In mice, the ubiquitin ligase RLIM/Rnf12 is a critical survival factor for mammary milk-producing alveolar cells, but little is known about how its activity is regulated. It is shown here that RLIM shuttles between the nucleus and cytoplasm in a phosphorylation-dependent manner, and shuttling is important for its alveolar survival function.
Abstract
The X-linked gene Rnf12 encodes the ubiquitin ligase really interesting new gene (RING) finger LIM domain–interacting protein (RLIM)/RING finger protein 12 (Rnf12), which serves as a major sex-specific epigenetic regulator of female mouse nurturing tissues. Early during embryogenesis, RLIM/Rnf12 expressed from the maternal allele is crucial for the development of extraembryonic trophoblast cells. In contrast, in mammary glands of pregnant and lactating adult females RLIM/Rnf12 expressed from the paternal allele functions as a critical survival factor for milk-producing alveolar cells. Although RLIM/Rnf12 is detected mostly in the nucleus, little is known about how and in which cellular compartment(s) RLIM/Rnf12 mediates its biological functions. Here we demonstrate that RLIM/Rnf12 protein shuttles between nucleus and cytoplasm and this is regulated by phosphorylation of serine S214 located within its nuclear localization sequence. We show that shuttling is important for RLIM to exert its biological functions, as alveolar cell survival activity is inhibited in cells expressing shuttling-deficient nuclear or cytoplasmic RLIM/Rnf12. Thus regulated nucleocytoplasmic shuttling of RLIM/Rnf12 coordinates cellular compartments during mammary alveolar cell survival.
INTRODUCTION
Really interesting new gene (RING) finger LIM domain–interacting protein (RLIM)/RING finger protein 12 (Rnf12) and Rnf6 define a class of RING finger ubiquitin ligases that is widely expressed during mouse embryogenesis (Tursun et al., 2005; Ostendorff et al., 2006). Whereas Rnf6 is detected in the nucleus and cytoplasm in cells (Tursun et al., 2005; Xu et al., 2009), RLIM/Rnf12 is detected mainly in the nuclear compartment, where it serves as a cofactor to promote or inhibit transcription factor activity in a context-dependent manner (Bach et al., 1999; Johnsen et al., 2009). Via proteasomal targeting, RLIM/Rnf12 regulates cellular levels of various mostly nuclear proteins, including CLIM/NLI/Ldb, LMO, HDAC2, TRF1, Smad4, and Rex1 (Ostendorff et al., 2002; Kramer et al., 2003; Her and Chung, 2009; Gontan et al., 2012; Zhang et al., 2012), thereby affecting the formation of transcriptional multiprotein complexes (Gungor et al., 2007). Moreover, functions of RLIM in promoting cell migration in vitro have recently been reported (Huang et al., 2011). RLIM/Rnf12 is encoded by the X-linked gene Rnf12 (Ostendorff et al., 2000), which plays major roles in sex-specific epigenetic regulation of female nurturing tissues in mice. Analysis of a conditional Rnf12-knockout (KO) mouse model identified Rnf12/RLIM as an important epigenetic regulator of female nurturing tissues. RLIM/Rnf12 expressed from the maternal allele is crucial for imprinted X chromosome inactivation (XCI) and the development of extraembryonic trophoblast cells in the placenta early during female embryogenesis (Shin et al., 2010). In adult female mice RLIM/Rnf12 expressed from the paternal allele functions as a critical survival factor specifically for milk-producing alveolar cells in the mammary gland. Indeed, alveolar cells lacking RLIM undergo apoptosis as soon as they differentiate in pregnant females, and evidence suggests that weaning-induced down-regulation of RLIM triggers involution (Jiao et al., 2012). Male mice with a germline Rnf12 knockout appear normal, however, and are fertile, suggesting that Rnf12 does not play general developmental roles in this species (Shin et al., 2010). This is in contrast to zebrafish, in which important roles of RLIM for embryonic morphogenesis and stem cell fate have been reported (Zhang et al., 2012).
Nucleocytoplasmic shuttling can convey signals that coordinate crucial cellular processes, including gene expression, cell motility, and survival (Hervy et al., 2006). Because RLIM/Rnf12 contains putative nuclear localization sequences (NLS) and nuclear export sequences (NES), to illuminate mechanisms controlling RLIM, we examined their relevance with respect to cellular localization and function of RLIM/Rnf12 protein. Whereas RLIM/Rnf12 protein is abundantly detected in the nuclei of cells, we find efficient nucleocytoplasmic shuttling of this protein and provide strong evidence that this process is regulated by phosphorylation of S214. We show that shuttling is critically involved in at least some of RLIM's biological activities, including cell migration and mammary alveolar cell survival.
RESULTS
Nuclear and cytoplasmic localization of RLIM
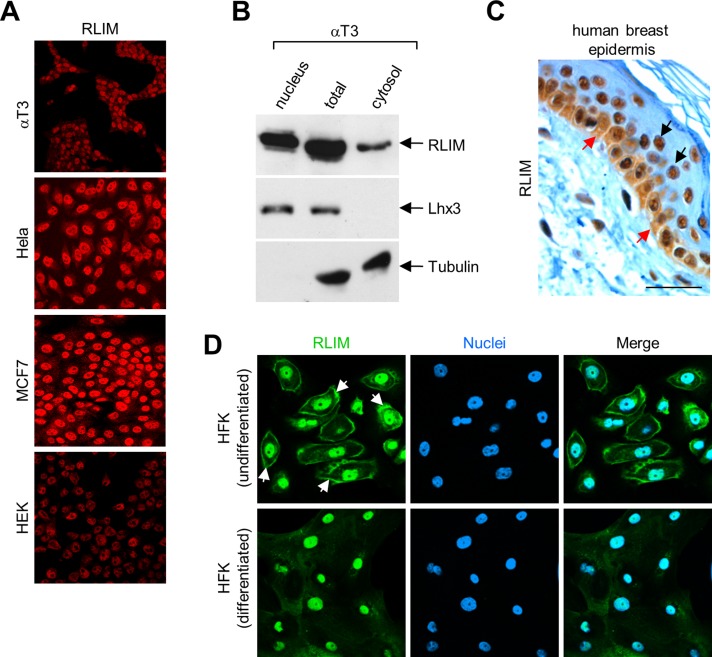
Known substrate proteins targeted for proteasomal degradation by RLIM/Rnf12 are mainly localized in the nucleus. In agreement with published results that showed predominant nuclear localization of RLIM in cells in culture (Ostendorff et al., 2002) and in many cell types during mouse embryogenesis, including neurons (Tursun et al., 2005; Ostendorff et al., 2006), immunocytochemical experiments showed high levels of endogenous RLIM protein in nuclei of all cell lines investigated (αT3, CHO, Cos7, HeLa, HEK293T, MCF7, and MDA-MB-231; Figure 1A and data not shown). Using Western blots on nuclear and cytoplasmic fractions of αT3 cell extracts, however, we found a significant portion of cellular RLIM localized in the cytoplasm (Figure 1B). Examining cell types that exhibit cytoplasmic expression in vivo, we identified high levels of RLIM expression in the basal layer of human breast epidermis, where undifferentiated keratinocytes actively divide and which displayed significant RLIM staining in nuclear and cytoplasmic compartments (Figure 1C). In differentiating keratinocytes that had migrated away from the basal layer, however, RLIM expression was mainly detectable in the nucleus. To further address this issue, we examined the distribution of RLIM in undifferentiated and differentiated primary human foreskin keratinocytes (HFKs). Staining of undifferentiated HFK cells showed a significant portion of RLIM in the cytoplasm adjacent to the cell membrane, whereas the differentiation of these cells in 10% serum for 48 h resulted in a specific dispersal and disappearance of the cytoplasmic RLIM staining along the plasma membrane (Figure 1D). Combined, these results suggest a regulated cellular distribution of RLIM during epithelial differentiation.
FIGURE 1:
RLIM localizes in the nuclear and cytoplasmic compartments in cells. (A) RLIM is found predominantly in the nuclei of cells in culture. αT3, HeLa, MCF7, and HEK293 cells stained with antibodies directed against RLIM. (B) Western blot on total, nuclear, and cytoplasmic fractions prepared from αT3 cells using antibodies directed against RLIM, Lhx3, and tubulin. (C) Expression of RLIM in human breast epidermis. Note that RLIM is expressed in the nucleus and cytoplasm in the basal layer in undifferentiated human breast epithelial cells (red arrows), whereas differentiating cells (black arrows) express RLIM mainly in the nucleus. Scale bar, 20 μm. (D) Immunocytochemical staining of undifferentiated and differentiated primary HFK cells with RLIM antibodies. Specific staining along the plasma membrane in undifferentiated cells is indicated by arrows.
RLIM shuttles between nucleus and cytoplasm
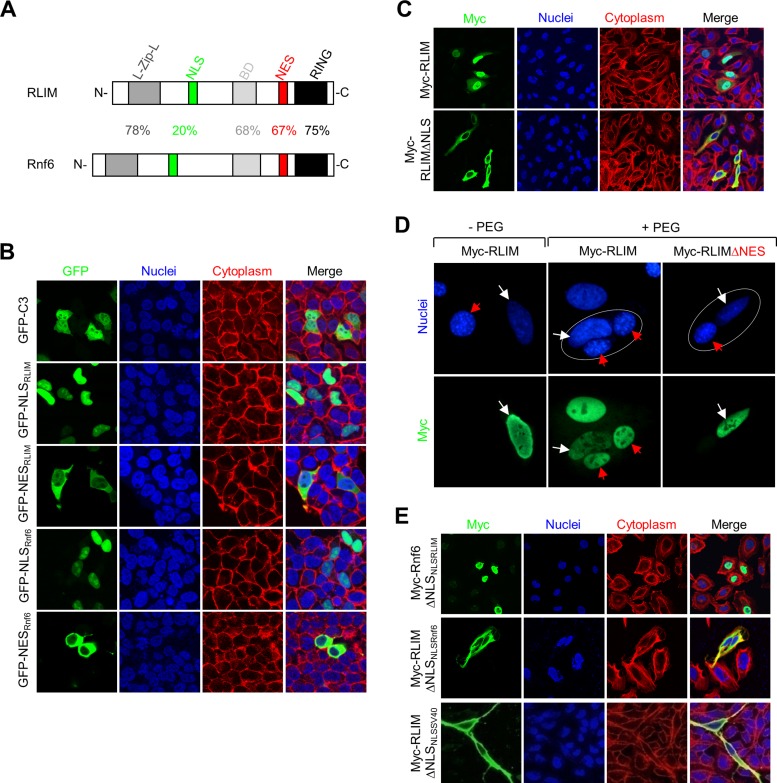
Sequence comparison between RLIM and the closely related ubiquitin ligase Rnf6 revealed that both proteins display significant amino acid homologies over large parts of their sequence, including the RING finger, a basic domain (BD), and a leucine-zipper-like domain (L-Zip-L; Figure 2A). Furthermore, both proteins contained putative NLS and NES regions. However, whereas the putative NES was highly conserved between both proteins, the predicted NLS regions contained much less sequence homology (Figure 2A). We investigated the functionality of the putative NLS and NES in RLIM and Rnf6 by generating NLS–green fluorescent protein (GFP) fusion proteins. Whereas transfected GFP protein localizes to both the cytoplasm and nucleus, fusing the NLS domain of RLIM or Rnf6 to GFP resulted in strictly nuclear localization of the respective NLS-GFP fusion proteins (GFP-NLSRLIM and GFP-NLSRnf6, respectively; Figure 2B). Similarly, the GFP-NESRLIM and GFP-NESRnf6 fusion proteins localized to the cytoplasm. Furthermore, an RLIM-deletion mutant protein lacking the putative NLS was strictly localized to the cytoplasm (Figure 2C). These results show that the NLS is required for the nuclear localization of RLIM and that the NLS and NES are functional, suggesting that RLIM shuttles between compartments. To address this question, we noted that the NES of RLIM and Rnf6 displayed high sequence homology to the NES of the Rev protein from HIV, which is exported via exportin 1 (CRM1 receptor; Pollard and Malim, 1998). Given that leptomycin B (LMB) inhibits protein export via CRM1 (Kudo et al., 1999), we tested whether LMB affects the cytoplasmic localization of GFP-NESRLIM and GFP-NESRnf6 proteins. Consistent with protein shuttling, LMB treatment of GFP-NESRLIM–and GFP-NESRnf6–transfected cells resulted in nuclear localizations of both proteins, similar to the GFP-NESRev/HIV protein used as positive control (Supplemental Figure S1). To obtain definitive evidence that RLIM is a shuttling protein, we performed a heterokaryon assay and transfected human HeLa cells with Myc-RLIM or as control Myc-RLIMΔNES and cultured transfected cells together with untransfected mouse NIH3T3 cells. Both cell types can be distinguished by 4′,6-diamidino-2-phenylindole staining, in which nuclei of NIH3T3 cells appear smaller and display a more speckled staining pattern than the nuclei of HeLa cells (Katahira et al., 1999). After mixing of transfected and untransfected cells, heterokaryons were generated using polyethylene glycol (PEG). We observed Myc staining in the nuclei of untransfected NIH3T3 cells only in PEG-treated heterokaryons that contained Myc-RLIM transfected HeLa cells and not in heterokaryons containing HeLa cells transfected with Myc-RLIMΔNES (Figure 2D). Indeed, Myc-RLIM staining in NIH3T3 cells was observed as early as 20 min, indicating rapid RLIM shuttling. We next examined protein localization in which the endogenous NLS was replaced in the context of the full-length protein. Replacing the endogenous NLS from Rnf6 with that of RLIM generated a protein Rnf6ΔNLSNLSRLIM that localized to the nucleus, whereas replacing the NLS in RLIM with the one of Rnf6 (RLIMΔNLSNLSRnf6) was not sufficient for nuclear localization and targeted the resulting protein to the cytoplasm (Figure 2E). In addition, replacement of the endogenous NLS in RLIM with the NLS from simian virus 40 (SV40) failed to target the resulting protein to the nucleus. The functionality of the SV40 NLS was verified in GFP fusion experiments (data not shown). A similar replacement of NES domains in the context of full-length protein did not lead to a change in the localization of the proteins RLIMΔNESNESRnf6 and Rnf6ΔNESNESRLIM (data not shown). Taken together, these results demonstrate the nucleocytoplasmic shuttling of RLIM in cells and show that the NLS in RLIM is uniquely responsible for its nuclear localization and function.
FIGURE 2:
RLIM is a nucleocytoplasmic shuttling protein. (A) Structure of RLIM and Rnf6 proteins showing a leucine-zipper-like domain (L-Zip-L), putative NLS (green), NES (red), BD (gray), and RING finger (RING, black). The percentage of amino acid identity between each region is indicated. (B) The NLS and NES in RLIM and Rnf6 are functional. Plasmids expressing GFP-NLS and GFP-NES sequences of RLIM (NLSRLIM and NESRLIM) and Rnf6 (NLSRnf6 and NESRnf6) were overexpressed in HeLa cells, and the fusion proteins were visualized using anti-GFP antibodies (green). (C) The putative NLS of RLIM is required for nuclear localization. Staining of Myc-tagged full-length or mutant protein deleted of the putative NLS domain (Myc-RLIMΔNLS) was overexpressed in HeLa cells and stained with anti-Myc antibodies (green). Nuclei were stained with Toto 3 dye (blue), and the cytoplasm was stained for actin with rhodamine–phalloidin (red). (D) RLIM shuttles between the nucleus and cytoplasm in heterokaryon assays. Human HeLa cells were transfected with Myc-RLIM or Myc-RLIMΔNES lacking the NES (white arrows) and fused with untransfected mouse NIH3T3 cells (red arrows). Arrows in PEG-treated cells point at nuclei in heterokaryons (circled). Note Myc-RLIM in the nuclei of NIH3T3 cells in a heterokaryon, indicating protein shuttling, whereas Myc-RLIMΔNES is retained in the nuclei of transfected HeLa cells. (E) The unique NLS of RLIM confers nuclear localization of RLIM. Cells transfected with a Myc-Rnf6 construct in which the NLS was replaced with that of RLIM (Myc-Rnf6ΔNLSNLSRLIM) and a Myc-RLIM construct in which the NLS was replaced with that of Rnf6 (Myc-RLIMΔNLSNLSRnf6) or SV40 (Myc-RLIMΔNLSNLSSV40). Cells were stained with anti-Myc antibody (green), Toto 3 dye (blue), and rhodamine-phalloidin (red).
The NLS of RLIM is phosphorylated in conserved RSRSP domains
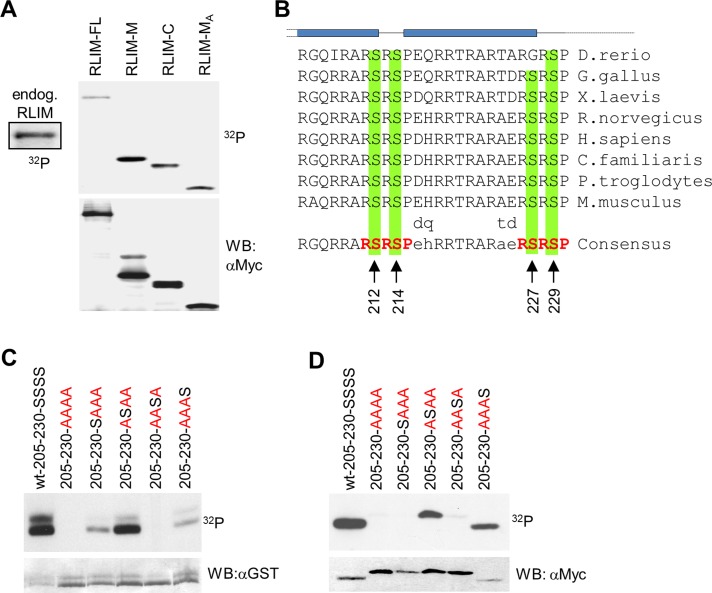
Because ubiquitin ligases are often regulated via posttranslational modifications such as phosphorylation (Pickart, 2001; Bach and Ostendorff, 2003), we examined whether RLIM is phosphorylated in cells. Indeed, immunoprecipitation of endogenous RLIM from cell extracts of metabolically labeled cells using specific antibodies directed against RLIM revealed that RLIM is phosphorylated in cells (Figure 3A). Mapping experiments showed that RLIM full-length and RLIM mutants RLIM-M (208–403) and RLIM-MA (208–305), containing the NLS, and RLIM-C (403–600), containing the C-terminal portion of RLIM, including the RING finger, are all phosphorylated (Figure 3A).
FIGURE 3:
The NLS of RLIM is phosphorylated at conserved Ser residues. (A) Metabolic labeling of transfected Myc-tagged RLIM protein mutants RLIM-M (amino acids [aa] 206–423), RLIM-C (aa 403–600), and RLIM-MA (aa 206–305) in cells. Note strong phosphorylation of RLIM-M and RLIM-MA deletion mutants, both containing the NLS. (B) Comparison of NLS of RLIM in vertebrates (205–230). Note the high conservation of the NLS. Serines at positions 212, 214, 227, and 229 that may serve as potential phosphorylation sites are indicated by arrows. RSRSP motifs are indicated in red. Predicted α-helices (GOR IV, PSIPRED, and Jnet) are indicated in blue. (C) In vitro phosphorylation of the mouse RLIM-NLS (205–230) fused to GST and RLIM-NLS Ser-to-Ala mutant proteins in which all serine residues were replaced by alanine residues (AAAA, in red) or containing only one serine residue at the indicated position. Note the strong serine phosphorylation at position 214. As loading control the same membrane was hybridized with an antibody directed against GST. (D) Metabolic labeling of Myc-tagged mouse RLIM-NLS (205–230) Ser-to-Ala mutations after transfection of corresponding expression plasmids. Note strong serine phosphorylation at positions 214 and 229. As a loading control the same membrane was hybridized with an antibody directed against Myc.
Owing to the very high degree of conservation of the NLS and secondary structure among different vertebrate species and the high frequency of sites for arginine- and/or proline-directed serine/threonine kinases within two RSRSP repeats (Figure 3B), we sought to determine whether these motifs are targets of phosphorylation and what functional role they might play in the regulation of RLIM. This was initially tested in vitro using purified glutathione S-transferase (GST) fusion proteins of the wild-type RLIM-NLS (amino acids 205–230), which contains all four serines (SSSS) as substrates for a kinase-enriched protein fraction from Hs578T human breast cancer cells. Of importance, mutation of all four serine residues to alanine (205-230-AAAA) resulted in complete inhibition of RLIM-NLS phosphorylation in vitro (Figure 3C). RLIM-NLS mutants containing the individual serine residues displayed strong phosphorylation of Ser-214 (205-230-ASAA), which is located between two predicted α-helices (Figure 3B), and weak phosphorylation of Ser-212 (205-230-SAAA) and Ser-229 (205-230-AAAS). To verify the results of the in vitro kinase assays, we transfected expression vectors for myc-epitope–tagged fusion proteins of the various RLIM-NLS mutant proteins into HEK293 cells and tested their phosphorylation by metabolic labeling. As shown in Figure 3D and similar to the in vitro kinase data, Ser-214 (205-230-ASAA) and Ser-229 (205-230-AAAS) in the RLIM-NLS are highly phosphorylated in cells. These data identify RLIM as a protein phosphorylated in cells at many sites, including the NLS.
Phosphorylation at S214 occurs during differentiation of human epithelial tissues
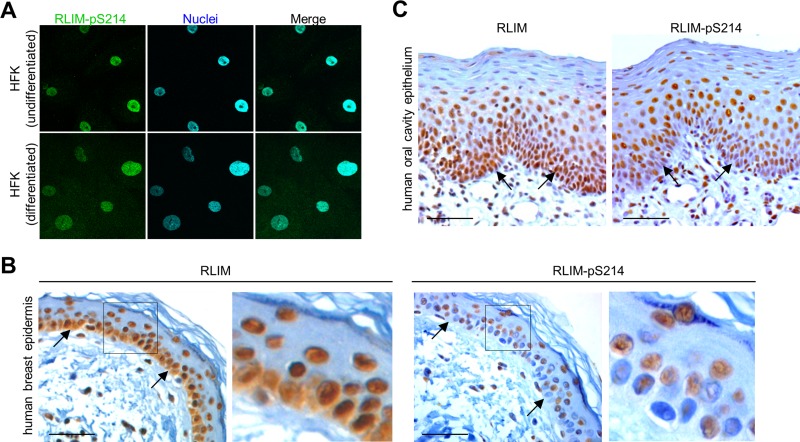
It has been shown that CLIM cofactors are widely expressed in epithelial tissues, in particular in breast epithelia, where, in conjunction with LMO4, they are believed to contribute to mammary gland differentiation and the occurrence of breast cancer (Visvader et al., 2001; Wang et al., 2004). Given that CLIM cofactors and LMOs serve as substrate proteins for RLIM-mediated ubiquitinations (Ostendorff et al., 2002) and RLIM is expressed in breast epithelial cells and human estrogen receptor–positive breast cancer (Johnsen et al., 2009), we further examined RLIM expression in human epithelial tissues and tested whether S214 in RLIM is phosphorylated in vivo. To test this, we generated a polyclonal phospho-specific antibody recognizing phosphorylated S214 (RLIM-pS214) in the RLIM-NLS (205–230) and, as a control, phosphorylation-independent antibodies recognizing the same region of RLIM. The specificity of the RLIM-pS214 antibody was demonstrated using competition experiments in immunohistochemical stainings. We hybridized sections of human microinvasive squamous cell carcinomas or mammary gland with RLIM-pS214 and the phosphorylation-independent antiserum, adding excess amounts of synthetic RLIM-NLS peptides containing a phosphorylated S214 (RLIM-NLS-205-230-pS214) or an unphosphorylated RLIM-NLS peptide (RLIM-NLS-205-230). Whereas the signals of the non–phospho-specific antiserum were inhibited in the presence of RLIM-NLS-205-230-pS214 and RLIM-NLS-205-230 peptide (Supplemental Figure S2A), signals obtained by the phospho-specific RLIM-pS214 antibody were competed by adding excess amounts of the phosphorylated but not the unphosphorylated peptide (Figure S2, B and C), confirming the specificity of the antibodies.
Based on use of a non–phospho-specific RLIM antibody on HFK cells, our results showed that in undifferentiated HFK cells a significant portion of RLIM localizes in the cytoplasm, whereas in differentiated cells RLIM mainly localizes in the nucleus (Figure 1D). To find the cellular compartment(s) in which the S214-phosphorylated RLIM localizes, we first performed immunocytochemistry using our antisera on HFK cells. In contrast to the non–phospho-specific RLIM antibody (Figure 1D), we detected exclusively nuclear RLIM staining using the RLIM-pS214 antibodies in both undifferentiated and differentiated cells (Figure 4A), consistent with a role of S214 phosphorylation for nuclear localization. Similarly, in HEK293 cells we also detected strictly nuclear signals using the phospho-specific antibody, whereas the phosphorylation-independent antibody yielded weak but detectable staining in the cytoplasm of cells (Supplemental Figure S2D). Next we tested RLIM S214 phosphorylation in human breast epidermis and human oral cavity epithelium specimens. The basal epithelia layer of normal breast epidermis showed strong staining using the phosphorylation-independent RLIM antibodies, whereas the phospho-specific RLIM-pS214 antibodies showed less staining of this layer (Figure 4B). In addition, whereas the RLIM-pS214 antibody showed only nuclear staining, we also observed cytoplasmic staining in cells of the basal layer using the phosphorylation-independent RLIM antibody (Figures 1C and 4B), corroborating results obtained in HFK cells (compare with Figures 1D and 4A). Similarly, in stainings of human oral cavity epithelium using the phosphorylation-independent RLIM antibody, we observed strong RLIM expression in oral epithelia, including basal layers (Figure 4C). Staining a parallel section with the phospho-specific RLIM-pS214 antibody, however, revealed significantly less staining in the basal layer, whereas suprabasal layers that contain differentiating keratinocytes displayed signal strengths similar to those obtained with the phosphorylation-independent RLIM antibody. Combined, these results indicate that pS214 phosphorylation occurs during the differentiation of epithelial tissues.
FIGURE 4:
Nuclear RLIM is phosphorylated at S214. (A) Immunocytochemical staining of undifferentiated and differentiated primary HFK cells with phospho-specific peptide antibodies specifically recognizing the NLS of RLIM phosphorylated on S214 (RLIM-pS214). Note exclusively nuclear staining in cells. (B, C) Immunohistochemical sections of adult human epithelia were stained with non–phospho-specific RLIM antibodies or phospho-specific RLIM-pS214 antibodies. Human breast epidermis is shown in B. Right, magnification of boxed area. Human oral cavity epithelium is shown in C. Note weak staining of basal layer (arrows) in sections stained with the phospho-specific RLIM-pS214 antibodies. Scale bars, 25 μm.
Phosphorylation at S214 is required for nuclear localization
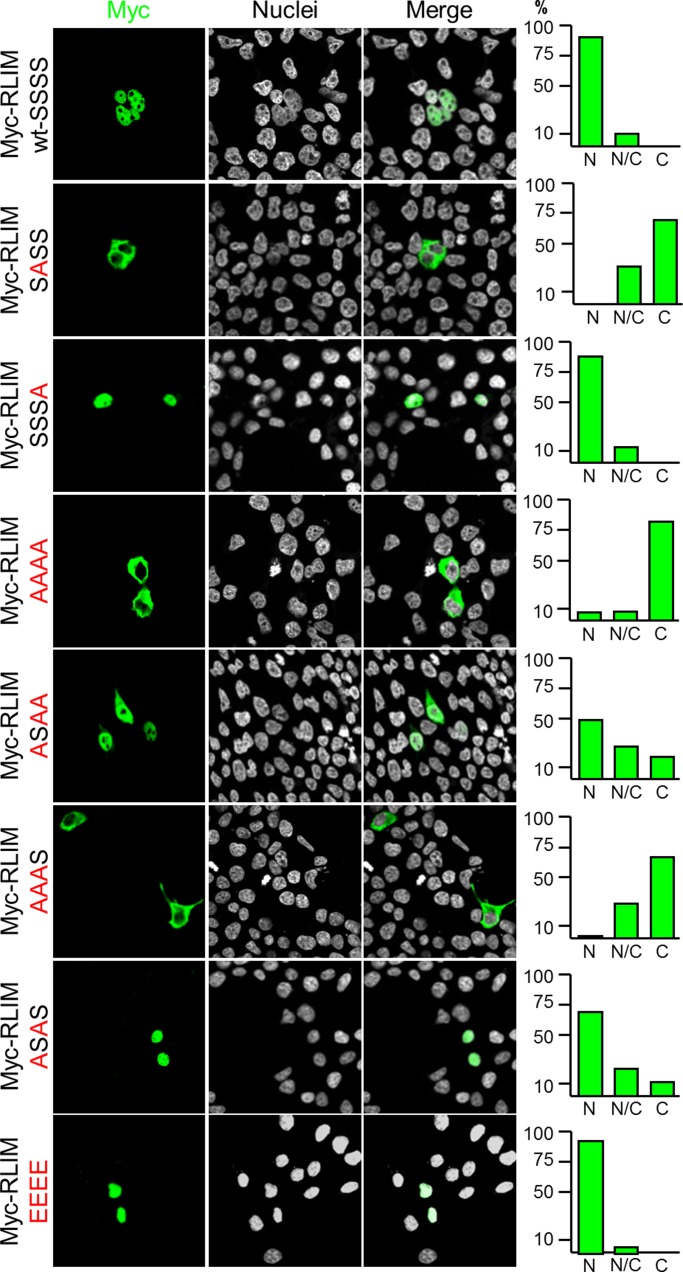
We first examined whether NLS phosphorylation affects the cellular localization of RLIM, using NLS mutant proteins in the context of the isolated NLS. Comparing the localization of the wild-type GFP-NLS fusion protein with a GFP-NLS fusion protein in which all four serine residues in the NLS were replaced with alanine (AAAA) showed more cytoplasmic localization for the AAAA mutant protein, indicating that serine residues in the NLS are involved in the regulation of nuclear localization (Supplemental Figure S1B). We next generated several Ser-to-Ala mutations in the context of the full-length RLIM protein and tested their distribution in transfected cells. Results of these experiments showed that the S214 is required for nuclear localization, as the S214A mutant protein (SASS) localized mainly in the cytoplasm, whereas mutating the analogous serine (S229) in the second RSRSP motif (SSSA) had little effect (Figure 5). Conversely, nuclear localization was partially restored when only S214 was present (ASAA) when compared with the AAAA mutant, which localized mainly in the cytoplasm. Efficient nuclear localization, similar to SSSS-wild type, was detected in the ASAS mutant protein, which contained both S214 and S229. A mutant RLIM protein in which the serine residues were replaced by glutamic acid (EEEE) to mimic phosphorylation showed a strong bias toward nuclear localization. In this context, mutating both prolines in the RSRSP motifs to alanines mildly decreased the nuclear localization, and, not surprisingly, removing the first RSRSP motif (Δ211–215) completely abolished nuclear localization of mutant proteins (data not shown). Combined, these results show that S214 of RLIM is most crucial for nuclear localization, and other residues in the RSRSP motifs play additional roles.
FIGURE 5:
The RLIM-NLS regulates localization of RLIM in a phosphorylation-dependent manner. Expression of Myc-tagged RLIM containing various Ser-to-Ala mutations at positions 212, 214, 227, and 229 in their NLS in the context of the full-length protein. Left, representative images of transfected cells; right, statistical analyses of at least 100 transfected cells. Note that replacement of Ser-214 with alanine (SASS) strongly inhibits nuclear localization and RLIM containing S214 and S229 (ASAS) is mainly localized in the nucleus, similar to wild-type RLIM (SSSS).
Nucleocytoplasmic shuttling regulates the functional activity of RLIM/Rnf12
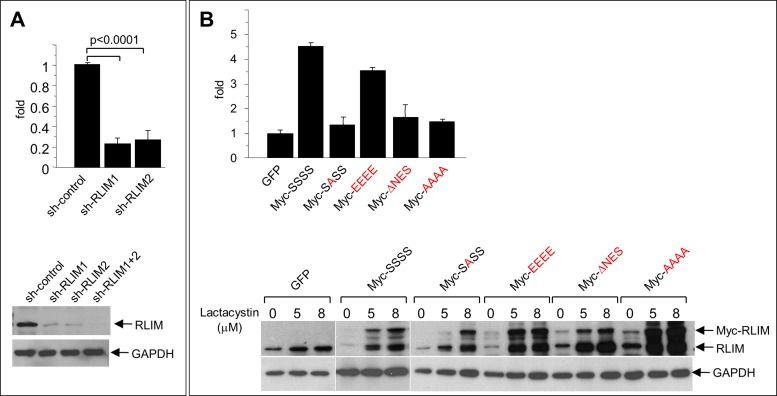
We first investigated the functional importance of RLIM nucleocytoplasmic shuttling for cell migration. Indeed, in agreement with previously published results (Huang et al., 2011), knocking down endogenous RLIM levels in MCF7 cells using two different short hairpin RNAs caused cells to migrate significantly slower in Transwell migration assays (Figure 6A), and knocking down RLIM in MDA-MB-231 breast cancer cells via small interfering RNAs (siRNAs) yielded similar results (Supplemental Figure S3). To examine the importance of RLIM localization for its ability to promote cell migration, we tested the migration potential of MCF7 cell lines that stably expressed RLIM mutant proteins in the context of the full-length protein via lentiviral infections. Whereas overexpression of wild-type (SSSS) and EEEE proteins promoted cell migration, the NES-deleted RLIM or NLS mutant SASS and AAAA proteins were no longer able to enhance cell migration (Figure 6B). Expression of Myc-tagged wild-type and mutant proteins was verified in Western blots. To better assess and compare cellular levels of endogenous and induced exogenous RLIM, we added increasing concentrations of the proteasome inhibitor lactacystin. Because the ΔNES and both NLS mutants, which locate in the nucleus (Figure 3D) and cytoplasm (Figure 6A), respectively, fail to promote cell migration, these results link protein shuttling with cell migration.
FIGURE 6:
Shuttling of RLIM is important for its function of promoting cell motility. (A) Knockdown of RLIM inhibits cell motility. MCF7 cells were infected with lentivirus containing control or short hairpin RNA against RLIM. Top, motility of infected MCF7 cells as measured in Transwell migration assays. Averages ± SEM for five independent measurements. Bottom, representative Western blot of infected cell extracts. The same blot was hybridized with antibodies against RLIM and GAPDH. (B) Function of RLIM to promote cell motility is dependent on nucleocytoplasmic shuttling. MCF7 cells were infected with lentivirus containing RLIM, various RLIM-NLS mutants, and, as control, GFP. Top, motility of infected MCF7 cells as measured in Transwell migration assays. Averages ± SEM for four independent measurements. Note that overexpression of Myc-RLIM wild type but not shuttling-deficient Myc-RLIM mutant proteins promotes cell migration. Bottom, representative Western blot of infected cell extracts. To compare cellular levels of endogenous with lentivirus-mediated expression of RLIM, cells were treated with various concentrations of the proteasome inhibitor lactacystin for 6 h. The same blot was hybridized with antibodies against RLIM recognizing exogenous and endogenous protein and GAPDH.
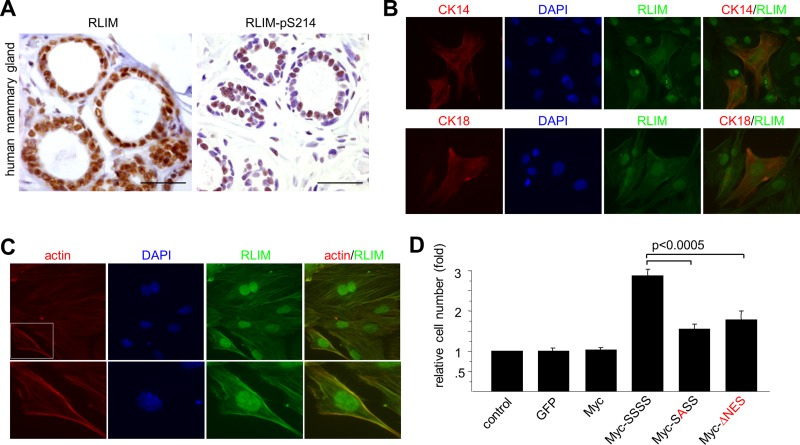
We next investigated the importance of RLIM S214 phosphorylation and nucleocytoplasmic shuttling for its functions in mammary glands. Hybridizing sections with phospho-specific RLIM-pS214 and phosphorylation-independent RLIM antibodies revealed that, similar to other epithelial cell types (Figure 4), RLIM is phosphorylated in nuclei of mammary epithelial cells (Figure 7A). Immunocytochemical costaining of primary mammary epithelial cells showed that RLIM is localized in both nucleus and cytoplasm of basal CK14-positive and luminal CK18-positive cell types (Figure 7B). Moreover, RLIM costainings with rhodamine–phalloidin revealed that in the cytoplasm RLIM colocalizes with actin filaments (Figure 7C). This was confirmed in costainings using actin antibodies (data not shown), suggesting that RLIM shuttling occurs between sites on the actin cytoskeleton and the nucleus. Using a conditional Rnf12-KO mouse model, we recently reported a crucial function of RLIM in the survival of mammary milk-producing alveolar cells (Jiao et al., 2012). Indeed, alveolar cells lacking RLIM undergo apoptosis when differentiated in culture, and cell death can be partially rescued by expressing exogenous RLIM via lentiviral infections. Thus, because these cells require RLIM expression for their survival, we tested the rescue potential of exogenous RLIM shuttling mutants in primary mammary epithelial cells lacking RLIM. To carry out these experiments, we isolated primary mammary epithelial cells of adult virgin female mice in which the floxed Rnf12 alleles are targeted to the mammary gland using transgenic mice that express Cre recombinase (Cre) under the control of the mouse mammary tumor virus (MMTV) long terminal repeat (Wagner et al., 1997; Jiao et al., 2012; Rnf12fl/fl x MMTV-Cre). Cells were then differentiated in culture and infected with lentivirus expressing the shuttling-deficient RLIM mutants or, as control, wild-type RLIM. Indeed, whereas the number of cells did not change upon lentiviral expression of GFP or a Myc tag alone when compared with uninfected cells, physiological levels of exogenous Myc-RLIM (SSSS) resulted in significantly increased cell numbers (Figure 7D). Because RLIM expression does not affect proliferation (Jiao et al., 2012), these results reflect a rescue from apoptosis. The shuttling-deficient Myc-SASS or Myc-ΔNES mutant RLIM, however, displayed a markedly lower rescue potential than wild-type RLIM (Figure 7D and Supplemental Figure S4). Combined, these results indicate an important contribution of nucleocytoplasmic shuttling of RLIM in its ability to promote alveolar cell survival.
FIGURE 7:
Nucleocytoplasmic shuttling regulates RLIM's ability to act as alveolar survival factor. (A) RLIM is phosphorylated in nuclei of mammary epithelial cells. Immunohistochemical sections of adult human mammary gland were stained with non–phospho-specific antibodies recognizing RLIM (left) or phospho-specific peptide RLIM-pS214 antibodies (right). Scale bars, 25 μm. (B) Dual localization of RLIM in the nucleus and cytoplasm in basal and luminal primary mammary cells. Primary mammary epithelial cells were isolated from an adult wild-type virgin female mouse and costained with antibodies directed against RLIM and basal marker CK14 or luminal marker CK18. (C) RLIM is associated with the actin cytoskeleton in primary mammary cells. Primary mammary epithelial cells were isolated from an adult wild-type virgin female mouse and costained with antibodies directed against RLIM and rhodamine–phalloidin. Boxed area is shown in higher magnification. (D) Primary mammary epithelial cells that lack RLIM were isolated from adult Rnf12fl/fl x MMTV-Cre virgin female mice. Cells were infected with lentivirus expressing Myc-tagged wild-type (SSSS) and shuttling-deficient RLIM mutant proteins (SASS) and ΔNES proteins. Cells infected with lentivirus expressing GFP and Myc-tag and uninfected cells were used as control. Averages ± SEM for four independent measurements.
DISCUSSION
In female mice, RLIM/Rnf12 appears to play essential, sex-specific roles in developmental processes such as XCI (Shin et al., 2010) and mammary alveolar cell survival and involution (Jiao et al., 2012). Other roles that RLIM might play are nonessential in mice because male mice carrying a germline Rnf12 KO appear healthy and are fertile (Shin et al., 2010). Thus, because RLIM is widely expressed in many epithelial and nonepithelial tissues in mouse embryos and adult animals (Bach et al., 1999; Ostendorff et al., 2000, 2006), the biological importance of RLIM in most of these tissues is unclear. Nevertheless, in cells RLIM reportedly contributes to apparently nonessential cellular functions such as cell migration (Huang et al., 2011), adjustment of levels of transcription factors/cofactors, assembly of multiprotein complexes, and transcriptional activity of several classes of transcription factors (Bach et al., 1999; Ostendorff et al., 2002; Gungor et al., 2007; Johnsen et al., 2009). Our finding that this protein is localized in both nucleus and cytoplasm in cells and shuttles between these cellular compartments is likely relevant for all of these functions.
RLIM phosphorylation
The NLS in RLIM containing two core RSRSP motifs is highly conserved among vertebrate species (Figure 3B). Secondary structural predictions place two α-helices in the NLS region, which are interrupted by the RSRSP motifs. A BLAST analysis of the RSRSP motif revealed many nuclear proteins that contain tandem RSRSP motifs (Supplemental Figure S5). Some of these proteins belong to the Ser/Arg-rich (SR) class of proteins known to contain many RS repeats (Graveley, 2000), whereas many others are not considered SR proteins. Of interest, nucleocytoplasmic shuttling of several of these proteins has been reported. The RNA-binding protein 1 (Lykke-Andersen et al., 2001) and the SR protein SF2/ASF both shuttle between the cytoplasm and nucleus, and, similar to RLIM, this shuttling is phosphorylation dependent for SF2 (Sanford et al., 2005). Thus the RSRSP motifs likely serve as a prototypic sequence for phosphorylation-dependent nuclear localization. Because Rnf6 contains functional NLS and NES (Figure 2B), it is likely that these also mediate nucleocytoplasmic shuttling. However, the RLIM NLS is unrelated to that of Rnf6, indicating divergent mechanisms that regulate protein shuttling.
In most tissues during mouse development (Ostendorff et al., 2006) and in all cell lines tested, cellular RLIM levels appear highest in the nucleus, where it is phosphorylated at S214. This argues that the kinase responsible for the NLS phosphorylation is widely expressed and/or that many different kinases may phosphorylate the RSRSP motif. Indeed, >50 kinases are predicted to phosphorylate the S214 by the PhosphoNET kinase predictor (www.phosphonet.ca), and phosphorylation was confirmed in human RLIM using quantitative phosphoproteomics (Olsen et al., 2010). This, combined with the finding that S214 phosphorylation is required for nuclear import, suggests that RLIM is phosphorylated at S214 in the cytoplasm, and this phosphorylation event triggers efficient binding to import proteins/nuclear import. However, the exact mechanisms regulating S214 phosphorylation and dephosphorylation and the nuclear import mechanisms remain unclear.
Our results show that besides the RSRSP motifs in the NLS, RLIM is phosphorylated at several different sites in cells (Figure 3A), suggesting that activities of RLIM other than its cellular localization also may be regulated by posttranslational phosphorylation. Indeed, a common theme emerging in recent years is that the activity of many ubiquitin ligases is regulated via posttranslational modifications, including phosphorylation (Hunter, 2007).
Relevance of RLIM shuttling for its biological functions
In this article, we provide evidence that protein shuttling occurs between sites of cytosolic actin filaments and the nucleus. Indeed, we show important functions of RLIM shuttling for alveolar cell survival and cell migration. The process of shuttling appears to be of high importance to regulating these functions, as expression of wild-type protein promotes both functions, but expression of cytoplasmic or nuclear shuttling-deficient RLIM mutant protein is much less efficient in mediating these processes. These results indicate that RLIM serves as a signaling molecule, which carries signals between these compartments. The fact that cell migration is mediated by restructuring of the actin cytoskeleton with which RLIM is associated (Figure 7B) suggests that RLIM/Rnf12 exerts its function on proteins localized with the actin cytoskeleton that mediate this process. In this context it is interesting to note that the RLIM-related protein Rnf6 regulates local actin dynamics in axonal growth cones in neurons by targeting LIM kinase 1 (LIMK1) for proteasomal degradation (Tursun et al., 2005). However, although RLIM can interact with the LIM domain of LIMK1 (Bach et al., 1999) and mediate its ubiquitination in vitro (Tursun et al., 2005), our results obtained by Western blotting and immunostainings in cells depleted of RLIM indicate that cellular LIMK1 is not regulated by RLIM (data not shown). The finding that cytoplasmic and nuclear shuttling-deficient RLIM mutant proteins (RLIM-SASS and RLIMΔNES, respectively) are much less efficient in promoting cell migration and alveolar cell survival indicates that both cytoplasmic and nuclear functions of RLIM are important. Thus RLIM acts as a signaling molecule that synchronizes functions in cellular compartments, and our results provide strong evidence that this is important to carry out these functions. Indeed, dual cytoplasmic and nuclear distribution has been observed for several proteins, including cytoplasmic LIM proteins, which are known to shuttle and signal between cellular adhesion sites and the nucleus. Similar to RLIM, these proteins often act as transcriptional cofactors in the nucleus to modulate transcription factor activity (Bach, 2000; Kadrmas and Beckerle, 2004). In this context, it is interesting to note that RLIM is detected along the plasma membrane in cells located in the epidermal basal cell layer and to a lower extent in mammary cell types (Figures 1, C and D, and 7, B and C).
Besides its crucial role in alveolar cell survival, it has been suggested that RLIM triggers involution via targeted proteasomal degradation by weaning-induced autoubiquitination (Jiao et al., 2012). This scenario would be analogous to the signal-induced apoptosis in thymocytes mediated by the RING finger ubiquitin ligase inhibitor of apoptosis (IAP), which also serves as a survival factor in this cell type. Of interest, IAP proteins localize to the cytoplasm, where they interact with a variety of proteins, including caspases, leading to their inhibition and thereby promoting cell survival (Vucic et al., 2011). Consistent with nucleocytoplasmic shuttling involved in RLIM's survival function, our results show RLIM localization in both compartments. Because RLIM rapidly shuttles (Figure 2D and data not shown) and RLIM levels in alveolar cells drop dramatically within 6 h upon forced weaning (Jiao et al., 2012), it is conceivable that to deplete cellular RLIM levels during involution it would be sufficient to target RLIM for degradation in only one of the compartments (e.g., the cytoplasm). Hence, to further decipher the role of RLIM in promoting alveolar cell survival and trigger involution, it will be interesting to elucidate the protein network with which RLIM is associated in the cytoplasm of mammary epithelial cells. Whether nucleocytoplasmic shuttling is important also for RLIM's functions during XCI (Jonkers et al., 2009; Shin et al., 2010; Barakat et al., 2011) is not clear. Because RLIM/Rnf12 does not appear enriched at inactive X chromosomes and X chromosomes that undergo XCI, however, the colocalization of RLIM with actin in the cytoplasm (Figure 7B and data not shown) is intriguing, as actin exists also in the nucleus and may therefore be involved in the nuclear distribution of RLIM. Indeed, nuclear β-actin/actin-related proteins participate in chromatin-remodeling and histone acetyltransferase) complexes, thereby influencing long-range chromatin organization (Pederson, 2008; Visa and Percipalle, 2010). Moreover, the two highly conserved core RSRSP motifs are also found in the zebrafish RLIM/Rnf12, and functions of this ubiquitin ligase during embryonic stem cell fate have been reported via proteasomal targeting of Smad4, which is localized in both cytoplasmic and nuclear compartments (Zhang et al., 2012). Thus the regulated nucleocytoplasmic shuttling of RLIM is likely to have important developmental functions in this organism.
In summary, we propose that phosphorylation-dependent nucleocytoplasmic shuttling of RLIM functionally coordinates cellular compartments during cell migration and for the survival of alveolar cells in the mammary gland.
MATERIALS AND METHODS
Plasmids/lentiviral constructs
Plasmids used in this work were RLIMΔNLS (Δ206–226) in pCS2MT and pGEX-KGK, RLIMΔNES (Δ502–513) in pCS2-MT and pGEX-KGK, GFP-NLSRnf6 (186–208) in pEGFP-C3 (Clontech), GFP-NESRnf6 (566–577) in pEGFP-C3, GFP-NLSRLIM (206–226) in pEGFP-C3, GFP-NESRLIM (501–513) in pEGFP-C3, pCS2-MT-Rnf6ΔRING (Δ589–667), pCS2-MT-Rnf6ΔRINGΔNES (Δ566–577; Δ589–667), pCS2-MT-RLIMΔRINGΔNLS (Δ206–226; Δ544–600), and previously described cDNA clones RLIM RLIMΔRING, N-RLIM, M-RLIM, and C-RLIM in eukaryotic and bacterial expression vectors (Bach et al., 1997, 1999; Ostendorff et al., 2002). RLIM serine point mutations were generated in the context of the pCS2-MT expression plasmid. Lentiviral infections were performed using lentiviral vectors (pLenti CMV/TO Puro) expressing NLS/NES mutant RLIM as described (Campeau et al., 2009). Correct sequence was verified via DNA sequencing.
Antibodies
Polyclonal antisera raised in rabbit and guinea pig against mouse RLIM and Lhx3 have been described (Ostendorff et al., 2002, 2006). Phospho-specific and phosphorylation-independent S214-RLIM antibodies (α-RLIM-pS214 and NP-8043, respectively) were generated and verified by enzyme-linked immunosorbent assay by Biogenes (Berlin, Germany). Anti-Myc polyclonal (Santa Cruz Biotechnology, Santa Cruz, CA) and monoclonal 9E10 (Roche, Mannheim, Germany) antibodies and anti-HA.11 monoclonal antibody (BAbCO, Richmond, CA) were used to detect/immunoprecipitate Myc- and hemagglutinin (HA)-tagged proteins, respectively. CK14 and CK18 antibodies (Abcam, Cambridge, MA) were used in immunostainings, and anti-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were purchased from Sigma-Aldrich (St. Louis, MO) and Chemicon (Temecula, CA), respectively. Secondary antibodies were horseradish peroxidase–anti-mouse and horseradish peroxidase–protein A (Bio-Rad, Munich, Germany). Biotinylated secondary antibodies for the immunohistochemistry were goat anti-guinea pig (Vector Laboratories, Burlingame, CA) and goat anti-rat (MoBiTec, Göttingen, Germany). For immunocytochemistry we used Alexa 488 goat anti-mouse (MoBiTec), Alexa 488 goat anti–guinea pig (MoBiTec), Cy3 goat anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove, PA), Cy5 goat anti-rabbit (MoBiTec), and Cy5 goat anti-rat (MoBiTec). Rhodamine–phalloidin and Toto-3 dye (Molecular Probes, Eugene, OR) were used to visualize actin filaments and nuclei, respectively.
Cell culture, cell transfection, heterokaryon assay, and immunocytochemistry
Cell lines used in this work were CHO, Cos-7, HEK293T, HeLa, Hs578T, MCF7, MDA-MB-231, NIH3T3, and the pituitary gonadotrope cell line αT3 (Windle et al., 1990). HeLa and Hs578T cells were grown in DMEM or DMEM/F12, respectively (PAA Laboratories, Pasching, Austria), containing 10% (vol/vol) fetal bovine serum (FBS; PAA Laboratories) and 1× antibiotic-antimycotic solution (Invitrogen). Cells were seeded in 24-well or 10-cm plates and transfected at 80% confluence with various combinations of plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. Transient transfections, cotransfections, and immunocytochemical experiments were carried out as described (Ostendorff et al., 2002; Tursun et al., 2005). Cells were treated with leptomycin B (a kind gift of M. Yoshida, University of Tokyo) as reported previously (Kudo et al., 1999). The heterokaryon shuttling assay was performed essentially as described (Katahira et al., 1999). Briefly, HeLa cells expressing Myc-RLIM or Myc-RLIMΔNES were cocultured with NIH3T3 cells on coverslips for 4 h. After treatment with 10 μM cycloheximide for 30 min, the cells were fused by 50% PEG 1500 and incubated in the presence of cycloheximide for 1 h. The cells were fixed, permeabilized, and incubated with anti-Myc monoclonal antibody, followed by incubation with Alexa 488–labeled anti-mouse immunoglobulin G. HFK cells (a kind gift of E. Androphy, University of Massachusetts Medical School) were cultured in serum-free keratinocyte media (Gibco/Life Technologies, Carlsbad, CA) supplemented with epidermal growth factor and pituitary extract. Cells were differentiated by adding fetal calf serum to 10% final concentration. For transient transfections of siRNA, cells were transfected twice in a 12-h period using FuGENE (Roche) according to the manufacturer's instructions. siRNAs against human RLIM have been described (Ostendorff et al., 2002).
Western blotting
Cell extracts were harvested in RIPA buffer (phosphate-buffered saline, 1% [wt/vol] Nonidet P-40, 0.5% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS) containing 100 μg/ml phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml aprotinin, 10 μg/ml leupeptin, and 500 μM sodium orthovanadate. Protein was separated on a SDS–10% (wt/vol) polyacrylamide gel and blotted onto Protran nitrocellulose membranes (Schleicher and Schuell, Keene, NH). Primary antibodies were detected by enhanced chemiluminescence (GE Healthcare Life Sciences, Piscataway, NJ) using horseradish peroxidase–conjugated protein A (Bio-Rad).
In vitro kinase assays
A cellular fraction enriched for RLIM-NLS kinase activity was obtained by performing a 40–60% ammonium sulfate precipitation. Briefly, Hs578T breast cancer cells were grown to confluence and harvested in RIPA buffer containing protease and phosphatase inhibitors. Cell lysate was initially precipitated by adding saturated ammonium sulfate to a concentration of 40% (vol/vol), and the insoluble fraction was removed by centrifugation. After centrifugation, the active fraction was obtained by further centrifugation after an increase in the ammonium sulfate concentration to 60% (vol/vol). This fraction was then resuspended in kinase buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 10 mM MgCl2, 25 mM NaCl, 10 mM sodium β-glycerophosphate, 0.1 mM Na3VO4, 0.2 mM PMSF, 1 mM dithiothreitol, 10 μM ATP), aliquoted, and stored at −80°C until use in in vitro kinase assays. For in vitro kinase assays 1 μg of GST fusion protein was incubated with 1 μl of kinase-enriched Hs578T cell extract in a total volume of 30 μl of kinase buffer containing 1 μCi of [32P]γ-ATP (220 TBq/mmol). After 20 min of incubation at 30°C, Laemmli buffer was added and samples were boiled for 5 min, separated by SDS–PAGE, and blotted onto nitrocellulose membranes. The blots were then analyzed by autoradiography for the presence of phosphorylated proteins. Equal loading of GST fusion proteins was demonstrated by anti-GST Western blot analysis.
Metabolic labeling
HEK293 cells were grown in 6-cm plates and transfected with 10 μg of myc-fusion protein expression plasmids using the polyethanolamine procedure. Briefly, 10 μg of DNA was incubated with 500 μl of PBS containing 100 μM polyethanolamine. After 20 min of incubation at room temperature, transfection mixes were added to cells and incubated overnight. At 16 h after transfection, cells were incubated in serum-free/phosphate-free medium for 30 min and another 4 h in 2.5 ml of phosphate-free medium containing 0.25 mCi of [32P]orthophosphate. Protein was harvested in 1 ml of RIPA buffer containing phosphatase and protease inhibitors. Myc-fusion proteins were recovered by immunoprecipitation overnight with 1.5 μg of anti–myc 9E10 mouse monoclonal antibody (Roche). Immune complexes were recovered with Protein A/G Agarose (Santa Cruz Biotechnology), washed three times with RIPA buffer containing phosphatase and protease inhibitors, boiled in Laemmli buffer, subjected to SDS–PAGE analysis, and blotted onto nitrocellulose membranes. The blots were then analyzed by autoradiography for the presence of phosphorylated proteins. Equal expression and loading of myc-fusion proteins was demonstrated by anti-myc Western blot analysis with a polyclonal anti-myc antibody. Phosphorylation of endogenous RLIM protein was similarly investigated using a specific anti-RLIM monoclonal antibody and metabolically labeled protein extracts from MDA-MB-231 breast cancer cells, which express high levels of RLIM protein.
Human tissue samples and immunohistochemistry
The paraffin-embedded primary breast and head and neck cancer tissue specimens were obtained from the Institute of Pathology, University Medical Center Hamburg-Eppendorf, in an anonymous manner. Sections from Formalin-fixed, paraffin-embedded tissue were deparaffinized and subsequently subjected to heat pretreatment in antigen decloaker chamber (Decloaker; BioCarta, Hamburg, Germany) for 5 min at 120°C. After blocking of endogenous peroxidase activity, RLIM expression was visualized by using polyclonal rabbit antibodies NP-8043 (non–phospho-specific) antibody (1:500 dilution, 1 μg/ml), α-RLIM-pS214 (1:250 dilution, 1.3 μg/ml), and α-RLIM (1:1000 dilution; Ostendorff et al., 2002). Slides were incubated with the primary antibodies for 16–18 h at 4°C in a humidified chamber. Application of the primary antibody was followed by incubation with biotinylated goat anti-mouse/anti-rabbit immunoglobulins and streptavidin conjugated to horseradish peroxidase (ChemMate Detection Kit, Peroxidase/DAB, Rabbit/Mouse; DakoCytomation, Hamburg, Germany). 3,3′-Diamonobenzidine tetrahydrochloride chromogen solution and a substrate buffer containing hydrogen peroxide served as substrate system. Sections were counterstained with Mayer's hemalaun solution (Merck, Darmstadt, Germany) and permanently mounted. For negative control, addition of the primary antibody was omitted. Specificity of the immunoreaction was tested by blocking the antibodies with the appropriate peptides used for immunization. Therefore the primary antibody at the concentration used for immunostaining was preabsorbed with 5 μg/ml blocking peptide for 2 h at room temperature.
Cell migration assays
Cell migration assays were performed essentially as described (Chen et al., 2007). Briefly, cells were rinsed in serum-free media and plated into migration chambers (BD Biosciences, San Diego, CA) at a density of 5 × 104 cells/insert. Media containing 10% FBS as a chemoattractant was placed in the lower chamber. After 20 h, cells were fixed, stained with Giemsa reagent, and counted at 10× magnification. Five fields were counted at this magnification for each migration insert and the sum of these fields determined. The migration activity of control cells was normalized to 1 and the data plotted relative to the controls.
Alveolar cell survival assays
Alveolar survival assays were performed on primary mammary epithelial cells prepared from adult virgin females lacking mammary RLIM/Rnf12 (Rnf12fl/fl x MMTV-Cre) essentially as described (Jiao et al., 2012). Briefly, fourth inguinal mammary glands were isolated from 2-mo-old virgin mice and digested using collagenase/hyaluronidase for 3-6 h. The suspension was incubated with trypsin/dispase/DnaseI, followed by filtration through a 40-μm cell strainer. Cells were cultured overnight with differentiation medium (DMEM/F12 containing 10% FBS and 800 ng/ml prolactin). Fibroblasts were removed as described (Langdon, 2004). To test for apoptosis, primary mammary epithelial cells were isolated and differentiated for 1 d in FBS before lentiviral infections. Lentiviral infections of primary cells or MCF7 cells were performed using lentiviral vectors (based on pLenti CMV/TO Puro) as described (Campeau et al., 2009) expressing GFP, 6xMyc tag (Myc), Myc-RLIM, and Myc-RLIM mutants SASS, EEEE, AAAA, and ΔNES. Two days after infections, cells were aliquoted and seeded on 96-well plates for counting using the CellTiter 95R Aqueous One Solution Cell Proliferation Assay Kit (Promega, Madison, WI). Results were pooled from eight individual wells per measuring point. Three independent experiments were carried out. Wt and Rnf12 mutated mice (Jiao et al., 2012) were bred in the animal facility of the University of Massachusetts Medical School according to National Institutes of Health guidelines established by the Institute of Animal Care and Usage Committee.
Supplementary Material
Acknowledgments
We are grateful to J. Benanti, T. Fazzio, E. Luna, A. Mercurio, and L. Shaw for advice and helpful discussions; E. Androphy for the HFK primary human foreskin keratinocytes; M. Yoshida for the gift of leptomycin B; and T. Loening for providing the paraffin-embedded human tissue material. I.B. is a member of the UMass DERC Morphology Core (DK32520). This work was supported by the National Institutes of Health (R01CA131158 to I.B.) and the Deutsche Krebshilfe (109088 to S.A.J.).
Abbreviations used:
- GFP
green fluorescent protein
- HFK
human foreskin keratinocyte
- IAP
inhibitor of apoptosis
- KO
knockout
- LIMK1
LIM kinase 1
- NES
nuclear export sequence
- NLS
nuclear localization sequence
- RING
really interesting new gene
- RLIM
RING finger LIM domain–interacting protein
- Rnf6
RING finger protein 6
- Rnf12
RING finger protein 12
- XCI
X chromosome inactivation
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0239) on July 31, 2013.
*These authors contributed equally.
Present addresses: †Kunming Institute of Zoology, Chinese Academy of Science, Kunming, Yunnan 650223, China.
‡Department of General, Visceral and Thoracic Surgery, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany.
§Roche Pharma AG, 79639 Grenzach-Wyhlen, Germany.
||National Institute of Cancer Research, National Health Research Institutes, Miaoli, Taiwan.
#Department of Bioengineering, Osaka City University, Osaka 558-8585, Japan.
Author contributions: I.B. designed the study. B.J., N.T.-I., C.G., M.A.P., Y.-W.C., S.R., A.D., L.G.A., J.S., R.P., T.T., and S.A.J designed and performed the experiments. All authors analyzed and discussed the results. I.B. wrote the manuscript with input from B.J., B.C.L., and S.A.J.
REFERENCES
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Bach I, Ostendorff HP. Orchestrating nuclear functions: ubiquitin sets the rhythm. Trends Biochem Sci. 2003;28:189–195. doi: 10.1016/S0968-0004(03)00055-0. [DOI] [PubMed] [Google Scholar]
- Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, Glass CK, Andersen B, Izpisua Belmonte JC, Rosenfeld MG. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat Genet. 1999;22:394–399. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- Barakat TS, Gunhanlar N, Pardo CG, Achame EM, Ghazvini M, Boers R, Kenter A, Rentmeester E, Grootegoed JA, Gribnau J. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 2011;7:e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, Klimstra DS, Mongeau ME, Tatem JL, Boyartchuk V, Lewis BC. Loss of p53 and Ink4a/Arf cooperate in a cell autonomous fashion to induce metastasis of hepatocellular carcinoma cells. Cancer Res. 2007;67:7589–7596. doi: 10.1158/0008-5472.CAN-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IJcken W, Grootegoed JA, Gribnau J. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungor C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc Natl Acad Sci USA. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her YR, Chung IK. Ubiquitin ligase RLIM modulates telomere length homeostasis through a proteolysis of TRF1. J Biol Chem. 2009;284:8557–8566. doi: 10.1074/jbc.M806702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yang Y, Gao R, Yang X, Yan X, Wang C, Jiang S, Yu L. RLIM interacts with Smurf2 and promotes TGF-beta induced U2OS cell migration. Biochem Biophys Res Commun. 2011;414:181–185. doi: 10.1016/j.bbrc.2011.09.053. [DOI] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Jiao B, et al. Paternal RLIM/Rnf12 is a survival factor for milk-producing alveolar cells. Cell. 2012;149:630–641. doi: 10.1016/j.cell.2012.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen SA, et al. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 2009;69:128–136. doi: 10.1158/0008-5472.CAN-08-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Gribnau J. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer OH, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon SP. Isolation and culture of ovarian cancer cell lines. Methods Mol Med. 2004;88:133–139. doi: 10.1385/1-59259-406-9:133. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;12:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Bossenz M, Mincheva A, Copeland NG, Gilbert DJ, Jenkins NA, Lichter P, Bach I. Functional characterization of the gene encoding RLIM, the corepressor of LIM homeodomain factors. Genomics. 2000;69:120–130. doi: 10.1006/geno.2000.6311. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Peirano RI, Peters MA, Schlüter A, Bossenz M, Scheffner M, Bach I. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Tursun B, Cornils K, Schluter A, Drung A, Gungor C, Bach I. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev Dyn. 2006;235:786–791. doi: 10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- Pederson T. As functional nuclear actin comes into view, is it globular, filamentous, or both. J Cell Biol. 2008;180:1061–1064. doi: 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci USA. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, et al. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 2005;19:2307–2319. doi: 10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O'Reilly L, White D, Williams R, Armes J, Lindeman GJ. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Kudryavtseva E, Ch'en IL, McCormick J, Sugihara TM, Ruiz R, Andersen B. Expression of an engrailed-LMO4 fusion protein in mammary epithelial cells inhibits mammary gland development in mice. Oncogene. 2004;23:1507–1513. doi: 10.1038/sj.onc.1207288. [DOI] [PubMed] [Google Scholar]
- Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- Xu K, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell. 2009;15:270–282. doi: 10.1016/j.ccr.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Mol Cell. 2012;46:650–661. doi: 10.1016/j.molcel.2012.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.