Ypt1 GTPase, in the context of an autophagy-specific module, regulates ER-phagy. Because Ypt1 is a known regulator of ER-to-Golgi transport, this means that a single Ypt/Rab can regulate two alternative transport steps from one compartment, the ER, to two different destinations, the Golgi and the autophagy pathway.
Abstract
Accumulation of misfolded proteins on intracellular membranes has been implicated in neurodegenerative diseases. One cellular pathway that clears such aggregates is endoplasmic reticulum autophagy (ER-phagy), a selective autophagy pathway that delivers excess ER to the lysosome for degradation. Not much is known about the regulation of ER-phagy. The conserved Ypt/Rab GTPases regulate all membrane trafficking events in eukaryotic cells. We recently showed that a Ypt module, consisting of Ypt1 and autophagy-specific upstream activator and downstream effector, regulates the onset of selective autophagy in yeast. Here we show that this module acts at the ER. Autophagy-specific mutations in its components cause accumulation of excess membrane proteins on aberrant ER structures and induction of ER stress. This accumulation is due to a block in transport of these membranes to the lysosome, where they are normally cleared. These findings establish a role for an autophagy-specific Ypt1 module in the regulation of ER-phagy. Moreover, because Ypt1 is a known key regulator of ER-to-Golgi transport, these findings establish a second role for Ypt1 at the ER. We therefore propose that individual Ypt/Rabs, in the context of distinct modules, can coordinate alternative trafficking steps from one cellular compartment to different destinations.
INTRODUCTION
At the cellular level, neurodegenerative diseases are associated with accumulation of aggregated proteins termed neurodegenerative-related (NDR) proteins, such as α-synuclein in Parkinson, amyloid precursor protein in Alzheimer, and PrP in prion-related diseases (Uversky et al., 2009). The cause of the toxicity of these aggregates is not well understood. One common feature of NDR proteins is that they are all integral or membrane-associated proteins (Fantini and Yahi, 2010). On synthesis, all integral membrane proteins are inserted into the endoplasmic reticulum (ER), a membrane network that spans the whole cell. Exit of membrane proteins from the ER to other cellular compartments depends on their proper folding, a process that is not 100% efficient, yielding a subset of misfolded proteins. One mechanism for clearance of misfolded proteins is shuttling to the lysosome for degradation via the autophagy pathway (Schroder, 2008; Vembar and Brodsky, 2008).
Transport of membranes and proteins through cellular trafficking pathways connects membrane-bound intracellular compartments with the plasma membrane (PM) and the cell milieu. In the exocytic pathway, transport flows from the ER through the Golgi toward the PM, whereas in the endocytic pathway, transport flows from the PM through endosomes toward the lysosome. In addition, selective and nonselective autophagic pathways deliver proteins and membranes, from various intracellular compartments or bulk cytoplasm, for degradation in the lysosome. ER-phagy is a selective autophagy pathway, which targets aberrant ER (Bernales et al., 2007; Deegan et al., 2013). Misfolded or excess proteins at the ER can be shuttled for degradation either to the proteosome or, via the ER-phagy pathway, to the lysosome (Bernales et al., 2007; Vembar and Brodsky, 2008). Disruption of this shuttling and subsequent accumulation of misfolded proteins at the ER results in ER stress and induction of the unfolded protein response (UPR; Schroder, 2008).
All types of autophagy start with the formation of the preautophagosomal structure (PAS; Nakatogawa et al., 2009; Yang and Klionsky, 2009). The PAS is required for the formation of the double-membrane autophagosome, which engulfs parts of the cytoplasm and then fuses with the lysosome. Much research has been done on elucidating the signaling pathways that induce or suppress autophagy (He and Klionsky, 2009; Wang and Levine, 2010) and identifying autophagy-specific machinery components (Nakatogawa et al., 2009; Yang and Klionsky, 2009). The current consensus is that the autophagosomal membrane originates from the ER and other cellular compartments, such as mitochondria or the Golgi–endosomal system (Yang and Klionsky, 2009; Tooze and Yoshimori, 2010). It is not clear, however, how autophagy machinery intersects with membrane trafficking machinery to generate the autophagosome (Rubinsztein et al., 2012).
Ypt/Rabs are conserved monomeric GTPases that regulate trafficking between cellular compartments. These GTPases are stimulated by guanine-nucleotide exchange factors (GEFs) to recruit multiple effectors, which mediate all vesicular transport events, from vesicle formation to their targeting and fusion (Segev, 2001a, b; Stenmark, 2009). Recently Ypt/Rab GTPases have also emerged as candidates for coordination of individual transport steps in the same pathway (Segev, 2011). Here we address the question of whether Ypt/Rabs can coordinate distinct intracellular trafficking pathways and, specifically, regulate the intersection between the exocytic and autophagic pathways.
In yeast, Ypt1 is required for ER-to-Golgi transport (Segev et al., 1988; Jedd et al., 1995) and autophagy (Segev and Botstein, 1987; Lynch-Day et al., 2010). The Ypt31/32 functional pair also plays a role in two transport steps: Golgi to PM and endosomes to Golgi (Jedd et al., 1997; Chen et al., 2005). One possible scenario is that a single Ypt can play different roles in distinct locations. Evidence in favor of this model has been presented for Ypt1, which in addition to its established roles in ER-to-Golgi transport and autophagy, was suggested to play a role in endosome-to-Golgi transport (Sclafani et al., 2010). An alternative explanation is that Ypt1 has different roles at a single location. Our new data favor the latter model, which may establish a new paradigm for the action of Ypt/Rab GTPases.
The GEF for Ypt1 and Ypt31/32 is the multisubunit modular TRAPP complex, which is found in at least three configurations. TRAPPI acts as a GEF for Ypt1 (Jones et al., 2000; Wang et al., 2000) and is required for ER-to-Golgi transport (Sacher et al., 1998). TRAPPIII, which contains the TRAPPI subunits plus Trs85, acts as a Ypt1 GEF in autophagy (Lynch-Day et al., 2010). TRAPPII, which contains Trs120 and Trs130 in addition to TRAPPI subunits, acts at the trans-Golgi (Sacher et al., 2001). The GEF specificity of TRAPPII has differing reports (Morozova et al., 2006; Cai et al., 2008).
We recently showed that a Ypt/Rab module consisting of the autophagy-specific Trs85-contaning TRAPPIII as a GEF, Ypt1, and Atg11 as an effector (Trs85-Ypt1-Atg11) is required for PAS formation (Lipatova et al., 2012). It was not clear, however, from which compartment this Ypt module shuttles membranes and proteins to the autophagic pathway. Here we show that the Trs85-Ypt1-Atg11 module functions at the ER to clear excess and/or misfolded proteins. Our results establish that Ypt1 acts at the ER to coordinate two different transport steps: ER-phagy and ER-to-Golgi transport. Because all of the players are conserved from yeast to humans, elucidation of such coordination is pertinent to tackling neurodegenerative disorders.
RESULTS
Accumulation of membrane proteins in aberrant ER of ypt1 mutant cells
Ypt1 is essential for both ER-to-Golgi transport and autophagy (Segev and Botstein, 1987; Segev et al., 1988; Jedd et al., 1995; Lynch-Day et al., 2010; Lipatova et al., 2012). We hypothesized that, like cargo-containing vesicles destined for the Golgi apparatus, Ypt1-mediated flow of cellular components to autophagy also originates from the ER. To test this idea, we used two ypt1 mutations that do not exhibit an ER-to-Golgi transport defect but confer an autophagy-specific block: ypt1-T40K (ypt1-1) and ypt1-T40A.
Cells expressing the ypt1‑1 mutation from the endogenous locus are sensitive to cold and, mildly, to elevated temperatures. At the permissive temperature, this mutation does not cause a vegetative growth defect or an ER-to-Golgi block (Segev and Botstein, 1987; Segev et al., 1988; Jedd et al., 1995). The second allele is ypt1-T40A, which is a substitution of the same amino acid as in the ypt1-1 allele, T40K, but to alanine. The ypt1-T40A allele, when expressed from a plasmid as the sole copy of YPT1, does not exhibit defects in the secretory pathway (Sclafani et al., 2010). To compare the two alleles, we constructed them in a CEN plasmid with the promoter and terminator of YPT1 and expressed in a ypt1∆ background.
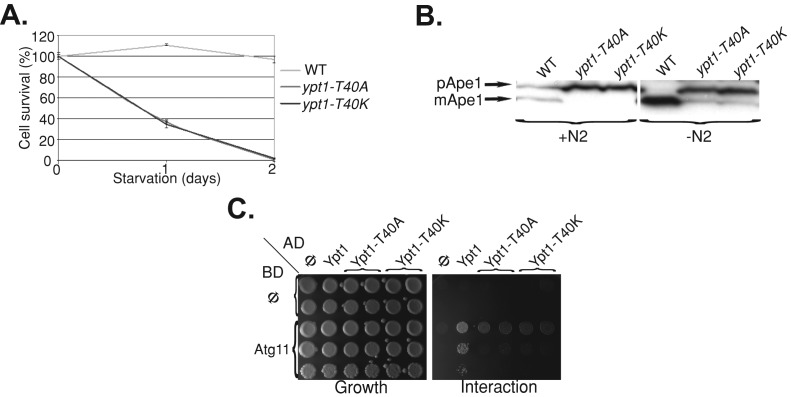
We previously showed that the ypt1‑1 chromosomal mutation confers severe selective and nonselective autophagy blocks (Segev and Botstein, 1987; Lipatova et al., 2012). In contrast, the ypt1-T40A allele was suggested to confer an endosome-to-Golgi transport block (Sclafani et al., 2010). First, we determined whether, like ypt1-1, the two mutations ypt1-T40A and ypt1-T40K expressed from a plasmid over the null confer an autophagy defect. Nonselective autophagy was determined by survival under nitrogen starvation; the selective autophagy cytosol-to-vacuole pathway (CVT) was determined by processing of Ape1. Like ypt1-1, both ypt1-T40A and ypt1-T40K alleles, when expressed from a plasmid over the null, confer a block in selective and nonselective autophagy (Figure 1, A and B). Second, we tested the interaction of Ypt1 and Atg11 using the yeast two-hybrid assay. We recently showed that, whereas the Ypt1 wild-type protein interacts with its autophagy-specific effector Atg11, the Ypt1-T40K mutant protein does not (Lipatova et al., 2012). Like Ypt1-T40K, Ypt1-T40A is defective in the Ypt1-Atg11 interaction (Figure 1C). Therefore the ypt1-T40A mutation appears to confer the same autophagy defects as the ypt1-T40K.
FIGURE 1:
The YPT1-T40A mutation, like YPT1-T40K, confers autophagy and Atg11-binding defects. (A) Similar to ypt1-T40K (ypt1-1), ypt1-T40A mutant cells are defective in nonselective autophagy. Cells were deleted for the YPT1 gene on the chromosome and express one of the following alleles of YPT1 from a CEN plasmid under its own promoter and terminator: YPT1 (WT), ypt1-T40K, or ypt1-T40A. The viability of the cells was determined before and after a shift to medium without nitrogen. Shown is percentage cell survival after the shift (100% before the shift); error bars, SD). Whereas wild-type cells remained viable, both ypt1 mutant strains lost their viability after 2 d of nitrogen starvation. (B) Similar to ypt1-T40K (ypt1-1), ypt1-T40A mutant cells are defective in CVT. Processing of Ape1 in the three strains (as in A) was determined using immunoblot analysis with anti-Ape1 antibodies before and 4 h after a shift to medium without nitrogen. Whereas wild-type cells process pApe1 to mApe1 (mature), both ypt1-T40K and ypt1-T40A mutant cells are defective in this processing. (C) The Ypt1-T40A mutant protein, like Ypt1-T40K, does not interact with Atg11 in the yeast two-hybrid (Y2H) assay. Interaction was determined using a mating assay with two Y2H plasmids. Activation domain (AD): Φ, Ypt1, Ypt1-T40K, and Ypt1-T40A (left to right). Binding domain (BD): Φ or Atg11 (top to bottom). Growth of the diploids carrying the two plasmids is shown on SD-Ura-Leu (left), and interaction is shown on SD-Ura-Leu-His (right). Whereas wild-type Ypt1 interacts with Atg11, both mutant proteins are defective in this interaction. Results represent at least two independent experiments.
To further characterize the autophagy-specific ypt1 mutations, we tested their effect on the localization of membrane proteins. One such membrane protein is Snc1, a vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptor that recycles through endosomes for multiple rounds of function between the Golgi and the PM. Intracellular accumulation of GFP-tagged Snc1 has been used as a marker of an endosome-to-Golgi transport block (e.g., in ypt31∆/ypt32ts mutant cells; Lewis et al., 2000; Chen et al., 2005) and was used as an indication for such a block in ypt1-T40A mutant cells (Sclafani et al., 2010). We recently showed, however, that intracellular accumulation of green fluorescent protein (GFP)–tagged Snc1 also occurs as a result of an ER-to-Golgi block (e.g., in ypt1-A136D temperature-sensitive mutant cells; Zou et al., 2012) and, as shown later, also as a result of a defect in ER-phagy.
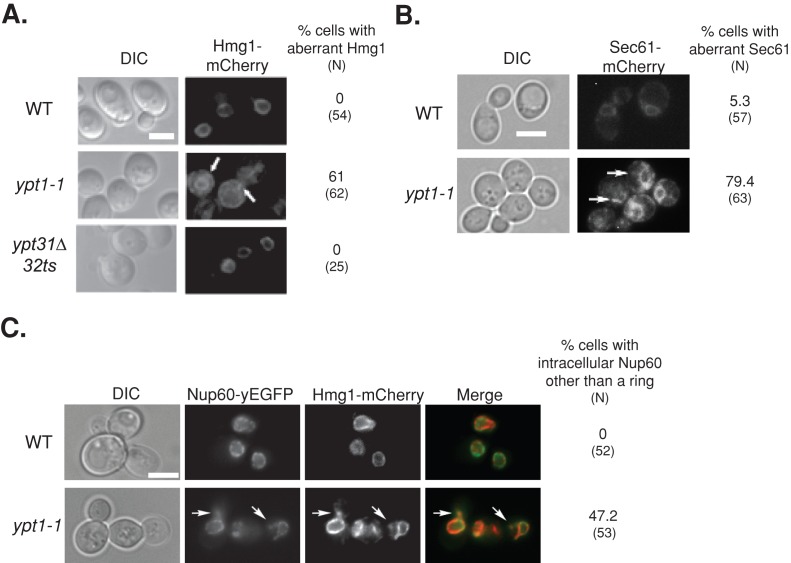
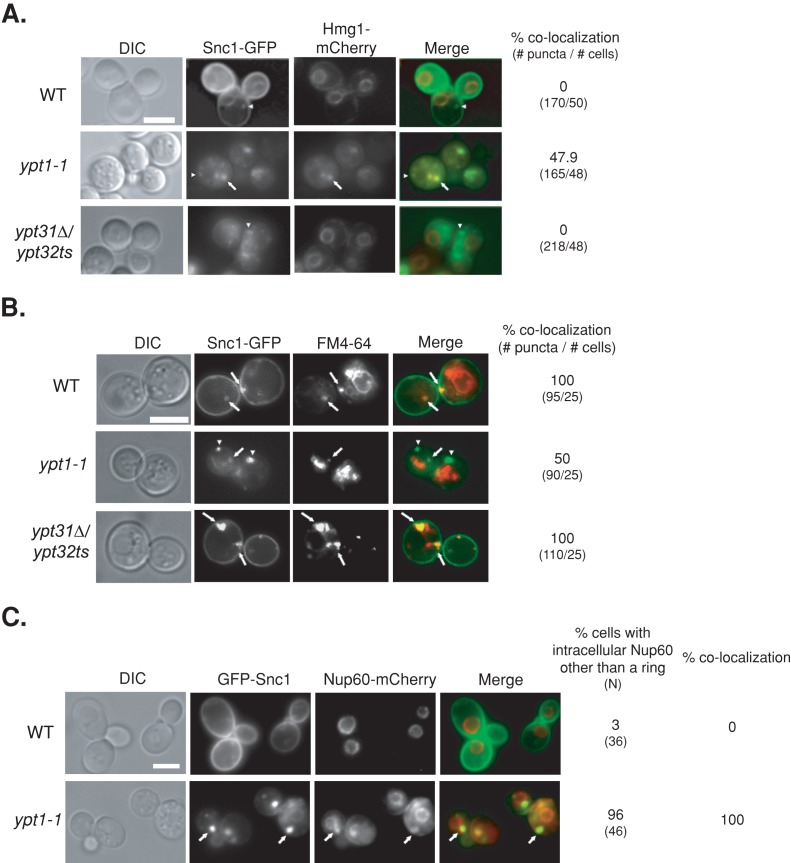
First, we tested the effect of the ypt1-1 mutation on the localization of Snc1-GFP. We determined the extent of colocalization of intracellular Snc1-GFP with an ER marker, Hmg1, and with endosomes (using a pulse and short chase with the membrane fluorescent dye FM4-64). Endogenous Hmg1 was tagged with mCherry in wild-type and ypt1-1 and ypt31∆/ypt32ts mutant cells (without expressing Snc1-GFP). Whereas in wild-type and ypt31∆/ypt32ts mutant cells Hmg1-mCherry localizes to rings around nuclei (Huh et al., 2003), the majority of ypt1-1 mutant cells contain aberrant structures in addition to the rings (Figure 2A). This was true also for another ER protein, the translocon subunit Sec61, and a nuclear pore subunit, Nup60 (Figure 2, B and C; Huh et al., 2003). In wild-type cells, Snc1-GFP localizes to the PM and to a few small puncta that colocalize with endosomes and not with the ER. In ypt31∆/ypt32ts mutant cells, which are defective in endosome-to-Golgi transport (Chen et al., 2005), there is intracellular accumulation of Snc1-GFP, which localizes to endosomes but not to the ER (Figure 3, A and B). We find that ypt1-1 mutant cells also accumulate intracellular Snc1-GFP as both small and very large puncta. Whereas ∼50% of the intracellular Snc1-GFP puncta in ypt1-1 mutant cells localize to endosomes (smaller puncta), ∼50% colocalize with the ER marker (larger puncta; Figure 3, A and B). This result suggests that transport of Snc1-GFP from the ER of ypt1-1 mutant cells is hindered but that some Snc1-GFP reaches the PM and can be recycled through endosomes.
FIGURE 2:
Excess ER accumulates in ypt1-1 mutant cells expressing fluorescently tagged ER proteins. (A) Excess ER accumulates in ypt1-1 mutant cells expressing Hmg1-mCherry. The ER protein Hmg1 was tagged on the chromosome with mCherry in wild-type and ypt1‑1 and ypt31∆/32ts mutant cells. Cells were visualized by live-cell fluorescence microscopy. Left to right, differential interference contrast (DIC), mCherry, and percentage of cells with aberrant Hmg1 structures (N, number of cells visualized). In wild-type and ypt31∆/32ts mutant cells, mCherry-tagged Hmg1 localizes to rings around nuclei, a typical ER staining pattern (Huh et al., 2003). On the other hand, excess ER structures accumulate in the majority of ypt1‑1 mutant cells. (B) Excess ER accumulates in ypt1-1 mutant cells expressing Sec61-mCherry. Sec61 was tagged on the chromosome with mCherry in wild-type and ypt1‑1 mutant cells. Cells were visualized by live-cell fluorescence microscopy. Left to right, DIC, mCherry, and percentage of cells with aberrant Sec61 structures (N, number of cells visualized). In wild-type cells, mCherry-tagged Sec61 localizes to rings around nuclei and underneath the PM, as previously seen for Sec61-GFP (Huh et al., 2003). In addition, excess ER structures accumulate in the majority of ypt1‑1 mutant cells. (C) Excess ER accumulates in ypt1-1 mutant cells expressing Nup60–yeast enhanced GFP (yEGFP) and Hmg1-mCherry. The nuclear pore protein Nup60 was tagged on the chromosome with yEGFP in wild type and ypt1‑1 mutant cells expressing Hmg1-mCherry. Cells were visualized by live-cell fluorescence microscopy. Left to right, DIC, GFP, mCherry, merge, percentage of cells with Nup60 other than the ring (N, number of cells visualized). In wild-type cells, Nup60-GFP localizes to rings around nuclei (Huh et al., 2003). About half of the ypt1-1 mutant cells (47%) contain structures other than the rings in which Nup60-GFP colocalizes with Hmg1-mCherry (92%). Arrows point to aberrant ER structures; bars, 5 μm. Results represent at least two independent experiments.
FIGURE 3:
GFP-tagged Snc1 accumulates in the ER of ypt1-1 mutant cells. (A) Colocalization of Snc1-GFP with the ER marker Hmg1 in ypt1-1, but not in wild-type or ypt31∆/32ts mutant cells. Wild-type and ypt1-1 and ypt31∆/32ts mutant cells expressing Hmg1-mCherry from its endogenous locus and Snc1-GFP from a 2 μ plasmid were visualized by live-cell fluorescence microscopy. Left to right, DIC, GFP, mCherry, merge, percentage colocalization of Snc1-GFP puncta with Hmg1-mCherry (number of GFP puncta/number of cells visualized). Arrows point to GFP puncta that colocalize with mCherry; arrowheads point to GFP puncta that do not colocalize with mCherry. In wild-type and ypt31∆/32ts mutant cells, Hmg1-mCherry localizes to rings around nuclei (Huh et al., 2003), which do not overlap with Snc1-GFP puncta. In contrast, in ypt1-1 mutant cells, about half of the Snc1-GFP puncta colocalize with Hmg1. (B) Only half of the intracellular Snc1-GFP is in endosomes of ypt1-1 mutant cells. Endosomes of cells expressing Snc1-GFP, as in Figure 1A, were labeled with FM4-64 and chased for 5 min. Cells were visualized by live-cell microscopy for GFP and FM4-64. Left to right, DIC, GFP, FM4-64, merge, and percentage colocalization of GFP with FM4-64 (number of GFP puncta/number of cells visualized). Arrows point to GFP puncta that colocalize with FM4-64; arrowheads point to GFP puncta that do not colocalize with FM4-64. Whereas all intracellular Snc1-GFP puncta overlap with endosomes in wild-type and ypt31∆/32ts mutant cells, only half localize to endosomes in ypt1-1 mutant cells. (C) The nuclear pore marker Nup60-mCherry colocalizes with intracellular GFP-Snc1. Wild-type and ypt1-1 mutant cells expressing endogenously tagged Nup60-mCherry and GFP-Snc1 from a 2 μ plasmid were visualized by live-cell microscopy. Left to right, DIC, GFP, mCherry, merge, percentage of cells with intracellular aberrant Nup60-mCherry (N, number of cells visualized), and percentage colocalization (percentage of cells in which intracellular GFP-Snc1 colocalizes with Nup60-mCherry). Arrows point to GFP puncta that colocalize with mCherry. In wild-type cells, Nup60-mCherry localizes to rings around nuclei (Huh et al., 2003). Almost all ypt1-1 mutant cells (96%) contain aberrant structures other than the rings in which Nup60-mCherry colocalizes with GFP-Snc1. Bars, 5 μm. Results represent at least two independent experiments.
Both Snc1 and Hmg1 are integral membrane proteins. To determine whether membrane-associated proteins also accumulate on membranes in ypt1-1 mutant cells, we tagged endogenous Nup60 with mCherry in wild-type and mutant cells overexpressing GFP-Snc1. We verified that GFP-Snc1 (tagged at the N-terminus), like Snc1-GFP (used earlier), accumulates in the ER of ypt1-1 mutant cells (Supplemental Figure S1). In wild-type cells expressing both tagged proteins, intracellular GFP-Snc1 does not colocalize with the Nup60-mCherry rings. In ypt1-1 mutant cells, however, Nup60-mCherry accumulates in puncta in addition to the rings. The Nup60-mCherry puncta colocalize with Snc1-GFP (Figure 3C). Together these results show that ypt1-1 mutant cells accumulate overexpressed membrane and membrane-associated proteins in aberrant ER structures.
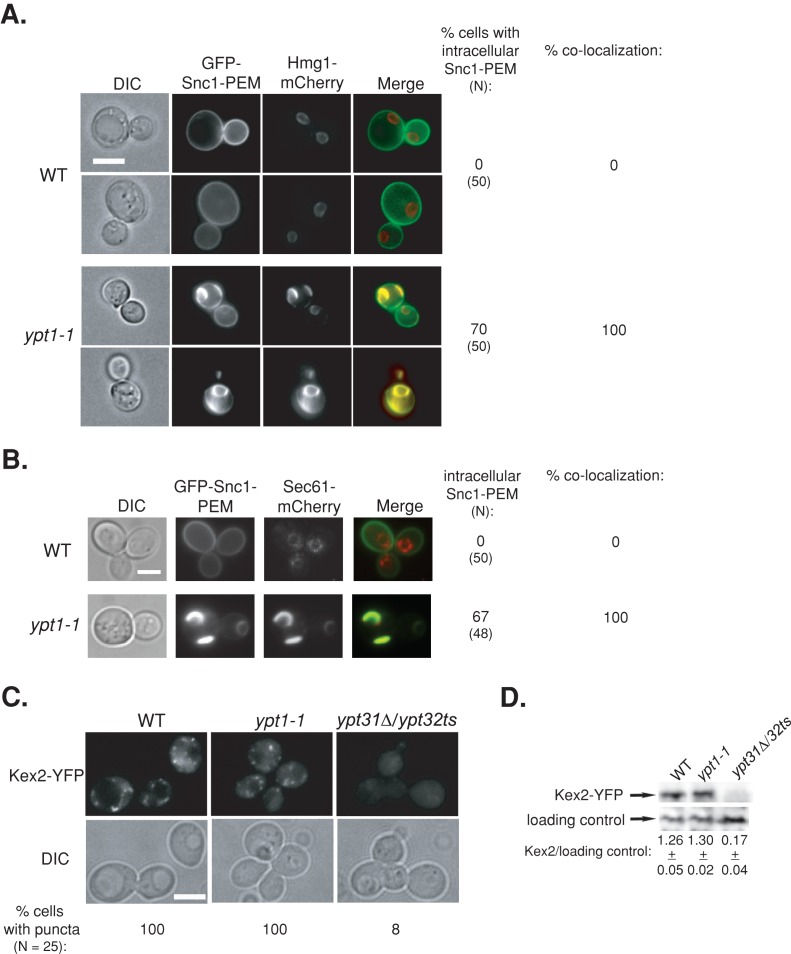
Because a subset of Snc1 accumulations in ypt1-1 mutant cells colocalize with endosomes, a phenomenon also observed in wild-type cells, we reasoned that these puncta arise through the normal process of Snc1 recycling from the PM to endosome. To establish that the larger accumulations in ypt1-1 mutant cells, which do not colocalize with endosomes, are not due to a PM recycling defect, we used GFP-Snc1-PEM, a mutant that cannot be internalized once it reaches the PM (Lewis et al., 2000). Thus intracellular accumulation of GFP-Snc1-PEM would be a result of a block in its transport to the PM and not PM recycling. We previously showed that ypt31∆/32ts mutant cells, which are defective in endosome-to-Golgi transport, do not accumulate intracellular GFP-Snc1-PEM (Chen et al., 2005). Accumulation of intracellular GFP-Snc1-PEM and its colocalization with two ER markers, Hmg1 and Sec61, was determined in wild-type and ypt1-1 mutant cells. In wild-type cells, all the GFP-Snc1-PEM localizes to the PM and does not colocalize with Hmg1-mCherry or Sec61-mCherry rings. In ypt1-1 mutant cells GFP-Snc1-PEM is found on the PM; however, ∼70% of the mutant cells also demonstrate intracellular accumulation of GFP-Snc1-PEM. Of importance, this intracellular GFP-Snc1-PEM completely colocalizes with Hmg1-mCherry and Sec61-mCherry (Figure 4, A and B). These results indicate that in ypt1-1 mutant cells GFP-Snc1-PEM transport is blocked at the ER before it reaches the PM. Moreover, intracellular accumulation of Snc1-PEM incapable of being internalized in ypt1-1 mutant cells indicates that the defect causing abnormal accumulation is not in the endosome-to-Golgi transport step.
FIGURE 4:
Accumulation of tagged proteins in ypt1-1 mutant cells is not due to an endosome-to-Golgi recycling defect. (A, B) GFP-Snc1-PEM, which cannot be internalized, accumulates in the ER in ypt1-1 mutant cells. Wild-type and ypt1-1 mutant cells expressing Hmg1-mCherry (A) or Sec61-mCherry (B) from their endogenous loci and GFP-Snc1-PEM from a 2 μ plasmid were visualized by live-cell fluorescence microscopy. left to right: DIC, GFP, mCherry, merge, percentage of cells with intracellular GFP-Snc1-PEM (N, number of cells visualized), and percentage colocalization (percentage of cells in which intracellular GFP-Snc1-PEM colocalizes with Hmg1 or Sec61). In wild-type cells all the GFP-Snc1-PEM is on the PM and does not colocalize with the Hmg1 or Sec61 rings. In ypt1-1 mutant cells, intracellular GFP-Snc1-PEM accumulates in ∼70% of the cells and completely colocalizes with Hmg1 and Sec61. (C, D) Endosome-to-Golgi recycling is not defective in ypt1-1 mutant cells. Endogenous Kex2-YFP was expressed in wild-type and ypt1-1 and ypt31∆/32ts mutant cells. Cells were visualized by live-cell fluorescence microscopy (C). Left to right, DIC, YFP, and percentage of cells with GFP puncta (N, number of cells visualized). Lysates from wild-type and ypt1-1 and ypt31∆/32ts mutant cells expressing Kex2-YFP were subjected to immunoblot analysis using anti-GFP antibodies (D). Bands were quantified, and the ratio of Kex2 over the loading control is shown; ±, SD. In wild-type and ypt1-1 mutant cells, the level of Kex2-YFP is similar, and Kex2-YFP puncta were observed. In contrast, in ypt31∆/32ts mutant cells, which are defective in endosome-to-Golgi transport, the level of Kex2-YFP is significantly lower, and almost no Kex2-YFP puncta were seen. Bar, 5 μm. Results represent at least two independent experiments.
To further verify that ypt1-1 mutant cells are not defective in endosome-to-Golgi transport, we examined a second cargo, Kex2, which cycles between the trans-Golgi and endosomes. In cells defective in endosome-to-Golgi transport, Kex2 is shuttled to the vacuole for degradation. We previously used Kex2-YFP to establish the endosome-to-Golgi transport defect of ypt31∆/32ts mutant cells (Chen et al., 2005). In ypt31∆/32ts mutant cells Kex2-YFP does not show the typical Golgi puncta and is degraded in the vacuole. In contrast, in wild-type and ypt1-1 mutant cells, Kex2-YFP localizes to puncta and is stable (Figure 4, C and D). Together the Snc1-PEM and Kex2 results support the idea that ypt1-1 mutant cells are not defective in endosome-to-Golgi transport. Therefore the intracellular accumulation of GFP-tagged Snc1 in ypt1-1 is not due to a PM recycling defect but instead is due to a transport block at the ER.
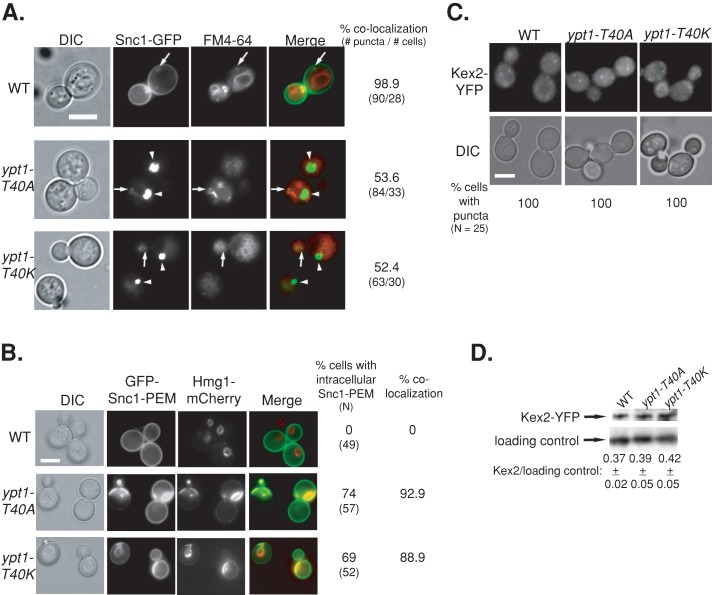
Next we determined whether, like ypt1-1, ypt1-T40A and ypt1-T40K expressed from a plasmid over the null confer accumulation of GFP-tagged Snc1 in the ER. Whereas all intracellular Snc1-GFP localizes to endosomes in wild-type cells, only ∼50% does so in ypt1-1 (Figure 3B), ypt1-T40K, and ypt1-T40A mutant cells (Figure 5A). In addition, whereas all the GFP-Snc1-PEM protein localizes to the PM of wild-type cells, ∼70% of the ypt1-1, ypt1-T40K, and ypt1-T40A mutant cells accumulate intracellular GFP-Snc1-PEM, which completely colocalizes with the ER marker Hmg1 (Figures 4A and 5B). Finally, like ypt1-1 (Figure 4, C and D), neither ypt1-T40K nor ypt1-T40A mutant cells are defective in recycling of Kex2-YFP (Figure 5, C and D). These results show that the two YPT1 mutant alleles ypt1-T40A and ypt1-T40K, which are defective in autophagy and not in the exocytic pathway, accumulate overexpressed GFP-tagged Snc1 in the ER. Moreover, this accumulation is not due to an endosome-to-Golgi transport defect.
FIGURE 5:
The YPT1-T40A mutation, like YPT1-T40K, confers Snc1-GFP accumulation in the ER but not endosome-to-Golgi transport defects. The three strains used here are YPT1 (WT), ypt1-T40K, and ypt1-T40A allele (as in Figure 1). (A) Half of the intracellular Snc1-GFP localizes to endosomes in ypt1-T40A and ypt1-T40K mutant cells. The endosomes of cells expressing Snc1-GFP were labeled with FM4-64, and cells were visualized using live-cell microscopy (as in Figure 3B). Left to right, DIC, GFP, FM4-64, merge, percentage colocalization of Snc1-GFP with endosomes (number of GFP puncta/number of cells visualized). Arrows point to GFP puncta that colocalize with FM4-64; arrowheads point to GFP puncta that do not colocalize with FM4-64. About 50% of the intracellular Snc1-GFP colocalizes with FM4-64 in ypt1-T40A and ypt1-T40K, as compared with ∼100% colocalization in wild-type cells. (B) Intracellular GFP-Snc1-PEM localizes to the ER in both ypt1-T40K and ypt1-T40A mutant cells. Cells expressing GFP-Snc1-PEM and the endogenously tagged ER marker Hmg1-mCherry (as in Figure 3A) were visualized by live-cell fluorescence microscopy. Left to right, DIC, GFP, mCherry, merge, percentage of cells with intracellular GFP-Snc1-PEM (N, number of cells visualized), and percentage colocalization (percentage of cells in which intracellular GFP-Snc1-PEM colocalizes with Hmg1). In wild-type cells all the GFP-Snc1-PEM is on the PM and does not colocalize with Hmg1 rings. In ypt1-T40A and Ypt1-T40K mutant cells, ∼90% of the intracellular GFP-Snc1-PEM colocalizes with Hmg1. (C) Kex2 recycling is not defective in ypt1-T40K and ypt1-T40A mutant cells. Endogenous Kex2-YFP was expressed in wild-type and ypt1-T40K and ypt1-T40A mutant cells. Top, cells were visualized by live-cell fluorescence microscopy (as in Figure 4C). Left to right, DIC, YFP, and percentage of cells with intracellular GFP puncta (N, number of cells visualized). (D) Immunoblot analysis of Kex2-YFP. Lysates from wild-type and ypt1-T40K and ypt1-T40A mutant cells expressing Kex2-YFP were subjected to immunoblot analysis using anti-GFP antibodies (as in Figure 4D). Bands were quantified and the ratio of Kex2 over the loading control is shown; ±, SD. Like wild-type cells, both ypt1-T40K and ypt1-T40A mutant cells show Kex2-YFP puncta and similar level of Kex2-YFP. Bar, 5 μm (A–C). Results represent at least two independent experiments.
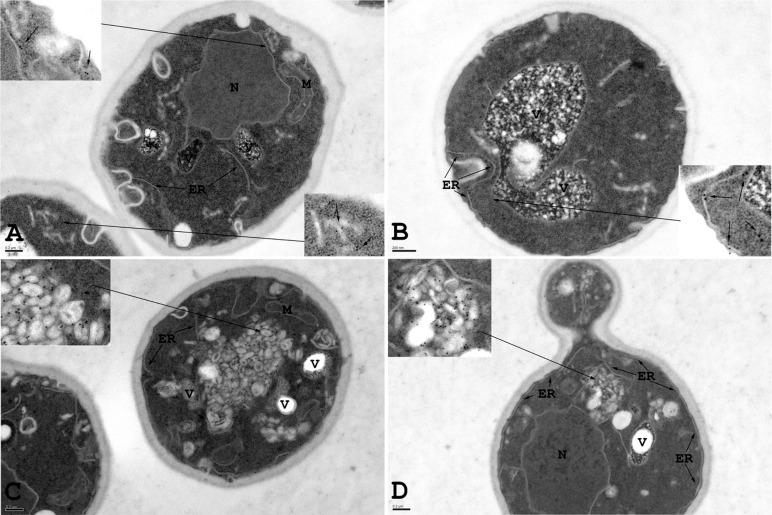
The accumulation of large puncta of fluorescently tagged Snc1 with Hmg1, Sec61, and Nup60 in ypt1-1 mutant cells suggests that these membrane or membrane-associated proteins accumulate on aberrant ER structures. To support this idea, we used immuno–electron microscopy (EM) of wild-type and ypt1-1 mutant cells expressing Snc1-GFP. Anti-Hmg1 antibodies were used to mark the ER (Hmg1 was not tagged). The elongated membrane structures identified as ER in wild-type cells are Hmg1 positive, and their morphology is consistent with ER membranes identified with other protein markers in earlier immuno-EM studies using similar methods (Preuss et al., 1991). In ypt1-1 mutant cells, there is an accumulation of aberrant structures that appear tubular-vesicular in nature and form aggregates. Unlike lipid droplets, these structures are membrane bound and contain electron-dense material. Of interest, these structures are decorated with anti-Hmg1 antibodies, indicative of their ER origin. We observed an >20-fold increase in membrane-associated Hmg1 in ypt1-1 compared with wild-type cells (Figure 6 and Supplemental Table S1). These Hmg1-positive aggregates of aberrant membranes mostly likely represent the fluorescent Hmg1-mCherry puncta observed in the fluorescence microscopy experiments in ypt1-1 mutant cells. Together these results indicate that ypt1-1 mutant cells accumulate overexpressed fluorescently tagged proteins on aberrant ER structures.
FIGURE 6:
Aberrant ER accumulation in ypt1-1 mutant cells overexpressing Snc1-GFP. Immuno-EM analysis was done with wild-type and ypt1-1 mutant cells expressing Snc1-GFP. Wild-type and ypt1-1 mutant cells expressing Snc1-GFP were grown to log phase, fixed, and processed for immuno-EM using anti-Hmg1 antibodies. Insets show enlarged view of ER structures labeled with gold particles. (A, B) In wild-type cells, Hmg1, an ER marker, is present exclusively on elongated ER membranes. (C, D) In ypt1-1 mutant cells Hmg1 is present on aberrant membrane structures. ER, endoplasmic reticulum; M, mitochondria; N, nucleus; V, vacuole. Bar, 200 nm. Results represent two independent experiments; quantification is shown in Supplemental Table S1.
A role for Ypt1 in ER-phagy
We hypothesized that in wild-type cells, excess and misfolded membrane proteins are shuttled from the ER for degradation in the vacuole (the yeast lysosome) through the autophagy pathway. In contrast, because ypt1-1 mutant cells are defective in autophagy, they accumulate aberrant ER structures filled with excess proteins, which have stalled before reaching the vacuole. Moreover, such accumulation should induce the UPR in the ER.
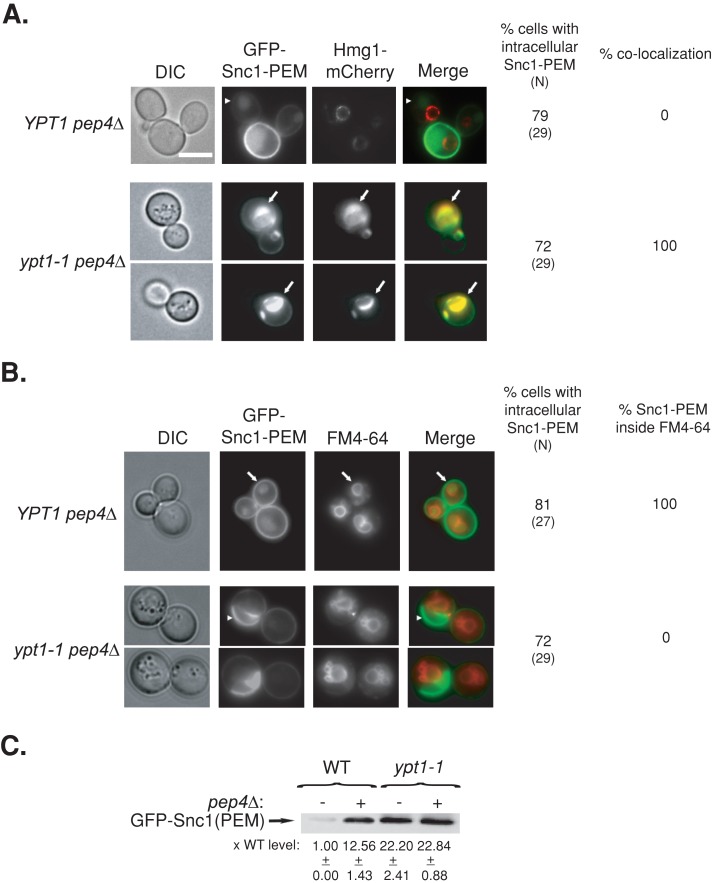
To test the first part of this idea, we analyzed the intracellular accumulation of the nonrecyclable GFP-Snc1-PEM protein in pep4∆ mutant cells, which are compromised for protein degradation in the vacuole (Jones et al., 1982). Because GFP-Snc1-PEM cannot get to the vacuole through endosomes, its accumulation in the vacuole in pep4∆ mutant cells is due to shuttling from the exocytic pathway. Whereas no intracellular GFP-Snc1-PEM accumulates in wild-type cells (Figure 4A), ∼80% of the pep4∆ mutant cells accumulate intracellular GFP-Snc1-PEM. Moreover, this internal GFP-Snc1-PEM accumulates inside the vacuole (labeled by FM4-64) and does not colocalize with the ER marker Hmg1 (Figure 7, A and B). In contrast, deletion of PEP4 in ypt1-1 mutant cells does not affect the localization of GFP-Snc1-PEM, which accumulates in the ER of ∼70% of the cells and not in the vacuole (compare Figures 4A and 7, A and B). To further support this microscopy observation, the level of GFP-Snc1-PEM in the four yeast strains was determined using an immunoblot analysis. In wild-type cells (YPT1), deletion of PEP4 results in accumulation of GFP-Snc1-PEM. In contrast, in ypt1-1 mutant cells, the level of GFP-Snc1-PEM is higher than in wild-type cells and is similar in PEP4 and pep4∆ cells (Figure 7C). These results support the idea that in wild-type cells, excess GFP-Snc1-PEM is shuttled to the vacuole for degradation, and this transport is defective in ypt1-1 mutant cells.
FIGURE 7:
GFP-Snc1-PEM accumulates in the vacuole of pep4∆, but not ypt1-1 pep4∆, mutant cells. (A) Accumulation of intracellular GFP-Snc1-PEM outside the ER of YPT1 (WT) pep4∆ cells defective in vacuolar degradation. GFP-Snc1-PEM was expressed from a 2 μ plasmid and mCherry-Hmg1 from its endogenous locus (as in Figure 4A) in YPT1 pep4∆ and ypt1‑1 pep4∆ mutant cells. Cells were visualized by live-cell fluorescence microscopy. Left to right, DIC, GFP, mCherry, merge, percentage of cells with intracellular GFP-Snc1-PEM (N, number of cells visualized), and percentage colocalization (percentage of cells in which intracellular GFP-Snc1-PEM colocalizes with Hmg1-mCherry). In YPT1 (WT) pep4∆ cells, intracellular GFP-Snc1-PEM does not colocalize with the Hmg1 rings (arrowheads). In ypt1-1 pep4∆ double mutant cells, intracellular GFP-Snc1-PEM colocalizes with Hmg1 (arrows). (B) Intracellular GFP-Snc1-PEM accumulates inside the vacuole of YPT1 pep4∆ but not ypt1-1 pep4∆ mutant cells. The vacuolar membrane of YPT1 pep4∆ and ypt1‑1 pep4∆ mutant cells expressing GFP-Snc1-PEM was labeled with FM4-64. Cells were visualized by live-cell microscopy. Left to right, DIC, GFP, FM4-64, merge, percentage of cells with intracellular GFP-Snc1-PEM (N, number of cells visualized), and percentage localization of intracellular GFP-Snc1-PEM inside the vacuole. Arrows point to GFP localized inside FM4-64-labled vacuoles YPT1 (WT) pep4∆ cells; arrowheads point to GFP that does not localize inside FM4-64-labled vacuoles in ypt1-1 pep4∆ double mutant cells. (C) Accumulation of GFP-Snc1-PEM in ypt1-1 mutant cells is not dependent on the vacuolar Pep4 protease. The protein level of GFP-Snc1-PEM in lysates of wild-type and ypt1-1 mutant cells with or without pep4∆ was determined using immunoblot analysis and anti-GFP antibodies. Bands were quantified, and the ratio of GFP-Snc1-PEM in mutant versus wild-type cells is shown at the bottom; ±, SD. The level of GFP-Snc1-PEM is higher in pep4∆ mutant cells than in wild-type cells, indicating that the protein is degraded in the vacuole. In contrast, the high level of GFP-Snc1-PEM In ypt1-1 mutant cells is not dependent on pep4∆, indicating that the protein does not reach the vacuole. Bar, 5 μm. Results represent at least two independent experiments.
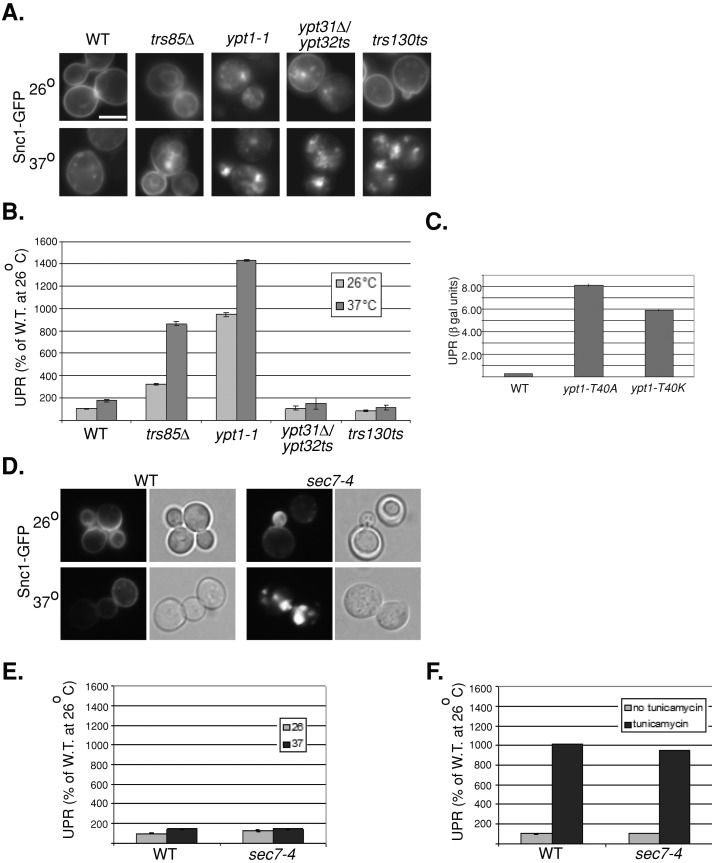
Overexpression and accumulation of fluorescently tagged ER proteins might generate a greater population of misfolded proteins. We reasoned that the UPR would be induced in cells that accumulate such proteins. To test this idea, we determined the degree of UPR induction in wild-type and ypt1-1 mutant cells expressing Snc1-GFP. Cells were transformed with a second plasmid encoding the lacZ gene behind four repeats of a UPR element, and the level of β-galactosidase (βgal) in cell lysates was determined (Kruse et al., 2006). UPR induction was 10-fold greater in ypt1-1 mutant cells than in wild-type cells. In contrast, no such induction was observed in ypt31∆/32ts mutant cells under conditions in which they accumulate intracellular Snc1-GFP (Figure 8, A and B). Furthermore, UPR is induced in both ypt1-T40K and ypt1-T40A mutant cells overexpressing Snc1-GFP (Figure 8C). These results suggest that ypt1‑1, ypt1-T40K, and ypt1-T40A, but not ypt31∆/32ts, mutant cells accumulate misfolded Snc1-GFP in their ER. Therefore these ypt1 mutant cells, which are not defective in ER-to-Golgi transport, are defective in ER-phagy.
FIGURE 8:
UPR is induced in ypt1 and trs85∆ but not ypt31∆/32ts, trs130ts, and sec7-4 mutant cells expressing Snc1-GFP. (A) Snc1-GFP accumulates in internal puncta in ypt1-1, trs85∆, ypt31∆/32ts, and trs130ts mutant cells. Cells expressing Snc1-GFP from a 2 μ plasmid were grown at 26°C (top), shifted to 37°C for 90 min (bottom), and visualized by live-cell microscopy. Representative cells are shown (a minimum of 24 cells were visualized for each panel). All mutant cells accumulate internal Snc1-GFP at 37°C and to a lesser degree at 26°C. (B) UPR is induced in ypt1-1 and trs85∆ but not ypt31∆/32ts and trs130ts mutant cells. Cells expressing Snc1-GFP (as in A) were transformed with a second plasmid carrying the LacZ gene under a UPR-inducible promoter (Kruse et al., 2006). Cells were grown at 26°C (light bars) and shifted to 37°C for 90 min (dark bars), and the induction of the UPR response was determined using the βGAL assay. UPR is expressed as percentage of the response of wild type at 26°C. The UPR response in trs85∆ and ypt1-1 mutant cells is induced at 26°C and is even higher at 37°C. No such induction is observed in ypt31∆/32ts or trs130ts mutant cells even under conditions in which these cells accumulate intracellular Snc1-GFP. (C) UPR is induced in ypt1-T40K and ypt1-T40A mutant cells expressing Snc1-GFP. UPR was determined in wild type and ypt1 mutant cells expressing LacZ under a UPR-inducible promoter and Snc1-GFP (as in B). (D) Intracellular Snc1-GFP accumulates in sec7-4 mutant cells. The experiment was done as described in A (50 cells were visualized for each panel). sec7-4 mutant cells accumulate internal Snc1-GFP at 37°C and to a much lesser degree at 26°C. (E) UPR is not induced in sec7-4 mutant cells. The experiment was done as described in B. No UPR induction is observed in sec7-4 mutant cells even under conditions in which these cells accumulate intracellular Snc1-GFP. (F) UPR can be induced in wild-type and sec7-4 mutant cells by tunicamycin. Cells were grown without tunicamycin (light bars) or with 5 μg/ml tunicamycin for 90 min (dark bars), and the induction of the UPR response was determined using the βGAL assay. Bar, 5 μm; error bars, SD (B, C, E, F). Results represent at least two independent experiments.
To verify that the ER-phagy pathway is independent of the Golgi, we determined the effect of Snc1-GFP overexpression on UPR in sec7-4 mutant cells. Sec7 is a GEF for Arf GTPases, which functions in the Golgi (Franzusoff et al., 1991). The sec7-4 mutation is in the GEF domain of Sec7 (Jones et al., 1999). Snc1-GFP accumulates in sec7-4 mutant cells, especially at 37°C (Figure 8D). Even though the UPR response can be induced by tunicamycin, however, it is not induced in sec7-4 mutant cells overexpressing Snc1-GFP (Figure 8, E and F).
A role for the Trs85-Ypt1-Atg11 module in ER-phagy
If Ypt1 regulates shuttling of excess GFP-tagged Snc1 through the autophagic pathway to the vacuole for degradation, we expect that it does so in the context of the autophagy-specific module, including the specific GEF and effector. Recently we showed that the GEF/Trs85-Ypt1-effector/Atg11 module regulates the onset of selective autophagy (Lipatova et al., 2012). We therefore tested the role of the Trs85-Ypt1-Atg11 module in ER-phagy.
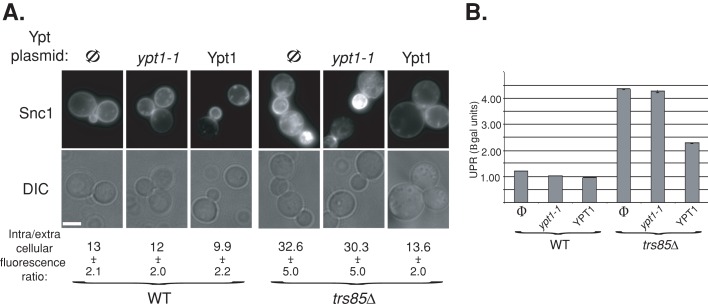
GEF mutations are expected to result in phenotypes similar to those of mutations in their Ypt substrates. We proposed that Trs85-containing TRAPPIII and Trs130-containing TRAPPII function as GEFs for Ypt1 and Ypt31, respectively (Morozova et al., 2006; Zou et al., 2012). Both trs85∆ and trs130ts mutant cells accumulate intracellular Snc1-GFP (Zou et al., 2012). The effect of overexpression of Snc1-GFP in trs85∆ and trs130ts mutations on UPR was determined. Similar to ypt1-1 mutant cells, UPR is induced in trs85∆ mutant cells expressing Snc1-GFP. In contrast, similar to ypt31∆/32ts mutant cells, UPR is not induced in trs130ts mutant cells under conditions in which they accumulate intracellular Snc1-GFP (Figure 8, A and B). These results show that the nature of Snc1-GFP accumulation in ypt1-1 and trs85∆ mutant cells is distinct from its accumulation in ypt31∆/32ts and trs130ts, which are defective in endosome-to-Golgi transport (Chen et al., 2005; Zou et al., 2012). Moreover, because both ypt1-1 and trs85∆ are defective in autophagy, these results suggest that accumulation of excess proteins in the ER of these mutant cells is due to a block in the autophagic pathway.
We showed that overexpression of Ypt1 can suppress the CVT phenotype of trs85∆ mutant cells (Lipatova et al., 2012). We tested whether it can also suppress the ER-phagy phenotypes of trs85∆ mutant cells. Overexpression of Ypt1 but not Ypt1-T40K mutant protein can suppress both the intracellular accumulation of Snc1-GFP and UPR induction in trs85∆ mutant cells (Figure 9, A and B, respectively). These results suggest that Trs85 functions upstream of Ypt1 in clearing out excess ER proteins through autophagy.
FIGURE 9:
A role for the Trs85-Ypt1 interaction in ER-phagy. (A) Overexpression of Ypt1 suppresses the Snc1-GFP accumulation defect of trs85∆ mutant cells. Top, wild-type (left) and trs85∆ mutant cells (right) expressing Snc1-GFP from a 2 μ plasmid were transformed with a second plasmid expressing Ypt1-T40K (mutant) or Ypt1 (wild-type) proteins (Φ, empty vector control). Cells were visualized by live-cell microscopy; representative cells are shown: GFP (top) and DIC (bottom). Bar, 5 μm. Bottom, quantification of intracellular/extracellular fluorescence ratio. The ratio of fluorescence inside and outside cells was determined using ImageJ (40 cells for each strain). The level of intracellular fluorescence in trs85∆ mutant cells is three times higher than in wild-type cells, and this increase is suppressed if Ypt1 but not Ypt1-T40K mutant protein is overexpressed. (B) Overexpression of Ypt1 suppresses UPR induction in trs85∆ mutant cells. Cells were transformed with three plasmids expressing LacZ under a UPR promoter, Snc1-GFP, and Ypt1 (Φ, Ypt1, or Ypt1-T40K). Induction of the UPR was determined in cell lysates using the βGAL assay (as in Figure 8B). UPR is induced in trs85∆ mutant cells (Φ) when compared with wild type, and this induction is partially suppressed by overexpression of Ypt1 but not Ypt1-T40K protein. Results represent at least two independent experiments; error bars, SD.
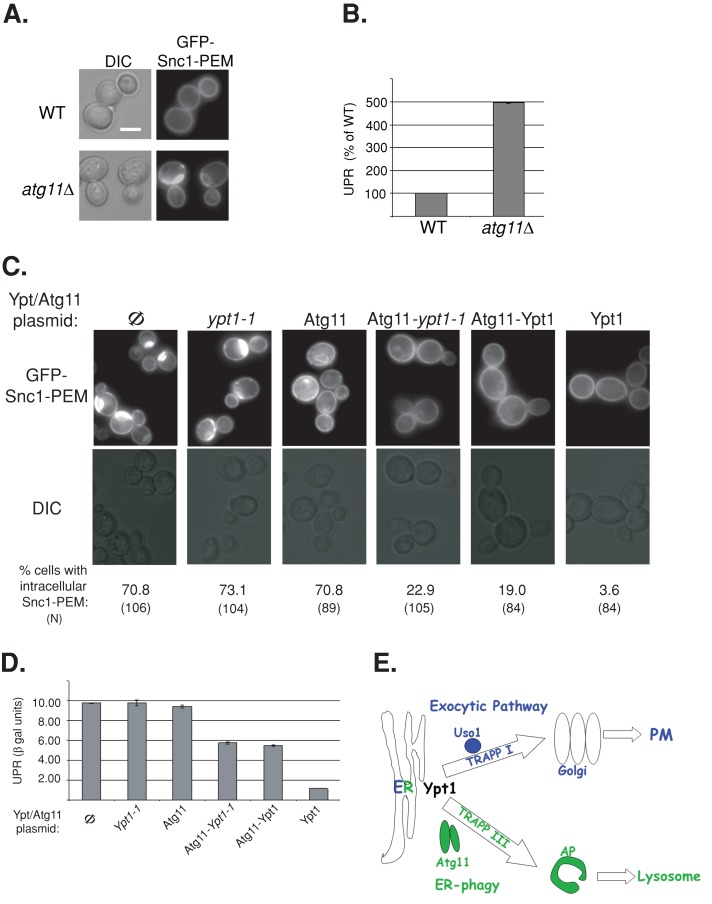
We also previously showed that Atg11 acts as an autophagy-specific Ypt1 effector, and an Atg11-Ypt1-T40K fusion protein can suppress the autophagy defects of ypt1‑1 mutant cells (Lipatova et al., 2012). Here we tested whether this is also true for the ER accumulation of GFP-Snc1 and UPR induction. First, intracellular accumulation of GFP-Snc1-PEM is observed in atg11∆ mutant cells, and UPR is induced in these cells (Figure 10, A and B), implicating Atg11 in ER-phagy. Second, the Atg11-Ypt1‑T40K fusion protein can rescue both the GFP-Snc1-PEM intracellular accumulation and UPR induction of ypt1-1 mutant cells. In contrast, neither the Ypt1-T40K mutant protein (which cannot interact with Atg11) nor Atg11 alone can rescue the GFP-Snc1-PEM intracellular accumulation and UPR induction phenotypes of ypt1-1 mutant cells (Figure 10, C and D). Together these results support the role of the Trs85-Ypt1-Atg11 module in shuttling excess ER proteins to the autophagy pathway for degradation.
FIGURE 10:
A role for the Ypt1-Atg11 interaction in ER phagy. (A) GFP-Snc1-PEM accumulates in atg11∆ mutant cells. Wild-type and atg11∆ mutant cells expressing GFP-Snc1-PEM were visualized by live-cell microscopy. Representative cells are shown (25 cells for each strain), DIC (left) and GFP (right). (B) UPR is induced in atg11∆ mutant cells expressing GFP-Snc1. UPR was determined in wild-type and atg11∆ mutant cells expressing GFP-Snc1 and LacZ under a UPR-inducible promoter as in Figure 8. (C) The Atg11-Ypt1-T40K fusion protein complements the GFP-Snc1-PEM accumulation defect of ypt1-1 mutant cells. Ypt1-1 mutant cells expressing GFP-Snc1-PEM were transformed with a CEN plasmid to express the following proteins (left to right): Φ, empty vector control, Ypt1-1, Atg11, Atg11-Ypt1-T40K, Atg11-Ypt1, and Ypt1. Cells were visualized by live-cell microscopy, and representative cells are shown (GFP, top; DIC, bottom). Percentage of cells with intracellular GFP-Snc1-PEM is indicated at the bottom (N, number of cells visualized). Approximately 70% of ypt1-1 mutant cells accumulate intracellular GFP-Snc1-PEM (Φ, left). This defect can be suppressed by Ypt1 (right), as well as by the two fusion proteins, Atg11-Ypt1-T40K and Atg11-Ypt1, but not by Ypt1-T40K or Atg11 proteins. (D) Suppression of UPR induction in ypt1-1 mutant cells by the Atg11-Ypt1-T40K fusion protein. Ypt1-1 mutant cells were transformed with two plasmids expressing LacZ under a UPR-inducible promoter and Atg11/Ypt1. Induction of the UPR was determined using the βGAL assay. UPR is induced in ypt1‑1 mutant cells (Φ, left, and this induction is suppressed by overexpression of Ypt1 (right), as well as by the two fusion proteins, Atg11-Ypt1-T40K and Atg11-Ypt1, but not by Ypt1-T40K or Atg11 proteins. Bar, 5 μm; error bars, SD (B, D). Results shown in A–D represent at least two independent experiments. (E) Model depicting a role for Ypt1, together with its process-specific GEFs and effectors, in coordinating ER-to-Golgi and ER-phagy. Here we show that, whereas in wild-type cells some overexpressed GFP-tagged Snc1 is transported from the ER to the PM via the exocytic pathway, some is shuttled to the lysosome (vacuole in yeast) for degradation through the autophagic pathway. Ypt1 is required for both pathways. In the exocytic pathway Ypt1 is activated by TRAPP I and mediates ER-to-Golgi transport via vesicles; Uso1/p115 is an example of an ER-to-Golgi–specific effector (Cao et al., 1998; Allan et al., 2000). In autophagy, Ypt1 is activated by Trs85-containing TRAPP III to shuttle ER-derived membranes loaded with misfolded proteins to autophagosomes; Atg11 is an autophagy-specific effector (Lipatova et al., 2012). In ypt1-1, trs85∆, and atg11∆ mutant cells, shuttling of GFP-Snc1 from the ER to the autophagosome is defective, GFP-Snc1 accumulates in aberrant ER structures, and the ER-stress response, UPR, is induced.
DISCUSSION
On the basis of results presented here, we conclude that a Ypt/Rab GTPase module, consisting of the Trs85-containing TRAPPIII GEF, Ypt1, and the Atg11 effector, functions in ER-phagy. We recently showed that this module regulates the onset of selective autophagy (Lipatova et al., 2012). However, the cellular compartment from which membrane and cargo destined to autophagy originate was not known. We now show that the Trs85-Ypt1-Atg11 module regulates shuttling of tagged, overexpressed, and likely misfolded proteins from the ER to the autophagic pathway (Figure 10E). Two observations point to ER as the origin of the Ypt1-mediated autophagic branch: colocalization of the accumulated proteins with ER markers and induction of UPR in trs85, ypt1, and atg11 mutant cells overexpressing tagged membrane proteins. Regulation of ER-phagy is distinctive from the role of Ypt1 in ER-to-Golgi transport because the ypt1 mutants used here are not defective in ER-to-Golgi transport (Jedd et al., 1995; Sclafani et al., 2010). The trs85∆ and atg11∆ mutants are defective specifically in autophagy as well (Kim et al., 2001; Meiling-Wesse et al., 2005; Nazarko et al., 2005).
In addition, results presented here argue against a role for Ypt1 in endosome-to-Golgi transport. Although our cumulative evidence points to a role for Ypt1 in ER-to-Golgi transport (Segev et al., 1988; Segev, 1991; Jedd et al., 1995) and ER-phagy (the present results), Sclafani et al. (2010) suggested that Ypt1 also plays a role in endosome-to-Golgi transport. This inference was based on the isolation of ypt1 mutants that do not exhibit secretory defects but accumulate intracellular GFP-Snc1. This accumulation was taken as an indication for an endosome-to-trans-Golgi transport defect. Using one mutant isolated and characterized in the Sclafani study, ypt1-T40A, we show that the block in this mutant is not in endosome-to-Golgi transport but in ER-phagy. Therefore we conclude that Ypt1 does not regulate endosome-to-Golgi transport. We previously showed that that role is performed by the Ypt31/Ypt32 GTPases (Chen et al., 2005).
On the basis of the present study, we propose a new paradigm for how Ypt/Rab GTPases coordinate intracellular trafficking pathways. In this paradigm, a single Ypt/Rab GTPase can regulate independent transport processes from the same compartment to more than one destination. We show here that in addition to the established role of Ypt1 in the regulation of transport of ER vesicles to the cis-Golgi, this GTPase also regulates transport of ER membranes—loaded with excess proteins—to autophagy. The separation of these two functions of Ypt1 was possible by the use of mutations that affect only autophagy and not ER-to-Golgi transport. The mechanism that allows Ypt1 to do that is to function in separate modules that contain process-specific GEF and effectors. The two proposed process-specific Ypt1 modules are TRAPPI and Uso1 in ER-to-Golgi transport, and TRAPPIII and Atg11 in ER-phagy (Figure 10E). An intriguing question is the nature of the cues that allow recruitment of the right Ypt/Rab module to the right membrane domain.
On the basis of the conservation of Ypt/Rabs, their GEF and effectors, and the autophagic machinery, we propose that Ypt1-dependent ER-phagy is also conserved. The importance of intracellular trafficking and autophagy in human disease (Howell et al., 2006; Choi et al., 2013) and the role that Ypt/Rabs play in autophagy (Chua et al., 2011) have been recognized. Because Ypt/Rabs play essential roles in multiple processes, it is crucial to elucidate mechanisms that allow these regulators to function in trafficking intersections toward the goal of developing drugs that target a specific process without affecting the others.
MATERIALS AND METHODS
Strains, plasmids, and reagents
Strains and plasmids used in this study are summarized in Supplemental Tables S2 and S3, respectively. All chemical reagents were purchased from Fisher Scientific (Bridgewater, NJ), unless otherwise noted. Media components other than amino acids were purchased from US Biological (Swampscott, MA). ProtoGel for Western blots was purchased from National Diagnostics (Atlanta, GA). Amino acids, 2-nitrophenyl β-d-galactopyranoside, tunicamycin, and protease inhibitors were purchased from Sigma-Aldrich (St. Louis, MO). Glass beads were purchased from BioSpec Products (Bartlesville, OK). Restriction enzymes and buffers were purchased from New England BioLabs (Ipswich, MA). Dithiothreitol (DTT) was purchased from Invitrogen (Carlsbad, CA). FM4-64 was purchased from Molecular Probes (Eugene, OR). Antibodies used in this study include rabbit anti–GAL4-BD, mouse monoclonal anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-hemagglutinin (Covance, Madison, WI), goat anti-rabbit horseradish peroxidase (HRP) and goat anti-mouse-HRP (GE Healthcare UK, Chalfont St. Giles, United Kingdom), affinity-purified rabbit anti-Hmg1 (a gift from R. Wright, University of Minnesota), and anti-Ape1 (a gift from D. Klionsky, University of Michigan).
Plasmid and strain construction
Plasmids.
pRS317-ypt1-T40A was made by site-directed mutagenesis of pRS317-YPT1. pRS327-YPT1 and pRS327-ypt1-T40K were made by subcloning the BamHI–ClaI fragments from pRS317-YPT1 and pRS317-ypt1-T40K, respectively, into pRS327. pRS425-GFP-SNC1 and pRS425-GFP-SNC1 PEM were made by subcloning the BamHI—PspXI fragments from pRS406 GS and pRS406 GSSOM (Lewis et al., 2000), respectively, into pRS425.
Yeast strains.
Kex2 was tagged on the chromosome at the COOH terminus with yellow fluorescent protein (YFP) using PCR product from the Kex2-YFP from the Yeast Resource Center (University of Washington, Seattle, WA) and homologous recombination. Hmg1, Sec61, and Nup60 were tagged on the COOH termini according to the standard technique (Wach et al., 1997).
Yeast culture conditions
Media preparation and yeast culture growth for all experiments were done as described (Segev and Botstein, 1987). For yeast two-hybrid assays, yeast cultures were grown overnight at 26°C in minimal (SD) media, normalized to the same density by OD600, and spotted onto agar plates in serial dilutions. Plates for yeast two-hybrid assay, indicated in figure legends, were incubated at 26°C.
Protein expression analyses
To determine the level of Kex2-YPF and Ape1 proteins, lysates of exponentially growing or starved cells were prepared as described (Cheong and Klionsky, 2008) and subjected to immunoblot analysis using anti-GFP and anti-Ape1 antibodies, respectively. The expression level of yeast two-hybrid constructs was determined as previously described (Lipatova et al., 2012). The level of GFP-Snc1-PEM protein was determined similarly to that for the yeast two-hybrid proteins, except that lysates were prepared from 7 OD units of cells, and anti-GFP antibodies were used.
Microscopy
For live-cell microscopy, cells carrying constructs for expression of fluorescently tagged proteins were grown to mid log phase in appropriate media. Fluorescence microscopy observation was carried out using a deconvolution Axioscope microscope (Carl Zeiss, Thornwood, NY) with fluorescein isothiocyanate (GFP and YFP) and Texas red (mCherry) sets of filters. Labeling of endosome and vacuole membranes with FM4-64 was done using a 5-min pulse and 5- and 60-min chases, respectively, as previously described (Vida and Emr, 1995). Immuno–electron microscopy was done as previously described (Preuss et al., 1992).
UPR βgal assay
The βgal assay was done with cells transformed with a plasmid (pJC104) encoding the lacZ gene behind four repeats of a UPR element (Kruse et al., 2006). To induce UPR, cells were grown in the presence of 5 μg/ml tunicamycin for 90 min. The level of β-galactosidase in cell lysates was determined as previously described (Reynolds et al., 2001). To prepare cell lysates, 5 OD600 units of early-log-phase cells were spun down, washed in distilled H2O + 2 mM phenylmethylsulfonyl fluoride, resuspended in 100 μl of lysis buffer (20 mM 1,4-piperazinediethanesulfonic acid, pH 7.0, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 1 mM DTT, protease inhibitors), and vortexed with glass beads. The resulting lysate was transferred to a fresh Eppendorf tube, the beads were vortexed with additional 200 μl of lysis buffer, and the lysate was combined with the previous one. One volume of the lysate was combined with nine volumes of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0) and allowed to equilibrate at 28°C for 15 min. On addition of 4 mg/ml o-nitrophenyl-β-d-galactoside, timing was started. When the solution turned yellow, 1 M Na2CO3 was added to terminate the reaction.
Supplementary Material
Acknowledgments
We thank J. Goeckeler and J. Brodsky for the UPR plasmid, R. Wright for anti-Hmg1 antibody, D. Klionsky for anti-Ape1, X. Zhang for technical help, and D. Taussig for critical reading of the manuscript. This research was supported by National Institutes of Health Grant GM-45444 to N.S.
Abbreviations used:
- EM
electron microscopy
- ER
endoplasmic reticulum
- GEF
guanine-nucleotide exchange factor
- GFP
green fluorescent protein
- PAS
pre-autophagosomal structure
- PM
plasma membrane
- UPR
unfolded protein response
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0269) on August 7, 2013.
REFERENCES
- Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol Biol Cell. 2005;16:178–192. doi: 10.1091/mbc.E04-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Chua CE, Gan BQ, Tang BL. Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci. 2011;68:3349–3358. doi: 10.1007/s00018-011-0748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan S, Saveljeva S, Gorman AM, Samali A. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci 70, 2425–2441. 2013 doi: 10.1007/s00018-012-1173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J, Yahi N. Molecular insights into amyloid regulation by membrane cholesterol and sphingolipids: common mechanisms in neurodegenerative diseases. Expert Rev Mol Med. 2010;12:e27. doi: 10.1017/S1462399410001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell GJ, Holloway ZG, Cobbold C, Monaco AP, Ponnambalam S. Cell biology of membrane trafficking in human disease. Int Rev Cytol. 2006;252:1–69. doi: 10.1016/S0074-7696(06)52005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW, Zubenko GS, Parker RR. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics. 1982;102:665–677. doi: 10.1093/genetics/102.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics. 1999;152:1543–1556. doi: 10.1093/genetics/152.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci USA. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the CVT pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol. 2006;8:1263–1269. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Lundblad V, Dorris D, Keaveney M. Yeast vectors and assays for expression of cloned genes. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb1306s39. Chapter 13, Unit 13.6. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, 3rd, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani A, Chen S, Rivera-Molina F, Reinisch K, Novick P, Ferro-Novick S. Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic. 2010;11:520–532. doi: 10.1111/j.1600-0854.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science. 1991;252:1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol. 2001a;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- Segev N. Ypt/Rab GTPases: regulators of protein trafficking. Sci STKE 2001. 2001b:re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- Segev N. Coordination of intracellular transport steps by GTPases. Semin Cell Dev Biol. 2011;22:33–38. doi: 10.1016/j.semcdb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N, Botstein D. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol Cell Biol. 1987;7:2367–2377. doi: 10.1128/mcb.7.7.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;10(Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wang RC, Levine B. Autophagy in cellular growth control. FEBS Lett. 2010;584:1417–1426. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Liu Y, Zhang XQ, Chen Y, Ye M, Zhu X, Yang S, Lipatova Z, Liang Y, Segev N. Modular TRAPP complexes regulate intracellular protein trafficking through multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics. 2012;191:451–460. doi: 10.1534/genetics.112.139378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.