The CARMIL1–capping protein (CP) interaction is essential for lamellipodial function and the processes of ruffling and macropinocytosis. CARMIL1’s capping-binding region can inhibit CP in cells. Evidence suggests that CARMIL1’s ability to bind CP in cells is regulated.
Abstract
The regulation of free barbed ends is central to the control of dynamic actin assembly and actin-based motility in cells. Capping protein (CP) is known to regulate barbed ends and control actin assembly in cells. The CARMIL family of proteins can bind and inhibit CP in vitro, but the physiological significance of the interaction of CARMIL with CP in cells is poorly understood. Mammalian cells lacking CARMIL1 have defects in lamellipodia, macropinocytosis, cell migration, and Rac1 activation. Here we investigate the physiological significance of the CARMIL1–CP interaction, using a point mutant with a well-defined biochemical defect. We find that the CARMIL1–CP interaction is essential for the assembly of lamellipodia, the formation of ruffles, and the process of macropinocytosis. In contrast, the interaction of CARMIL1 with CP shows little to no importance for other functions of CARMIL1, including localization of CARMIL1 to the membrane, activation of Rac1, and cell migration. One implication is that lamellipodia are only marginally important for cell migration in a wound-healing model. The results also suggest that the ability of CARMIL1 to inhibit CP in cells may be regulated.
INTRODUCTION
Actin assembly is important for multiple cellular processes, including cytokinesis and cell migration (Pollard and Cooper, 2009). Actin polymerization in cells occurs primarily at free barbed ends of actin filaments, which makes the creation and regulation of barbed ends a critical determinant of actin assembly (Cooper and Sept, 2008). Barbed ends are also important in cells because they mediate the attachment of actin filaments to structures such as sarcomeric Z-lines and plasma membranes. Therefore the creation and regulation of free barbed ends in cells is critically important.
Cells have specific mechanisms to regulate the creation of free barbed ends. Barbed ends can be created by the nucleating action of Arp2/3 complex, formins, and spire proteins (Chesarone and Goode, 2009). In addition, new barbed ends can be created as a result of severing preexisting filaments by proteins such as cofilin (Bernstein and Bamburg, 2010). Finally, barbed ends can be generated by uncapping preexisting capped filaments (Cooper and Sept, 2008).
Capping protein (CP) is a highly conserved heterodimeric protein that binds to and functionally caps the barbed end of actin filaments (Cooper and Sept, 2008). Capping protein is a critical component of the dendritic nucleation model, which describes the generation of branched actin filament networks by Arp2/3 complex (Pollard, 2007). Decreasing the cellular concentration of CP in vertebrate cells inhibits lamellipodia formation and dramatically increases the size and number of filopodia on the cell surface (Mejillano et al., 2004). Understanding how CP is regulated in cells is critical to understanding how cells regulate barbed ends and therefore actin assembly.
The CARMIL family proteins are potential regulators of CP in cells. CARMILs are highly conserved, large, multidomain proteins discovered in amoeba such as Acanthamoeba Acan125 (Xu et al., 1995) and Dictyostelium p116/CARMIL (Jung et al., 2001). CARMIL homologues are present in all metazoans. Most mammalian genomes have three CARMIL genes, and those genes encode three isoforms whose sequences are conserved across species (Liang et al., 2009). CARMILs can interact with CP in cells, and the CARMIL1 isoform colocalizes with CP at the leading edge of migrating cells (Yang et al., 2005; Liang et al., 2009). The capping protein–binding region (CBR) of CARMIL proteins is a potent inhibitor of CP in vitro. Of greatest note, the CBR fragment is able to rapidly uncap CP-capped actin filaments in vitro by binding and inducing an allosteric change in the conformation of the actin-binding surface of CP (Kim et al., 2012).
The potent anti-CP activity of the CARMIL CBR in vitro suggests that CARMILs may be key regulators of CP function in cells. The physiological significance and the role of the CARMIL-CP interaction have been studied to a limited extent (Yang et al., 2005; Liang et al., 2009). Open questions include whether CARMIL1 binds to CP in living cells and, if so, whether CARMILs function to inhibit CP or to target active CP to certain locations in cells (Yang et al., 2005; Hernandez-Valladares et al., 2010; Zhao et al., 2013). In addition, there is conflicting evidence as to the activity of full-length CARMIL compared with that of the CBR fragment in vitro (Yang et al., 2005; Uruno et al., 2006).
In this study, we address the physiological significance of the interaction of CARMIL1 with CP in human cultured cells, using expression of a point mutant with defined biochemical defects. We find that certain cellular functions of CARMIL1 depend heavily on the interaction, whereas others, including localization of CARMIL1, do not. The results are relevant to the important issue of the physiological role of lamellipodia in cell migration.
RESULTS
CARMIL1 point mutant deficient in binding of capping protein
The physiological significance of the CARMIL1–capping protein interaction in cells has been examined, but only to a limited extent. A previous study tested a 122–amino acid residue internal deletion of CARMIL1 (Yang et al., 2005). Here we generated a point mutant form of CARMIL1 with two amino acid changes chosen based on sequence conservation and a cocrystal structure.
CARMIL1 contains a CP-binding motif, LxHxTxxRPK(6X)P (Bruck et al., 2006). A cocrystal structure of CARMIL1 with CP revealed that conserved residues in this motif, called CPI for capping-protein interaction, make up the primary binding site between CARMIL1 and CP (Hernandez-Valladares et al., 2010). CARMIL1 and CP have a second site of interaction, revealed in the cocrystal structure, which involves a second conserved motif, called CSI for CARMIL-specific interaction.
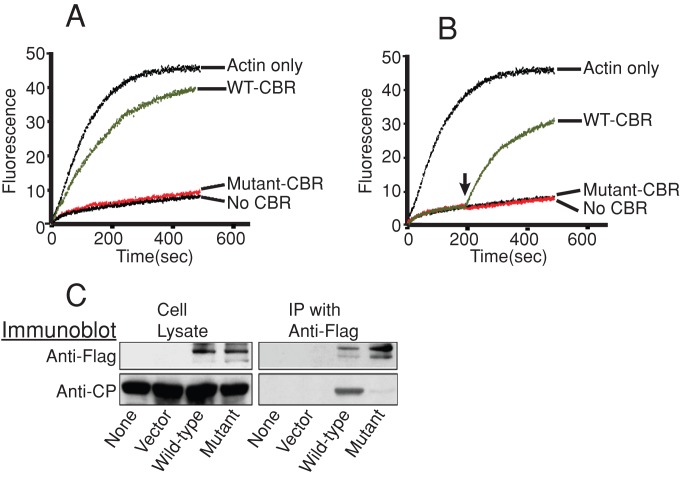
We tested the activity of mutations affecting CARMIL1 residues in the CPI motif. We changed two amino acid residues, K987 and R989, to alanine. These residues are highly conserved, and they make close contacts in the cocrystal structure. The KR987/989AA mutant had very little to no activity in two biochemical assays for the CARMIL1–CP interaction. In pyrene–actin polymerization assays, the CARMIL1 mutant, in the context of the CBR fragment of CARMIL1, had little to no ability to prevent CP from capping the barbed ends of growing actin filaments (Figure 1A). In addition, the mutant form of the CBR fragment had little to no ability to reverse the capping activity of CP (Figure 1B).
FIGURE 1:
Activity of the CARMIL1 CP-binding mutant KR987/989AA. (A) Inhibition of capping activity. Pyrene–actin subunits polymerized over time, adding to barbed ends of filaments nucleated by spectrin–actin seeds. The CBR fragment of CARMIL1 CBR was added to CP at time zero. Solutions contained 10 nM CP and either 10 nM wild-type CBR (green) or 4500 nM mutant CBR (red). A representative experiment is shown; n = 3. (B) Reversal of capping. CP was added at time zero, and CBR was added at 200 s. Concentrations of CBR and CP were the same as in A. A representative experiment is shown; n = 3. (C) Lack of association of the CARMIL1 mutant with CP in cell lysates. Full-length FLAG-CARMIL1 expressed in cells was immunoprecipitated from whole-cell lysates, and the precipitates were probed with antibodies to CP and FLAG.
In addition, we tested the ability of the CARMIL1 KR987/989AA mutant to bind CP in cells, by immunoprecipitation from whole-cell lysates. Here we tested full-length CARMIL1, not the CBR fragment. The amount of endogenous CP that precipitated with the mutant form of epitope-tagged full-length CARMIL1 was severely decreased compared with wild-type (wt) CARMIL1 (Figure 1C).
We used this CARMIL1 mutant, KR987/989AA, to test the physiological significance and the role of the CARMIL1-CP interaction in cells. We expressed the mutant form of CARMIL1 in cells as a full-length protein or the CBR fragment.
Localization of the CARMIL1 mutant
First, we asked whether the ability to bind CP is required for the localization of CARMIL1, which is found at cell edges in association with dynamic actin and Arp2/3 complex (Liang et al., 2009). CARMIL1 is most concentrated at free cell edges, where cells are not in contact with other cells. These cell edges contain dynamic lamellipodia with high ruffling activity. In cells that are migrating, their leading edges are free and show this activity.
We generated yellow fluorescent protein (YFP)–tagged constructs of full-length versions of the CARMIL1 CP-binding mutant and wt CARMIL1. These constructs were expressed at low levels in HT1080 cells. The expression of endogenous CARMIL1 was not targeted for inhibition in these experiments. An empty vector expressing YFP served as a negative control.
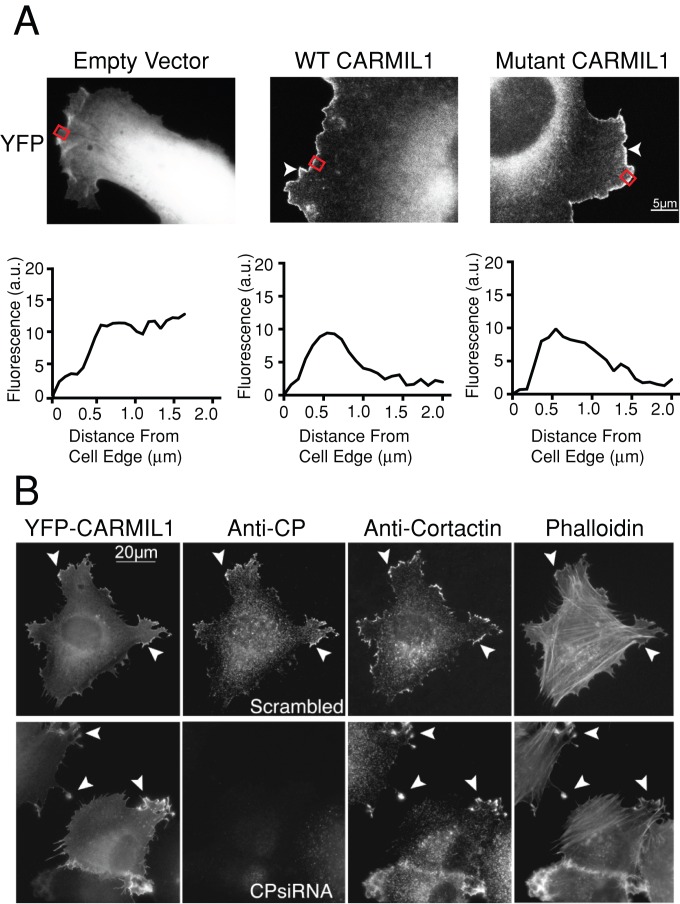
The CP-binding mutant localized normally to the leading edge of cells (Figure 2A). The low levels of expression of CARMIL1 used here had no observable effect on the cells. The localization pattern for the mutant was indistinguishable from that of wt CARMIL1. The YFP empty-vector control showed a diffuse localization pattern. This was supported by line-scan analysis of cells, with fluorescence intensity peaking at around 0.5 μm from the cell edge in cells transfected with YFP CARMIL1 constructs. There was no corresponding peak in YFP empty-vector-transfected cells. Thus CARMIL1 localization to the leading edge does not depend on its ability to bind CP.
FIGURE 2:
(A) CARMIL1 localization does not depend on the ability to bind CP. YFP-tagged fusions of wild-type CARMIL1 or the CP-binding mutant KR987/989AA were expressed in cells at low levels. Representative images are shown; n = 15 cells. The pEYFPC-1 vector, expressing YFP alone, was used as a control. Arrowheads indicate the leading edge of cells. Red rectangles indicate the region of the cell analyzed in the line scan below the image. YFP-CARMIL1 appears at the actin-rich cortex. The expression levels here were lower than the levels needed to induce changes in cell shape and actin distribution, described later. (B) The CARMIL1 localization phenotype does not depend on CP. Cells overexpressing YFP-CARMIL1 were treated with siRNA targeting CP. Cell edges show abnormal protrusions (arrowheads), which are rich in YFP-CARMIL1, cortactin, and F-actin. Loss of CP had no noticeable effect on the localization of CARMIL 1 or the formation or molecular composition of the protrusions. Scale bar, 20 μm. Representative images are shown; n = 11 cells.
To further explore the relationship between CARMIL1 and CP localization in cells, we depleted CP from cells and localized wild-type full-length CARMIL1. The CARMIL1 was expressed in these cells as a YFP fusion at relatively low levels; YFP-CARMIL1 was still concentrated at the leading edge despite the loss of CP (Figure 2B, arrowheads). This result provides further evidence that CARMIL1 localization does not depend on its ability to bind to CP.
Rescue of CARMIL1 loss-of-function phenotypes by CP-binding mutant
To investigate whether and how the biochemical interaction of CARMIL1 with CP is important for the function of CARMIL1 in cells, we asked whether expression of the CARMIL1 CP-binding mutant could rescue the knockdown phenotypes characteristic of CARMIL1. We expressed short hairpin RNA (shRNA)–resistant versions of cDNAs expressing mutant and wt CARMIL1. We used a pFLRu lentiviral expression plasmid that simultaneously expressed shRNA to knock down endogenous CARMIL1 along with an shRNA-resistant, YFP-tagged form of the CARMIL1 cDNA being tested for rescue. In these experiments, immunoblots with anti-CARMIL1 antibodies showed that the protein levels for wild-type and mutant shRNA-resistant YFP-CARMIL1 were similar to each other and to the level of endogenous CARMIL1 protein in control cells (data not shown).
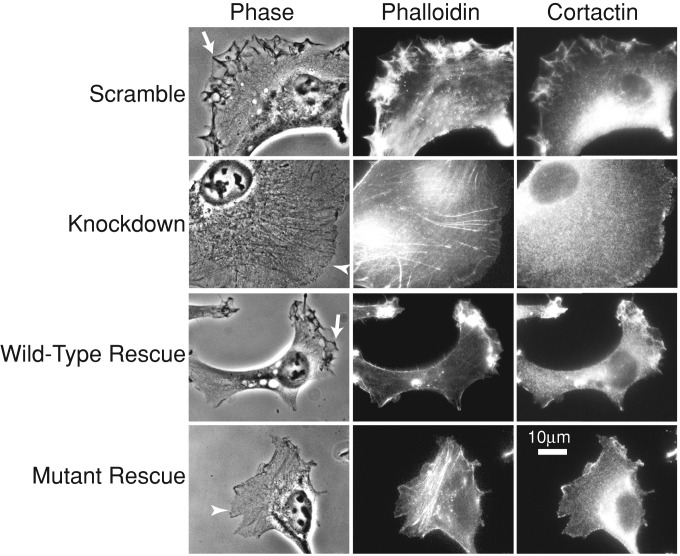
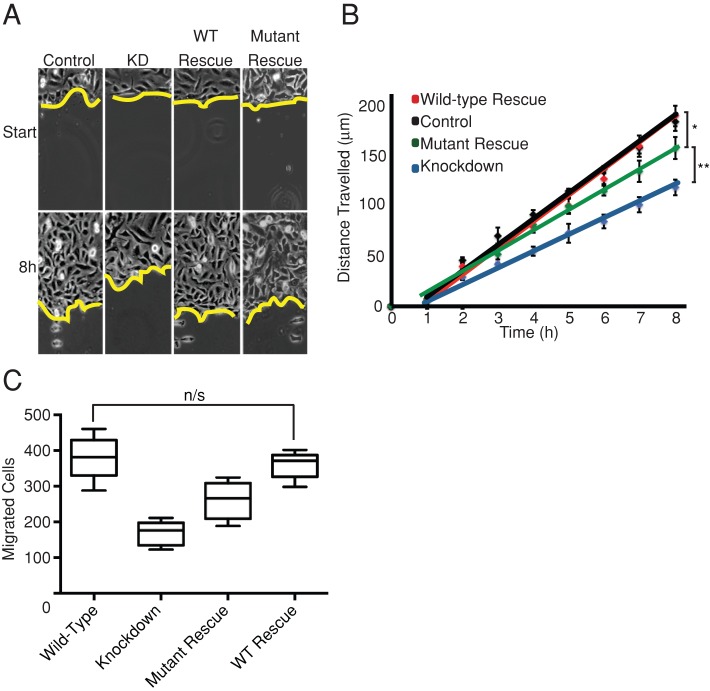
First, we examined lamellipodial assembly and dynamics, which are deficient in CARMIL1-knockdown cells (Liang et al., 2009). Staining with fluorescent phalloidin revealed decreased lamellipodial F-actin in the knockdown cells (Figure 3 and Supplemental Figure S4). Expression of the CP-binding mutant failed to rescue this defect, which was rescued nearly completely by expression of wild-type CARMIL1 (Figure 3 and Supplemental Figure 4).
FIGURE 3:
CARMIL1 CP-binding mutant fails to rescue the actin assembly defect of CARMIL1-knockdown cells. Representative images are shown; n = 30 cells. Cells on fibronectin-coated coverslips were fixed and stained for F-actin (phalloidin) and cortactin. Ruffles at cell edges (arrows) are prominent in control (scrambled) and wild-type rescue cells but not knockdown or mutant rescue cells. Cell edges in knockdown and mutant rescue cells show decreased staining for F-actin and cortactin (arrowheads) compared with control (scrambled) and wild-type rescue cells.
Phase-contrast movies of living cells revealed that knockdown cells had greatly decreased numbers of protrusions and ruffles at their free edges compared with control cells (Figure 3 and Supplemental Movies S1/S5 and S2/S6). This was supported by quantitative kymograph analysis of protrusion rates at the leading edge (Supplemental Figure S2). CARMIL1 knockdowns showed a dramatic reduction in the rate of protrusions formed at the leading edge of cells. Expression of mutant CARMIL1 failed to rescue this phenotype (Figure 3, Supplemental Figure S2, and Supplemental Movies S4 and S8), which was rescued nearly completely by expression of wild-type CARMIL1 (Figure 3, Supplemental Figure S2, and Supplemental Movie S3). The rate of protrusions increased slightly upon expression of the mutant rescue construct (Supplemental Figure S2), although the difference was not statistically significant.
Together these results show that the CARMIL1–CP interaction is important for lamellipodial assembly and function. These effects most likely involve the dynamic assembly of actin that occurs in lamellipodia.
Next we examined ruffling and macropinocytosis. Ruffling at the cell edge often leads to macropinocytosis, which results in the formation of intracellular vesicles filled with extracellular fluid. These vesicles—macropinosomes—are bright in phase-contrast optics because of the low density of their contents. CARMIL1-knockdown cells show a loss of ruffling and macropinocytosis (Liang et al., 2009). We tested the ability of the CP-binding mutant to rescue these phenotypes.
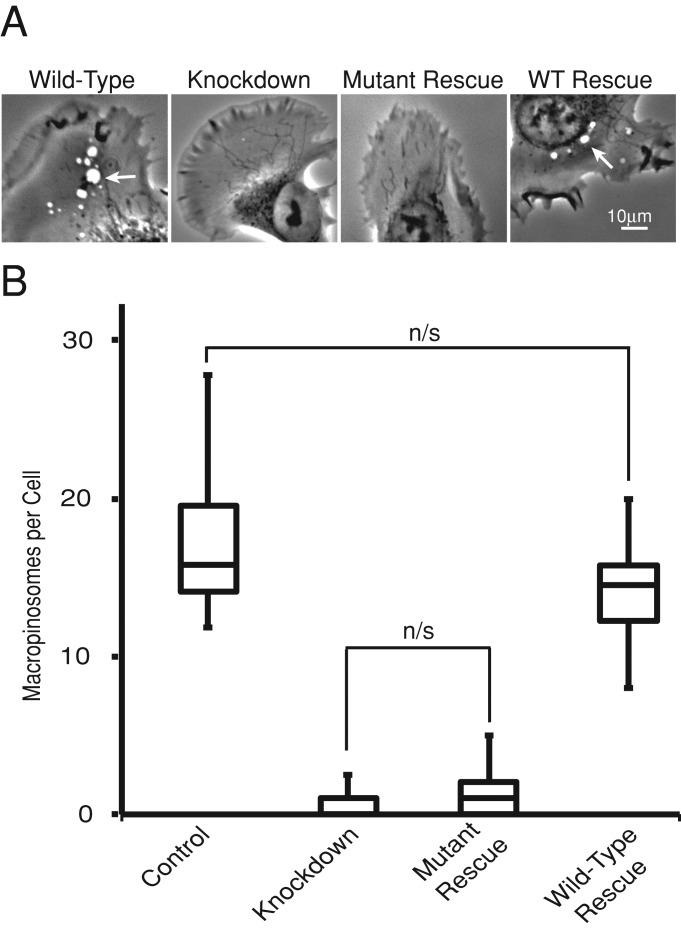
We quantitatively analyzed macropinocytosis by counting macropinosomes that formed during time-lapse movies of HT-1080 cells (Figure 4). CARMIL1-knockdown cells showed a nearly complete loss of macropinosome formation. Expression of the CP-binding mutant essentially failed to rescue this defect. The number of macropinosomes increased by only a very small extent, and the difference was not statistically significant. Expression of wild-type CARMIL1 restored the number of macropinosomes to a normal level, indistinguishable from control. These effects on macropinosome formation can also be appreciated by viewing the phase-contrast movies (Supplemental Movies S1–S8). Thus CARMIL1’s role in macropinocytosis depends completely on its ability to bind CP.
FIGURE 4:
CARMIL1 CP-binding mutant fails to rescue the macropinocytosis defect of CARMIL1-knockdown cells. (A) Representative frames taken from Supplemental Movies S1–S4. Arrowheads indicate macropinosomes. (B). The number of macropinosomes per cells in a box-and-whisker format showing the median, the interquartile range, and the extremes. Macropinocytosis was scored by analyzing 10 movies. All differences were significant (p < 0.01), unless otherwise indicated on the plot.
The CARMIL1–CP interaction has only marginal importance for cell migration
In several studies, lamellipodia and Arp2/3-based actin assembly have not been found to be important for overall rates of cell migration (Gupton et al., 2005; Wu et al., 2012). In previous studies CARMIL1-knockdown cells were found to display decreased rates of cell migration in wound-healing assays (Yang et al., 2005; Liang et al., 2009). Because we found that the CARMIL1–CP interaction is important for lamellipodia formation and dynamics, we were able to ask whether lamellipodia are important for cell migration in this setting, by attempting to rescue the cell migration defect with the CP-binding mutant of CARMIL1.
We measured cell migration in wound-healing assays, examining CARMIL1-knockdown cells that expressed wild-type or CP-binding mutant rescue constructs (Figure 5, A and B). The CARMIL1-knockdown cells closed the wound more slowly than did control cells, consistent with previous results (Liang et al., 2009). This defect was almost completely rescued by expression of wild-type CARMIL1. The CP-binding mutant was able to rescue the wound-healing defect. The level of rescue was not complete, but it was substantial: greater than half.
FIGURE 5:
Rescue of the wound-healing defect of CARMIL1-knockdown cells by the CP-binding mutant. Cells were infected with virus-carrying plasmids expressing control shRNA, CARMIL1-knockdown shRNA, knockdown plus wild-type rescue, or knockdown plus mutant rescue. (A) Images from representative experiments. (B) Distance traveled by the edge of the wound. The mean of six independent experiments is plotted; error bars, SEM. The CP-binding mutant shows an intermediate level of rescue. Distances traveled after 8 h between control and mutant rescue * and mutant rescue and knockdown ** were significant (p < 0. 01). (C) Transwell assay data showing the total number of cells migrating through 8-μm Transwell pores to the lower chamber after 4 h. Data are plotted in a box-and-whisker format showing the median, the interquartile range, and the extremes, n = 6. All differences were significant (p < 0.01), unless otherwise indicated on the plot.
We tested the migration of individual cells using Transwell assays, to further assess the role of the CARMIL–CP interaction in cell migration. In a wound-healing model, cell migration is heavily influenced by cell–cell contacts. The dynamics of migrating as a sheet of cells might potentially reduce the dependence on lamellipodia-driven migration. In a Transwell assay, cells migrate independent of cell–cell contacts, through pores, toward a chemoattractant in the lower chamber. The results of this assay were consistent with the wound-healing data (Figure 5C). Expression of the CARMIL1 CP-binding mutant showed substantial rescue (>50%) of the migration defect observed in CARMIL1-knockdown cells. Migration was completely restored to normal levels by expressing wild-type CARMIL1.
Because cells expressing the CARMIL1 CP-binding mutant have very few lamellipodia but migrate at rates near normal in both assays, we conclude that lamellipodia are not critical for cell migration in these settings.
The CARMIL1–CP interaction is not required for Rac1 activation
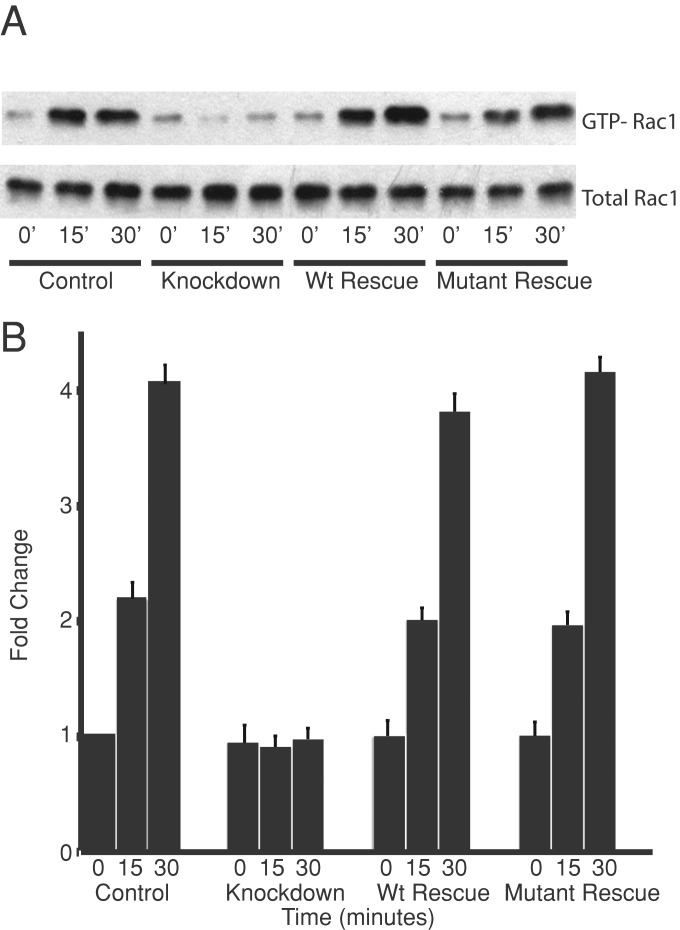
We investigated the role that the CARMIL1–CP interaction plays in the activation of Rac1 that occurs when cells spread on fibronectin. Spreading-induced Rac1 activation is lost, to an essentially complete extent, when CARMIL1 is knocked down (Liang et al., 2009). Here we found that wt CARMIL1 and the CP-binding mutant both fully rescued the Rac1 activation defect seen in CARMIL1-knockdown cells (Figure 6). Therefore, in striking contrast to the situation for lamellipodia and ruffles, the CARMIL1–CP interaction is not needed for spreading-induced Rac1 activation.
FIGURE 6:
Rac1 activation in spreading cells requires CARMIL1 but not the interaction between CARMIL1 and CP. Cells were infected with lentivirus-carrying plasmids to simultaneously knock down endogenous CARMIL1 with shRNA and express rescue forms of full-length CARMIL1. Cells were allowed to spread on fibronectin for the indicated times. (A) Immunoblot showing the result of a representative GTP-Rac1 assay. (B) The fold change in GTP-Rac1 levels relative to the control at time zero. Mean of four independent assays; error bars, SEM.
Overexpression phenotypes of the CP-binding mutant form of CARMIL1
We investigated whether increasing the level of CARMIL1 would lead to inhibition of CP activity in cells. Expressing CARMIL1 at high levels is known to induce phenotypes that include effects on the actin-rich cortex (Liang et al., 2009), so we asked whether those phenotypes were the result of the interaction of CARMIL1 with CP. On one hand, because CP localizes to the actin-rich cortex and CP is important for actin dynamics, one might expect that the CARMIL1–CP interaction would be necessary for the CARMIL1 overexpression phenotypes. CARMIL1 is a large protein with multiple domains of unknown function, however, so the overexpression phenotypes might be due to biochemical interactions of CARMIL1 with molecules other than CP. In addition, the extent to which CARMIL1 is able to bind to CP in cells is an open question.
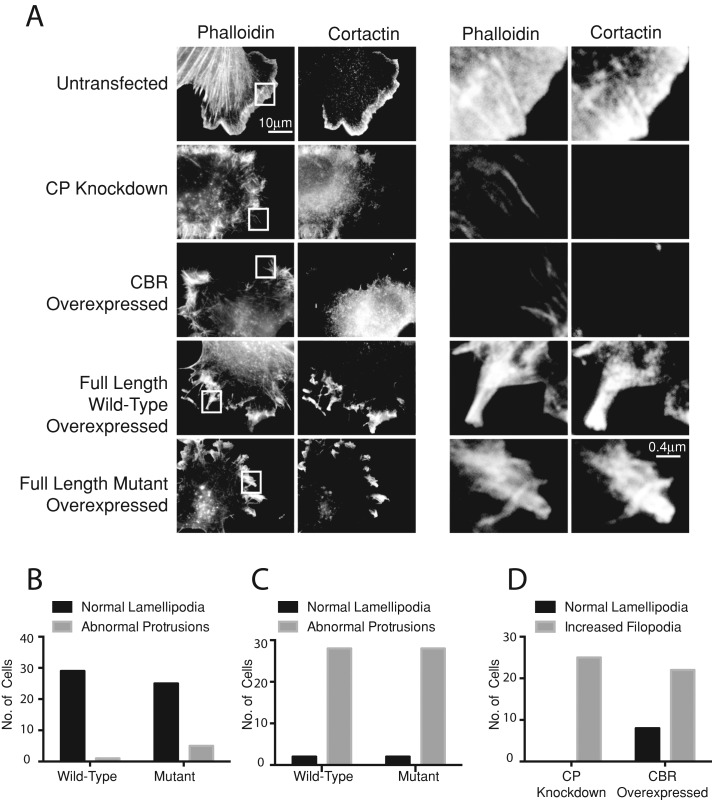
First, we asked whether the CP-binding mutant form of CARMIL1 was able to induce the overexpression phenotypes. Wild-type CARMIL1 was expressed to medium levels in HT1080 cells, in which the expression of endogenous CARMIL1 was not inhibited. This level of overexpression led to the formation of abnormal actin-rich protrusions at the cell edge (Figure 7, A and C), consistent with previous observations (Liang et al., 2009). Expression of the CARMIL1 CP-binding mutant caused a similar appearance of abnormal protrusions. In both cases, wild type and mutant, the abnormal protrusions were rich in F-actin and cortactin. Overall the effects of mutant CARMIL1 were indistinguishable from those of the wild type (Figure 7A). Higher levels of expression showed even greater distortions of shape at the cell edge, again with no difference between wild-type CARMIL1 and the CP-binding mutant.
FIGURE 7:
The role of CP in the effects of CARMIL1 overexpression. (A) Comparison of the effects of CP knockdown in cells with those of overexpression of full-length CARMIL1, the CBR fragment of CARMIL1, and the CP-binding mutant form of full-length CARMlL1. Cells deficient in CP were stained for cortactin and F-actin (phalloidin). Boxed regions in the fluorescent phalloidin channel are magnified in the columns on the right. Representative images; n = 30 cells. (B) Effects of wt and mutant CARMIL1 overexpression on lamellipodia at low expression levels. (C) Effects of wt and mutant CARMIL1 overexpression on lamellipodia at high expression levels. (D) Effects of CP knockdown vs. CARMIL1 CBR overexpression on lamellipodia and filopodia formation at the leading edge.
To address this issue further, we reasoned that if the overexpression phenotypes induced by CARMIL1 are due to inhibition of CP, one might expect that loss of CP would produce effects similar to those of CARMIL1 overexpression or that loss of CP would block the effects of CARMIL1 overexpression. To examine these possibilities, we first inhibited CP by shRNA knockdown. Loss of CP was confirmed by immunoblots (Supplemental Figure S1), which also revealed a small increase in the level of total cellular actin, consistent with previous studies in other cell types (Hug et al., 1995; Canton et al., 2005).
We observed that the loss of CP led to a loss of lamellipodia, with an increase in the number of fine filopodial surface projections, consistent with previous findings in other cell types (Hug et al., 1995; Mejillano et al., 2004). In contrast, overexpression of CARMIL1 caused the formation of abnormal protrusions shaped like clubs and spikes, distinct from the fine filopodial projections seen in CP-knockdown cells. The molecular composition of the abnormal protrusions in the CARMIL1 overexpressers revealed them to be lamellipodial in nature, in that they stained for the Arp2/3 regulators WAVE2 (not shown) and cortactin (Figure 7A). Overall, protrusions induced by overexpression of CARMIL1, both wild type and mutant, were strikingly different from those induced by loss of CP in terms of morphology and molecular markers. These results indicate that the overexpression effects of CARMIL1 are not due to inhibition of CP.
The fact that expression of CARMIL1 did not mimic the loss of CP raised the question of whether, or to what extent, CARMIL1 is capable of inhibiting CP in cells. To address this issue, we asked whether expression of the 115–amino acid CBR fragment of CARMIL1, which is known to potently inhibit CP in vitro in biochemical assays, might mimic the loss of CP in cells. Indeed, we found this to be the case (Figure 7, A and D). Expression of the CBR fragment induced filopodial protrusions identical to those resulting from the knockdown of CP in 24 of 30 cells observed (Figure 7D). One would expect that the interaction of the CBR fragment with CP would be necessary for the overexpression effects of CBR. Again, this was the case. Expression of the CP-binding mutant form of CBR, carrying the KR987/989AA changes, produced essentially no effect on cells (data not shown). Thus CARMIL1 CBR, expressed on its own as an active fragment, is able to inhibit CP in cells. These results suggest that CARMIL1 may be able to inhibit CP activity in cells and raise the possibility that full-length CARMIL1 may be regulated in its ability to inhibit CP in cells.
In addition, we asked whether the phenotypes caused by overexpression of full-length CARMIL1 required the presence of CP. We depleted cells of CP by small interfering RNA (siRNA) treatment and then expressed wild-type CARMIL1. We observed abnormal lamellipodial protrusions (Figure 2B, arrowheads) in these cells identical to those produced upon overexpression of CARMIL1 in wild-type cells (Figure 2B). Therefore the effects resulting from overexpression of CARMIL1 do not depend on CP.
Localization of CP in CARMIL1-knockdown cells
We tested whether the CARMIL1-CP interaction was important for the localization of CP to the leading edge. CP localized to lamellipodia at the leading edge of migrating cells (Supplemental Figure S3), where it is presumably involved with dynamic actin assembly and lamellipodia formation (Zhao et al., 2013). CP fails to localize to the leading edge in CARMIL1-knockdown cells (Supplemental Figure S3). Expression of the CARMIL1 CP-binding mutant did not restore CP localization to the leading edge (data not shown). CARMIL1-knockdown cells have a significant defect in lamellipodia formation (Liang et al., 2009; Figures 3 and S4), making it impossible to determine whether the effect we observe is due to direct interaction of CP with CARMIL or indirectly to interaction of CP with actin filaments or other components of the lamellipodia.
DISCUSSION
In this study we investigated the physiological significance of the CARMIL1–CP interaction. Our most important discovery is that the ability of CARMIL1 to bind capping protein is required for the ability of CARMIL1 to contribute to lamellipodial assembly and function, which is the basis for ruffling and macropinocytosis. In contrast, the localization of CARMIL1 and the ability of CARMIL1 to activate Rac1 do not depend on the CARMIL1–CP interaction.
The physiological role of the CARMIL1–CP interaction in cells
In this study we investigated the relevance and function of the CARMIL1–CP interaction in cells. We found the interaction to be important for lamellipodial assembly and ruffling of the cell edge, which are the basis for macropinocytosis. These processes require a dynamic network of actin filaments at the cell cortex (Chhabra and Higgs, 2007), implying that the CARMIL1–CP interaction contributes to dynamic actin assembly.
CARMIL1 localizes in close proximity to the plasma membrane. We found that the localization of CARMIL1 does not depend on its ability to bind capping protein. Therefore CARMIL1 must be localized via some other biochemical interaction with another cellular component. This result suggests that free CP, diffusing about the cytoplasm, might be recruited to the plasma membrane by CARMIL1, similar to what was found recently for the membrane adaptor CD2AP (Zhao et al., 2013). If CARMIL1 does recruit CP to the membrane, then one might speculate that the bound CP would not be active to bind actin. This mechanism would serve to promote barbed-end growth near the membrane. Alternatively, in vitro biochemical assays show that the ability of CP to bind actin is greatly inhibited, but not abolished, by its interaction with CARMIL. This raises the possibility that CP bound to CARMIL does remain active for capping barbed ends (Yang et al., 2005; Uruno et al., 2006), so that the net effect of CARMIL is to promote barbed-end capping near the membrane.
Another possible mechanism of action for CARMIL1 with respect to dynamic actin assembly is that actin filament barbed ends created by Arp2/3 or other nucleators and then capped by CP might subsequently become uncapped when they encounter CARMIL1 near the membrane. This mechanism might contribute to the disassembly of actin filaments, depending on other factors that influence barbed-end growth and shrinkage.
Does CARMIL1 inhibit CP in cells?
Other results in our study address the question of whether CARMIL1 inhibits CP in cells. First, we found that the CBR fragment of CARMIL1, which is a potent inhibitor of CP in actin polymerization assays in vitro, is able to inhibit CP in cells. Expression of the CBR fragment alone had effects on cells that closely resembled the effects of the loss of CP from shRNA-mediated knockdown. In striking contrast, we found that expression of full-length CARMIL1 did not mimic the loss of CP. One possible interpretation of this result is that CARMIL1 has other biochemical functions in addition to binding CP, which makes sense because CARMIL1 is large and has multiple domains of unknown function. Another possible interpretation of this result is that full-length CARMIL1 is inhibited from binding CP in cells by either an autoinhibition mechanism or the action of another molecule.
Previous biochemical studies differ on the issue of autoinhibition for CARMIL. For Acanthamoeba CARMIL, a compelling biochemical analysis provided strong evidence for autoinhibition (Uruno et al., 2006). In that study, a fragment of Acanthamoeba CARMIL, produced by limited proteolysis, was a far more potent inhibitor of CP than was full-length CARMIL. In contrast, a study of mouse CARMIL1 found that a fragment containing the CP-interacting region had the same ability to inhibit CP as did full-length CARMIL1 (Yang et al., 2005). This discrepancy might be due to differences in the experimental systems and designs. The question is open and important in the context of a cell.
Functions of CARMIL1 that do not require its interaction with CP
CARMIL1 appears to have functions that affect the architecture of the actin-rich core but do not require CP. At the molecular level, the abnormal protrusions induced by CARMIL1 overexpression appear to be lamellipodial in nature, which is not what one would expect from inhibition of CP. More important, the overexpression effects from full-length CARMIL were not affected when the ability of CARMIL to interact with CP was abolished by mutation. In addition, CARMIL1 localization to the cortex also does not require interaction to CP.
Finally, the activation of Rac1 observed when cells spread on fibronectin also does not require CARMIL1–CP interaction. Rac1 plays a well-documented role in lamellipodia formation (Ridley and Hall, 1992; Ridley et al., 1992). Because the abnormal protrusions induced by CARMIL1 overexpression are lamellipodial in nature, one might speculate that the ability of CARMIL1 to activate Rac1 has physiological significance.
Lamellipodia are not important for cell migration
The results of our rescue experiments with the CARMIL1–CP interaction mutant suggest that lamellipodia are not required for cell migration in wound-healing and Transwell migration models. The CARMIL1-mutant rescue cells had very few lamellipodia but migrated almost as well as control cells, which have prominent lamellipodia. Our cell migration result contrasts with that in a previous study (Yang et al., 2005), although that study examined a CARMIL1 mutant with a large internal deletion. Here the CP-binding mutant carried only two amino acid changes. Thus the failure of the CARMIL1 mutant to rescue cell migration in the previous study may have been due to effects on biochemical functions other than binding to CP.
Our findings support a growing body of evidence indicating that the lamellipodium is not critical for cell migration (Gupton et al., 2005; Suraneni et al., 2012; Wu et al., 2012). Other studies concluded that the actin networks in the lamella or the filopodia, not the lamellipodia, generate the force that moves the cell forward (Gupton et al., 2005; Suraneni et al., 2012). Lamellipodia are important for ruffling at the cell edge and the process of macropinocytosis, in which ruffles close back onto the cell body and engulf extracellular fluid. Macropinocytosis allows cells to sample their environment and take up antigens for processing (von Delwig et al., 2006).
MATERIALS AND METHODS
Antibodies and reagents
Reagents and materials were from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) unless stated otherwise. To detect capping protein, we used mouse monoclonal antibody (mAb) clone 2A3 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) for immunoblots (Schafer et al., 1996) and rabbit polyclonal antibody (pAb) R26 against the C-terminus of β2 for immunostaining (Schafer et al., 1994). Other antibodies and sources were as follows: ARPC2/p34 (rabbit pAb), cortactin (mouse mAb 4F11), WAVE2 (rabbit pAb), and actin (mouse mAb C4) from Millipore (Billerica, MA); α-tubulin (mouse mAb) and FLAG (mouse mAb M2) from Sigma-Aldrich; and anti–green fluorescent protein (GFP; rabbit, pAb), Dynabeads M-280 sheep anti-rabbit immunoglobulin G, and horseradish peroxidase– and Alexa-conjugated secondary antibodies from Invitrogen (Carlsbad, CA). Chicken antibodies to human CARMIL1 were produced and characterized as described (Liang et al., 2009).
Cell culture, transfection, knockdown, and rescue of CARMIL1
Human HT1080 cells and HEK-293 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) in an atmosphere containing 5% CO2. Cells were transfected using Transit LT1 (Mirus, Madison, WI). For overexpression, cells were transfected with 5 μg of DNA/106 cells and fixed 48 h after transfection.
For knockdown of human CARMIL1, an shRNA construct in lentivector pFLRu-FH-GFP was used as described (Liang et al., 2009). Target sequence was ATGCCATTGTTCATCTGGAT for CARMIL1, with CAGTCGCGTTTGCGACTGG as a nontargeting control. For rescue by expression, site-directed mutagenesis was used to construct a pFLRu shRNA–resistant CARMIL1 lentiviral-based expression plasmid. Resistance to shRNA was conferred by the following three codon-silent nucleotide changes: GCC to GCT, GTT to GTG, and CTG to CTC.
The CP-binding mutant form of CARMIL1 was generated by changing two individual amino acid residues, lysine 987 and arginine 989, to alanine by site-directed mutagenesis (Stratagene, La Jolla, CA). We refer to this mutant as KR987/989AA.
To knock down CP, we expressed an shRNA in the lentiviral vector PLKO.1 or treated cells with siRNA targeting the coding region of the CP β subunit, AAGGATTACCTTTTGTGTGAC. siRNA was purchased from Dharmacon (Lafayette, CO). The nontargeting sequence GCCTGGTAGAGGACATGGAAA was used as a control for both siRNA- and shRNA-based knockdown. siRNA and shRNA constructs both target all isoforms of CP β because the isoforms are produced by alternative splicing from one gene and the mature mRNAs of all the isoforms contain the target sequence. We purchased the shRNA construct targeting CP, developed by the RNAi Consortium at the Broad Institute, from the Children's Discovery Institute/Genome Sequencing Center at Washington University (St. Louis, MO).
Immunofluorescence and live-cell imaging
HT1080 cells grown on glass coverslips coated with 15 μg/ml fibronectin (Sigma-Aldrich) were fixed in paraformaldehyde and processed as described (Mejillano et al., 2004). Immunostaining was performed with the primary and secondary antibodies listed. Cells were imaged with 100×/1.4 numerical aperture (NA) and 40×/0.75 NA objectives on an Olympus IX70 inverted microscope (Olympus, Melville, NY) equipped with a cooled charge-coupled device camera. Images were collected and initially processed with QED In Vivo software (Media Cybernetics, Silver Spring, MD).
For time-lapse movies, cells were grown on glass-bottom culture dishes (MatTek, Ashland, MA) coated with fibronectin (15 μg/ml). Cells were adapted to L-15 medium (Invitrogen) supplemented with 10% FBS (Sigma-Aldrich) at 37°C. Temperature was maintained at 37°C, and images were acquired every 6 s for 10 min. Movie files were processed with ImageJ (National Institutes of Health, Bethesda, MD).
Macropinocytosis was quantified from time-lapse movies of migrating single cells (N = 10) not in contact with other cells. The observer counted each discrete macropinocytotic vesicle formed at a ruffling edge. The number of vesicles was normalized to the number of cells.
Kymography
Kymographs were generated in ImageJ from a 2-μm linear region of interest (ROI) at the leading edge of 1-h time-lapse movies.
Line scans
Fluorescence intensity was measured in ImageJ from a 2-μm rectangular ROI at the edge of the cell. Measurements were background subtracted and plotted.
Coimmunoprecipitations and immunoblots
Immunoprecipitation with anti-FLAG M2 affinity beads (Sigma-Aldrich) and anti-GFP beads (Invitrogen) was performed as described (Liang et al., 2009). The beads were washed and boiled with 2× SDS loading buffer and then analyzed by SDS–PAGE and immunoblotting. Immunoblots were developed with electrochemiluminescence (ECL; PerkinElmer-Cetus, Boston, MA) and exposed to autoradiography film.
Rac1 activation assays
Rac1 activation assays were performed on spreading cells as described (Liang et al., 2009). Rac1-GTP levels were assayed via pull down with glutathione S-transferase (GST) fused to the p21PAK1 Rac1/Cdc42 (p21)–binding domain of human p21-activated kinase 1 (Cell Biolabs, San Diego, CA). Immunoblots with anti-Rac1 were developed using ECL (PerkinElmer-Cetus). Rac1 band intensity levels on the immunoblots were measured by densitometry with a ChemiDoc MP System (Bio-Rad, Hercules, CA). First, Rac1-GTP levels were measured and found to be similar in all cell samples—control, knockdown, and rescue. Second, Rac1-GTP levels were measured at 15 and 30 min of cell spreading. The fold increase of Rac1-GTP was calculated, compared with the level of Rac1-GTP in control cells at time zero.
Protein expression and purification
The CBR fragments of human CARMIL1a (GenBank FJ009082) Glu-964–Ser-1078 (pBJ 1841), described elsewhere (Liang et al., 2009), were amplified from cDNAs by PCR and cloned into pGEX-6P-3 (GE Healthcare, Piscataway, NJ). Complete DNA sequencing of the insert and junctions verified the plasmids. The mutant CARMIL1-CBR KR987/989AA was made using QuikChange site-directed mutagenesis (Stratagene). GST fusion proteins were expressed in BL21 (DE3) Escherichia coli and purified with glutathione Fast-Flow Sepharose resin (GE Healthcare). Cultures were grown and induced with isopropyl-β-d-thiogalactoside at 23°C. After elution from the glutathione resin, GST-CBR was mixed with PreScission protease (GE Healthcare). The mixture was dialyzed into S-Sepharose buffer A (10 mM Tris, pH 8.0, 10 mM KCl, 0.1 mM EDTA, 0.5 mM dithiothreitol [DTT], 1 mM NaN3) overnight, applied to an S-Sepharose column, and eluted with a KCl gradient (10–700 mM). For storage, CBR was dialyzed into 10 mM Tris, pH 8.0, 40 mM KCl, 0.1 mM EDTA, 0.5 mM DTT, and 1 mM NaN3 and kept on wet ice. The concentration of CBR was calculated from A280, based on predicted extinction coefficients, and confirmed by SDS–PAGE with Coomassie blue staining.
Actin polymerization assays
Actin was purified from rabbit skeletal muscle as described (Wear et al., 2003). Pyrene–actin polymerization assays, including inhibition and uncapping of by CARMIL, were performed as described (Wear et al., 2003; Kim et al., 2012). The actin concentration was 1.5 μM, with 7% pyrene label. For capping assays, 10 nM CP was added to a mixture of pyrene-labeled actin and spectrin–actin seeds at time zero. To assay for reversal of capping, pyrene–actin was polymerized from seeds in the presence of 10 nM CP. After 200 s, the CBR fragment of CARMIL was added, and polymerization was followed for 300 s.
Cell migration in a wound-healing model
HT-1080 cells were infected with lentivirus-carrying plasmids for knockdown or knockdown/rescue of CARMIL1. The cells were grown to a monolayer and starved for serum for 12 h. The monolayer was wounded with a pipette tip, and the culture medium was changed to fresh medium with serum (10% FBS). Images were collected every hour for 8 h. We used ImageJ to measure the distance traveled by the edge of the wound.
Transwell cell migration assay
HT-1080 cells were infected with lentivirus-carrying plasmids for knockdown or knockdown/rescue of CARMIL1. The cells were grown to a monolayer and starved for serum for 12 h. Then 1 × 104 cells were added to the upper chamber of an 8-μm Transwell (Corning, Tewksbury, MA). Cells were allowed to migrate toward a 10% FBS gradient in the lower chamber at 37°C for 4 h. Inserts were then cut out and crystal violet stained. The number of cells that migrated through the pores to the underside of the insert was scored under a light microscope.
Statistical analysis
We used Tukey's multiple comparison tests to test significance of our results when comparing knockdowns to controls and rescue experiments. We performed Student's t test on population means to determine whether the total distances traveled after 8 h in our wound-healing assays were significant between the indicated populations. In all cases p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank our lab colleagues for their comments and assistance with the project and the manuscript. This work was supported by National Institutes of Health Grants GM 38542 and GM 95509 to J.A.C. M.E. was supported by National Institutes of Health Training Grant 5T90DA02287104.
Abbreviations used:
- CARMIL
capping protein Arp2/3 myosin I linker
- CBR
capping protein binding region
- CP
capping protein
- CPI
capping protein interaction
- CSI
CARMIL-specific interaction
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0270) on July 31, 2013.
REFERENCES
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck S, Huber TB, Ingham RJ, Kim K, Niederstrasser H, Allen PM, Pawson T, Cooper JA, Shaw AS. Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J Biol Chem. 2006;281:19196–19203. doi: 10.1074/jbc.M600166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton DA, Olsten ME, Kim K, Doherty-Kirby A, Lajoie G, Cooper JA, Litchfield DW. The pleckstrin homology domain-containing protein CKIP-1 is involved in regulation of cell morphology and the actin cytoskeleton and interaction with actin capping protein. Mol Cell Biol. 2005;25:3519–3534. doi: 10.1128/MCB.25.9.3519-3534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, et al. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Valladares M, Kim T, Kannan B, Tung A, Aguda AH, Larsson M, Cooper JA, Robinson RC. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat Struct Mol Biol. 2010;17:497–503. doi: 10.1038/nsmb.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, Cooper JA. Capping protein levels influence actin assembly and cell motility in Dictyostelium. Cell. 1995;81:591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA 3rd. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Ravilious GE, Sept D, Cooper JA. Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. J Biol Chem. 2012;287:15251–15262. doi: 10.1074/jbc.M112.345447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Niederstrasser H, Edwards M, Jackson CE, Cooper JA. Distinct roles for CARMIL isoforms in cell migration. Mol Biol Cell. 2009;20:5290–5305. doi: 10.1091/mbc.E08-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harb Symp Quant Biol. 1992;57:661–671. doi: 10.1101/sqb.1992.057.01.072. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T, Remmert K, Hammer JA 3rd. CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J Biol Chem. 2006;281:10635–10650. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- von Delwig A, Hilkens CM, Altmann DM, Holmdahl R, Isaacs JD, Harding CV, Robertson H, McKie N, Robinson JH. Inhibition of macropinocytosis blocks antigen presentation of type II collagen in vitro and in vivo in HLA-DR1 transgenic mice. Arthritis Res Ther. 2006;8:R93. doi: 10.1186/ar1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA. How capping protein binds the barbed end of the actin filament. Curr Biol. 2003;13:1531–1537. doi: 10.1016/s0960-9822(03)00559-1. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zot AS, Zot HG. Identification of Acan125 as a myosin-I-binding protein present with myosin-I on cellular organelles of Acanthamoeba. J Biol Chem. 1995;270:25316–25319. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]
- Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bruck S, Cemerski S, Zhang L, Butler B, Dani A, Cooper JA, Shaw AS. CD2AP links cortactin and capping protein at the cell periphery to facilitate formation of lamellipodia. Mol Cell Biol. 2013;33:38–47. doi: 10.1128/MCB.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.