Abstract
Tumor cell motility and invasion rely on actin cytoskeleton rearrangements mediated by the activation of RhoGTPase signaling pathways. Invadopodia are membrane-degrading protrusions that mediate extracellular matrix degradation. Here, we provide procedures for imaging RhoGTPase biosensors in tumor cells during the formation of invadopodia and matrix degradation.
Keywords: RhoGTPase, FRET, Biosensors, Invadopodia
1 Introduction
The use of fluorescent biosensors in systems that are highly significant to understanding disease processes including breast adenocarcinomas [1] has opened new avenues to address signal transduction studies in contexts that were not possible before. One such process recently described is how the Rho family of GTPases regulates tumor invasive protrusions [1]. When plated on two-dimensional matrices, tumor cells form actin-rich membrane degrading protrusions on their ventral surfaces called invadopodia [2]. In transverse section, invadopodia display a dot-like shape and are rich in actin and actin binding proteins such as cofilin, cortactin, and Arp2/3, which mediate actin polymerization.
Invadopodia formation is a multistep process that begins with the formation of an invadopodium precursor structure that matures into a matrix degrading invadopodia [3]. Invasive tumor cells including the rat adenocarcinoma cell line MTLn3 or the human MDA-MB-231 cell line have been used as model systems to study the different stages of invadopodia formation [3, 4]. RhoGTPases have been shown to be directly involved and regulate the formation of invadopodia [1], and thus are ideal targets to elucidate the regulatory mechanisms and dynamics of tumor cell invasion. As such, the use of the FRET (Forster Resonance Energy Transfer)-based biosensors in these systems requires careful considerations of the methods in order to ascertain that the biosensor readout is physiologically relevant. Here we provide methods to image RhoGTPase activation by using FRET based biosensors [1, 5] during invadopodia formation in tumor cells.
2 Materials
All stock solutions and buffers should be prepared in distilled water unless otherwise mentioned.
Cell lines: MTLn3 [6], MDA-MB-231, LinXE 293T and LinXA 293T cells (www.bioxys.com).
α-MEM: Supplemented with 5 % Fetal Bovine Serum (FBS) and 0.5 % penicillin/streptomycin.
D-MEM: Supplemented with 10 % FBS and 0.5 % penicillin/streptomycin.
L-15 media (Gibco): Store at 4 °C, protected from light.
Opti-MEM: Store at 4 °C, protected from light.
Ham's F-12K without phenol-red.
Trypsin.
Lipofectamine 2000 reagent (Invitrogen).
Lipofectamine and Plus reagents (Invitrogen).
Nucleofection Kit V (Lonza).
PBS.
Gelatin from Porcine skin, Type A (Sigma-Aldrich).
Glutaraldehyde solution: Grade I, 25 % in distilled water (Sigma-Aldrich).
Poly-l-lysine: 5 mg/mL stock in distilled water. Prepare a 1:10 dilution in PBS.
Hexadimethrine bromide (Polybrene; Sigma-Aldrich): 8 μg/μL stock.
Oxyfl uor reagent (Oxyrase).
Mouse EGF (Gibco) and human EGF (Invitrogen): 50 μM stock. Store mouse EGF at −80 °C in 5 μL aliquots and human EGF at −20 °C in 5 μL aliquots.
NaBH4 (Sodium Borohydrate): Store in a desiccated environment.
Paraformaldehyde (PFA) 16 % stock in distilled water.
Bovine serum albumin (BSA): Store at 4 °C.
dl-Lactate.
Triton X-100: 0.1 % stock in PBS.
Blocking solution: 1 % BSA, 1 % FBS in PBS.
1 % BSA in PBS.
MatTek dishes: 14 mm microwell (MatTek corporation).
G-418/neomycin: 100 μg/μL stock.
Puromycin: 10 μg/μL stock.
Doxycycline (Dox): 10 μg/mL stock.
Penicillin/streptomycin: 10,000 units of penicillin and 10,000 μg of streptomycin per mL. Use 0.5 % of this stock for cell culture.
Alexa 568 carboxylic acid, succinimidyl ester (Molecular Probes).
TagRFP-Cortactin plasmid [3].
pREV-tet-OFF advanced (Clontech), p-Babe-sin-puro-tet-CMV-RhoA biosensor [5] (https://www.addgene.org), pCL-ECO and pCL-AMPHO (www.imgenex.com).
Antibodies: Cortactin (ab81208, Abcam), Tks5 (Santa Cruz Biotechnology, sc-30122).
Beckman Coulter MoFlo XDP FACS System.
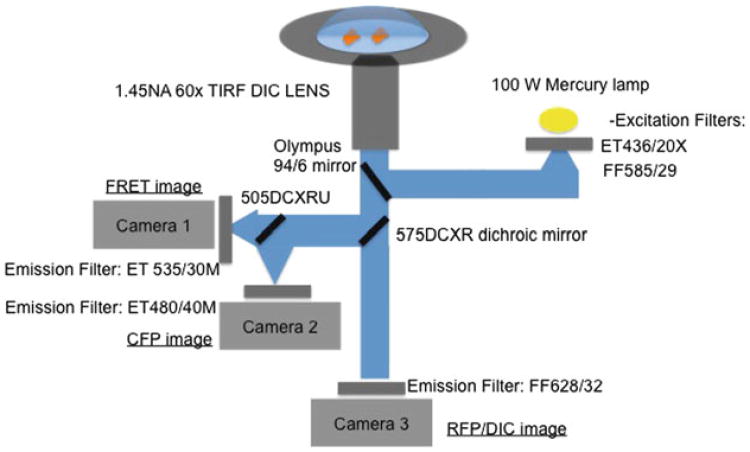
Microscope Filters: The filter sets used for ratiometric imaging were (excitation, emission, respectively): ECFP/mCerulean: ET436/20X, ET480/40M (Chroma Technology, Rockingham, VT); FRET: ET436/20X, ET535/30M (Chroma Technology); and TagRFP: FF585/29, FF628/32 (Semrock, Rochester, NY). The main fluorescence turret utilized a 10/90 (Reflection/Transmittance) mirror (Olympus, Center Valley, PA) that provided compatibility with all of the band pass filters used. Additionally, a 575DCXR dichroic mirror (Chroma Technology, Rockingham, VT) was installed to replace the internal prism of the microscope, enabling beamsplitting of CFP-YFP FRET emissions to the left port of the microscope and all the longer wavelengths to the bottom port of the microscope. 505DCXRU dichroic mirror (Chroma Technology, Rockingham, VT) was used to split the CFP and YFP-FRET emissions in the external beamsplitter attached to the left side port of the microscope. A 94:6 mirror from Olympus.
Microscope Lens: Olympus 60× Plan Apo N 1.45 NA UIS2 DIC lens.
3 Methods
3.1 Preparation of Gelatin Matrices
Place 25 mm round coverslips in a 6 well plate (see Note 1).
Treat 25 mm round coverslips or MatTek dish glass wells with 50 μg/mL of poly-L - lysine in PBS. Add 400 μL of poly-l-lysine to the inner MatTek dish glass well or 2 mL to completely cover the 25 mm coverslips (see Note 2).
Leave the coverslips/MatTek dishes at room temperature for 20 min.
Wash three times with 2 mL PBS.
Dissolve the gelatin powder in PBS to 0.2 %. Adjust the volume needed depending on the number of coverslips/MatTek dishes to prepare.
Preheat the PBS with the gelatin in a microwave for 2–3 min. Make sure all the gelatin is dissolved. Let it cool at 37 °C in a water bath.
Add 500 μL of 0.2 % gelatin (see Subheading 3.1.1 for different labeling options) to the MatTek dishes or 2 mL to the covrslips in the 6 well dishes. Incubate for 10 min at room temperature.
Wash three times with 2 mL PBS.
Treat MatTek dishes with 300 μL (or coverslips with 2 mL) of 0.5 % glutaraldehyde diluted in PBS for 15 min at room temperature. Glutaraldehyde cross-links the gelatin.
Wash three times with 2 mL PBS.
Dissolve NaBH4 in PBS to make a 5 mg/mL solution. Bubbles will form after adding the powder. Add the solution immediately to the dishes. Treat dishes/coverslips with 2 mL of NaBH4 for 15 min at room temperature (see Note 3). This treatment will quench autofluorescence.
Wash three times with 2 mL PBS.
Prepare PBS with 5 % penicillin/streptomycin. Add 2 mL per dish/coverslip to sterilize and keep at 4 °C in a humidified chamber (see Note 4).
Before plating cells wash with 2 mL PBS three times and add 2 mL of appropriate media (depending on the cell type). Leave at 37 °C for 15 min.
3.1.1 Preparing Labeled Gelatin
Gelatin can be prepared in two different ways: un-labeled or fluorescently labeled, depending on the requirements of the experiment. Un-labeled gelatin can be mixed with labeled gelatin in order to produce a fluorescent matrix to study the processes of matrix degradation by invadopodia as follows:
Conjugate gelatin with Alexa dyes following manufacturer's instructions (Invitrogen). Alexa 568 can be used for these assays.
Prepare 0.2 % gelatin stock solution in PBS.
After heating the gelatin, let it cool at room temperature. Mix 0.2 % gelatin with Alexa- conjugated gelatin in a 40:1 dilution ratio.
After this step follow the protocol in Subheading 3.1 starting from step 8.
Place the dishes in a humidified chamber, covered with aluminum foil (see Note 4).
3.2 EGF Preparation for Tumor Cell Stimulation
Prepare 1 μM EGF by diluting the mouse/human EGF 50 μM stock in 245 μL of starvation medium (see Subheading 3.7). Keep at 4 °C on ice during the experiment.
Prepare 5 nM (MTLn3 cells) or 10 nM (MDA-MD-231 cells) EGF by diluting 1 μM solution in starvation imaging medium (see Subheading 3.8).
Keep at 37 °C.
3.3 Viral Production and Stable tet-OFF Cell Lines
Coat 10 cm dishes with poly-l-lysine by adding 5 mL of diluted poly-l-lysine (diluted 1:10 in PBS from 5 mg/mL stock) for 20 min and aspirate prior to plating cells.
Wash with PBS three times.
Plate 6 × 106 LinXE 293 or LinXA 293 cells overnight in a 10 cm dish coated with poly-l-lysine (see Note 5).
- The following day prepare the transfection mixture.
- Eppendorf tube 1: 10 μg pREV tet-OFF, 6 μg of pCL-ECO, 120 μL of Lipofectamine Plus reagent; all mixed in 800 μL of Opti-MEM. For MDA-MB-231 use 6 μg pCL-Ampho instead of pCL-ECO (see Note 5).
- Eppendorf tube 2: 100 μL of Lipofectamine diluted in 800 μL of Opti-MEM.
- Incubate both tubes for 15 min at room temperature.
Combine the solutions of the two eppendorf tubes into one tube and incubate for 15 min at room temperature.
Replace the growth media in the LinXE or LinXA plates with 5 mL of Opti-MEM during the DNA–Lipofectamine incubation time. Add the transfection mixture to the cells following the 15 min incubation.
After 6 h stop the transfection by changing the media to α-MEM for MTLn3 or DMEM for MDA-MB-231.
Transfer the plates to a 32 °C incubator. At this temperature virus production is more efficient [7].
Collect the viral supernatant 48 h after transfection. Centrifuge for 5 min at 660 × g to remove any debris. Add polybrene at 8 μg/mL.
The day before infection plate 2–5 × 105 MTln3 or MDA-MB-231 cells on a 10 cm dish (see Note 6).
Add the supernatant from LinXE or LinXA cells containing the virus to the MTLn3 or MDA-MD-231 cells. We usually infect four times, once every 12 h before starting the selection.
Select for stable incorporation of the tet-OFF tetracycline trans-activator using G418 at 1 mg/mL.
3.4 Viral Production and Stable Biosensor Cell Line
Virus production can be performed as described in Subheading 3.3 by using the viral carrier construct under the tet-CMV promoter, such as p-Babe-sin-puro-tet-CMV-RhoA biosensor [5] containing the biosensor expression cassette, instead of pREV tet-OFF.
Transduction of MTLn3 or MDA-MB-231 cells expressing pREV tet-OFF plasmid can be performed as described in Subheading 3.3, steps 9 – 11. The transductions should be performed in the presence of 1 μg/mL Dox in order to suppress the biosensor expression during infection.
The selection of stably transduced cells is performed by adding gradually the selection reagent (puromycin) starting at 1 μg/mL, and eventually reaching the concentration of 10 μg/mL (see Note 7).
Once stable populations of biosensor expressing cells are obtained, induce the biosensor expression by removing the Dox for 72 h. At this point, cells should be FACS sorted to obtain uniform expression levels (see Note 8).
- For routine imaging experiments, biosensor expression through Dox removal should follow the following steps.
- Wash cells with PBS and briefly trypsinize to lift cells.
- Resuspend in growth medium without Dox, and centrifuge at 300 × g for 5 min to pellet the cells.
- Suction out as much of the medium as possible following the centrifugation, then resuspend in fresh growth medium without Dox.
- Plate cells at sparse density, routinely 1–2 × 105 cells per 10 cm dish (see Note 9).
3.5 Transfection of Tumor Cells with Invadopodia Markers
3.5.1 MTLn3
Plate 2 × 105 MTLn3 cells in a 6 well plate the day before transfection.
- Prepare transfection mixture in two eppendorf tubes (the volumes are for a single transfection in one well of a 6 well plate; for multiple transfections scale up the volumes).
- Eppendorf 1: 4 μL of Lipofectamine 2000 reagent in 250 μL of Opti-MEM.
- Eppendorf 2: 0.5–1 μg of DNA (invadopodium marker such as TagRFP-cortactin (red- shifted fluorescence proteins can be used in combination with a CFP/YFP biosensor)) in 250 μL of Opti-MEM.
- Incubate for 5 min at room temperature.
Mix the two solutions and incubate for 20 min at room temperature.
During the incubation, wash the cells with Opti-MEM and add 500 μL of Opti-MEM.
Add the transfection mixture to the cells and leave for 45 min (see Note 10).
After the transfection wash three times with growth medium.
8 h following the transfection, wash cells with PBS and trypsinize. Plate 2 × 105 cells per gelatin coated coverslip/MatTek dish.
3.5.2 MDA-MB-231
For MDA-MB-231 we recommend using Lonza Nucleofection Kit V.
Use the kit following manufacturer's instructions by using 1 μg of DNA per 1 × 106 cells.
Plate 2 × 105 cells per gelatin coated coverslips/MatTek dish.
3.6 Biosensor Induction and Transfection for Invadopodium Precursor Imaging
Induce biosensor expression in MTLn3 or MDA-MB-231 cells 72 h before the start of imaging (see Note 11). Wash the cells two times with PBS. Trypsinize and centrifuge for 5 min at 300 × g. Make sure to remove all media containing the Dox.
24 h before the experiment perform transfection with the invadopodium marker Cortactin-RFP [3] as described in Subheading 3.5.
Plate 2 × 105 cells on un-labeled gelatin coverslips.
3.7 Tumor Cell Starvation
3.7.1 MTLn3 Cells
Prepare starvation medium: L -15 containing 0.35 % BSA. Filter it to sterilize.
Wash cells two times with L-15 medium.
Wash cells once with the starvation medium and keep them in starvation medium.
Place cells for 3 h at 37 °C in a CO2-free incubator in starvation medium (see Note 12).
3.7.2 MDA-MB-231 Cells
MDA-MB-231 cells should be starved overnight with starvation medium: DMEM, 0.5 % FBS, 0.8 % BSA.
Wash cells two times with L-15 medium the next day. 10 min before the experiment, change the medium to L-15 with 0.35 %BSA.
Proceed with the EGF stimulation as in Subheading 3.9.
3.8 Starvation Imaging Medium
6 mL of Ham's F-12K without phenol-red [8, 9] is warmed to 37 °C to release any dissolved gases. Ham's F-12K reduces the background autofluorescence in the channels used for CFP/YFP FRET.
Bubble argon gas into the medium for 1 min to displace the oxygen.
BSA is added at 0.35 % and the medium is aliquoted into 2 mL tubes together with Oxyfl uor reagent (Oxyrase) at 1:100 dilution along with 5 mM DL -lactate (see Note 13).
The mixture is then incubated at 37 °C for 1 h and spun for 1 min at 24 °C, 20,000 × g to remove any debris from the Oxyfluor treatment prior to imaging. Filter it to sterilize.
3.9 EGF Stimulation and Image Acquisition
Mount the coverslips onto an imaging chamber as previously described [9] (Fig. 1) by using silicone vacuum grease. Add 400 μL of starvation imaging medium to the coverslips and place onto the microscope heating stage.
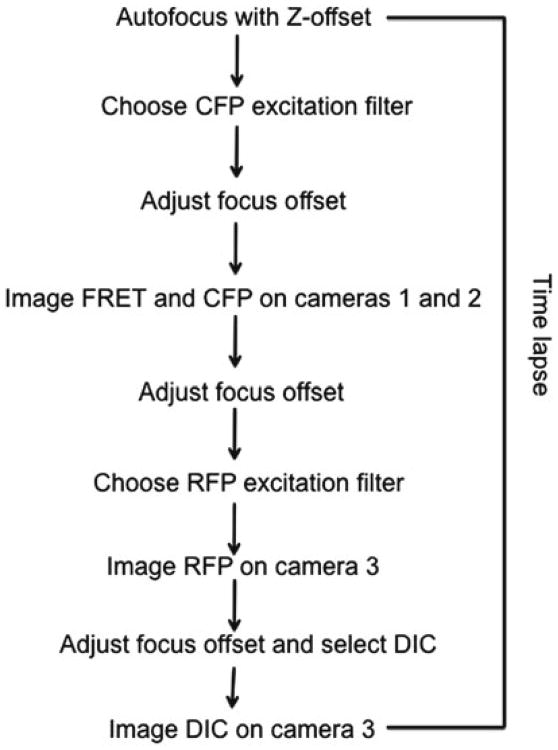
- Start the imaging following the diagram in Fig. 2. For sequential acquisition mode of four channels (CFP, FRET, RFP, DIC) we used a microscope set up [9] with a side port with two cameras for simultaneous acquisition of CFP/FRET and a bottom port for Red/DIC images (Fig. 3).
- First acquire CFP and FRET images simultaneously for 500–700 ms exposure time with binning 2 × 2 (see Note 14).
- Acquire TagRFP-Cortactin image by exposing for 200–400 ms at 2 × 2 binning.
- Acquire DIC image.
After 1–2 min add 400 μL of EGF at 10 nM for MTLn3 cells or 5 nM for MDA-MB-231 cells (the final concentration of EGF on the coverslips is 5 nM for MTLn3 and 2.5 nM for MDA-MB-231). Mix the volumes gently.
Continue imaging for 10–15 min.
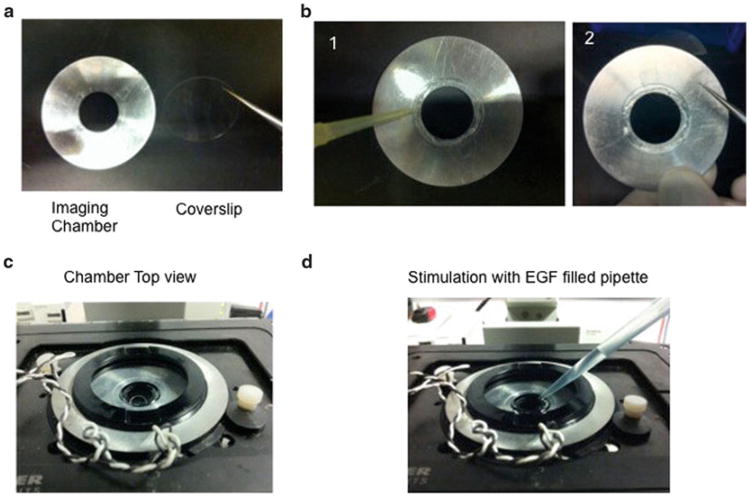
Fig. 1.
Mounting the imaging chamber. (a) Front view of the metal chamber and the coverslip. (b) Add grease to the inner part of the metal chamber (1) and place the coverslip on top of the greased area with the cell facing the grease (2). (c) Cover the coverslips with media. (d) Example of EGF stimulation. Approximately 800 μL can be added to the imaging chamber without spilling the imaging solution
Fig. 2. Flow diagram for the dual-camera for single-chain biosensor imaging. The red fluorescence acquisition and DIC acquisition using a third camera is also shown. The autofocus is achieved by an Olympus ZDC system [9].
Fig. 3. The light path diagram for the dual-camera, single-biosensor mode imaging. The third camera will enable acquisitions of red and DIC images.
3.10 Fixed Cell Imaging of RhoGTPase Biosensors
Alternatively, tumor cells expressing Rho GTPase biosensors can be fixed after EGF stimulation or in steady state conditions and immunofluorescence can be performed to localize endogenous proteins at invadopodia and visualize matrix degradation.
The day before the experiment plate 2 × 105 MTLn3 or MDA-MB-231 cells on Alexa 568 gelatin matrices.
Prepare 3.7 % PFA in PBS by diluting the 16 % stock.
After tumor cell stimulation or under steady state conditions, remove the media and add 2 mL of 3.7 % PFA and fix for 20 min at room temperature.
Wash two times with PBS for 5 min each.
Permeabilize the cells with 0.1 % Triton X-100 for 5 min.
Wash three times with PBS for 5 min each.
Incubate in blocking solution for 1 h.
Incubate with the primary antibody (anti-cortactin at 1:400 dilution or anti-TKs5 at 1:100 dilution) in blocking solution for 1 h at room temperature.
Wash three times with 1 % BSA in PBS.
Incubate with secondary antibody (use a far-red labeled secondary antibody) at room temperature for 1 h.
Wash two times with PBS and store at room temperature before imaging (see Note 15).
4 Notes
25 mm coverslips need to be washed in ethanol to sterilize before starting the poly-l-lysine treatment. We recommend keeping the coverslips in 100 % ethanol and flame sterilize before starting the coating.
We recommended using 25 mm coverslips for live cell imaging. When incorporated in the imaging chamber (see Subheading 3.9) grease sealing provides for better stability when doing EGF stimulations. For immunofluorescence we recommend using MatTek dishes since the volumes of antibodies can be minimized by adding 100 μL to the inside well.
NaBH4 should be kept in a desiccated environment with no air. Use the Desi-vac system (Fisher).
Gelatin-coated coverslips and MatTek dishes can be stored at 4 °C for 1–2 weeks.
MDA-MB-231 cells have to be transduced by using virus generated in cells expressing pCL-Ampho (LinXA) to allow infection of human cell lines and MTLn3 cells have to be transduced with virus generated in cells expressing pCL-ECO (LinXE).
When growing MTLn3 cells, it is very important to use very low passage (between 19 and 25) and not let them grow beyond 30–50 % confluence. MDA-MB-231 cells should not be kept for more than a month in culture.
Suggested increments; 1, 2, 4, 8, and 10 μg/mL. Add the puromycin at the time of cell splitting and allow cells to reach the correct subconfluency.
It is useful to obtain several differently gated populations in FACS sorting, representing low, medium and high biosensor expressors. Also, maintain frozen cell aliquots of unsorted cells for later analysis of effects of various expression levels of the biosensor, as necessary.
Here, sparse plating is important for induction. In order to enhance the levels of biosensor expression, trypsinization and direct replating at 24 and 48 h time points during induction can enhance biosensor expression.
In order to express transiently fluorescent proteins in MTLn3 cells, Lipofectamine 2000 transfection gives the best transfecion efficiency. Do not leave the transfection mixture more than 45 min as this causes significant cell death.
Induction times may vary depending on the biosensor used and the cell line in which the biosensor is being expressed. For Rho biosensors we usually get the best expression levels 72 h after induction.
L-15 media must be used in CO2-free conditions.
Treatment of the medium with argon gas and Oxyfluor/Lactate will control the production of oxygen radicals during imaging.
Imaging conditions must be controlled to minimize photodamage [8]. Furthermore, signal to noise ratio of 2:1–3:1 should be targeted between the cell fluorescence and the background at the dimmest part of the cell to achieve optimal results [8].
Gelatin coated dishes with stained cells can be stored at 4 °C for a week.
Acknowledgments
We thank the Condeelis, Cox, Hodgson, and Segall laboratories for helpful discussions. This work was funded by GM093121 (J.J.B-C, Y.M., L.H.), T32 GM007491 (Y.M.), and CA150344 (J.C, J.J.B-C).
References
- 1.Bravo-Cordero JJ, et al. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr Biol. 2011;21:635–644. doi: 10.1016/j.cub.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oser M, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oser M, et al. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J Cell Sci. 2010;123:3662–3673. doi: 10.1242/jcs.068163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 6.Segall JE, et al. EGF stimulates lamellipod extension in metastatic mammary adenocarcinoma cells by an actin-dependent mechanism. Clin Exp Metastasis. 1996;14:61–72. doi: 10.1007/BF00157687. [DOI] [PubMed] [Google Scholar]
- 7.Kaptein LC, Greijer AE, Valerio D, van Beusechem VW. Optimized conditions for the production of recombinant amphotropic retroviral vector preparations. Gene Ther. 1997;4:172–176. doi: 10.1038/sj.gt.3300373. [DOI] [PubMed] [Google Scholar]
- 8.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr Protoc Cell Biol. 2010;Chapter 14(Unit 14.11):11–26. doi: 10.1002/0471143030.cb1411s46. Editorial board, Juan S. Bonifacino … [et al.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spiering D, Hodgson L. Multiplex imaging of Rho family GTPase activities in living cells. Methods Mol Biol. 2012;827:215–234. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]