Abstract
Mammalian cochlear supporting cells remain quiescent at postnatal ages and age-dependent changes in supporting cell proliferative capacity are evident. Ectopic Atoh1 expression in neonatal supporting cells converts only a small percentage of these cells into hair cell-like cells. Despite tremendous potential for therapeutics, cellular reprogramming in the mammalian inner ear remains a slow inefficient process that requires weeks, with most cells failing to reprogram. Cellular reprogramming studies in other tissues have shown that epigenetic inhibitors can significantly improve reprogramming efficiency.
Very little is known about epigenetic regulation in the mammalian inner ear, and almost nothing is known about the histone modifications. Histone modifications are vital for proper transcriptional regulation, and aberrant histone modifications can cause defects in the regulation of genes required for normal tissue development and maintenance. Our data indicate that cofactors of repressive complexes such as NuRD and PRC2 are present in the neonatal organ of Corti. These NuRD cofactors are present throughout most of the organ of Corti from E18.5 until P4. By P6, these NuRD cofactors are mostly undetectable by immunofluorescence and completely lost by P7, but are detectable again at P8 and continue to be present through P21. The PRC2 enzymatic subunit, EZH2 is also highly present from E18.5 to P0 in the organ of Corti, but lost between P2 and P4. However, EZH2 staining is evident again throughout the organ of Corti by P6 and persists through P21. Our data provide evidence that HDACs, DNA methyltransferases, histone methyltransferases, and histone demethylases are expressed postnatally within the organ of Corti, and may be targets for drug inhibition to increase the capacity, speed, and efficiency of reprogramming a supporting cell into a hair cell.
Keywords: epigenetics, NuRD, PRC2, HDAC, KDM1A, EZH2, inner ear, cochlea
1.1 Introduction
Cellular reprogramming studies have tremendous potential for therapeutic development, disease studies, and developmental processes (Soldner et al., 2009; Yamanaka, 2009). However, direct reprogramming through ectopic expression of defined transcription factors is a slow and inefficient process with most cells failing to reprogram (Huangfu et al., 2008; Mikkelsen et al., 2008). Additionally, the efficiency and yield of reprogrammed cells was shown to decline with increasing age and differentiation status of the donor cell (Hanna et al., 2010; Kim et al., 2010; Lister et al., 2011). Studies using induced pluripotent stem cells (iPSCs) have shown that inhibitors of epigenetic events such as DNA methylation, histone deacetylation, and histone methylation are able to improve reprogramming efficiency (Hanna et al., 2010; Huangfu et al., 2008; Kim et al., 2010; Lister et al., 2011; Mikkelsen et al., 2008).
In the auditory field, ectopic expression of the bHLH transcription factor, Atoh1, in neonatal cochlear supporting cells converts 6-11% of cochlear supporting cells into auditory hair cell-like cells that expressed multiple auditory hair cell markers and survive for at least 2 months in vivo (Kelly et al., 2012; Liu et al., 2012). However, the newly generated hair cell-like cells do not express the outer hair cell terminal differentiation marker Prestin, and lack mature hair cell morphology (Liu et al., 2012). Additionally, ectopic Atoh1 expression at P30 is unable to convert supporting cells into hair cell-like cells (Kelly et al., 2012; Liu et al., 2012). These data suggest that cochlear supporting cells may lose their cellular plasticity and capacity for cellular reprogramming through epigenetic modifications that occur during early postnatal inner ear development (Walters and Zuo, 2013).
Epigenetics encompass a broad variety of biological processes but may best be described as a change in phenotype that is not caused by a change in DNA sequence. The two most well understood mechanisms of epigenetic alterations that lead to these phenotypic changes are DNA methylation and histone modifications. Posttranslational histone modifications alter the histones interaction with DNA and nuclear proteins. Histone H3 and H4 can be covalently modified at several sites, and modifications on specific lysine residues play a fundamental role in transcriptional regulation. The enzymes responsible for maintaining proper histone acetylation states include histone acetyltransferases (HATs) and histone deacetylases (HDACs), while the enzymes responsible for maintaining proper histone methylation status are histone methyltransferases and histone demethylases (Figure 1).
Figure 1.
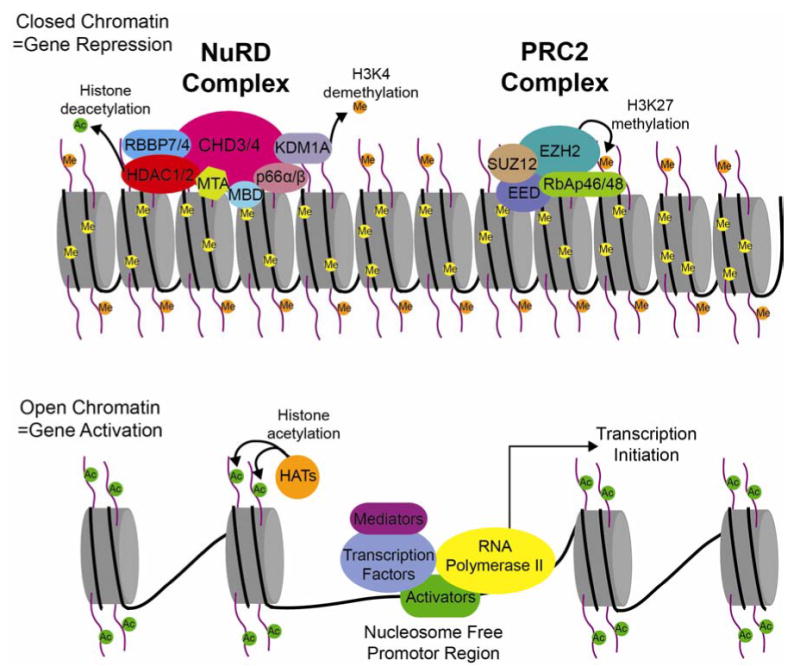
Cartoon diagram depicting the nucleosome dense, compacted chromatin associated with gene repression (upper portion), compared to the open more relaxed chromatin of an actively transcribed gene (bottom portion). The NuRD and PRC2 complexes modify the chromatin structure through histone modifications to initiate and maintain gene repression.
Class I HDACs, HDAC1 and HDAC2 are found in three distinct multiprotein complexes including the nucleosome remodeling and deacetylation (NuRD), CoREST, and Sin3 complexes (Cunliffe, 2008; Hayakawa and Nakayama, 2011). These complexes are highly conserved and function in distinct cellular processes. The NuRD complex is a transcriptional co-repressor essential for developmental transitions. The NuRD complex comprises at least two enzymatic subunits and several non-enzymatic subunits (Lai and Wade, 2011). Lysine-specific histone demethylase 1 (KDM1A), is also associated with the NuRD complex to demethylate di- and tri-methylated histone H3K4 (Wang et al., 2009). Histone deacetylation by the NuRD complex has been shown to recruit the polycomb repressive complex 2 (PRC2) to specific NuRD target genes (Reynolds et al., 2012). PRC2 is a transcriptional repressor complex required for gene silencing during multiple developmental processes. The histone methyltransferase activity of the PRC2 complex is done by the enzymatic subunit, enhancer of zeste homolog 2 (EZH2). EZH2 acts to repress transcription by di- and tri-methylating histone H3K27. Indicating that silencing of some NuRD target genes may require multiple repressive complexes for proper gene silencing during development.
Very little is known about epigenetic regulation in the mammalian inner ear. Although some DNA methylation studies in the mammalian inner ear have been reported (Mutai et al., 2009; Waldhaus et al., 2012), almost nothing is known about histone modifications that take place during mammalian inner ear development. Histone modifications are vital for proper transcriptional regulation, and aberrant histone modifications can cause defects in the regulation of genes required for normal tissue development and maintenance. A better understanding of the types of histone modifications made during mammalian inner ear development may provide information vital for future research toward complete cellular reprogramming and ultimately hearing regeneration. Here we report our comprehensive analysis of the distribution of several vital epigenetic factors involved in histone modification during late embryonic and early postnatal development of the auditory sensory epithelium (organ of Corti). Our results provide a framework for future studies of molecular mechanisms underlying epigenetic changes in the developing cochlea.
2.1 Material and Methods
2.2 Mice
FVB/NJ and C57BL/6J wild type mice were obtained from the Jackson Laboratory. Mice are housed with a 12/12h dark/light cycle and fed ad libitum. Each neonatal litter was divided into at least two neonatal time points, and a minimum of 3 littermates from at least 2 litters was assayed at each neonatal time point for both FVB/NJ and C57BL/6J to control for intra and inter litter variation. Timed pregnancies were established for both FVB/NJ and C57BL/6J wild type mice, and the morning of plug identification designated as E0.5. Embryos were collected following dam euthanization by CO2 asphyxiation and hysterectomy, and washed briefly in PBS. All procedures were approved by St. Jude Children’s Research Hospital Animal Care and Use Committee (ACUC).
2.3 Immunofluorescence
Embryonic (E18.5) and neonatal (P0-P7) ears from FVB/NJ and C57BL/6J wild type littermates were fixed in 4% paraformaldehyde overnight at 4°C. P8-P21 FVB/NJ and C57BL/6J wild type littermates were anesthetized with 250 mg/kg body weight tribromoethanol and perfusion fixed with 4% paraformaldehyde. P8-P21 ears were removed and placed in 4% paraformaldehyde overnight at 4°C, then incubated in 100mM EDTA for 2 days. All ears were treated with 30% sucrose protection overnight at 4°C, then flash frozen in TFM freezing medium (Triangle Biomedical Sciences, Durham, NC) for cryosectioning at 12μm. Following cryosectioning, sections were processed for immunofluorescence with antibodies against HDAC1 (1:500; Abcam, Cambridge, MA), HDAC2 (1:1000; Abcam), KDM1A (1:500; Abcam), CHD4 (1:200; Abcam), EZH2 (1:100; Cell Signaling, Danvers, MA), RCOR2 (1:200; Millipore, Temecula, CA), Histone H4ac (pan-acetylation) (1:1000; Active Motiff, Carlsbad, CA), and PVALB (1:500; Sigma, St. Louis, MO). Secondary antibodies were used at 1:200 and conjugated with Alexa 488, Alexa 568, Alexa 647 (Invitrogen, Carlsbad, CA) or HRP and then labeled with Tyramide Signal Amplification Kit (Vector Laboratories, Burlington, CA). Images were captured on a confocal microscope (Zeiss LSM 700 confocal microscope) using the Zen 2011 software then processed in LSM Image Browser and Photoshop CS3. Laser intensity was consistent between ages and samples.
2.4 RNA Isolation and Real-Time PCR
Each neonatal litter was divided into at least two neonatal time points, and a minimum of 3 littermates from at least 2 litters was assayed at each neonatal time point for both FVB/NJ and C57BL/6J. Pups were euthanized by decapitation, ears removed by gross dissection, placed in ice cold HBSS, and then microdissected for the organ of Corti. Organ of Corti RNA was isolated using the RNAqueous-Micro RNA Isolation Kit (Ambion, Austin, TX). Isolated RNA was treated with DNase I prior to cDNA synthesis. cDNA was generated using Superscript First-Strand cDNA Synthesis system for RT-PCR (Invitrogen) with random primers.
Relative expression levels were assayed utilizing TaqMan Gene Expression Master Mix and TaqMan probes (Applied Biosystems, Foster City, CA) for Hdac1, Hdac2, Ezh2, Chd4, Kdm1a, Rcor2, Prestin, Atoh1, mTert, Dnmt3a, Dnmt3b, 18S, GAPDH, and Actb. Reactions were run in triplicate in an Eppendorf Realplex2 Mastercycler System. The level of 18S, GAPDH, and Actb were used as internal controls and were run as a multiplex reaction with each assayed gene. The difference in CT between the assayed gene and 18S, GAPDH, and Actb for any given sample was defined as ∆CT(X). The difference in ∆CT(x) between two samples was defined as ∆∆CT(X), which represents a relative difference in expression of the assayed gene. The fold change of the assayed gene relative to 18S, GAPDH, and Actb was defined as 2-∆∆CT (Livak and Schmittgen, 2001). DataAssist software (Applied Biosystems) was used for statistical analysis and to confirm ∆CT(X) calculation.
3.1 Results
3.2 Histone deacetylases are present in the neonatal organ of Corti
Histone deacetylation plays a vital role in cell cycle regulation, cellular differentiation, and tissue development in mammals. Hdac1 and Hdac2 are typically co-expressed and often show functional redundancy in many tissues and cultured cell lines (LeBoeuf et al., 2010; Wilting et al., 2010; Yamaguchi et al., 2010). HDAC1 and HDAC2 play a crucial role in cellular proliferation and differentiation by regulating the expression of genes such as Cdkn1a (p21), Cdkn1b (p27), Cdkn2a (p16), and Trp53 (LeBoeuf et al., 2010; Wilting et al., 2010; Yamaguchi et al., 2010). Since mammalian cochlear supporting cells remain quiescent at postnatal ages and age-dependent changes in supporting cell proliferative capacity are evident (White et al., 2006), we analyzed late embryonic and postnatal wild type organ of Corti for the presence of histone deacetylases, HDAC1 and HDAC2, by immunofluorescence. Since staining for HDAC1 and HDAC2 was identical for each age in wild type mice, HDAC1 and HDAC2 will be referred to as HDAC1/2. We found that at E18.5 (FVB n=6; B6 n=6) HDAC1/2 are broadly found throughout the organ of Corti including the OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 2A and A’). At P0 (FVB n=8; B6 n=6), HDAC1/2 remain present in the OHCs, IHC, DCs, OP, LER, GER but is variably present in the IPC and IPh (Figure 2B and B’). At P2 (FVB n=8; B6 n=6) and P4 (FVB n=8; B6 n=6), HDAC1/2 appear reduced but present in the OHCs, IHC, DCs, OPC, GER and is variably present in the IPC, IPh, and LER (Figure 2C, C’, D and D’). However by P6 (FVB n=8; B6 n=6), HDAC1/2 are largely reduced but present in the GER and variably present in IHC, IPC, and IPh (Figure 2E and E’), while by P7 (FVB n=8; B6 n=6) HDAC1/2 staining is no longer apparent within the organ of Corti (Figure 2F and F’). Surprisingly, HDAC1/2 reappear in the organ of Corti in the OHCs, IHC, DCs, OPC, IPC, and IPh at P8 (FVB n=6; B6 n=6) (Figure 2G and G’), and continues to be present from P9-P14 (data not shown) and past hearing onset at P21 (FVB n=6; B6 n=6) (Figure 2H and H’).
Figure 2.
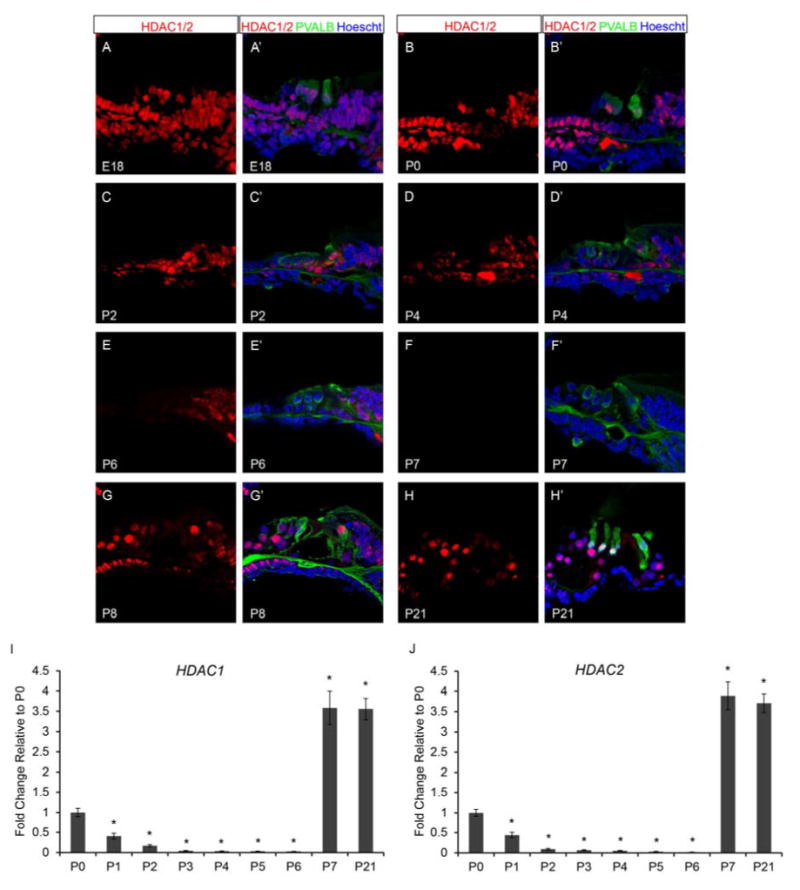
Neonatal murine organ of Corti expresses histone deacetylases, HDAC1 and HDAC2. Immunofluorescence of wild type neonatal mouse cochlea was performed using antibodies against HDAC1 and 2 (red), Parvalbumin (PVALB) (green), and Hoechst (blue). Taqman gene expression assays were done on microdissected organ of Corti. Expression levels are relative to endogenous gene controls 18S, GAPDH, and Actb. Fold changes are shown relative to P0. (A & A’) HDAC1/2 are found throughout the organ of Corti at E18.5. (B & B’) HDAC1/2 are still present in a majority of cell types in the organ of Corti at P0. (C-D’) Staining for HDAC1/2 appear reduced at P2 and P4 compared to E18.5 and P0. (E & E’) P6 organ of Corti retains some HDAC1/2 label in the OPCs, IPCs, IPhs, and IHCs. (F & F’) HDAC1/2 staining is no longer apparent in the organ of Corti by P7. (G-H’) HDAC1/2 reappears in the organ of Corti by P8 and continues to be detected at P21 (I) Expression of Hdac1 and Hdac2 are very similar at each time point analyzed. Hdac1 and Hdac2 expression steadily decreases from P0 to P6, then at P7 and P21 expression is increased compared to the P0 time point. All representative images were taken from the middle turn of the cochlea. *P<0.05 by Data Assist Software.
To determine whether HDAC1/2 affect histone acetylation levels, we analyzed late embryonic and postnatal wild type organ of Corti for the presence of histone H4 acetylation using antibodies against histone H4 pan-acetylation (H4ac). We found an inverse correlation between HDAC1/2 and H4ac in the organ of Corti. HDAC1/2 staining steadily decreases from E18.5 to P7, while H4ac levels increase from E18.5 to P7 (FVB n=8; B6 n=6) in the organ of Corti (Figure 3A-F’). While at P8 when HDAC1/2 reappear in the organ of Corti, H4ac at P8 appears reduced (FVB n=6; B6 n=6) (Figure 3G and G’) and remains low through P21 (FVB n=6; B6 n=6) in the organ of Corti (Figure 3H and H’).
Figure 3.
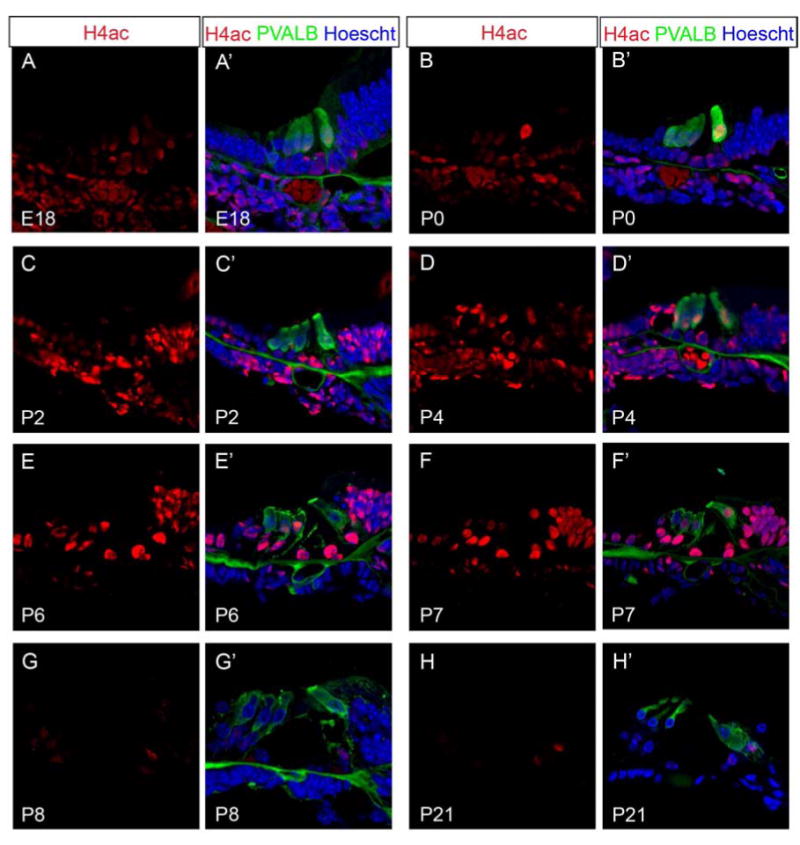
Histone H4 pan-acetylation staining appears inversely correlated with HDAC1/2 staining during postnatal inner ear development. Immunofluorescence of wild type postnatal mouse organ of Corti was performed using antibodies against H4ac (red), PVALB (green), and Hoechst (blue). Confocal laser intensity and gain were kept consistent between samples and ages. (A-H’) H4ac staining increases from E18.5 to P7, while at P8 H4ac staining is reduced and remains low through P21 in the organ of Corti. All representative images were taken from the middle turn of the cochlea.
Since the half-life of a protein can widely vary within a cell from minutes to several days, we analyzed Hdac1 and Hdac2 mRNA levels in the micro-dissected neonatal organ of Corti by quantitative real-time PCR (qPCR) using Taqman probes. Hdac1 and Hdac2 mRNA levels were analyzed at P0, P1, P2, P3, P4, P5, P6, P7, and P21 (for each time point FVB n=6; B6 n=6), values are presented as relative to P0 (each time point FVB n=6; B6 n=6). We found that Hdac1 and Hdac2 follow a similar pattern of expression within the neonatal organ of Corti. Hdac1 and Hdac2 mRNA levels steadily decrease from P0 to P6, then at P7 and P21, both Hdac1 and Hdac2 mRNA levels increase approximately 4 fold compared to the P0 time point (Figure 2I and J). The apparent spike in Hdac1 and Hdac2 mRNA levels precedes HDAC1/2 protein detection by 1 day in the organ of Corti, suggesting that HDAC1 and HDAC2 may be subject to post-transcriptional and/or translational regulation (Spriggs et al., 2010; Vogel and Marcotte, 2012).These data together with the immunofluorescence data indicates that both Hdac1 and Hdac2 are expressed and may have a functional role during postnatal organ of Corti maturation.
3.3 Neonatal organ of Corti expresses lysine-specific histone demethylase 1 (KDM1A)
KDM1A is a lysine-specific histone demethylase that acts to repress gene transcription by removing methylation sites from H3K4me1/2 (Wang et al., 2007; Wang et al., 2009). However, KDM1A typically functions with additional co-factors such as HDAC1 and HDAC2 in repressive complexes including the Co-REST and NuRD complexes to control transcription factor access to target genes by altering chromatin structure thereby regulating gene transcription (Wang et al., 2007; Wang et al., 2009). To test whether KDM1A is present in a similar pattern to HDAC1/2, we analyzed wild type organ of Corti for the presence of KDM1A by immunofluorescence. We found that at E18.5 (FVB n=6; B6 n=6) KDM1A is present throughout the organ of Corti including the OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 4A and A’). At P0 (FVB n=8; B6 n=6), KDM1A is retained throughout the organ of Corti including the OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 4B and B’). At P2 (FVB n=8; B6 n=6), KDM1A appears reduced but present in the OHCs, DCs, OPC, IPC, IPh, GER and is variably present in the IHC and LER (Figure 4C and C’). By P4 (FVB n=8; B6 n=6), remains present in the OHC, IPh, GER and is variably present in DC, OPC, IPC, and IHC (Figure 4D and D’). At P6 (FVB n=8; B6 n=6), KDM1A is greatly reduced but remains present in the GER and appears variably in IPC and IPh (Figure 4E and E’). By P7 (FVB n=8; B6 n=6), KDM1A staining is no longer apparent within the organ of Corti (Figure 4F and F’). Similar to HDAC1/2 staining, KDM1A is apparent again at P8 (FVB n=6; B6 n=6) in the OHCs, IHC, DCs, OPC, IPC, and IPh (Figure 4G and G’), and remains detectable in the organ of Corti from P9 to P14 (data not shown) and beyond hearing onset at P21 (FVB n=6; B6 n=6) (Figure 4H and H”).
Figure 4.
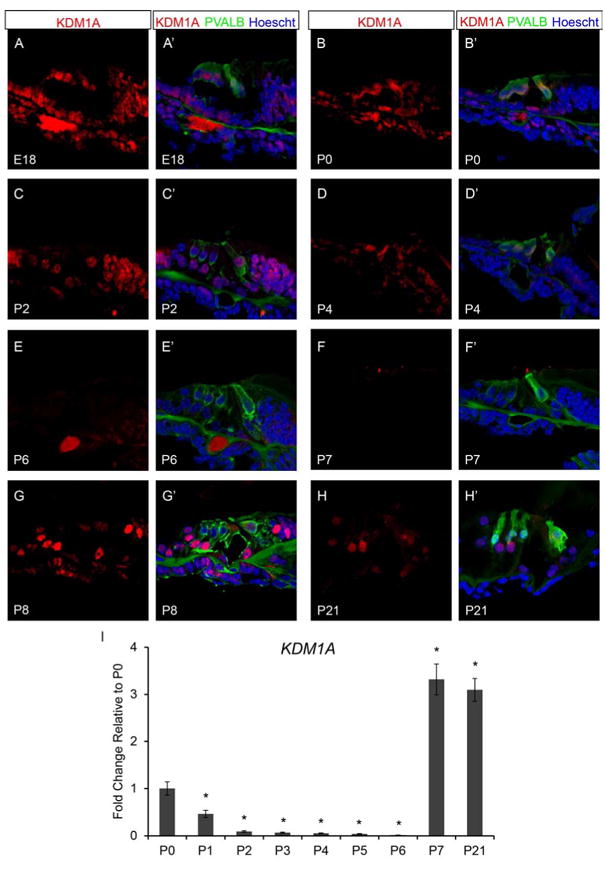
Histone demethylase, KDM1A is detected in the neonatal mouse cochlea. Immunofluorescence of wild type neonatal mouse organ of Corti was performed using antibodies against KDM1A (red), PVALB (green), and Hoechst (blue). Taqman gene expression assays were done on microdissected organ of Corti. Expression levels are relative to endogenous gene controls 18S, GAPDH, and Actb. Fold changes are shown relative to P0. (A & A’) KDM1A are found throughout the organ of Corti at E18.5. (B & B’) KDM1A is still present in most cell types in organ of Corti at P0. (C & C’) Staining is present but less apparent in the organ of Corti by P2. (D & D’) Staining for KDM1A is variable in IHCs, DCs, OPCs, and IPCs but is retained in OHCs, IPhs, and GER cells at P4. (E-F’) Very little KDM1A label remains apparent in the organ of Corti at P6, and is undetectable by P7. (G-H’) KDM1A is detected again in the organ of Corti by P8 and continues to be present through P21. (I) Kdm1a expression levels steadily decrease from P0 to P6, but is increased at P7 and P21 compared to the P0 time point. All representative images were taken from the middle turn of the cochlea. *P<0.05 by Data Assist Software.
We analyzed the wild type neonatal organ of Corti by qPCR using TaqMan probes for the mRNA expression levels Kdm1a (each time point FVB n=6; B6 n=6). Similar to Hdac1 and Hdac2 mRNA levels, Kdm1a expression levels decrease from P0 to P6 (Figure 4I). Kdm1a expression at both P7 and P21 increases by approximately 3 fold compared to the P0 time point (Figure 4I), which coincides with Hdac1 and Hdac2 expression levels at P7 and P21. The KDM1A immunofluorescence and qPCR data taken together with the HDAC1 and HDAC2 data suggests that these epigenetic factors may have a combinatorial role in regulating maturation of the postnatal organ of Corti.
3.4 The NuRD complex is present within the neonatal inner ear
The NuRD complex is required for lineage-specific gene expression by regulating the chromatin state through histone modifications and chromatin remodeling (Lai and Wade, 2011; Reynolds et al., 2012; Wang et al., 2009). The core component of the NuRD complex is the ATP-dependent chromatin remodeling protein, chromodomain helicase DNA binding protein 4 (CHD4) (Lai and Wade, 2011). To test whether CHD4 is present in the neonatal organ of Corti, we analyzed wild type organ of Corti for the presence of CHD4 by immunofluorescence. We found that at E18.5 (FVB n=8; B6 n=6) CHD4 is present in organ of Corti including the OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 5A and A’). From P0 (FVB n=8; B6 n=6) to P2 (FVB n=8; B6 n=6), CHD4 continues to be detected in the OHCs, IHC, DCs, OPC, IPC, IPh, GER and is variably detectable in the LER (Figure 5B-C’). While at P4 (FVB n=8; B6 n=6), CHD4 appears reduced but present in the OHCs, IHC, DCs, OPC, IPC, IPh, GER and remains variably detectable in the LER (Figure 5D and D’). By P6 (FVB n=8; B6 n=6), CHD4 is only found in the IHC and is undetectable within the organ of Corti by P7 (FVB n=8; B6 n=6) (Figure 5E-F’). CHD4 is apparent again in the organ of Corti in the OHCs, IHC, DCs, OPC, IPC, and IPh at P8 (FVB n=6; B6 n=6) (Figure 5G and G’), and continues to be present from P9-P14 (data not shown) and past hearing onset at P21 (FVB n=6; B6 n=6) (Figure 5H and H’).
Figure 5.
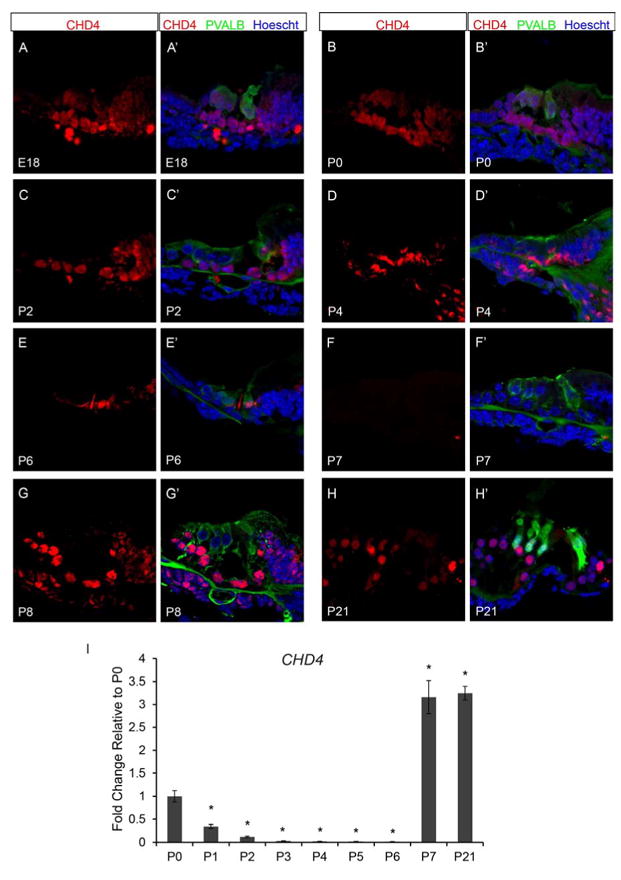
The core subunit of the NuRD repressive complex, CHD4, is detected in a similar pattern of expression as HDAC1, HDAC2, and KDM1A. Immunofluorescence of wild type organ of Corti was performed using antibodies against CHD4 (red), PVALB (green), and Hoechst (blue). Taqman gene expression assays were done on microdissected organ of Corti. Expression levels are relative to endogenous gene controls 18S, GAPDH, and Actb. Fold changes are shown relative to P0. (A & A’) CHD4 is highly present throughout the organ of Corti at E18.5. (B-C’) From P0 to P2, CHD4 continues to be detected in the vast majority of cells in the organ of Corti. (D & D’) While at P4, CHD4 continues to be detected throughout the organ of Corti but appears reduced in intensity. (E & F’) By P6, CHD4 staining is only found in the IHCs of the organ of Corti, and is no longer apparent by P7. (G-H’) KDM1A is detected again in the organ of Corti by P8 and continues to be present through P21. (I) Similar to Hdac1, Hdac2, and Kdm1a, Chd4 mRNA levels steadily decrease in expression from P0 to P6, and then at P7 and P21 expression is increased compared to the P0 time point.. All representative images were taken from the middle turn of the cochlea. *P<0.05 by Data Assist Software.
To determine the temporal expression pattern of Chd4, we analyzed Chd4 mRNA levels in the micro-dissected neonatal organ of Corti by qPCR using Taqman probes (each time point FVB n=6; B6 n=6). We found a similar pattern of Chd4 mRNA expression within the neonatal organ of Corti as was detected for Hdac1, Hdac2, and Kdm1a, with expression levels steadily decreasing from P0 to P6, then at P7 and P21 expression is increased approximately 3 fold compared to the P0 time point (Figure 5I). Altogether, our data suggest that the NuRD complex functions to regulate proper temporal and cell-type specific gene expression within the organ of Corti to achieve terminal cellular differentiation during postnatal development.
Since HDAC1, HDAC2, and KDM1A are known to be co-factors in both the CoREST and NuRD repressive complexes (Lai and Wade, 2011; LeBoeuf et al., 2010; Qureshi et al., 2010; Reynolds et al., 2012; Wang et al., 2007; Wang et al., 2009), we analyzed wild type organ of Corti for the presence of the core component of these repressive complexes by immunofluorescence and qPCR (FVB n=8; B6 n=6). The core component of the murine CoREST complex is RCOR2. We were unable to detect the presence or mRNA expression of RCOR2 by immunofluorescence or qPCR, indicating that the CoREST complex is unlikely to have a functional role during neonatal organ of Corti maturation.
3.5 PRC2 is present in the neonatal cochlea and may function in combination with NuRD
Precise control of gene expression is vital for both cellular differentiation and development. During development lineage-specific genes require a precise temporal and spatial pattern of expression, while the expression of genes such as pluripotency genes must be repressed. Achieving proper gene expression during developmental transitions requires multiple levels of gene regulation from transcription factor availability to the chromatin state. NuRD has been shown to recruit additional repressive complexes such as PRC2 to specific target genes (Reynolds et al., 2012). Similar to NuRD, PRC2 is essential for proper developmental transitions. The enzymatic subunit of the PRC2 repressive complex is the histone methyltransferase, EZH2. To test whether PRC2 may work in conjunction with NuRD during neonatal organ of Corti maturation, we examined wild type organ of Corti for the presence of EZH2 by immunofluorescence. We found that at E18.5 (FVB n=6; B6 n=6) EZH2 can be detected throughout the organ of Corti including OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 6A and A’). EZH2 continues to be present in the OHCs, IHC, DCs, OPC, IPC, IPh, GER, and variably in the LER at P0 (FVB n=8; B6 n=6) (Figure 6B and B’). However, by P2 (FVB n=8; B6 n=6) EZH2 is no longer detected within the organ of Corti and remains absent through P4 (FVB n=8; B6 n=6) (Figure 6C-D’). Then at P6 (FVB n=8; B6 n=6), EZH2 is found present again within IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 6E and E’). By P7 (FVB n=8; B6 n=6), EZH2 is once again found throughout the organ of Corti including OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER (Figure 6F and F’). EZH2 remains detectable in the organ of Corti at P8 (FVB n=6; B6 n=6) (Figure 6G and G’), and is still apparent from P9-P14 (data not shown) and past hearing onset at P21 (FVB n=6; B6 n=6) (Figure 6H and H’).
Figure 6.
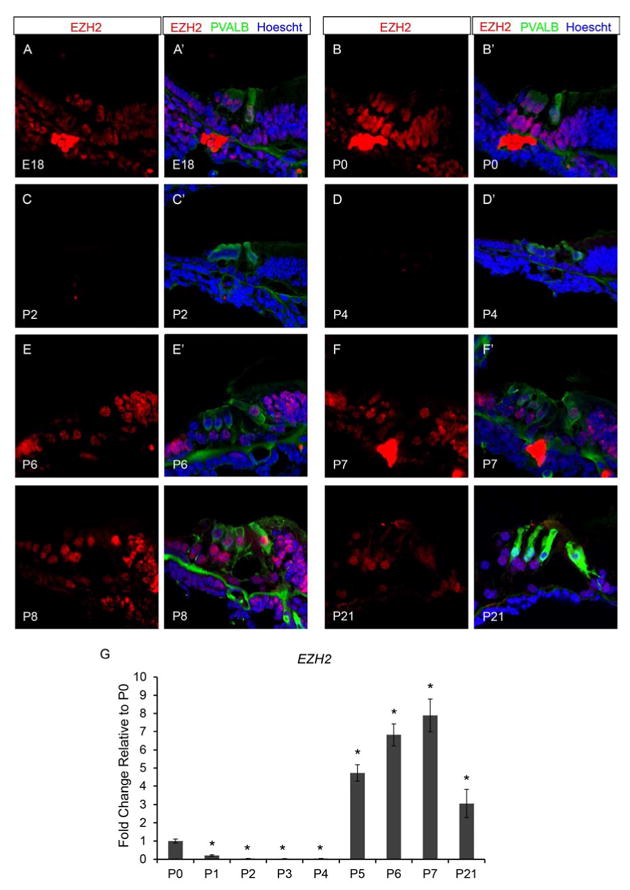
Neonatal murine organ of Corti expresses the PRC2 enzymatic subunit, the histone methyltransferase, EZH2. Immunofluorescence of wild type neonatal mouse cochlea was performed using antibodies against EZH2 (red), PVALB (green), and Hoechst (blue). Taqman gene expression assays were done on microdissected organ of Corti. Expression levels are relative to endogenous gene controls 18S, GAPDH, and Actb. Fold changes are shown relative to P0. (A-B’) EZH2 is detected throughout the organ of Corti at both E18.5 and P0 time points. (C-D’) Staining for CHD4 is no longer apparent in the organ of Corti from P2 to P4. (E & E’) At P6, EZH2 staining is detected again in a majority of cell types in the organ of Corti. (F & H’) EZH2 is present throughout the organ of Corti by P7 and remains detectable from P8 to P21. (I) Ezh2 expression rapidly declines from P0 to P1, and expression remains very low through P4. From P5-P7 and at P21, Ezh2 expression is increased in the organ of Corti compared to the P0 time point. All representative images were taken from the middle turn of the cochlea. *P<0.05 by Data Assist Software.
The temporal expression pattern of Ezh2 was measured by qPCR using Taqman probes in micro-dissected neonatal organ of Corti (each time point FVB n=6; B6 n=6). Ezh2 expression rapidly declines from P0 to P1, expression remains very low until P5 at which point Ezh2 expression increases approximately 5 fold compared to P0 time point (Figure 6I). From P5 until P7, Ezh2 expression increases with approximately an 8 fold increase in expression at P7 and 3 fold increase at P21 compared to the P0 time point. The transient temporal pattern of HDAC1, HDAC2, KDM1A, CHD4, and EZH2 staining and mRNA levels suggests that both NuRD and PRC2 may be required for specific developmental transitions within the neonatal organ of Corti and may function alone or in combination at lineage-specific target genes.
3.6 Repressive histone modifications correspond with altered gene expression
During development, the expression of lineage-specific genes must be up-regulated, while genes required for maintaining cellular plasticity and proliferation need to be down-regulated in a precise temporal pattern. NuRD and PRC2 are required during these developmental transitions for proper gene regulation. Genes such as telomerase reverse transcriptase(mTert) are known to be highly expressed in cells that have greater capacity for proliferation, mTert then becomes increasingly down-regulated during differentiation as cells become more committed to a cellular lineage (Armstrong et al., 2000). We found by qPCR using Taqman probes that during neonatal organ of Corti maturation mTert expression steadily decreases between P0 and P7 (each time point FVB n=6; B6 n=6) (Figure 7A), which correlates with previous work showing loss of postnatal proliferative capacity in vivo (Ruben, 1967). We also analyzed by qPCR using Taqman probes the expression of Atoh1, which is thought to be one of the earliest determinants of hair cell fate. Although Atoh1 expression is highest in the embryonic inner ear (Driver et al., 2013), we analyzed Atoh1 expression levels by qPCR during neonatal maturation (each time point FVB n=6; B6 n=6). We found that Atoh1 mRNA levels continuously decline from P0 to P7 (Figure 7B), with P7 Atoh1 levels being nearly undetectable. To determine whether lineage-specific genes become up-regulated during neonatal inner ear maturation, we analyzed by qPCR using Taqman probes mRNA expression levels of the outer hair cell-specific gene, Prestin, by qPCR. Prestin expression levels were not consistently detectable in the neonatal organ of Corti until P3, thus expression levels are relative to P3 instead of P0 as described above. Prestin mRNA levels steadily increase from P3 to P6, then at P7 Prestin expression increases by approximately 17 fold compared to the P3 time point (each time point FVB n=6; B6 n=6) (Figure 7C).
Figure 7.
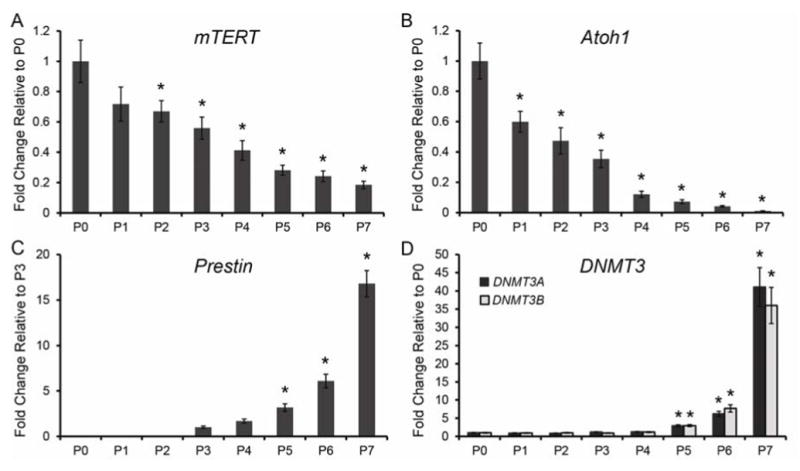
Organ of Corti maturation coincides with alterations gene during neonatal development. TaqMan gene expression assays were done on microdissected organ of Corti. Expression levels are relative to endogenous gene controls 18S, GAPDH, and Actb. Fold changes are shown relative to P0 for all genes except Prestin which is relative to the P3 time point. (A and B) Expression of both mTert and Atoh1 steadily decrease from P0 to P7. (C) Prestin expression was not readily detectable in the organ of Corti until P3, and then Prestin expression levels continue to increase from P4 to P7. (D) Dnmt3a and Dnmt3b have low levels of expression between P0 and P6, and then expression of Dnmt3a and Dnmt3b increases dramatically at P7. *P< 0.05 by Data Assist Software.
Age-related DNA methylation studies have found a positive correlation between aging and increased DNA methylation, which results in gene silencing (Hernandez et al., 2011; Maegawa et al., 2010). Studies in the mammalian inner ear show a positive correlation between increased DNA methylation on Sox2 enhancers and supporting cell differentiation and loss of stemness (Waldhaus et al., 2012). NuRD binding specificity relies upon the methyl binding properties of their methyl-CpG binding domain protein subunits, MBD2 and MBD3 (Lai and Wade, 2011). DNA methylation plays an important role during development and changes in the DNA methylation pattern serve as a key epigenetic process in transcriptional regulation (Jurkowska et al., 2011). Two DNA methyltransferase enzymes, DNMT3A and DNMT3B, are responsible for establishing de novo DNA methylation patterns during development (Jurkowska et al., 2011). Additionally, NuRD and PRC2 were shown to recruit DNMTs to target genes to stabilize the silencing of target genes (Cai et al., 2013; Vire et al., 2006). To determine whether Dnmt3a and Dnmt3b are expressed during neonatal organ of Corti maturation, we measured the expression levels of Dnmt3a and Dnmt3b by qPCR using Taqman probes (each time point FVB n=6; B6 n=6). We found that Dnmt3a and Dnmt3b have a relatively low level of expression from P0 to P4, then both Dnmt3a and Dnmt3b expression increases approximately 7 fold by P5, and an approximate 35-40 fold increase in expression by P7 compared to the P0 time point (Figure 7D). These data taken together with the NuRD and PRC2 data suggest that a key developmental transition occurs in the organ of Corti after the first week of postnatal development, and that this developmental transition requires multiple levels of epigenetic regulation.
4.1 Discussion
Epigenetic events such as histone modifications and DNA methylation are essential for proper development. Developmental patterns of epigenetic events in the mammalian organ of Corti may help elucidate targets toward developing new therapeutics in the treatment of hearing loss and hearing regeneration. Our data indicates that cofactors of repressive complexes such as NuRD and PRC2 are present in both FVB/NJ and C57BL/6J wild type organ of Corti during the first postnatal week. HDAC1, HDAC2, KDM1A, and CHD4 are all highly present in OHCs, IHC, DCs, OPC, IPC, IPh, LER, and GER just prior to birth at E18.5. These NuRD cofactors remain present in the OHCs, IHC, DCs, OPC, IPC, IPh, and GER from P0 until P4. At P6, these cofactors of the NuRD complex are mostly undetectable by immunofluorescence within the organ of Corti, completely lost by P7, returns at P8, and remain present through hearing onset at P21. Surprisingly, there was no discernible difference in the expression or staining pattern between the two wild type strains tested, which suggests that these epigenetic factors are fundamentally required for postnatal organ of Corti maturation.
Although some variability in the presence of HDAC1, HDAC2, KDM1A, and CHD4 exists, the variability in staining may be explained by the location within the organ of Corti, basal to apical. Immunofluorescence for HDAC1, HDAC2, KDM1A, and CHD4 was lost progressively within the organ of Corti from base to apex. During development, the organ of Corti matures in a similar pattern (base to apex) (Ruben, 1967), which may explain some of the variability seen in the HDAC1, HDAC2, KDM1A, and CHD4 staining pattern for specific cell types. Differential rates of protein degradation between cell types within the organ of Corti may also contribute to variability in the staining pattern, since immunofluorescence visualization is dependent upon a threshold level of protein availability. However, the large degree of overlap in HDAC1, HDAC2, KDM1A, and CHD4 staining that exists both temporally and spatially indicates that the NuRD repressive complex functions to help regulate proper gene expression during postnatal inner ear development.
The disparity between mRNA and protein detection at P7 for components of the NuRD complex is somewhat confounding. However, producing too little or too much of a single subunit of a protein complex can compromise the proper assembly of the entire complex. Many regulatory mechanisms occur after mRNAs are produced to precisely time proper complex assembly including cap-dependent translation initiation, ribosomal entry, and mRNA sequestration by miRNAs and RNA binding proteins (Spriggs et al., 2010; Vogel and Marcotte, 2012). Recent work has shown that mRNA from the myogenic determination gene Myf5 is sequestered by miR-31 in mRNP granules to precisely regulate cellular differentiation. Altogether these data suggest that the precise regulation of both terminal cellular differentiation and NuRD complex assembly may contribute to the 1 day discordance between mRNA expression levels and protein detection.
During development, NuRD may recruit additional repressive complexes such as PRC2 to specific target genes (Reynolds et al., 2012). Our data show that the PRC2 enzymatic subunit, EZH2, is present at E18.5 and P0 in a similar pattern of expression as the NuRD subunit, HDAC1, HDAC2, KDM1A, and CHD4. Interestingly, EZH2 is not detectable from P2 to P4 in the organ of Corti but returns throughout the organ of Corti by P6 and remains present through P21. These data suggest that NuRD and PRC2 may have a combinatorial role for regulating gene expression at specific developmental time points, but NuRD and PRC2 are differentially present within the organ of Corti from P2 to P7. These data together suggest that although NuRD may recruit PRC2 to specific target genes during postnatal inner development, both NuRD and PRC2 also have independent functional roles for regulating the chromatin state.
Histone modifications made by NuRD and PRC2 are typically repressive marks that silence the expression of specific-target genes during cellular transitions including cell fate specification, cellular differentiation, and tissue development (Hernandez et al., 2011; Lande-Diner et al., 2007; LeBoeuf et al., 2010; Maegawa et al., 2010; McCabe et al., 2012; Miranda and Jones, 2007; Wang et al., 2007; Wang et al., 2009; Yamaguchi et al., 2010). Genes required for proliferation (mTert) and cell fate specification (Atoh1) were increasingly down regulated between P0 and P7 in the organ of Corti, which is consistent with reports related to organ of Corti quiescence and maturation during neonatal development. However, these developmental time points also correlate with increased expression of genes such as Prestin, Dnmt3a, and Dnmt3b which are required for terminal cellular differentiation. Although NuRD and PRC2 are unlikely to directly regulate genes whose expression is elevated, the increased expression of these genes corresponds with the role of NuRD and PRC2 in terminal cellular differentiation during mammalian tissue development.
An age-related decline in proliferative and reprogramming capacity in the mammalian cochlea has been shown both in vitro and in vivo (Driver et al., 2013; Izumikawa et al., 2005; Kelly et al., 2012; Liu et al., 2012; Malgrange et al., 2002; Oshima et al., 2007; Ruben, 1967; Sinkkonen et al., 2011; Waldhaus et al., 2012; Zheng and Gao, 2000). In vivo studies of hair cell regeneration found that both transient and irreversible induction of ectopic Atoh1 in supporting cells during the first postnatal week, leads to the formation of hair cell-like cells (Kelly et al., 2012; Liu et al., 2012). However, induction of Atoh1 at later postnatal ages (P8-P14) showed a dramatic decrease in the capacity of supporting cells to form hair cell-like cells, and by P30, induction of Atoh1 no longer leads to the formation of hair cell-like cells (Kelly et al., 2012; Liu et al., 2012). These data are consistent with our findings that during the first postnatal/neonatal week, epigenetic alterations are occurring within the organ of Corti. The presence of repressive complexes such as NuRD and PRC2 suggests that silencing histone marks may change the chromatin state such that transcription factors no longer have adequate access to target genes required for supporting cell to hair cell conversion. Additionally, these data correlate with our gene expression profiles showing a dramatic increase in Hdac1, Hdac2, Kdm1a, Chd4, Ezh2, Dnmt3a, and Dnmt3b mRNA levels at P7. These gene products are all associated with a global repression of gene transcription (Cheng et al., 2003; McCabe et al., 2012; Palmieri et al., 2009; Reynolds et al., 2012) and likely play a role in altering the reprogramming capacity of the mammalian cochlea.
Cellular reprogramming has extraordinary potential as a therapeutic for hearing loss. Epigenetic effects on cellular reprogramming have been characterized in iPSCs. Similar to cellular reprogramming in the inner ear, reprogramming somatic cells to a pluripotent state is a long inefficient process with most cells failing to reprogram (Cohen and Melton, 2011; Hanna et al., 2010; Huangfu et al., 2008; Kim et al., 2010; Li et al., 2009; Mikkelsen et al., 2008; Yamanaka, 2009). Studies have found that under normal reprogramming condition (transcription factor induction) somatic cells induced to become iPSCs retain the epigenetic memory of their donor cell (Cohen and Melton, 2011; Hanna et al., 2010; Huangfu et al., 2008; Kim et al., 2010; Li et al., 2009; Mikkelsen et al., 2008; Yamanaka, 2009). The epigenetic memory of the donor cell greatly influences the iPSCs capacity to become other cell types but iPSCs do retain their ability to be easily converted back to the original donor cell type (Cohen and Melton, 2011; Hanna et al., 2010; Huangfu et al., 2008; Kim et al., 2010; Li et al., 2009; Mikkelsen et al., 2008; Yamanaka, 2009). The speed and efficiency of cellular reprogramming to an iPSC state can be enhanced by epigenetic inhibitor treatment. Donor cells treated with inhibitors against HDACs, DNA methyltransferases, histone methyltransferases and/or histone demethylases have a faster more efficient reprogramming capacity (Huangfu et al., 2008; Kim et al., 2010; Papp and Plath, 2011). Our data provides evidence that HDACs (Hdac1 and Hdac2), DNA methyltransferases (Dnmt3a and Dnmt3b), histone methyltransferases (Ezh2), and histone demethylases (Kdm1a) are expressed postnatally within the organ of Corti, and may be targets for drug inhibition to increase the capacity, speed, and efficiency of reprogramming a supporting cell into a hair cell.
Highlights.
Histone deacetylases are present in the neonatal organ of Corti
Neonatal organ of Corti expresses lysine-specific histone demethylase 1 (KDM1A)
The NuRD complex is present within the neonatal inner ear
PRC2 is present in the neonatal cochlea and may function in combination with NuRD
Repressive histone modifications correspond with altered gene expression
Acknowledgments
This work was supported by grants from the National Institutes of Health (DC006471, DC008800, and CA21765 to J.Z.), the Office of Naval Research (N000140911014, N000141210775, and N000141210191 to J.Z.), and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. J.Z. is a recipient of The Hartwell Individual Biomedical Research Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Wanda S. Layman, Email: wanda.layman@stjude.org.
Mario A. Sauceda, Email: mario.sauceda@stjude.org.
Jian Zuo, Email: jian.zuo@stjude.org.
References
- Armstrong L, Lako M, Lincoln J, Cairns PM, Hole N. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mechanisms of development. 2000;97:109–116. doi: 10.1016/s0925-4773(00)00423-8. [DOI] [PubMed] [Google Scholar]
- Cai Y, Geutjes EJ, de Lint K, Roepman P, Bruurs L, Yu LR, Wang W, van Blijswijk J, Mohammad H, de Rink I, et al. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene. 2013 doi: 10.1038/onc.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- Cohen DE, Melton D. Turning straw into gold: directing cell fate for regenerative medicine. Nat Rev Genet. 2011;12:243–252. doi: 10.1038/nrg2938. [DOI] [PubMed] [Google Scholar]
- Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr Opin Genet Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver EC, Sillers L, Coate TM, Rose MF, Kelley MW. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Developmental biology. 2013;376:86–98. doi: 10.1016/j.ydbio.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, Ben-Porath I, Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G, Cedar H. Role of DNA methylation in stable gene repression. J Biol Chem. 2007;282:12194–12200. doi: 10.1074/jbc.M607838200. [DOI] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Developmental cell. 2010;19:807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, Ayrault O, Zindy F, Gan L, Roussel MF, Zuo J. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters' cells to immature hair cells by Atoh1 ectopic expression. J Neurosci. 2012;32:6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgrange B, Belachew S, Thiry M, Nguyen L, Rogister B, Alvarez ML, Rigo JM, Van De Water TR, Moonen G, Lefebvre PP. Proliferative generation of mammalian auditory hair cells in culture. Mechanisms of development. 2002;112:79–88. doi: 10.1016/s0925-4773(01)00642-6. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Mutai H, Nagashima R, Sugitani Y, Noda T, Fujii M, Matsunaga T. Expression of Pou3f3/Brn-1 and its genomic methylation in developing auditory epithelium. Dev Neurobiol. 2009;69:913–930. doi: 10.1002/dneu.20746. [DOI] [PubMed] [Google Scholar]
- Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, Geleoc GS, Edge A, Holt JR, Heller S. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. Journal of the Association for Research in Otolaryngology : JARO. 2007;8:18–31. doi: 10.1007/s10162-006-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri D, Lockman PR, Thomas FC, Hua E, Herring J, Hargrave E, Johnson M, Flores N, Qian Y, Vega-Valle E, et al. Vorinostat inhibits brain metastatic colonization in a model of triple-negative breast cancer and induces DNA double-strand breaks. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6148–6157. doi: 10.1158/1078-0432.CCR-09-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Plath K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell research. 2011;21:486–501. doi: 10.1038/cr.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Gokhan S, Mehler MF. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta oto-laryngologica. 1967;(Suppl 220):221–244. [PubMed] [Google Scholar]
- Sinkkonen ST, Chai R, Jan TA, Hartman BH, Laske RD, Gahlen F, Sinkkonen W, Cheng AG, Oshima K, Heller S. Intrinsic regenerative potential of murine cochlear supporting cells. Scientific reports. 2011;1:26. doi: 10.1038/srep00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhaus J, Cimerman J, Gohlke H, Ehrich M, Muller M, Lowenheim H. Stemness of the organ of Corti relates to the epigenetic status of Sox2 enhancers. PLoS One. 2012;7:e36066. doi: 10.1371/journal.pone.0036066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Zuo J. Postnatal development, maturation and aging in the mouse cochlea and their effects on hair cell regeneration. Hear Res. 2013;297:68–83. doi: 10.1016/j.heares.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J, DePinho RA, Dannenberg JH. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C, Matthias P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]


