes, such as TFF-1 and GREB-1, were down-modulated indicating that ER signaling does not regulate TRPS-1 gene expression (supplementary Figure S3, available at Annals of Oncology online). In a Kaplan–Meier survival analysis, patients with a TRPS-1 QS ≥ 4 had significantly greater OS (log-rank, P = 0.001 with 95% power) and DFS (log-rank, P = 0.003 with 89% power) durations than did patients with a TRPS-1 QS of <4 (Figure 3E and F). Univariate Cox regressive analysis carried out on this subgroup demonstrated that only TRPS-1 QS ≥ 4 and small tumor size (T1 or T2) were significantly associated with longer OS and DFS durations; using a forward selection method for the multivariate model, only TRPS-1 QS ≥ 4 was included at the α = 0.05 significance level (Figure 3G).
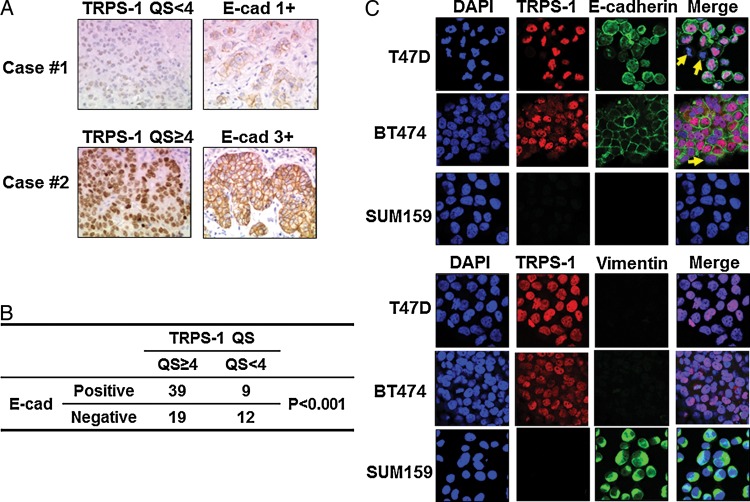
correlation of E-cadherin and TRPS-1 expression in a clinical set of ER+ invasive ductal carcinoma samples
Because evidence has mounted that TRPS-1 is involved in the process of MET [9, 10, 14, 18, 35], we conducted IHC staining in 79 ER+ invasive ductal carcinoma sample sets to analyze the relationship between expression of E-cadherin, an epithelial marker, and TRPS-1. Figure 4A shows two examples in which there was a positive correlation of TRPS-1 expression with E-cadherin at ×200 magnification. Overall, there was a strong significant association between TRPS-1 expression and E-cadherin expression in the ER+ BC cases (P < 0.001, Figure 4B). This was confirmed by immunofluorescence staining in T47D and BT474, the ER+ BC cell lines and SUM159, a Claudin-low mesenchymal BC cell line characterized by the low to absent expression of tight junction proteins including Claudin 3 and E-cadherin and high expression of markers associated with EMT [36]. As shown in Figure 4C, T47D and BT474 co-expressed TRPS-1 and E-cadherin, did not express vimentin, a mesenchymal marker. In contrast, SUM159 over-expressed vimentin, but did not express TRPS-1 and E-cadherin.
Figure 4.
TRPS-1 expression is positively correlated with E-cadherin (E-cad). (A) Examples of two cases by IHC, one with low TRPS-1 (QS < 4) and E-cad (1+) levels, and one with higher TRPS-1 (QS ≥ 4) and high E-cad (3+) levels. (B) Table summarizing analysis of 79 ER+ BC cases showing a significant association between TRPS-1 and E-cadherin expression (P < 0.001). (C) Co-staining and immunofluorescence microscopy of TRPS-1 with E-cadherin, TRPS-1 with vimentin in T47D, BT474, and SUM159 BC cell lines. Most T47D and BT474 cells co-expressed TRPS-1 and E-cadherin, and did not express vimentin, a mesenchymal marker, while a few cells which did not express TRPS-1, also did not express E-cadherin (yellow arrows in merged images). In contrast, SUM159 over-expressed vimentin, but did not express TRPS-1 and E- cadherin.
gene profiling for EMT-associated genes in different subtypes of BC
We then were interested in how TRPS-1 was expressed in different subtypes of BC, especially in basal and Claudin-low subsets enriched in mesenchymal-like cells that are members of the so-called ‘triple-negative’ BC subset. Hierarchical clustering analysis of EMT-associated gene expression data from ArrayExpress showed differences in TRPS-1 expression levels across 54 different BC cell lines originally profiled by Neve et al. [29]. TRPS-1 had a significantly positive correlation with CDH1 (P < 0.05, r2 = 0.3, Spearman's correlation coefficient). TRPS-1, as well as other epithelial markers (for example, CDH1, KRT18, KRT8, and GATA3), were higher in ER+ cell lines, but were absent in Claudin-low cell lines, such as SUM159 [36], and basal-like BC cell lines, such as SUM149 [37] (supplementary Figure S4A, available at Annals of Oncology online). Hierarchical clustering analysis of Claudin-low primary BC surgical samples from the UNC microarray database also found that TRPS-1 expression was conspicuously absent (supplementary Figure S4B, available at Annals f Oncology online).
discussion
GATA transcription factors have been found to be critical in mediating normal breast ductal epithelial cell differentiation [11, 38]. One GATA factor in particular, GATA-3, is a prevalent transcription factor facilitating ductal epithelial cell differentiation associated with ER expression in both normal ducts and in BC cells [39, 40]. The role of GATA-3 has also be explored as a prognostic marker in BC [41–43 ] and found in some studies to be predictive of improved OS and DFS [41, 42, 44]. Our study here sheds further light on the role of GATA family in BC by showing that another prevalent member of this critical transcription factor family expressed in BC, TRPS-1, has an independent prognostic value. We observed that TRPS-1 expression over a certain threshold (QS ≥ 4 using qIHC) was significantly associated with longer OS and DFS by both univariate and multivariate Cox regressive analysis in BC patients, especially in early-stage BC patients who received endocrine therapy alone. Our data also suggest that BC tumors with lower TRPS-1 may have more tumor-initiating and mesenchymal characteristics and thus may be more resistant to the treatments [45, 46].
In our initial IHC study, we found that ∼90% of BC samples expressed TRPS-1 at differential intensities, ranging from low to highly intense staining [1]. We found that traditional IHC scoring methods using subjective cut-offs for the degree of positive staining was not useful in revealing the role of TRPS-1 expression and its association with clinical and pathological parameters. We have also found that some microarray platforms do not accurately reflect the extent of TRPS-1 gene expression due to shortcomings in the probes used and TRPS-1 expression was not consistent between the mRNA level and the protein level in some BC cell lines (unpublished observations). Because of these issues, TRPS-1 may have been ‘missed’ as a potentially important prognostic or predictive marker in BC in many microarray-based studies. In addition, Venet et al. have recently found that most published BC outcome gene signatures could not significantly predict better outcomes [47]. Due to this issue, we previously developed a quantitative IHC method to accurately and reproducibly measure TRPS-1 protein expression, which could then be compared with pathologic tumor characteristics and other clinical variables associated with prognosis [2, 3]. In this study, we focused on the ER+ BC subset to determine whether TRPS-1 may be involved in the response to anti-hormone therapy. We found that TRPS-1 was a stronger independent predictor of OS and DFS than percentage of ER and Ki-67 expression, the method widely used by pathology laboratories currently across the world.
About 70% of BC cases express ER and are mostly dependent on estrogen for survival. Despite the effectiveness of treatment, approximately 30% of women eventually have relapses even after treatment with endocrine therapy [48, 49]. We observed that there was no significant difference in TRPS-1 expression between luminal A and B subtypes of BC [50] (data not shown), however, higher TRPS-1 expression when analyzed using univariate and multivariate models predicted better OS and DFS in a subgroup of ER+, stage I/II BC patients who received endocrine therapy alone. In this subgroup, TRPS-1 expression was not significantly correlated with ER expression. Moreover, we found that TRPS-1 expression was not regulated by ER signaling, since estrogen withdrawal using charcoal-stripped serum did not affect TRPS-1 gene or protein expression in ER+ BC cell lines. What regulates TRPS-1 expression currently is not known, although recent data suggests that bone morphogenetic factors may be involved [14]. Nevertheless, our results show that although TRPS-1 tends to be expressed more in ER+ BC overall, its prognostic value is not related to hormonal status in ER+ BC patients receiving endocrine therapy, and patients with low-expressing TRPS-1 tumors might be more resistant to the effects of hormonal manipulation and might therefore benefit from additional chemotherapy or newer targeted therapies. Although the power of this analysis in the subgroup was high enough (95% for OS and 89% for DFS), it is important to point out that the data should be validated using a much larger patient cohort.
TRPS-1 has been shown to regulate the process of MET and is a modulator in MET during the differentiation of chondrocytes, hair follicle cells, osteoblasts, and kidney tubular epithelium [9, 10, 35]. A recent study also found that TRPS-1 was a key regulator of EMT/MET in breast ductal epithelial cells and BC cells by repressing ZEB-1, Sox9, and other EMT-regulatory genes [18]. Our results are consistent with these findings and reveal TRPS-1 as a key regulator of EMT/MET. Our IHC studies on human BC samples found that TRPS-1 expression was positively associated with expression of E-cadherin, a marker of breast epithelial cell differentiation, while low expression of TRPS-1 is associated with vimentin expression (data not shown). This was also confirmed in immunofluorescence studies on BC cell lines. In addition, we analyzed the expression of a group of 29 genes associated with EMT (up- or down- regulated during EMT), including TRPS-1, in 54 different BC cell lines and Claudin-low (mesenchymal)BC samples and found that TRPS-1 expression was higher in ER+ BC cell lines, but conspicuously absent in Claudin-low tumors and basal subtype tumors enriched with mesenchymal features [46, 51, 52]. Creighton et al. [46] also confirmed that the remaining tumor cells in primary ER+ BC after anti-hormone therapy with letrozole had a gene expression signature enriched in mesenchymal markers including decreased TRPS-1 [46]. These results, as well as data showing that TRPS-1 can drive the differentiation of mesenchymal cells and prevent EMT in breast ductal epithelial cells [14, 18], suggest that TRPS-1 may be a new marker to discern the state of EMT, or degree of ‘mesenchymal-ness’, in BC. Low TRPS-1 expression signals a tumor with more mesenchymal characteristics that is more resistant to therapy and metastatic and vice versa [51]. However, this hypothesis needs to be tested in a large number of tumors and BC cell lines.
In conclusion, we have demonstrated that TRPS-1 has a significant association with outcome in early-stage BC. The qIHC test of TRPS-1 protein expression could be a useful prognostic tool in early-stage BC and may be a more objective tool than traditional ER, as measured by percentage of cells and non-quantitative IHC. TRPS-1 could be a powerful marker in addition to traditional ER, PR, and HER2/neu staining in guiding clinicians in the type of adjuvant therapy to carry out. However, validation studies need to be done prospectively in tumors from a large number of early-stage BC patients before TRPS-1 can be used for clinical decision-making.
funding
This work was supported in part by a research grant from Sanofi Pasteur Canada (LR), by Susan G. Komen for the Cure grant KG080061 (LR), and by the National Institutes of Health (NIH) grant 1 P50 CA 116199-01 (Breast Cancer SPORE grant), and NIH grant CA 16672 (Cancer Center Support Grant).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
We thank Sunita Patterson, Department of Scientific Publications, UT MD Anderson Cancer Center, for her editorial assistance with this manuscript. We thank Sanofi Pasteur Canada for providing anti-TRPS-1 antibody (clone 8D11) for IHC staining.
references
- 1.Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. doi:10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen JQ, Litton J, Xiao L, et al. Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer. 2010;1:21–33. doi: 10.1007/s12672-010-0008-8. doi:10.1007/s12672-010-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen JQ, Bao Y, Litton J, et al. Expression and relevance of TRPS-1: a new GATA transcription factor in breast cancer. Horm Cancer. 2011;2:132–143. doi: 10.1007/s12672-011-0067-5. doi:10.1007/s12672-011-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Momeni P, Glockner G, Schmidt O, et al. Mutations in a new gene, encoding a zinc-finger protein, cause tricho-rhino-phalangeal syndrome type I. Nat Genet. 2000;24:71–74. doi: 10.1038/71717. doi:10.1038/71717. [DOI] [PubMed] [Google Scholar]
- 5.Chang GT, van den Bemd GJ, Jhamai M, et al. Structure and function of GC79/TRPS1, a novel androgen-repressible apoptosis gene. Apoptosis. 2002;7:13–21. doi: 10.1023/a:1013504710343. doi:10.1023/A:1013504710343. [DOI] [PubMed] [Google Scholar]
- 6.Malik TH, Von Stechow D, Bronson RT, et al. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho-rhino-phalangeal syndromes. Mol Cell Biol. 2002;22:8592–8600. doi: 10.1128/MCB.22.24.8592-8600.2002. doi:10.1128/MCB.22.24.8592-8600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gai Z, Gui T, Muragaki Y. The function of TRPS1 in the development and differentiation of bone, kidney, and hair follicles. Histol Histopathol. 2011;26:915–921. doi: 10.14670/HH-26.915. [DOI] [PubMed] [Google Scholar]
- 8.Malik TH, Shoichet SA, Latham P, et al. Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. EMBO J. 2001;20:1715–1725. doi: 10.1093/emboj/20.7.1715. doi:10.1093/emboj/20.7.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piscopo DM, Johansen EB, Derynck R. Identification of the GATA factor TRPS1 as a repressor of the osteocalcin promoter. J Biol Chem. 2009;284:31690–31703. doi: 10.1074/jbc.M109.052316. doi:10.1074/jbc.M109.052316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wuelling M, Kaiser FJ, Buelens LA, et al. Trps1, a regulator of chondrocyte proliferation and differentiation, interacts with the activator form of Gli3. Dev Biol. 2009;328:40–53. doi: 10.1016/j.ydbio.2009.01.012. doi:10.1016/j.ydbio.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Kouros-Mehr H, Slorach EM, Sternlicht MD, et al. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. doi:10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavallaro U, Schaffhauser B, Christofori G. Cadherins and the tumour progression: is it all in a switch? Cancer Lett. 2002;176:123–128. doi: 10.1016/s0304-3835(01)00759-5. doi:10.1016/S0304-3835(01)00759-5. [DOI] [PubMed] [Google Scholar]
- 13.Di Croce L, Pelicci PG. Tumour-associated hypermethylation: silencing E-cadherin expression enhances invasion and metastasis. Eur J Cancer. 2003;39:413–414. doi: 10.1016/s0959-8049(02)00815-8. doi:10.1016/S0959-8049(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 14.Gai Z, Zhou G, Itoh S, et al. Trps1 functions downstream of Bmp7 in kidney development. J Am Soc Nephrol. 2009;20:2403–2411. doi: 10.1681/ASN.2008091020. doi:10.1681/ASN.2008091020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh S, Kanno S, Gai Z, et al. Trps1 plays a pivotal role downstream of Gdf5 signaling in promoting chondrogenesis and apoptosis of ATDC5 cells. Genes Cells. 2008;13:355–363. doi: 10.1111/j.1365-2443.2008.01170.x. doi:10.1111/j.1365-2443.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- 16.Napierala D, Garcia-Rojas X, Sam K, et al. Mutations and promoter SNPs in RUNX2, a transcriptional regulator of bone formation. Mol Genet Metab. 2005;86:257–268. doi: 10.1016/j.ymgme.2005.07.012. doi:10.1016/j.ymgme.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser FJ, Moroy T, Chang GT, et al. The RING finger protein RNF4, a co-regulator of transcription, interacts with the TRPS1 transcription factor. J Biol Chem. 2003;278:38780–38785. doi: 10.1074/jbc.M306259200. doi:10.1074/jbc.M306259200. [DOI] [PubMed] [Google Scholar]
- 18.Stinson S, Lackner MR, Adai AT, et al. TRPS1 Targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. doi:10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 19.Tomaskovic-Crook E, Thompson EW, Thiery JP. Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 2009;11:213. doi: 10.1186/bcr2416. doi:10.1186/bcr2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. doi:10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The world Health Organization histological typing of breast tumors—second edition. The World Organization. Am J Clin Pathol. 1982;78:806–816. doi: 10.1093/ajcp/78.6.806. [DOI] [PubMed] [Google Scholar]
- 22.Black MM, Speer FD. Nuclear structure in cancer tissues. Surg Gynecol Obstet. 1957;105:97–102. [PubMed] [Google Scholar]
- 23.Birner P, Oberhuber G, Stani J, et al. Evaluation of the United States Food and Drug Administration-approved scoring and test system of HER-2 protein expression in breast cancer. Clin Cancer Res. 2001;7:1669–1675. [PubMed] [Google Scholar]
- 24.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. doi:10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 25.Tan PH, Bay BH, Yip G, et al. Immunohistochemical detection of Ki67 in breast cancer correlates with transcriptional regulation of genes related to apoptosis and cell death. Mod Pathol. 2005;18:374–381. doi: 10.1038/modpathol.3800254. doi:10.1038/modpathol.3800254. [DOI] [PubMed] [Google Scholar]
- 26.Thike AA, Chng MJ, Fook-Chong S, et al. Immunohistochemical expression of hormone receptors in invasive breast carcinoma: correlation of results of H-score with pathological parameters. Pathology. 2001;33:21–25. [PubMed] [Google Scholar]
- 27.Bertolo C, Guerrero D, Vicente F, et al. Differences and molecular immunohistochemical parameters in the subtypes of infiltrating ductal breast cancer. Am J Clin Pathol. 2008;130:414–424. doi: 10.1309/J3QV9763DYPV338D. doi:10.1309/J3QV9763DYPV338D. [DOI] [PubMed] [Google Scholar]
- 28.Tan DS, Potts HW, Leong AC, et al. The biological and prognostic significance of cell polarity and E-cadherin in grade I infiltrating ductal carcinoma of the breast. J Pathol. 1999;189:20–27. doi: 10.1002/(SICI)1096-9896(199909)189:1<20::AID-PATH394>3.0.CO;2-2. doi:10.1002/(SICI)1096-9896(199909)189:1<20::AID-PATH394>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. doi:10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. doi:10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen PL, Taghian AG, Katz MS, et al. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol. 2008;26:2373–2378. doi: 10.1200/JCO.2007.14.4287. doi:10.1200/JCO.2007.14.4287. [DOI] [PubMed] [Google Scholar]
- 32.Gabos Z, Thoms J, Ghosh S, et al. The association between biological subtype and locoregional recurrence in newly diagnosed breast cancer. Breast Cancer Res Treat. 2010;124:187–194. doi: 10.1007/s10549-010-1135-1. doi:10.1007/s10549-010-1135-1. [DOI] [PubMed] [Google Scholar]
- 33.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. doi:10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. doi:10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suemoto H, Muragaki Y, Nishioka K, et al. Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev Biol. 2007;312:572–581. doi: 10.1016/j.ydbio.2007.10.001. doi:10.1016/j.ydbio.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. doi:10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. doi:10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. doi:10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 39.Hoch RV, Thompson DA, Baker RJ, et al. GATA-3 is expressed in association with estrogen receptor in breast cancer. Int J Cancer. 1999;84:122–128. doi: 10.1002/(sici)1097-0215(19990420)84:2<122::aid-ijc5>3.0.co;2-s. doi:10.1002/(SICI)1097-0215(19990420)84:2<122::AID-IJC5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 40.Eeckhoute J, Keeton EK, Lupien M, et al. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. doi:10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 41.Albergaria A, Paredes J, Sousa B, et al. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11:R40. doi: 10.1186/bcr2327. doi:10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciocca V, Daskalakis C, Ciocca RM, et al. The significance of GATA3 expression in breast cancer: a 10-year follow-up study. Hum Pathol. 2009;40:489–495. doi: 10.1016/j.humpath.2008.09.010. doi:10.1016/j.humpath.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Mehra R, Varambally S, Ding L, et al. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005;65:11259–11264. doi: 10.1158/0008-5472.CAN-05-2495. doi:10.1158/0008-5472.CAN-05-2495. [DOI] [PubMed] [Google Scholar]
- 44.Yoon NK, Maresh EL, Shen D, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum Pathol. 2010;41:1794–1801. doi: 10.1016/j.humpath.2010.06.010. doi:10.1016/j.humpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taube JH, Herschkowitz JI, Komurov K, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. doi:10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. doi:10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. doi:10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 49.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. doi:10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 50.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. doi:10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 51.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. doi:10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mani SA, Guo W, Liao MJ, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. doi:10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.