Misfolding of cystic fibrosis transmembrane conductance regulator protein (CFTR) causes the fatal lung disease cystic fibrosis. VX-809 was developed to suppress disease-related folding defects in CFTR. VX-809 suppresses folding defects in CFTR by modulating the conformation of membrane-spanning domain 1. VX-808 is thereby able to partially restore function to F508del-CFTR and other disease-related mutants.
Abstract
Cystic fibrosis (CF) is a fatal genetic disorder associated with defective hydration of lung airways due to the loss of chloride transport through the CF transmembrane conductance regulator protein (CFTR). CFTR contains two membrane-spanning domains (MSDs), two nucleotide-binding domains (NBDs), and a regulatory domain, and its channel assembly requires multiple interdomain contacts. The most common CF-causing mutation, F508del, occurs in NBD1 and results in misfolding and premature degradation of F508del-CFTR. VX-809 is an investigational CFTR corrector that partially restores CFTR function in people who are homozygous for F508del-CFTR. To identify the folding defect(s) in F508del-CFTR that must be repaired to treat CF, we explored the mechanism of VX-809 action. VX-809 stabilized an N-terminal domain in CFTR that contains only MSD1 and efficaciously restored function to CFTR forms that have missense mutations in MSD1. The action of VX-809 on MSD1 appears to suppress folding defects in F508del-CFTR by enhancing interactions among the NBD1, MSD1, and MSD2 domains. The ability of VX-809 to correct F508del-CFTR is enhanced when combined with mutations that improve F508del-NBD1 interaction with MSD2. These data suggest that the use of VX-809 in combination with an additional CFTR corrector that suppresses folding defects downstream of MSD1 may further enhance CFTR function in people with F508del-CFTR.
INTRODUCTION
Cystic fibrosis (CF) is caused by the loss of epithelial chloride transport due to mutations in the CF transmembrane conductance regulator (CFTR) gene that encodes the CFTR protein (Riordan et al., 1989a). The CFTR protein is a chloride channel that is normally present at the cell surface of epithelial cells, where it regulates salt, fluid, and pH balance in multiple organs (Quinton, 1983, 2008; Boucher, 2007). In people with CF, the loss of chloride transport due to defects in the CFTR protein results in the accumulation of thick, sticky mucus in the bronchi of the lungs, loss of exocrine pancreatic function, impaired intestinal absorption, reproductive dysfunction, and elevated sweat chloride concentration (Boucher, 2007; Castellani et al., 2008).
CFTR is a 1480–amino acid ATP-binding cassette transporter protein that contains two membrane-spanning domains (MSD1 and MSD2) that form the chloride channel pore, two nucleotide-binding domains (NBD1 and NBD2) that bind and hydrolyze ATP to open and close the channel pore (channel gating), and a regulatory domain with several protein kinase A phosphorylation sites (Ikuma and Welsh, 2000; Rowe et al., 2005). Formation of the CFTR channel requires the coordinated folding and assembly of the individual membrane and cytoplasmic domains (Cyr, 2005; Kim and Skach, 2012). Each MSD is composed of six transmembrane segments (TM1–6 and TM7–12) that are associated with long α-helical cytosolic extensions known as coupling helices, which are connected by intracellular loops (ICLs). Contact formation between the ICLs and NBDs is critical for the proper assembly and Cl− channel function of CFTR (Anderson et al., 1991; Riordan et al., 1989b; Riordan, 2008).
The most common CFTR mutation in patients with CF is the deletion of phenylalanine at position 508 (F508del) in NBD1. Normally, F508 is critical for folding and assembly of native CFTR, as its backbone and side chain participate in the formation of a network of interdomain contacts that stabilizes interactions between NBD1 and ICL1 of MSD1 and ICL4 of MSD2 (Mornon et al., 2008; Serohijos et al., 2008). In the absence of F508, folding of the individual CFTR subdomains is initiated, but channel assembly is arrested at an intermediate stage (Lukacs et al., 1994; Younger et al., 2006; Rosser et al., 2008; Serohijos et al., 2008). This leads to accumulation of an ensemble of nonnative F508del-CFTR biogenic intermediates that are rapidly targeted for proteasomal degradation by the endoplasmic reticulum quality control machinery (Meacham et al., 2001; Younger et al., 2006; Grove et al., 2011). As a result, little to no F508del-CFTR is delivered to the cell surface, and CFTR chloride transport activity is abolished.
A potential therapeutic strategy to treat CF would be the use of small-molecule compounds that increase the amount of F508del-CFTR delivered to the cell surface. Such compounds are known as CFTR correctors. The investigational CFTR corrector, VX-809, increases the delivery of functional F508del-CFTR to the cell surface, resulting in partial restoration of chloride transport in cultured human bronchial epithelial cells derived from people with CF who are homozygous for F508del-CFTR. In vitro, the amount of chloride transport can be further enhanced by combining VX-809 with other CFTR correctors, such as Corr-4a (Pedemonte et al., 2005) or VRT-325 (Van Goor et al., 2006, 2011), or with CFTR potentiators, such as Ivacaftor (also known as VX-770; Van Goor et al., 2011). In clinical trials, VX-809 improved clinical measures of CFTR function and lung function when combined with the CFTR potentiator Ivacaftor (Van Goor et al., 2011; Clancy et al., 2012). The aim of this in vitro study was to determine the mechanism by which VX-809 modulates the folding pathway of CFTR to suppress misfolding of F508del-CFTR and other disease-related CFTR mutants.
RESULTS
VX-809 stabilizes N-terminal CFTR fragments that contain MSD1
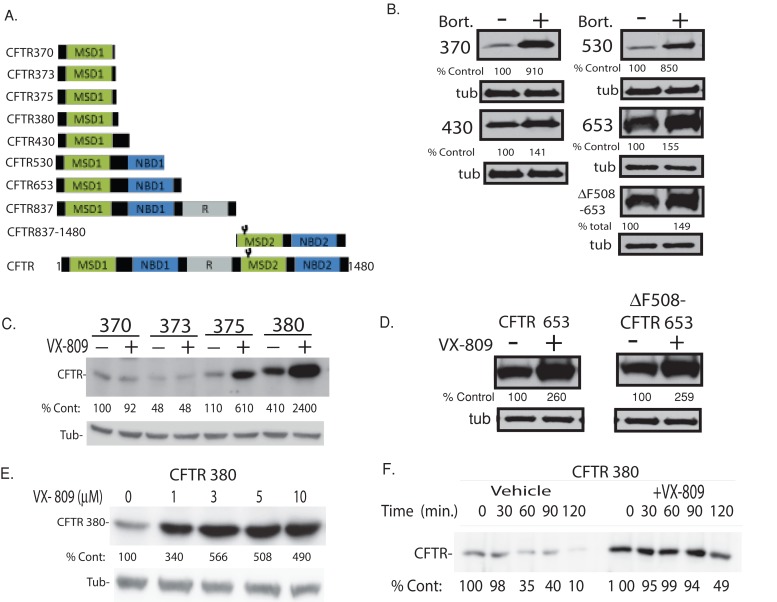
For identification of regions of CFTR required for VX-809 activity, CFTR fragments of different lengths were expressed in HEK-293 cells, and their steady-state accumulation and half-lives were quantified in biochemical assays (Figures 1, A and B, and Supplemental Figure S1). Because CFTR folding initiates cotranslationally and channel assembly is completed posttranslationally (Higgins, 1992), CFTR fragments, which may resemble folding intermediates, have been used to study CFTR biogenesis. The underlying rationale is that differences in accumulation or half-life between CFTR fragments of different lengths are believed to reflect differences in the stability of the protein conformation and resistance to endoplasmic reticulum–associated degradation (ERAD). Consistent with this, CFTR fragments 370 and 530, which accumulated at markedly lower levels compared with CFTR 430 and CFTR 653, were more susceptible to proteasome inhibition by bortezomib (Figure 1B). Differences in accumulation and half-life of CFTR fragments have been used successfully to identify Hsp70- and calnexin-dependent steps in CFTR folding and degradation pathways (Meacham et al., 1999; Okiyoneda et al., 2004; Farinha and Amaral, 2005; Lukacs and Verkman, 2012); identify defective interactions between MSD1, NBD1, and MSD2 as an underlying cause of premature degradation of F508del-CFTR (Younger et al., 2006; Cui et al., 2007; Du and Lukacs, 2009); and identify MSD2 as the site of action of the CFTR corrector Corr-4a. Coexpression of nonoverlapping N- and C-terminal CFTR fragments in cells led to an increase in chloride transport, suggesting these individual CFTR fragments were able to coassemble and form a functional CFTR channel (Csanady et al., 2000).
FIGURE 1:
Stabilization of N-terminal regions of CFTR by VX-809. (A) Domain boundaries of CFTR fragments used in this study. (B) The impact of bortezomib (Bort, 10 μM) on the steady-state levels of N-terminal CFTR fragments expressed in HEK293 cells. Indicated CFTR fragments were generated via introduction of a stop codon after the specified residue. Respective fragments were detected by Western blot with an antibody against the N-terminal tail of CFTR. (C and D) The impact of VX-809 (5 μM) on CFTR fragment levels. (E) Dose response of CFTR 380 to VX-809. (F) Pulse–chase analysis of the impact of VX-809 on the half-life of [35S]CFTR 380. Data are representative of three experiments. See Materials and Methods for details.
The shortest-length CFTR fragment affected by VX-809 was CFTR 375, which contains only MSD1 (Figure 1C). As expected, longer-length CFTR fragments were also increased by VX-809, including CFTR fragments that contained NBD1 with the F508del mutation (Figures 1D and S1). VX-809 did not increase the stability of CFTR 837–1480, which contains only MSD2 and NBD2 (Figure S1). In dose-response studies using CFTR 380, the maximal effective concentration of VX-809 was 3 μM, which is similar to that observed for full-length F508del-CFTR (Figure 1E; Van Goor et al., 2011). In pulse–chase studies, the half-lives of CFTR 375, CFTR 380, CFTR 430, and CFTR 653 were increased compared with untreated controls (Figures 1F and S1), indicating that the increase in steady-state accumulation caused by VX-809 is associated with an increase in the stability of the individual CFTR fragments. The inability of VX-809 to increase the accumulation of CFTR370 suggests that VX-809 does not act as a proteasome inhibitor, as bortezomib was able to increase the stability of this CFTR fragment (Figure 1B). Taken together, these data indicate that the shortest-length CFTR requirement for VX-809 action contains only MSD1. Moreover, these data suggest that VX-809 increases the stability of the protein conformation of MSD1 and, as a consequence, its resistance to ERAD.
VX-809 alters the protein conformation of MSD1
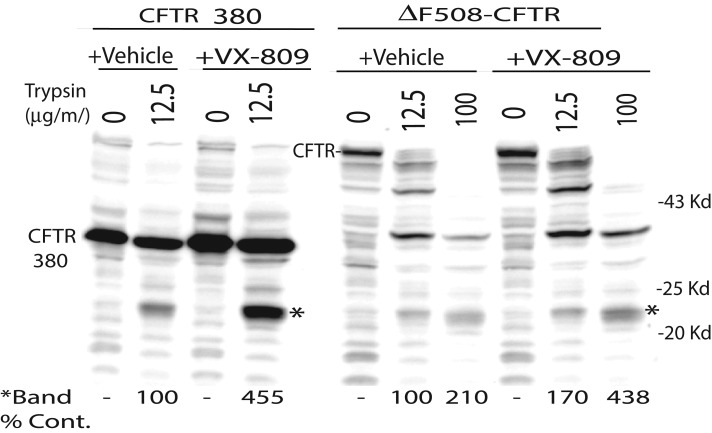
To test whether VX-809 alters the protein conformation of MSD1 to result in a more stable folded form that is resistant to ERAD, we tested the compound's ability to protect a subdomain of MSD1 from proteolytic digestion, using limited proteolysis of CFTR in cell extracts made with the nonionic detergent Triton X-100 (Figure 2). This technique, based on the premise that folded proteins are more compact and therefore typically more resistant to proteolytic digestion than unfolded or partially folded proteins, has been used to probe differences in protein folding between wild-type and F508del-CFTR, as well as between uncorrected and VX-809–corrected F508del-CFTR (Lukacs et al., 1994; Van Goor et al., 2011). For these studies, CFTR 380 was used, as it was the most stable CFTR fragment following treatment with VX-809. In cells expressing CFTR 380 or full-length F508del-CFTR, treatment with VX-809 increased the liberation of a protease-resistant species with an apparent molecular weight of 22 kDa that was detected with an antibody directed against the N-terminal tail of CFTR. Taken together, data presented in Figures 1 and 2 suggest that VX-809 alters the protein conformation of MSD1 to suppress folding defects in F508del-CFTR.
FIGURE 2:
Comparison trypsin-resistant fragments liberated from CFTR 380 and ΔF508-CFTR in the presence or absence of VX-809. CFTR 380 and ΔF508-CFTR were expressed in HEK293 cells in the absence or presence of VX-809 (5 μM). Cells were incubated 18 h in the presence of VX-809 and were lysed with PBS containing 1% Triton X-100. The indicated concentration of trypsin was added, and digestions were carried out on ice for 15 min. CFTR was detected via Western blot with an N-terminal–tail antibody.
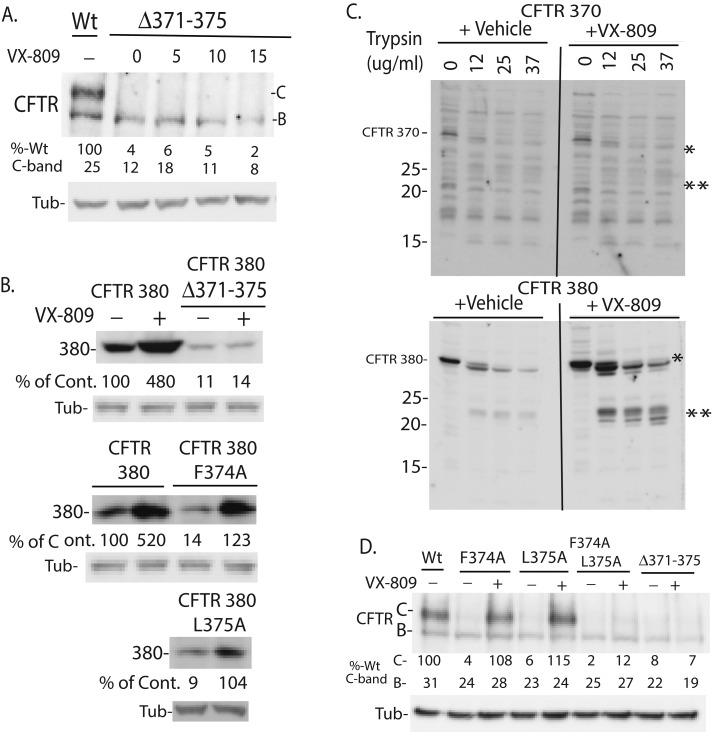
Residue 374 and 375 help MSD1 fold to a conformation that is stabilized by VX-809
The above data indicate that residues 374–375 are required for sensitivity of MSD1 fragments to VX-809, whereas residues 376–380 appear to aid in folding of MSD1 to a more stable form (Figure 1C). Deletion of residues 371–375 from full-length CFTR caused a severe folding defect, resulting in little to no mature form (C-band) and eliminating sensitivity to VX-809 (Figure 3A). Similarly, Δ371–375 CFTR 380 accumulated at 10% of CFTR 380 levels and was insensitive to VX-809 (Figure 3B). Thus residues 371–375 are important for proper folding of CFTR, and their deletion caused a biogenic defect that was not corrected by VX-809.
FIGURE 3:
CFTR amino acids 370–380 are required for folding of MSD1 to a conformation stabilized by VX-809. (A) Deletion of residues 371–375 causes a folding defect in CFTR that is insensitive to VX-809. (B) Mutation of residues 374 and 375 causes folding defects in CFTR 380 that are suppressed by VX-809. (C) CFTR 380, but not CFTR 370, folds to a trypsin-resistant conformation that is stabilized by VX-809. (D) F374A L375A CFTR exhibits a folding defect that is not efficaciously suppressed by VX-809. Levels of indicated forms of CFTR expressed in HEK293 cells were detected by Western blot with N-terminal–tail antibody. (C) Cells were lysed with PBS that contained 1% Triton X-100 and the indicated concentration of trypsin, and digestions were carried out on ice for 15 min. Unless otherwise noted, VX-809 was 5 μM.
To determine whether residues 370–380 are critical for folding of MSD1 to a conformation that is sensitive to VX-809, we compared the resistance of CFTR 370 and CFTR 380 to digestion by trypsin (Figure 3C). CFTR 370 was completely digested by trypsin and was not protected by VX-809, whereas CFTR 380 adopted a conformation that is partially resistant to trypsin digestion and was protected from digestion by VX-809. Thus residues 371–380 are important for folding of MSD1 to a biochemically stable conformation. Once translation of CFTR past residue 375 occurs, all the forms of CFTR examined are biochemically stabilized by VX-809 (Figures 1–3 and S1).
Given that the extension of CFTR 373 by two residues confers sensitivity of MSD1 fragments to VX-809, residues F374 and L375 might be involved in folding of CFTR to a conformation that is sensitive to VX-809. Despite the severe biogenic defects exhibited by F374A-CFTR, L375A-CFTR, the double mutation F374A/L375A-CFTR (Figure 3A), F374A-CFTR 380, and L375A-CFTR 380 (Figure 3B), VX-809 almost completely suppressed the biogenic defects caused by the F374A and L375A mutations at the 5-μM concentration that maximally suppressed folding defects in F508del-CFTR (Figure 3, A and B). Thus, while residues 374 and 375 are important for CFTR folding, mutation of F374 and L375 does not alter the potency or efficacy of VX-809, suggesting that neither of these residues was critical for binding of VX-809 to CFTR. Additional studies are now required to identify the VX-809 binding site in MSD1.
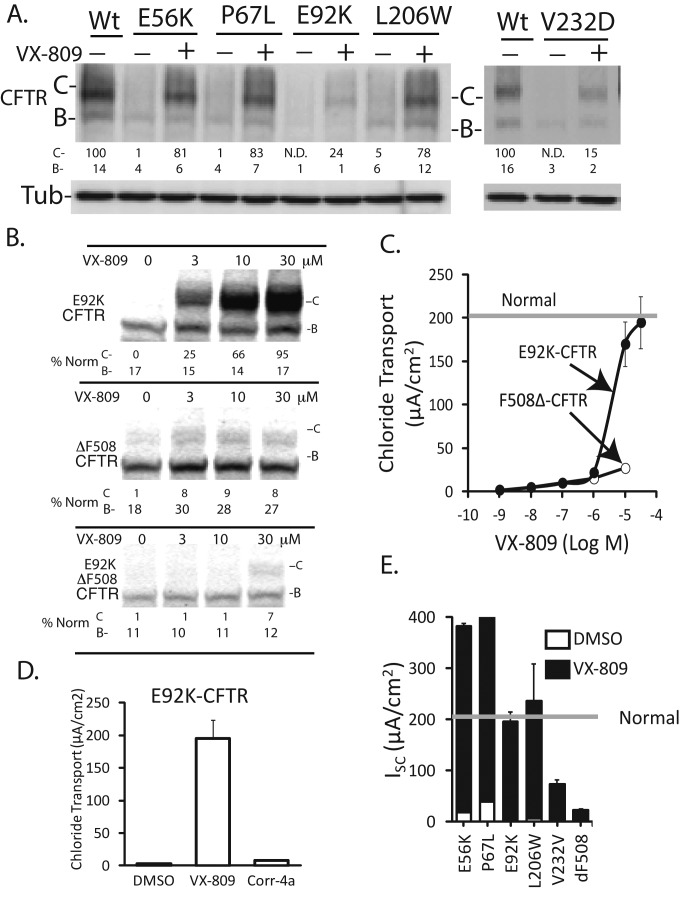
VX-809 suppresses folding defects in CFTR caused by disease-related mutations in MSD1
Data presented thus far suggest that MSD1 is the minimum-length fragment required for VX-809 action. There are several CF-associated mutations in MSD1 that cause defects in CFTR processing and function: N-terminal tail (E56K and P67L), TM1 (E92K), TM2 (L206W), and TM4 (V232D) (Figure 4, A–E). The severe folding (Figure 4, A–B) and functional (Figure 4E) defects exhibited by E56L, P67L and L206W were completely corrected by 5 μM VX-809. In contrast, 5 μM VX-809 only partially restored folding and function to E92K and V232D (Figure 4, A and E). Interestingly, VX-809 demonstrated reduced potency for E92K-CFTR relative to F508del-CFTR, for both correcting folding and function (Figure 4, B and C), yet was able to fully restore E92K-CFTR at 30 μM. However, Corr4a could not restore E92K-CFTR function (Figure 4D).
FIGURE 4:
Functional defects in CFTR caused by disease-related mutations in MSD1 are suppressed by VX-809. (A) Folding defects caused by mutation of CFTR's N-terminus are differentially suppressed by VX-809 (5 μM). (B) Dose-dependent correction of E92K-CFTR misfolding by VX-809. (C) Dose-dependent correction of E92K-CFTR function by VX-809. (D) VX-809 (30 μM) and Corr4a (15 μM) restore E92K-CFTR channel activity to different levels. (E) VX-809 is efficacious at restoration of Cl− channel function to forms of CFTR that have mutations in MSD1. Western blots of mutant CFTR levels in HEK293 are shown in (A) and (B). Results from electrophysiological measurements of forskolin-stimulated (10 μM) CFTR channel activity are shown in (A–C). CFTR was stably expressed in polarized FRT cells and cAMP-stimulated currents were measured in Ussing chambers. (E) VX-809 was 5 μM, except in the case of E92K-CFTR, for which it was 30 μM. (C–D): n = 3 ± SE.
V232D-CFTR was the least responsive to VX-809, and higher concentrations of the compound did not restore function beyond the 25% of normal CFTR observed in the presence of 5 μM VX-809. Taken together with the observation that Corr4a restored V232D-CFTR biogenesis and function to normal levels (Caldwell et al., 2011), these data suggest that correctors such as VX-809 and Corr4a can act to selectively suppress folding defects in CFTR caused by different disease-related mutations in MSD1.
Because E92K-CFTR was corrected to normal levels of function but F508del was corrected to ∼15% of normal function, the impact of VX-809 on the double mutation E92K/F508del-CFTR was tested. (Figure 4B). The effect of VX-809 on the C-band of E92K/F508del-CFTR was consistent with the dose response for E92K-CFTR, while the level of efficacy was consistent with that for F508del-CFTR. Thus the E92K mutation causes a folding defect in E92K/F508del-CFTR that requires a higher compound concentration, but the folding defects caused by F508del limit the efficacy of VX-809. These data support the concept that VX-809 action on MSD1 aids in suppression of some but not all of the folding defects caused by ΔF508.
VX-809 is highly efficacious at correction of folding defects in CFTR caused by some, but not all, of the missense mutations in MSD1 that were evaluated. E92K is unique among these mutants, as it alters the potency of VX-809, yet the biogenic defects caused by this mutation are completely corrected by VX-809. Mutational analysis of E92 suggested that mutation of this residue disrupts a salt bridge in MSD1 that is required for CFTR folding (Figure S3). E92 may therefore not be directly involved in binding VX-809 and instead appears to be required for folding of MSD1 to a conformation that binds VX-809 with high affinity.
Interdomain communication is required for VX-809 to enhance CFTR folding
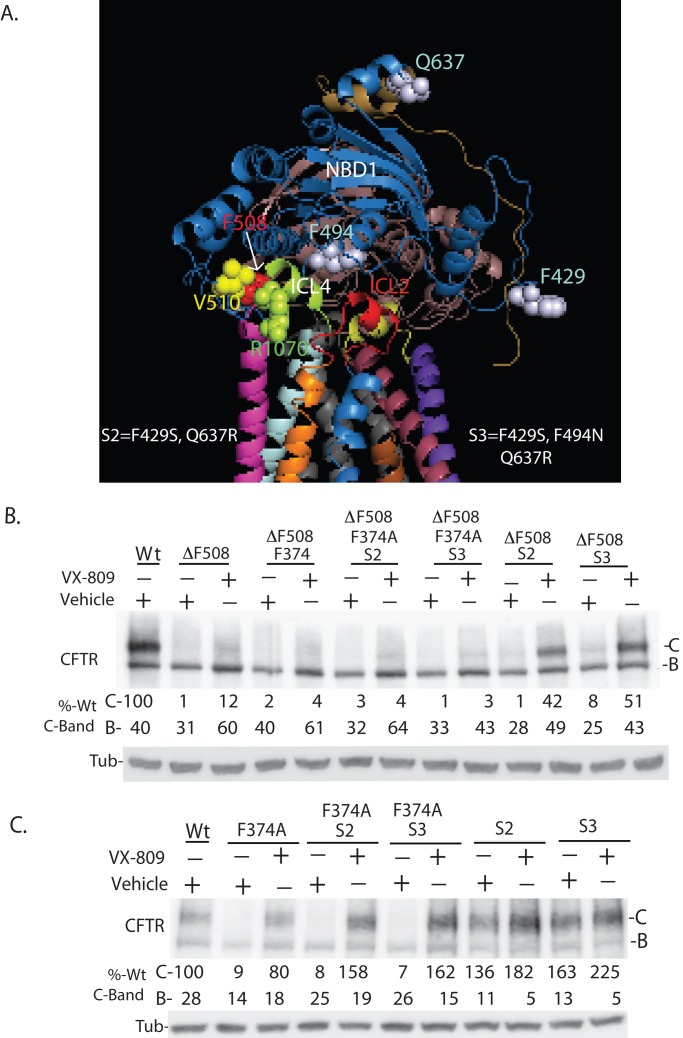
VX-809 was efficacious at correcting the folding defects caused by missense mutations in MSD1, and its ability to stabilize MSD1 appears to partially suppress the folding defects caused by F508del from NBD1. Yet VX-809 efficacy was limited by folding defects that appear to occur downstream of its effects on MSD1. Because VX-809 restored the function of some MSD1 CFTR mutants to normal levels in model cells, there is potential that it could act in concert with an additional corrector to restore F508del-CFTR function to normal levels (Van Goor et al., 2011; Mendoza et al., 2012; Rabeh et al., 2012). Understanding the nature of the defective folding step(s) that limit VX-809 efficacy on F508del-CFTR may aid in the development of such corrector combinations. The biogenic defects in F508del-CFTR that might limit VX-809 efficacy include 1) defective assembly caused by increased thermodynamic instability of F508del-NBD1 relative to NBD1 (Wang et al., 2010) or 2) defective assembly of F508del-NBD1 into a complex with ICL4 of MSD2 caused by loss of the F508 side chain (Serohijos et al., 2008).
To determine the extent to which thermodynamic instability of F508del-NBD1 impacts efficacy of VX-809 on F508del-CFTR, we examined the effect of VX-809 on a set of intragenic suppressor mutations that increase the solubility of purified NBD1 and thereby partially suppress biogenic defects in F508del-CFTR (Teem et al., 1993; Pissarra et al., 2008; Amaral and Farinha, 2013). These mutations are termed solubilizing (S) mutations and were introduced into NBD1 in different combinations (Figure 5A, S2 [F429S, Q637R] and S3 [F429S, F494N, and Q637R]). VX-809 increased accumulation of the C-band of F508del-CFTR to around 14% of normal CFTR, and the S mutations by themselves have little impact on accumulation of the C-band of F508del-CFTR (Figure 5B). Yet, in the presence of VX-809, the C-band of S2/F508del-CFTR and S3/F508del-CFTR accumulated at up to 45% of normal (wild type; Figure 5B). Interestingly, the positive effect of S2 and S3 on F508del-CFTR biogenesis was abolished by the F374A mutation, as VX-809 could not drive high-level accumulation of the folded C-band of F374A/S2/F508del-CFTR or F374A/S3/F508del-CFTR. In addition, the F374A mutation hindered VX-809 from suppressing folding defects in F508del-CFTR.
FIGURE 5:
Interdomain communication between MSD1 and NBD1 is required for VX-809 to enhance biosynthetic processing of CFTR. (A) Schematic of CFTR showing the position of indicated amino acid residues and domains. (B) Mutation of F374A hinders the ability of misfolding suppressor mutations S2 and S3 to increase the efficacy of VX-809 on F508del-CFTR. (C) Mutation of F374A hinders the ability of S2 and S3 to increase accumulation of the folded C-band of CFTR. Data in panels B and C are from Western blots of cell extracts. S2 (F429S, Q637R) and S3 (F429S, F494N, and Q637R) are mutations introduced into NBD1 to increase the thermodynamic stability of NBD1 and thereby increase CFTR and F508del-CFTR (B and C) folding efficiency (Pissarra et al., 2008; Teem et al., 1993). Panel are representations of three experiments. Quantitation is normalized to 100% of total C-band for CFTR detected for wild type. Bands B and C denote the ER-localized and plasma membrane–localized forms of CFTR.
In experiments with CFTR, the S2 and S3 mutations by themselves increased C-band accumulation almost twofold, and this effect was blocked by F374A (Figure 5C). Yet, in contrast to results with F508del-CFTR, VX-809 restored accumulation of the C-form of F374A/S2-CFTR and F374A/S3-CFTR to levels of S2 CFTR and S3 CFTR under control conditions.
Two important observations are to be drawn from these data. First, mutations in NDB1 that increase the thermodynamic stability of purified NBD1 (i.e., S2 or S3) also increase the efficacy of VX-809 on F508del-CFTR. Therefore the thermodynamic instability of F508del-NBD1 limits the efficacy of VX-809 on F508del-CFTR. Second, F374 is located in the cytosolic linker domain, positioned between TM6 and NBD1, required for folding of MSD1 to a conformation that can be modulated by VX-809 (Figures 2 and 3). The folding defect caused by F374A is efficaciously corrected by VX-809 (Figure 3), but the F374A mutation blocks VX-809 action on F508del-CFTR. In addition, F374A also blocks the positive effect of S mutations on CFTR and F508del-CFTR biogenesis. Thus F374 appears to facilitate interdomain communication between MSD1 and NBD1, and, further, allosteric communication of structural information between MSD1 and NBD1 appears critical for VX-809 correction of F508del-CFTR.
Stabilization of the NBD1:ICL4 interface increases VX-809 efficacy on F508del-CFTR
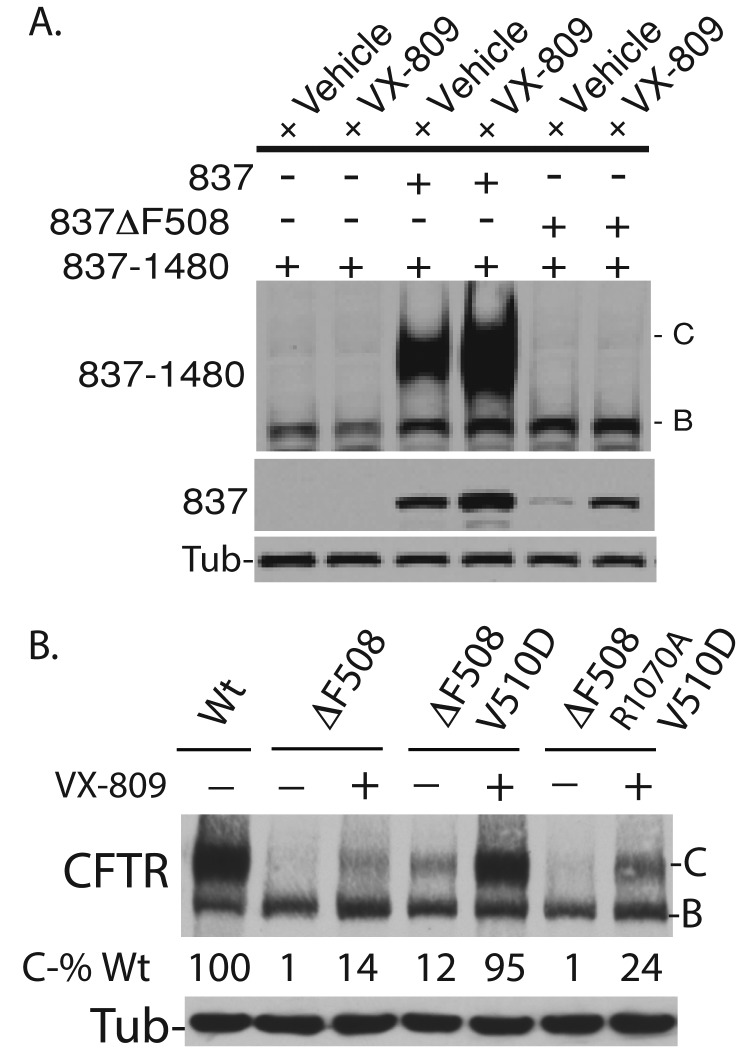
Defective assembly of NBD1 into a complex with solvent-exposed residues on ICL4 hinders F508del-CFTR folding (Serohijos et al., 2008). The extent to which this folding defect limits VX-809 efficacy (Figure 6) was tested by examining the ability of VX-809 to enhance the assembly of nonoverlapping N- and C-terminal CFTR fragments into a complex that can escape the ER (Figure 6A). Fragments of CFTR that contain the N- (1–837) and C- (837–1480) terminus assemble into a complex that folds, escapes the ER, and accumulates as a maturely glycosylated species (Rosser et al., 2008). Thus, if VX-809 acts on MSD1 to positively impact TM segment assembly, VX-809 would be expected to increase CFTR fragment assembly. Consistent with previous results (Figures 1, 2, and S1), VX-809 does not increase accumulation of CFTR 837–1480 alone (Figure 6A, lanes 1 and 2), but it did increase accumulation of CFTR 837. In addition, VX-809 stimulated the CFTR 837–dependent accumulation of C-band by greater than twofold (Figure 6A, lane 3 vs. lane 4).
FIGURE 6:
Restoration of contact between F508del-NBD1 and ICL4 increases the efficacy of VX-809 in repair of F508del-CFTR misfolding. (A) VX-809 enhances assembly of CFTR 837 into a complex with CFTR 837–1480, but cannot correct assembly defects caused by ΔF508 from CFTR 837. Bands B and C represent the ER-localized and plasma membrane–localized forms of CFTR 837–1480, respectively. (B) Introduction of the V510D suppressor mutation into NBD1 permits VX-809 to drive high-level folding of F508del-CFTR. B and C denote the ER-localized and plasma membrane forms of F508del-CFTR, respectively. (A and B) Western blots decorated with N-terminal–tail CFTR antibody. VX-809 was present at 5 μM.
Deletion of F508 prevents accumulation of F508del-CFTR 837 (Figure 6A, lane 3 vs. lane 5), and F508del-CFTR 837 does not productively interact with CFTR 837–1480 (i.e., the C-band does not accumulate in the presence of F508del-CFTR 837). VX-809 increased the accumulation of F508del-CFTR 837 by several fold (Figure 6A, lane 5 vs. lane 6) but did not promote interactions between F508del-CFTR 837 and CFTR 837–1480, as indicated by a lack of C-band (Figure 6A, lane 6).
Defective contacts between F508del-NBD1 and ICL4 that limit F508del-CFTR assembly have been partially restored by introduction of the V510D mutation into NBD1 by permitting the formation of a salt bridge between D510 and R1070 of ICL4 (Wang et al., 2007; Figure 6B). The V510D mutation partially corrects misfolding of F508del-CFTR, as shown by the C-band for V510D/F508del-CFTR (Figure 6B, lane 4), to levels similar to that for F508del-CFTR in the presence of VX-809. Remarkably, VX-809 stimulated the accumulation C-band of V510D/F508del-CFTR to the levels observed for normal CFTR.
To determine whether formation of a salt bridge between D510 and R1070 was important for this effect, we introduced the R1070A mutation into V510D/F508del-CFTR. In the presence of VX-809, the accumulation of the C-band of R1070A/V510D/F508del-CFTR was reduced by 75% relative to V510D/F508 -CFTR (Figure 6B, lane 7). Yet VX-809 was still able to increase folded R1070A/V510D/F508- CFTR to levels that were significantly higher than those for VX-809–treated F508del-CFTR. Because the V510D mutation can modestly improve the thermodynamic stability of purified NBD1 (Lewis et al., 2010; Wang et al., 2010), the residual VX-809 corrector function on R1070A/V501D/F508del-CFTR could result from thermodynamic stabilization of NBD1 that would occur in the absence of salt-bridge formation between D510 and R1070.
Collectively these data are consistent with the concept that VX-809 action on MSD1 partially restores F508del-CFTR function through modulation of the protein conformation of MSD1. This action of VX-809 appears to have global effects on the conformation of CFTR. However, to achieve higher levels of correction of F508del-CFTR biogenesis by VX-809 requires either further stabilization of NBD1 and/or restoration of contacts between F508del-NBD1 and MSD2.
DISCUSSION
Data presented reveal that VX-809 acts early in CFTR biogenesis to modulate the conformation of MSD1 and thereby suppress folding defects in ΔF508-CFTR and disease-related MSD1 mutants. Corrective conformational changes in MSD1 that are driven by VX-809 appear to be communicated to other domains in CFTR via a mechanism involving F374 in the linker domain that is located between TM6 and NBD1. VX-809 positively impacts the conformation of MSD1 to increase the efficiency of interactions between MSD1 with NBD1, MSD2, and NBD2, thereby restoring function to a portion of nascent F508del-CFTR.
VX-809 restores F508del-CFTR function to 15% of normal in lung cells from CF patients (Van Goor et al., 2011), and the data presented here suggest that the efficacy of the drug is impacted by the thermodynamic instability of ΔF508-NBD1 and defective interactions between ΔF508-NBD1 and MSD2. This conclusion is supported by observations that the use of VX-809 in combination with approaches that either enhance the stability of NBD1 or stabilize the NBD1:ICL4 interface permit folded F508del-CFTR to accumulate at levels that are 50–100% of normal CFTR. Interestingly, in the absence of VX-809, the thermodynamic stabilization of NBD1 or restoration of the NBD1:ICL4 interface has little impact on F508del-CFTR folding. Therefore VX-809 action on MSD1 primes F508del-CFTR for additional corrective events that permit high-level accumulation of its properly folded form. Identification of a small molecule that acts downstream of VX-809 to effectively stabilize F508del-NBD1 and/or restore contacts between ΔF508-NBD1 and MSD2 may lead to development of a corrector combination therapy to treat CF.
Evidence to support the conclusion that VX-809 partially corrects F508del-CFTR folding by altering the conformation of MSD1 is as follows: 1) VX-809 biochemically stabilizes CFTR fragments that contain only MSD1. 2) VX-809 alters the conformation of MSD1 to protect it from proteolytic digestion with trypsin. 3) CFTR fragments fold to a conformation that is stabilized by VX-809 after biosynthesis of amino acid F374. F374 is located in the linker between MSD1 and NBD1 and helps stabilize MSD1 in conformation that is sensitive to VX-809. 4) VX-809 cannot overcome folding defects caused by the combined mutation of F374A and L375A but efficaciously suppresses folding defects caused by each individual mutation. VX-809 can modulate the conformation of MSD1 in the vicinity of the interface between MSD1 and NBD1.
VX-809 is highly efficacious at correcting disease-causing mutations in CFTR MSD1, but also significantly restores folding of disease-related mutations in CFTR ICL4 (He et al., 2013). Therefore a possible explanation of these data is that VX-809 also acts directly on the NBD1/ICL4 interface (He et al., 2013). However, several observations from the data presented herein support the interpretation that VX-809 acts on MSD1 to allosterically suppress the assembly defects in CFTR caused by ICL4 mutations through interdomain communication: 1) VX-809 has no detectable effect on thermally induced unfolding of purified NBD1, so it does not appear to act directly on NBD1 (see Supplemental Figure S2); 2) VX-809 has no impact on the accumulation of a C-terminal CFTR fragment that contains MSD1 and NBD2, so it does not appear to directly modulate the conformation of MSD2; and 3) VX-809 enhances the assembly of CFTR 1–837 into a complex with CFTR 837–1480 and also increases the accumulation of F508del-CFTR 1–837 to the level of the normal CFTR control. Yet VX-809 was not able to suppress defects in F508del-CFTR 1–837 assembly into a complex with CFTR 837–1480.
Data presented are consistent with the concept that correctors such as VX-809 modulate the conformation of a specific region of CFTR to enhance global protein folding and assembly (Lukacs and Verkman, 2012; Moniz et al., 2013). Because CFTR folding involves cooperative domain assembly (Du and Lukacs, 2009), the action of VX-809 on MSD1 has potential to promote high-level functional correction of CFTR in people with CF who harbor mutations other than F508del (Bobadilla et al., 2002). Indeed, the data presented here demonstrate that VX-809 restores significant levels of function to a number of rare CFTR mutations. The development of personalized corrector therapies to treat people with CF will likely require the identification of individual CFTR genotypes whose functional defects can be suppressed by VX-809 monotherapy or combination therapy with additional correctors.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents
CFTR expression plasmids pcDNA3.1(+)-CFTR and pcDNA3.1(+)ΔF508-CFTR have been described elsewhere (Meacham et al., 2001; Younger et al., 2006). CFTR constructs representing CF disease–causing point mutants or truncated biogenic intermediates were made using the QuikChange protocol (Stratagene, Santa Clara, CA). The CFTR antibody used in this study was MM13-4 (N-terminal tail epitope) from Millipore, Darmstadt, Germany. Use of VX-809 in experiments with cultured cells was previously described (Van Goor et al., 2011).
Cell culture and transfection
HEK293 cells from the American Type Culture Collection (Manassas, VA) were maintained in DMEM (Gibco, Grand Island, NY) supplemented with 1% fetal bovine serum (Thermoscientific, Waltham, MA) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin; Gibco) at 37°C in an atmosphere of 5% CO2. Cell transfections were performed using Effectene reagent (Qiagen, Valencia, CA). The empty pcDNA3.1(+) vector was used to ensure equal microgram quantities of DNA were used in all transfection reactions.
Analysis of CFTR biogenesis
CFTR steady-state levels.
Steady-state levels of CFTR and its mutants were determined by Western blot analysis. HEK293 cells were transiently transfected with the indicated plasmids (Grove et al., 2011). The transfected cells were allowed to recover for ∼18 h before addition of DMEM and were then supplemented with VX-809 or dimethyl sulfoxide (DMSO; control). The cells were incubated with the correctors for 24 h before being isolated for Western blot analysis. The harvested cells were diluted with 2× SDS sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 0.05% bromophenol blue, 20% glycerol), sonicated, and heated at 37°C prior to the proteins being resolved on SDS–PAGE gels. The proteins were transferred to nitrocellulose membranes, and the membranes were probed with the designated antibodies. Tubulin levels were monitored as a loading control.
CFTR processing efficiency.
CFTR processing efficiency was measured by pulse–chase analysis (Grove et al., 2011). Transiently transfected HEK293 cells were allowed to recover for 18 h. The cells were then incubated with DMEM supplemented with VX-809 or DMSO for 2 h. Next, cells were starved in methionine-free MEM (Sigma-Aldrich, St. Louis, MO) for 30 min, pulse labeled for 30 min with [35S]methionine (100 μCi/six wells; 1200 Ci/mmol; ICN Biomedicals, Costa Mesa, CA), and then chased for the indicated amount of time. VX-809 was also included in the media during these steps of the pulse–chase reaction. Cells were then lysed in phosphate-buffered saline (PBS) buffer supplemented with 1% Triton (PBS-T (1%), 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor cocktail (Roche, Indianapolis, IN). Soluble lysates were obtained by centrifugation at 20,000 × g for 10 min. Lysates were normalized to contain the same total amount of protein. 35S-labeled CFTR was immunoprecipitated by incubation with a polyclonal α-CFTR antibody directed against the N-terminus; this was followed by addition of a 50% protein G bead slurry. The beads were washed with PBS-T (1%) supplemented with 0.2% SDS, the bound CFTR was eluted with 2× sample buffer, and the samples were heated at 55°C for 10 min. The samples were analyzed by SDS–PAGE and visualized by autoradiography.
Limited Proteolysis of CFTR.
The content of six wells of a six-well plate containing HEK293 cells were transfected with 1 μg of the indicated CFTR plasmid (Rosser et al., 2008). At 24 h posttransfection, the cells were harvested in citric saline and lysed in PBS-Tr (0.1%) for 1 h at 4°C. Lysates were cleared by centrifugation at 20,000 rpm for 10 min in a Beckman Allegra 64R centrifuge. Supernatants were removed, and total microgram quantities of protein were determined by the DC Bio-Rad protein determination assay (Hercules, CA). Cell lysates were then diluted to a concentration of 2 μg/ml, and trypsin was added at the indicated final concentrations. The cleavage reactions were incubated on ice for 15 min and were then quenched by addition of complete protease inhibitor (Roche) and trypsin inhibitor. Sample buffer was added to a final 1× concentration, and samples were run on 12.5% SDS–PAGE gels. Gels were transferred to nitrocellulose and probed with N-terminal–tail CFTR antibody (MM13-4 1:1000 dilution).
Coexpression of N-and C-terminal domains of CFTR.
Cells were transfected with pcDNA3.Then, 1-CFTR837X (1 μg) and pcDNA3.1-CFTR837–1480 (1 μg), individually or in combination, were used to evaluate the impact of VX-809 on the assembly of CFTR's membrane domains (Rosser et al., 2008). Reactions were balanced with pcDNA3.1 such that all transfections were performed with equal microgram quantities of DNA. At 24 h posttransfection, cells were harvested with citric saline, diluted in 2× sample buffer, sonicated for 10 s, and warmed to 37°C for 10 min prior to loading on 10% SDS–PAGE gels. Proteins were transferred to nitrocellulose using a Bio-Rad mini gel wet transfer apparatus. Blots were blocked in blocking buffer containing 10% fat-free milk and 0.1% Triton-X 100 in PBS and probed with α-CFTR monoclonal N-terminal tail (MM13-4 1:1000 dilution).
Electrophysiology
Ussing chamber techniques with Fisher rat thyroid (FRT) cells that were stably transfected with the indicated form of CFTR were used to record the transepithelial current resulting from CFTR-mediated chloride transport (Van Goor et al., 2006, 2011). Standard conditions were utilized for Ussing chamber measurements of forskilin (10 µM)-stimulated CFTR activity (Van Goor et al., 2006); for normal CFTR activity peaked at 205.5 µA/cm2.
Supplementary Material
Acknowledgments
D.M.C. is supported by National Institutes of Health (NIH) grants R01GM56981 and R01GM067785 and the North American Cystic Fibrosis Foundation (NACFF). D.E.G. was supported by a postdoctoral fellowship from the NACFF. S.A.H. is supported by a predoctoral fellowship from the NIH.
Abbreviations used:
- CF
cystic fibrosis
- CFTR
CF transmembrane conductance regulator
- DMSO
dimethyl sulfoxide
- ERAD
endoplasmic reticulum–associated degradation
- FRT
Fisher rat thyroid
- ICL
intracellular loop
- MSD
membrane-spanning domain
- NBD
nucleotide-binding domain
- PBS
phosphate-buffered saline
- S mutation
solubilizing mutation
- TM
transmembrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0240) on August 7, 2013.
*These authors contributed equally to this work.
†Present address: Parion Sciences, Durham, NC 27713.
REFERENCES
- Amaral MD, Farinha CM. Rescuing mutant CFTR: a multi-task approach to a better outcome in treating cystic fibrosis. Curr Pharm Des. 2013;19:3497–3508. doi: 10.2174/13816128113199990318. [DOI] [PubMed] [Google Scholar]
- Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- Bobadilla JL, Macek M, Jr, Fine JP, Farrell PM. Cystic fibrosis: a worldwide analysis of CFTR mutations—correlation with incidence data and application to screening. Hum Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- Boucher RC. Cystic fibrosis: a disease of vulnerability to airway surface dehydration. Trends Mol Med. 2007;13:231–240. doi: 10.1016/j.molmed.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Caldwell RA, Grove DE, Houck SA, Cyr DM. Increased folding and channel activity of a rare cystic fibrosis mutant with CFTR modulators. Am J Physiol Lung Cell Mol Physiol. 2011;301:L346–352. doi: 10.1152/ajplung.00044.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani C, et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros. 2008;7:179–196. doi: 10.1016/j.jcf.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy JP, et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012;67:12–18. doi: 10.1136/thoraxjnl-2011-200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanady L, Chan KW, Seto-Young D, Kopsco DC, Nairn AC, Gadsby DC. Severed channels probe regulation of gating of cystic fibrosis transmembrane conductance regulator by its cytoplasmic domains. J Gen Physiol. 2000;116:477–500. doi: 10.1085/jgp.116.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- Cyr DM. Arrest of CFTRΔF508 folding. Nat Struct Mol Biol. 2005;12:2–3. doi: 10.1038/nsmb0105-2. [DOI] [PubMed] [Google Scholar]
- Du K, Lukacs GL. Cooperative assembly and misfolding of CFTR domains in vivo. Mol Biol Cell. 2009;20:1903–1915. doi: 10.1091/mbc.E08-09-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha CM, Amaral MD. Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol Cell Biol. 2005;25:5242–5252. doi: 10.1128/MCB.25.12.5242-5252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove DE, Fan CY, Ren HY, Cyr DM. The endoplasmic reticulum–associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3–dependent degradation of nascent CFTRΔF508. Mol Biol Cell. 2011;22:301–314. doi: 10.1091/mbc.E10-09-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Kota P, Aleksandrov AA, Cui L, Jensen T, Dokholyan NV, Riordan JR. Correctors of ΔF508 CFTR restore global conformational maturation without thermally stabilizing the mutant protein. FASEB J. 2013;27:536–545. doi: 10.1096/fj.12-216119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Ikuma M, Welsh MJ. Regulation of CFTR Cl− channel gating by ATP binding and hydrolysis. Proc Natl Acad Sci USA. 2000;97:8675–8680. doi: 10.1073/pnas.140220597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Skach WR. Mechanisms of CFTR folding at the endoplasmic reticulum. Front Pharmacol. 2012;3:201. doi: 10.3389/fphar.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, et al. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2010;396:406–430. doi: 10.1016/j.jmb.2009.11.051. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs GL, Verkman AS. CFTR: folding, misfolding and correcting the ΔF508 conformational defect. Trends Mol Med. 2012;18:81–91. doi: 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, Feranchak AP, Brautigam CA, Thomas PJ. Requirements for efficient correction of ΔF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148:164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz S, Sousa M, Moraes BJ, Mendes AI, Palma M, Barreto C, Fragata JI, Amaral MD, Matos P. HGF stimulation of Rac1 signaling enhances pharmacological correction of the most prevalent cystic fibrosis mutant F508del-CFTR. ACS Chem Biol. 2013;8:432–442. doi: 10.1021/cb300484r. [DOI] [PubMed] [Google Scholar]
- Mornon JP, Lehn P, Callebaut I. Atomic model of human cystic fibrosis transmembrane conductance regulator: membrane-spanning domains and coupling interfaces. Cell Mol Life Sci. 2008;65:2594–2612. doi: 10.1007/s00018-008-8249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Harada K, Takeya M, Yamahira K, Wada I, Shuto T, Suico MA, Hashimoto Y, Kai H. ΔF508 CFTR pool in the endoplasmic reticulum is increased by calnexin overexpression. Mol Biol Cell. 2004;15:563–574. doi: 10.1091/mbc.E03-06-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedemonte N, Lukacs GL, Du K, Caci E, Zegarra-Moran O, Galietta LJ, Verkman AS. Small-molecule correctors of defective ΔF508-CFTR cellular processing identified by high-throughput screening. J Clin Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pissarra LS, Farinha CM, Xu Z, Schmidt A, Thibodeau PH, Cai Z, Thomas PJ, Sheppard DN, Amaral MD. Solubilizing mutations used to crystallize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem Biol. 2008;15:62–69. doi: 10.1016/j.chembiol.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Quinton PM. Chloride impermeability in cystic fibrosis. Nature. 1983;301:421–422. doi: 10.1038/301421a0. [DOI] [PubMed] [Google Scholar]
- Quinton PM. Cystic fibrosis: impaired bicarbonate secretion and mucoviscidosis. Lancet. 2008;372:415–417. doi: 10.1016/S0140-6736(08)61162-9. [DOI] [PubMed] [Google Scholar]
- Rabeh WM, et al. Correction of both NBD1 energetics and domain interface is required to restore ΔF508 CFTR folding and function. Cell. 2012;148:150–163. doi: 10.1016/j.cell.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JR. CFTR function and prospects for therapy. Annu Rev Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989a;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA [correction published in Science (1989). 245,1437] Science. 1989b;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rosser MF, Grove DE, Chen L, Cyr DM. Assembly and misassembly of cystic fibrosis transmembrane conductance regulator: folding defects caused by deletion of F508 occur before and after the calnexin-dependent association of membrane spanning domain (MSD) 1 and MSD2. Mol Biol Cell. 2008;19:4570–4579. doi: 10.1091/mbc.E08-04-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem JL, Berger HA, Ostedgaard LS, Rich DP, Tsui LC, Welsh MJ. Identification of revertants for the cystic fibrosis ΔF508 mutation using STE6-CFTR chimeras in yeast. Cell. 1993;73:335–346. doi: 10.1016/0092-8674(93)90233-g. [DOI] [PubMed] [Google Scholar]
- Van Goor F, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F, et al. Rescue of ΔF508 CFTR trafficking and gating in human cystic fibrosis airway primary cultures by small molecules. Am J Physiol Lung Cell Mol Physiol. 2006;290:L1117–L1130. doi: 10.1152/ajplung.00169.2005. [DOI] [PubMed] [Google Scholar]
- Wang C, et al. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 2010;19:1932–1947. doi: 10.1002/pro.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Loo TW, Bartlett MC, Clarke DM. Correctors promote maturation of cystic fibrosis transmembrane conductance regulator (CFTR)-processing mutants by binding to the protein. J Biol Chem. 2007;282:33247–33251. doi: 10.1074/jbc.C700175200. [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.