Abstract
Dysfunction of protein handling has been implicated in many neurodegenerative diseases and inhibition of the ubiquitin-proteasome system (UPS) has been linked to the formation of protein aggregates and proteinopathies in such diseases. While proteasomal inhibition could trigger an array of downstream protein handling changes including up-regulation of heat shock proteins (HSPs), induction of molecular chaperones, activation of the ER stress/unfolded protein response (UPR), autophagy and aggresome formation, little is known of the relationship of proteasomal inhibition to the sequence of activation of these diverse protein handling systems. In this study we utilized the reversible proteasome inhibitor MG132 and examined the activity of several major protein handling systems in the immortalized dopaminergic neuronal N27 cell line. In the early phase (up to 6 hours after proteasomal inhibition), MG132 induced time-dependent proteasomal inhibition which resulted in stimulation of the UPR, increased autophagic flux and stimulated heat shock protein response as determined by increased levels of phosphorylation of the eukaryotic translation initiation factor 2 alpha (eIF2α), C/EBP homologous protein (CHOP)/GADD153, turnover of autophagy related microtubule-associated protein 1 light chain 3 (LC3) and increased levels of Hsp70 respectively. After prolonged proteasomal inhibition induced by MG132, we observed the formation of vimentin-caged aggresome-like inclusion bodies. A recovery study after MG132-induced proteasomal inhibition indicated that the autophagy-lysosomal pathway participated in the clearance of aggresomes. Our data characterizes the relationship between proteasome inhibition and activation of other protein handling systems. These data also indicated that the induction of alternate protein handling systems and their temporal relationships may be important factors that determine the extent of accumulation of misfolded proteins in cells as a result of proteasome inhibition.
Keywords: Proteasome activity, ER stress, Autophagy, Heat shock protein responses, Aggresome, UPS dysfunction
1. Introduction
Neurodenegerative disorders including Parkinson’s disease (PD), Parkinsonian like syndromes such as Amyotrophic Lateral Sclerosis (ALS) and Progressive Supranuclear Palsy (PSP), Alzheimers Disease (AD) and poly Q disorders such as Huntington’s disease, are characterized by the accumulation of misfolded proteins, diagnostic protein aggregates and inclusion body formation which in turn leads to toxicity, loss of neurons and eventual loss of functional capacity [1–3]. Considerable attention has been focused on protein handling in Parkinson’s disease (PD) given the critical role of α-synuclein aggregation in disease etiology [4]. Although the molecular mechanism of loss of dopaminergic neurons is not fully understood, dysfunction of the ubiquitin proteasomal system (UPS) has been proposed as an important factor in the pathogenesis of both familial and sporadic PD [2,3,5,6]. In addition, we and others have also demonstrated that dopaminergic metabolites can cause inhibition of the proteasome leading to impaired protein handling [7,8]. Finally, a number of animal models have emphasized the role of the UPS and overall protein handling in the pathogenesis of Parkinson’s disease [9–11]. A comprehensive recent review has summarized the role of protein handling systems in Parkinson’s Disease [12].
The ubiquitin-proteasome pathway is the major proteolytic system in the cytosol and nucleus of all eukaryotic cells. Most substrates of this pathway are labeled for degradation by attachment of multiple molecules of ubiquitin, a small 8kDa protein [13,14]. The resulting ubiquitinated proteins are then recognized and degraded by the 26S proteasome complex, which contains chymotrypsin-like, caspase-like and trypsin-like active sites [15]. Functional blockade of this degradative system leads to enhanced aggresome formation [16] which have been defined as protein aggregates assembling near the microtubule-organizing center (MTOC) likely representing a cellular defense system for accumulation of aggregated proteins [17]. Perinuclear aggresome formation is characterized by colocalization of ubiquitin positive aggregates with the intermediate filament vimentin [18] and aggresomes have been implicated in proteinaceous inclusions found in neurodegenerative diseases [19]. Other cellular protein handling systems have been implicated in neurodegenerative diseases including the ER stress response/unfolded protein response (UPR), autophagy, and the heat shock protein response [20,21]. When challenged with misfolded proteins the ER stress sensor PERK is activated to phosphorylate and inhibit eIF2α which alleviates the burden of newly synthesized polypeptides by decreasing protein translation [22,23]. Heat shock proteins function as cellular chaperones that assist in the correct folding of many proteins and are often the first line of defense against misfolded proteins [24]. Autophagy is a highly regulated process that sequesters damaged organelles, long-lived proteins and potential toxic aggregates into an autophagosome, which subsequently fuses with lysosomes resulting in degradation of proteins and damaged organelles [25].
Although the ER stress response/UPR, autophagy, heat shock response and aggresome formation have all been implicated as compensatory mechanisms that occur downstream of proteasomal inhibition [26–30], little is known about the interrelationship between proteasomal inhibition and other cellular protein handling systems, especially with respect to the sequence of activation of these systems when stressed cells are challenged with accumulated proteins. The majority of studies to date have assessed one or two markers of protein handling which leads to potentially conflicting data regarding the involvement of protein handling mechanisms in toxicity. We therefore conducted this study using the proteasomal inhibitor MG132 to define in a comprehensive manner changes in all major protein handling systems subsequent to proteasomal inhibition. Disturbances in protein handling are considered to be crucial in neurodegenerative diseases in general but Parkinson’s disease (PD) in particular [2,3,5,6]. Since loss of dopaminergic neurons is an established characteristic the pathology of PD, we utilized dopaminergic cells in this study to define changes in protein handling systems downstream of proteasomal inhibition. We have also examined which system(s) participate in the clearance of ubiquitin protein aggregates under conditions of impaired proteasomal activity.
2. Materials and methods
2.1. Materials and Antibodies
Pepstatin A, leupeptin, tunicamycin, chloroquine diphosphate salt, ammonium chloride and 3-methyladenine were obtained from Sigma Chemical (St. Louis, MO, USA). MG132, mouse monoclonal anti-Hsp27, anti-Hsp70 antibodies and rabbit polyclonal anti-Hsp90α antibody were purchased from Enzo (Farmingdale, NY, USA). The fluorescently labeled proteasome substrate Suc-Leu-Leu-Val-Tyr-AMC was obtained from Bachem (Torrance, CA, USA). Both proteasome inhibitor and substrate were dissolved in dimethyl sulfoxide. Rabbit polyclonal anti-tyrosine hydroxylase (AB152) was purchased from Millipore (Temecula, CA, USA). Mouse monoclonal anti-vimentin (clone V-9) was obtained from Neomarkers (Thermo Scientific, USA). Rabbit polyclonal anti-LC3 (NB100–2331) was obtained Novus (Littleton, CO, USA). Rabbit polyclonal anti-phospho-eIF2α (Ser51, #9721), anti-eIF2α (#9722), anti-caspase-3 (#9662) and mouse monoclonal anti-CHOP (#2895) antibodies were purchased from Cell Signaling (Danvers, MA, USA). Rabbit polyclonal antibody targeting both the α and β subunits of the 20S proteasome were obtained from Abcam (Cambridge, MA, USA).
2.2. Cell Culture and treatment
Immortalized rat mesencephalic dopaminergic cells (N27) were obtained from Dr. Curt Freed, Department of Clinical Pharmacology and Toxicology, University of Colorado Anschutz Medical Campus, Aurora CO [31]. N27 cells were cultured in RPMI-1640 media (Cellgro, Manassas, VA, USA) containing 10% (v/v) fetal bovine serum, l00U/ml penicillin and 100μg/ml streptomycin (Cellgro, Manassas, VA, USA) in a humidified atmosphere of 5% CO2 at 37°C.
2.3. Proteasomal activity assay
Proteasomal activity was determined in N27 cells by measuring the fluorescence of cleaved substrate Suc-Leu-Leu-Val-Tyr-AMC (chymotrypsin-like activity) as described previously [32,33]. Following treatment with MG132 cells were lysed with proteasomal activity assay buffer (50mM Tris-HCl, pH 7.5, 250mM sucrose, 1mM dithiothreitol, 5mM MgCl2, 2mM ATP, 0.5mM EDTA and 0.025% (w/v) digitonin) and then centrifuged at 10,000g for 14min at 4°C. The supernatants were collected and protein concentrations were determined using the method of Lowry [34]. To determine proteasome activity 20μg of cell lysate was incubated with 100μM fluorogenic peptide probe in proteasomal activity assay buffer (200μl) at 37°C. After 30min the reactions were stopped by the addition of 200μ1 ice-cold ethanol and the samples were centrifuged at 10,000g for 4min. The supernatant was transferred to a 96-well plate and the fluorescence of liberated 7-amino-4-methylcoumarin (Ex: 380nm; Em: 460nm) was measured using a fluorescence micro-plate reader (Molecular Devices, Sunnyvale CA, USA).
2.4. Immunblot Analysis
Following treatment with MG132 cells were washed with phosphate buffered saline (PBS) and then lysed with RIPA buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 0.1% (w/v) SDS, 1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate) supplemented with protease inhibitors (Complete, Mini Protease Inhibitor Cocktail, Roche, Germany) and phosphatase inhibitors (Sigma Chemical, USA). Cells were sonicated briefly on ice and then centrifuged at 10,000g for 14 min at 4°C. The supernatants were collected and protein concentrations were determined by the method of Lowry [34]. Proteins were diluted in 2X Laemmli SDS sample buffer and heated to 70°C for 5 min. Proteins were separated using either a 7.5% (polyubiquitinated proteins) or 12% SDS-PAGE precast minigel (BioRad Laboratories, Hercules, CA, USA) Proteins were transferred to PVDF membranes in 25mM Tris, 192mM glycine containing 20% (v/v) methanol at 120mA for 4h at 4°C. Membranes were blocked for 1h at room temperature in 10mM Tris-HCl, pH 8.0, 150mM NaCl, 0.2% (v/v) Tween-20 (TBST) containing 5% (w/v) non-fat dry milk. Membranes were incubated with primary antibodies overnight at 4°C. Membranes were then washed extensively in TBST for 1h and then probed with HRP-conjugated goat anti-rabbit or HRP-conjugated goat anti-mouse IgG (1:5000; Jackson Immunoresearch Labs, West Grove, PA, USA) for 30min at room temperature. Membranes were washed three-times in TBST and protein bands were visualized using enhanced chemiluminescence (Thermo Scientific, USA).
2.5. Analysis of apoptosis
Apoptosis was measured using FITC-conjugated anti-annexin V antibody and propidium iodide (PI). The FITC-conjugated anti-annexin V antibody was purchased from Invitrogen (Carlsbad, CA, USA) and binding buffer was obtained from BioLegend (San Diego, CA, USA). After the indicated duration of treatment with MG132, both attached floating cells were collected and pelleted by centrifugation at 10,000g for 5 min. Cell pellets were gently resuspended in 500μl annexin V binding buffer and incubated with anti-annexin V (2μl) and PI (1μl of 100μg/ml stock in phosphate-buffered saline) for 10 min at 37°C in the dark. Samples were processed on ice and analyzed on a BD Biosciences FACS Calibur Flow Cytometer (San Jose, CA, USA) measuring fluorescence emission at 530nm (FL1, FITC) and PI at above 600nm (FL2). FL1 and FL2 were collected on log-scale with voltages of 423 and 376, respectively. Signal overlap was adjusted and compensated. Data was acquired and analyzed using Cellquest software (Becton-Dickenson, Mountainview, CA, USA).
2.6. Immunocytochemistry and quantification of aggresomes
Sub-confluent N27 cells were grown on glass coverslips and treated with MG132 for the indicated times. Cells were then washed three times with PBS and fixed in 3.7% (v/v) formaldehyde in PBS for 12min. After washing with PBS, cells were permeabilized with 0.1% (v/v) Triton X-100 in PBS for 10min, and then washed extensively with PBS and incubated in blocking buffer (RPMI1640 +10% (v/v) FBS) for 1h. Cells were incubated with anti-ubiquitin antibody (1:100); anti-vimentin antibody (1:1000); mouse anti-Hsp70/72 antibody (1:1000) overnight at 4°C. Cells were then washed three times in TBST and incubated with secondary antibodies Alexa Fluor 594 conjugated goat anti-rabbit IgG (1:1000) and Alexa Fluor 488 conjugated goat anti-mouse IgG (1:2000) for 30 min. DAPI (1μg/ml) was included with the secondary antibodies for nuclear staining. Coverslips were sequentially washed three times with TBST then once in deionized water and mounted on glass slides using SuperMount (BioGenex, USA). Cells were viewed on a Nikon TE2000 microscope with a Nikon C1 confocal imaging system. In control studies no significant immunostaining was observed in the absence of primary antibodies (data not shown). The number of aggresome (both ubiquitin and vimentin positive staining aggregates) was quantified by counting 100 cells in 20 random fields of each coverslip. Results shown are an average of at least 3 independent experiments for each treatment.
3. Results
3.1. Exposure to MG132 induced caspase 3 cleavage and toxicity in N27 cells
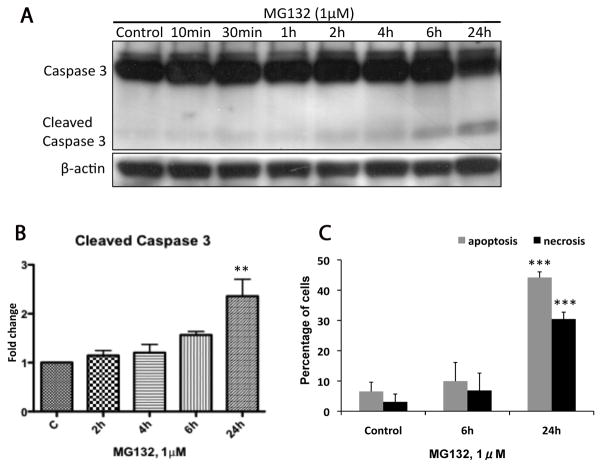
We first examined the potential toxicity of proteasomal inhibition in dopaminergic neuronal N27 cells by using the tripeptide aldehyde MG132. MG132 is a commonly used reversible proteasome inhibitor which enters cell rapidly [13]. A low concentration of MG132 (1μM) was used and this concentration was sufficient to inhibit specifically the chymotrypsin-like activity of the proteasome in cell culture without affecting other proteases [13,35]. We found MG132 induced time-dependent apoptosis in N27 cells. As shown in Figure 1A and B, treatment with MG132 resulted in a detectable increase of cleaved caspase 3 which reached a significant level at 24h. Using flow cytometric analysis, we further confirmed low dose MG132 induced significant apoptotic cell death at 24h with more than 40% of cells staining positive for Annexin V (Fig. 1C). Moreover, after 6h exposure to MG132, an increased level of PI positive cells (necrosis) was observed and the percentage of necrosis increased to 30% at 24h (Fig. 1C), suggesting that necrosis accompanied the apoptogenic activity of MG132. No significant induction of either apoptosis or necrosis after 2h and 4h exposure to MG132 was found. These results indicated that MG132 induced both time-dependent apoptosis and necrosis in N27 cells.
Figure 1. Time course of MG132 induced cytotoxicity.
The induction of apoptosis and necrosis was measured in N27 cells at 6 and 24h following treatment with MG132 (1μM). (A) The representative immunoblot shows caspase 3 cleavage increased to a significant level at 24h. (B) Fold changes of cleaved caspase 3 are normalized by control and estimated by densitometry. (C) Apoptosis was also measured in N27 cells using annexin V/PI cell staining in combination with flow cytometry. Significant apoptosis and necrosis were observed after 24h treatment. These data are presented as mean ± SD, (n=3–6); **p<0.01 and ***p<0.001 are considered significant by ANOVA using Dunnet’s multiple comparison test.
3.2. MG132 inhibited proteasomal activity and induced the accumulation of polyubiquitinated proteins in a time-dependent manner in N27 cells
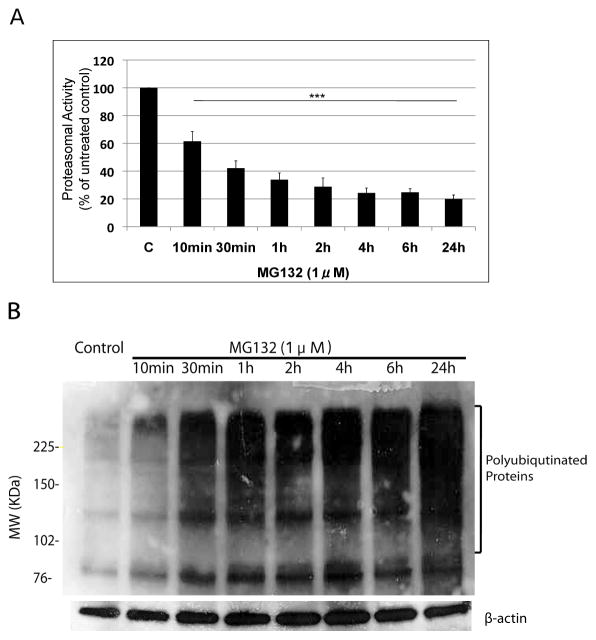
We next conducted a detailed time-course and measured proteasomal activity after treating N27 cells with MG132 from 10min up to 24h. The enzymatic activity of 20S/26S proteasome was determined by measuring the cleavage of the specific fluorogenic substrate Suc-LLVY-AMC at 380nm/460nm as previously described [33]. Proteasomal activity was rapidly decreased (by 40%) after a 10min exposure to MG132, and only 20% of proteasomal activity remained after 24h treatment (Fig. 2A). No significant differences in proteasomal activity were found between 2h and 24h treatments (Fig. 2A), indicating that a 2h exposure to low dose MG132 (1μM) was sufficient to impair the majority of proteasomal activity in N27 cells.
Figure 2. Time course of proteasome inhibition following treatment with MG132.
Proteasesome activity (A) and the accumulation of HMW polyubiquitinated proteins (B) were measured in N27 cells after treatment with MG132 (1μM). The proteasome activity was quantified by measuring fluorescence of Suc-Leu-Leu-Val-Tyr-AMC (chymotrypsin-like activity) cleavage at 380/460nm. The data is presented as mean ± SD, (n=3). ***p<0.001 is considered significant by one way analysis of variance (ANOVA) using Dunnet’s multiple comparison test. (B) The representative immunoblot blot shows time-dependent accumulation of higher molecular weight polyubiquitinated proteins and β-actin was included as a loading control.
Accumulation of high molecular weight (HMW) polyubiquitinated proteins has been used as an indicator of proteasomal inhibition [36]. To examine whether proteasome inhibition resulted in the accumulation of HMW polyubiquitinated proteins in N27 cells, we performed immunoblot analysis on whole cell lysates following MG132 treatment. HMW polyubiquitinated proteins could be detected after a 10min exposure to MG132 and increased to a more significant level at 24h (Fig. 2B), which correlated with proteasomal activity assays (Fig. 2A).
3.3. Rapid posphorylation of eIF2a in N27 cells after treatment with MG132. Induction of ER stress response/UPR is tightly associated with early proteasomal inhibition in N27 cells
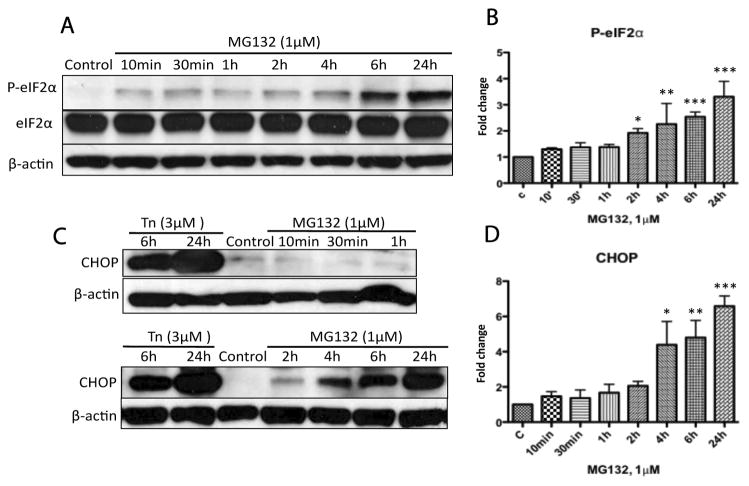
The mammalian endoplasmic reticulum (ER) is a crucial compartment for synthesis, folding and quality control of most secretory and transmembrane proteins. PERK is a rapidly activated ER stress sensor [23], and a recent study found that PERK and phosphorylated eIF2α co-localize with α-synuclein positive Lewy bodies in neurons of PD patients [37]. Therefore, we examined whether the PERK pathway was activated in response to proteasomal inhibition. As shown in Figure 3A and B, eIF2α was rapidly phosphorylated after 10min exposure to MG132 (1μM) and was significantly increased after 2h as determined by densitometry analysis. Induction of CHOP, a down-stream signal of phospho-eIF2α, could be detected after 2h and accumulated to a significant level, as indicated by densitometry analysis, after 4h exposure to MG132 (Fig. 3C, D). Tunicamycin (Tn) was included as a positive control for the induction of ER stress [38]. Our data demonstrates a close relationship between proteasomal inhibition and induction of ER stress response in dopaminergic cells.
Figure 3. Time course of ER stress activation following treatment with MG132.
N27 cells were treated with MG132 (1μM) then processed for immunoblot analysis for phospho-eIF2α (Ser-51), eIF2α and CHOP. Treatment with tunicamycin (Tn, 3μM) was included as a positive control for ER stress and β-actin was included as a loading control. (A, B) Phosphorylation of eIF2α; following treatment with MG132 increased to a significant level at 2h as indicated by densitometry. (C, D) Increases in CHOP reached a significant level at 4h as indicated by densitometry. (B) (D) Fold changes of the indicated protein levels are normalized by control and estimated by densitometry. These data are presented as mean ± SD, (n=3); *p<0.05, **p<0.01 and ***p<0.001 are considered significant by ANOVA using Dunnet’s multiple comparison test.
3.4. Proteasomal inhibition stimulated the turnover ofLC3 I/II and induced autophagy
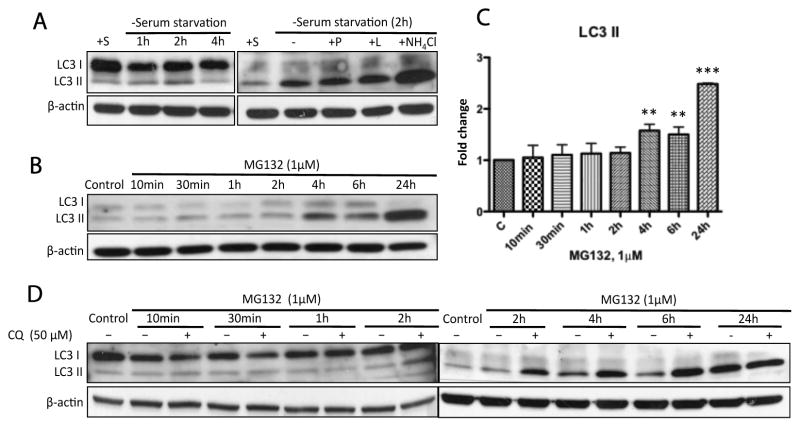
Since the autophagic flux in N27 cells has not been determined using lysosomal protease inhibitors in previous studies of autophagy, we performed initial experiments to define whether N27 cells were autophagy-competent. We conducted a serum starvation experiment with or without co-treatment with lysosomal protease inhibitors in N27 cells. The conversion of LC3 I to LC3 II, an indicator of autophagy [39,40], increased up to 2h following serum starvation,. However, after 4h of serum starvation the conversion of LC3 I to LC3 II decreased (Fig. 4A, left) suggesting that autophagic flux was an early event. In addition, serum starvation (2h) in combination with lysosomal inhibitors (pepstatin A, leupeptin and ammonium chloride) resulted in a further increase in LC3 II accumulation (Fig. 4A, right). These data indicate that N27 cells are autophagy competent.
Figure 4. Prolonged proteasomal inhibition by MG132 stimulated time-dependent autophagic flux.
(A) N27 cells were confirmed to be autophagy-competent. LC3 II accumulation is a marker for autophagosome formation. (B) The representative immunoblot shows autophagy induction following treatment with MG132 (1μM). (C) Significant accumulation of LC3 II could be detected at 4h and to a greater extent at 24h. Fold changes of LC3 II levels are normalized by control and estimated by densitometry. These data are presented as mean ± SD, (n=3); **p<0.01 and ***p<0.001 are considered significant by ANOVA using Dunnet’s multiple comparison test. (D) Induction of autophagy by MG132 was confirmed by co-incubation with chloroquine (CQ). Cells were exposed to MG132 (1μM) with or without the co-treatment of CQ for the indicated times and then processed for immunoblot analysis. Note that CQ is a specific lysosome inhibitor and could suppress the fusion of autophagosomes with lysosomes. The elevated levels of LC3 II in the presence of CQ (>4h) excluded the possibility that MG132 stimulated autophagic flux was due to the blockade of autophagic degradation in N27 cells.
To determine the relationship between proteasomal inhibition and autophagy, we performed the same time-course study using MG132 (1μM) and found the accumulation of LC3 II was not significantly increased until 4h following treatment (Fig. 4B, C). However, even though autophagy was induced and sustained at a high level as indicated by the elevated accumulation of LC3 II after exposure to MG132, this protective mechanism could not prevent cell death caused by prolonged proteasomal inhibition. To further distinguish whether the increased levels of LC3 II by MG132 treatment were due to either upregulation of autophagosome formation or blockage of autophagic degradation [39], we utilized chloroquine (CQ) as an inhibitor of lysosome-dependent autophagic degradation. Treatment of N27 cells with CQ (50μM) for 24h induced no observable cytotoxicity (data not shown) and treatment with MG132 had no effect on LC3 II accumulation at early time points (10min to 2h) in the absence or presence of CQ (Fig. 4D). After 4h exposure the levels of LC3 II accumulation were further elevated and increased to a much higher level in the presence of CQ (Fig. 4D). This demonstrates that MG132 induced increases in LC3 II were not a result of inhibition of lysosome-dependent autophagic degradation.
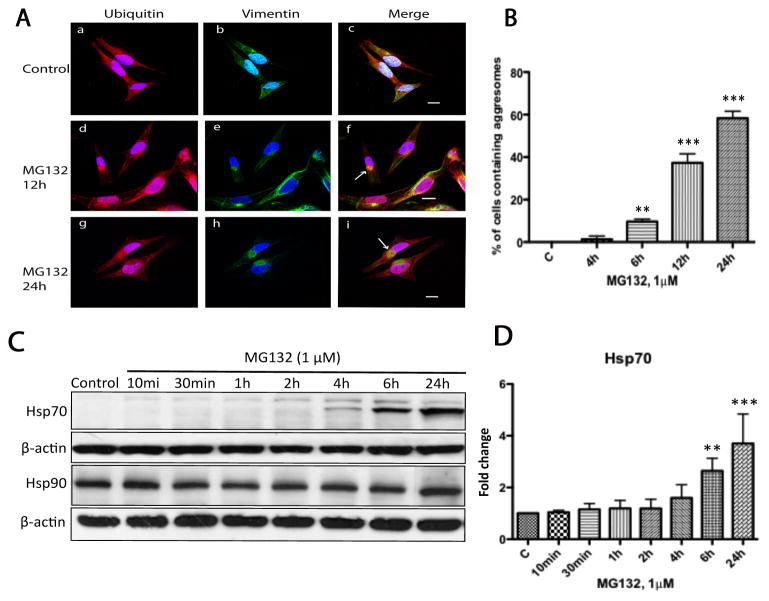
3.5. Prolonged proteasomal inhibition resulted in formation of aggresome-like inclusion bodies and induction of heat shock protein 70 (Hsp70) in N27 cells
There is an increasing body of evidence implicating aggresomes as a cytoprotective mechanism since they sequester misfolded proteins and potentially toxic protein aggregates [41]. Lewy bodies found in Parkinson’s Disease have found to display many similarities to aggresomes, particularly in the composition of ubiquitinated proteins [19,42]. Using double immunostaining in association with confocal microscopy we performed a time-course study in N27 cells treated with MG132 to examine the effect of proteasomal inhibition on the distribution of intracellular ubiquitinated proteins and to determine how early aggresomes are formed in N27 cells in response to proteasome inhibition. Aggresomes can be identified in cells by their characteristic perinuclear accumulation of ubiquitin positive and vimentin caged inclusion bodies [18]. In untreated cells ubiquitin was well distributed throughout the cytoplasm while vimentin filaments could be found transversing the cell body (Fig. 5A, a–c). In contrast, in cells that had been treated with MG132 for 24h (Fig. 5A), vimentin was completely collapsed and redistributed to form a cagelike structure surrounding ubiquitin inclusion bodies in the perinuclear region (Fig. 5A, g–i). Intermediate changes in vimentin and ubiquitin distribution could be detected after 12h of incubation with MG132 (Fig 5A, d–f). To further confirm the site of aggresome formation as the centrosome/microtubule-organizing center (MTOC) [43], we performed immunostaining to examine ubiquitin and γ-tubulin co-localization. In MG132 treated cells (1μM, 24h), γ-tubulin was found to co-localize with ubiquitin-positive inclusion bodies while in control cells no association could be observed (data not shown). To define the relationship between proteasomal inhibition and aggresome formation we performed the same time-course study as above using MG132 (1μM). In these studies aggresomes could be detected but only 6h after MG132 treatment (Fig. 5B), suggesting that aggresome formation was a relatively late event and was triggered by prolonged proteasomal inhibition in N27 cells.
Figure 5. Prolonged treatment with MG132 resulted in aggresome formation.
(A) Aggresome formation was examined by confocal microscopy in N27 cell treated with MG132 (1μM) for 24h. Aggresomes were characterized as vimentin-caged, ubiquitin-positive perinuclear inclusions. For these studies cells were either treated with DMSO (a–c) or MG132 (1μM, d–f) for 2h to 24h, then processed for confocal laser microscopy using double immuno-labeling with antibodies to ubiquitin (red) and vimentin (green). Nuclei were stained with DAPI (blue). Arrows (panels f, i) indicate aggresome-like inclusion bodies. In these studies no aggresomes could be found at early time points (2 and 4h) after treatment with MG132. (B) Quantitative determination of aggresome formation. Cells were treated with MG132 (1μM) and at the indicated times the percentage of aggresome containing cells were determined by counting 100 cells across 20 random fields. The data represents the means ± SD, (n=3). Scale bar 20μm. (**p<0.01 and ***p<0.001, ANOVA using Dunnet’s post test). (C) Induction of Hsp70 but not Hsp90 following treatment with MG132. N27 cells were exposed to MG132 (1μM) from 10min to 24h as indicated and cytosolic fractions were then analyzed by immunoblot analysis for Hsp70 and Hsp90. (D) Fold changes of Hsp70 levels are normalized by control and estimated by densitometry. These data are presented as mean ± SD, (n=3); **p<0.01 and ***p<0.001 are considered significant by ANOVA using Dunnet’s multiple comparison test.
Induction of heat shock proteins and molecular chaperones as a neuroprotective response and accordingly Hsp70 and Hsp90 have also been shown to colocalize with Lewy bodies [42]. Therefore, we performed a time-course study using MG132 (1μM) and examined the expression levels of Hsp70 and Hsp90. Consistent with previous findings Hsp70 was not detectable in untreated cells [24] whereas the level of Hsp70 increased after 4h exposure to MG132, and was further elevated to a more significant level at 6h and 24h (Fig. 5C, D). In addition, double immunolabeling with confocal microscopy confirmed colocalization of Hsp70 with ubiquitin containing aggresomes in N27 cell after treatment with MG132 (1μM) for 24h (data not shown). While Hsp90 was constitutively expressed in unstressed cells, it was not upregulated even after prolonged proteasomal inhibition (Fig. 5C). In addition to Hsp70 and Hsp90, Hsp27 and Hsp40 are also expressed in the central nervous system and it has been reported that Hsp27 expression is protective against α-synuclein-induced toxicity [44]. However, levels of Hsp27 and Hsp40 were not significantly altered in N27 cells by proteasomal inhibition as indicated by immunoblot analysis and confocal microscopy (data not shown).
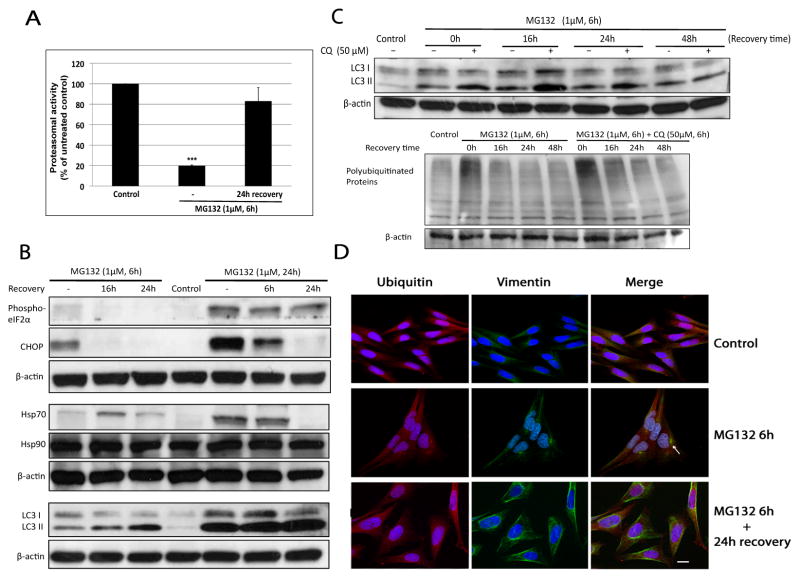
3.6. Involvement of autophagy but not the ER stress response/UPR or the heat shock response, in the clearance of aggresomes during recovery from proteasomal inhibition
Our data indicated the PR, autophagy, aggresome formation and Hsp70 were all activated in response to proteasomal inhibition prior to the earliest signs of the occurrence of cell death. After 6h exposure to MG132, it seems likely that all these protein-handling systems work cooperatively to deal with the accumulation of misfolded proteins and aggregates resulting from UPS impairment. To further elucidate which protein handling system played a major role in restoring intracellular protein homeostasis we conducted an MG132 recovery study. As shown in Figure 6A, proteasomal activity was inhibited by ~80% following treatment with MG132 for 6h and after 24h incubation with fresh medium in the absence of MG132, proteasomal activity was restored confirming the effect of MG132 on proteasome inhibition was reversible as previously reported [13]. Since cell death was detectable after 6h exposure to MG132 and increased to a more significant level at 24h (Fig. 1B), we performed the recovery experiment after 6h and 24h. Interestingly, the amount of LC3 II was found to be elevated during the 24h recovery period in both sets of recovery experiments whereas the level of p-eIF2α, CHOP and Hsp70 were decreased and the expression of Hsp27, Hsp40 (data not shown) and Hsp90 was not changed (Fig. 6B). These results demonstrated that the restoration of proteasome activity and cellular recovery was associated with stimulated autophagic flux, but not the misfolded protein response of the ER or Hsp chaperone activity.
Figure 6. Autophagy was induced and associated with aggresome clearance during the recovery period of proteasomal inhibition.
(A) Treatment of N27 cells with MG132 (1μM) inhibited proteasomal activity by greater than 80% after 6h and proteasome inhibition could be reversed following a 24h recovery in complete media in the absence of MG132. Proteasome activity was measured as described in Material and Methods. Results are presented as mean ± SD (n=3) (***p<0.001, by ANOVA using Dunnet’s post test). (B) Autophagic LC3 II levels were elevated during the recovery process whereas biomarkers of ER stress response/UPR and heat shock protein responses were attenuated. Cells were treated with MG132 (1μM) for 6h or 24h, and then exposed to fresh medium (minus MG132) for the indicated recovery period. (C) Induction of autophagy during the recovery process. N27 cells were exposed to MG132 (1μM) in the presence or absence of CQ (50μM). After 6h, the cultured medium was exchanged with drug free medium for the indicated recovery period. Note that aggresomes were removed as indicated by the disappearance of HMW polyubiquitinated proteins in the bottom immunoblot of (C) as well as the number of cells with of ubiquitin and vimentin positive perinuclear inclusions (D). Arrow in D pointed out an aggresome-like inclusion body. Scale bar 20μm.
To confirm the increase of LC3 II during the recovery period was not due to the blockage of lysosome-dependent autophagic degradation, we co-treated cells with CQ for 6h following by a 48h recovery incubation with fresh medium. Compared to MG132 treatment alone, a further elevation of LC3 II was observed in the presence of CQ within the 24h recovery period (Fig. 6C, top), which confirms activation of autophagy. Cells were essentially devoid of ubiquitin and vimentin positive aggresomes following a 24h recovery (Fig. 6D) indicating that autophagy may be involved in the clearance of polyubiquitinated aggregates resulting from proteasomal inhibition in N27 cells. Moreover, we detected a decrease in the levels of LC3 II and polyubiquitinated proteins at the end of a 48h recovery period (Fig. 6C) suggesting that the autophagy clearing process was completed.
4. Discussion
Our data indicates a potential link between proteasomal inhibition and alterations in other major intracellular protein handling systems. EIF2α was rapidly phosphorylated after MG132 exposure indicating that the induction of the ER stress response/UPR was tightly associated with early proteasomal inhibition. Accumulating evidence suggests a role for the ER stress response/UPR in both PD pathogenesis [37,45] and in response to disruption of the UPS [30,46]. ER associated protein degradation occurs via the proteasome and consequently proteasomal inhibition blocks degradation of unfolded proteins resulting in ER stress and induction of a protective UPR. Further aggravation of ER stress can trigger either apoptosis or necrosis [30,47,48], and in our study, we observed both apoptosis and necrosis in N27 cells after prolonged treatment with MG132.
Coupling of proteasomal inhibition to autophagy has also been suggested in neurodegenerative diseases [26,49]. Our results are supportive of this hypothesis demonstrating that proteasomal inhibition by MG132 activated autophagy in N27 dopaminergic cells. Interestingly, we found that activation of autophagy was a delayed response relative to activation of the ER stress response, and recent work examining the interrelationship of proteasomal inhibition and autophagy in prostate cancer cells has demonstrated similar findings [50]. Heat shock proteins in neuronal cells have a buffering capacity against protein misfolding disorders [24]. We were able to detect the induction of Hsp70 but not Hsp90, Hsp27 and Hsp40 in response to proteasomal inhibition, suggesting N27 cells rely on Hsp70 as a relatively early defense mechanism against protein misfolding triggered by UPS impairment.
Our results demonstrate that during the early phase of proteasomal inhibition, the UPR is activated to attenuate intracellular protein load, autophagy helps to facilitate clearance of misfolded protein aggregates and the molecular chaperone Hsp70 is upregulated to assist protein refolding. However, when the intracytosolic degradative capacity is exceeded by prolonged proteasomal inhibition, perinuclear aggresomes are formed. We observed the formation of aggresomes at later times after UPS impairment subsequent to accumulation of polyubiquitinated protein suggesting that aggresome formation may act as a protective mechanism to segregate the toxic intracellular aggregates and it also fits well with previous speculation that Lewy bodies are generated due to aggresome related processes and often occur as a late event in PD [19]. We were able to detect colocalization of Hsp70 with ubiquitin-positive aggresomes in N27 cells and aggresomes are known to recruit various chaperones to assist in folding and clearance [51,52].
Autophagy is able to protect against aggregated proteins and aggresomes are substrates for autophagy [53–55]. Importantly, our data demonstrates that autophagy, but not the ER stress response/UPR or the heat shock response, may be involved in the clearance of aggresomes during the proteasome recovery process in N27 cells. Although a previous study reported that proteasome subunits colocalized with aggresomes at the MTOC [51] and speculated that aggresomes might sequester misfolded proteins for proteasomal degradation, it has been shown that large aggregates are poor substrates for the proteasome because they can be excluded from the proteasome core particle [56,57].
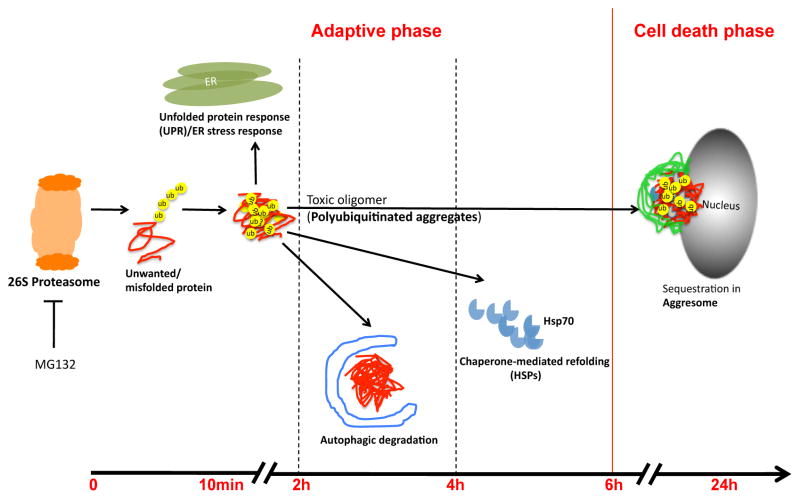
In summary, we have defined the sequence of activation of protein handling systems after inhibition of the proteasome in dopaminergic cells and our data suggests that the activation of diverse protein handling systems is a highly regulated process. After proteasomal inhibition (Fig. 7), the ER stress response/UPR is most rapidly activated followed by autophagy and Hsp70 induction with subsequent formation of aggresomes. We further defined the protective role of autophagy but not other protein handling systems during recovery of the cell from proteasomal inhibition. These results indicate that the mechanisms of induction of alternate protein handling systems and their temporal relationship may be important parameters determining the extent of accumulation of misfolded proteins in cells as a result of proteasomal inhibition. Determination of the relationship between different protein handling systems should provide a better understanding of accumulation of protein aggregates in neurodegenerative diseases associated with proteasomal inhibition.
Figure 7.
The schematic representation shows the activation sequence of major cellular protein handling systems.
Highlights.
We defined changes in major protein handling systems after proteasomal inhibition.
The ER stress response/UPR was most rapidly activated by proteasomal inhibition.
UPR, autophagy and Hsp70 were induced prior to the earliest signs of cell death.
The formation of vimentin-caged, ubiquitin rich aggresomes was a late event
Only autophagy was found to be involved in the clearance of aggresomes.
Acknowledgments
This work was supported by the National Institutes of Health Grant R01ES018943.
Abbreviations
- UPS
ubiquitin-proteasome system
- HSPs
heat shock proteins
- UPR
unfolded protein response
- eIF2α
eukaryotic translation initiation factor 2 alpha
- CHOP
C/EBP homologous protein
- LC3
autophagy related microtubule-associated protein 1 light chain 3
- CQ
chloroquine
- P
pepstatin A
- L
leupeptin
Footnotes
Conflict of interest statement
The authors declare there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 2.McNaught KSP, Olanow CW. Proteolytic stress: A unifying concept for the etiopathogenesis of Parkinson’s disease. Ann Neurol. 2003;53:S73–S86. doi: 10.1002/ana.10512. [DOI] [PubMed] [Google Scholar]
- 3.McNaught KSP, Belizaire R, Isacson O, Jenner P, Olanow CW. Altered proteasomal function in sporadic Parkinson’s disease. Experimental Neurology. 2003;179:38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- 4.Olanow CW. Etiology and pathogenesis of Parkinson’s disease. Annual Review of Neuroscience. 1999 doi: 10.1146/annurev.neuro.22.1.123. [DOI] [PubMed] [Google Scholar]
- 5.Chung KKK, Dawson VL, Dawson TM. New insights into Parkinson’s disease. Journal of Neurology. 2003;250 doi: 10.1007/s00415-003-1304-9. [DOI] [PubMed] [Google Scholar]
- 6.Olanow CW. Mov Disord. 2006. Ubiquitin–proteasome system and Parkinson’s disease - Olanow - 2006 - Movement Disorders-Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- 7.Zafar KS. A Potential Role for Cyclized Quinones Derived from Dopamine, DOPA, and 3,4-Dihydroxyphenylacetic Acid in Proteasomal Inhibition. Molecular Pharmacology. 2006;70:1079–1086. doi: 10.1124/mol.106.024703. [DOI] [PubMed] [Google Scholar]
- 8.Jinsmaa YY, Florang VRV, Rees JNJ, Mexas LML, Eckert LLL, Allen EMGE, et al. Dopamine-derived biological reactive intermediates and protein modifications: Implications for Parkinson’s disease. Chem Biol Interact. 2011;192:118–121. doi: 10.1016/j.cbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie W, Li X, Li C, Zhu W, Jankovic J, Le W. Proteasome inhibition modeling nigral neuron degeneration in Parkinson’s disease. Journal of Neurochemistry. 2010;115:188–199. doi: 10.1111/j.1471-4159.2010.06914.x. [DOI] [PubMed] [Google Scholar]
- 10.Vernon AC, Johansson SM. BMC Neuroscience | Full text | Non-invasive evaluation of nigrostriatal neuropathology in a proteasome inhibitor rodent model of Parkinson’s disease. BMC Neuroscience. 2010 doi: 10.1186/1471-2202-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukhatwa S, Iravani MM, Zeng BY, Cooper JD, Rose S, Jenner P. An immunohistochemical and stereological analysis of PSI-induced nigral neuronal degeneration in the rat. Journal of Neurochemistry. 2009;109:52–59. doi: 10.1111/j.1471-4159.2009.05956.x. [DOI] [PubMed] [Google Scholar]
- 12.Ebrahimi-Fakhari D, Wahlster L, McLean PJ. Acta Neuropathologica, Volume 124, Number 2 - SpringerLink. Acta Neuropathologica. 2012 doi: 10.1007/s00401-012-1004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chemistry & Biology. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg A. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003 doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 15.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Ikenoue T, Chen X, Li L, Inoki K, Guan KL. Rheb controls misfolded protein metabolism by inhibiting aggresome formation and autophagy. Proc Natl Acad Sci USa. 2009;106:8923–8928. doi: 10.1073/pnas.0903621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ron KR. Aggresomes, inclusion bodies and protein aggregation. Trends in Cell Biology. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 19.Olanow CW, Perl DP, DeMartino GN, McNaught KSP. Lewy-body formation is an aggresome-related process: a hypothesis. The Lancet Neurology. 2004;3:496–503. doi: 10.1016/S1474-4422(04)00827-0. [DOI] [PubMed] [Google Scholar]
- 20.Nishitoh H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & Development. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. The Journal of Cell Biology. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 23.Lin W, Popko B. Endoplasmic reticulum stress in disorders of myelinating cells. Nat Neurosci. 2009;12:379–385. doi: 10.1038/nn.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Brown IR. Neuronal expression of constitutive heat shock proteins: implications for neurodegenerative diseases. Cell Stress and Chaperones. 2007;12:51–58. doi: 10.1379/CSC-236R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinsztein D. Autophagy Induction Rescues Toxicity Mediated by Proteasome Inhibition 10.1016/j.neuron.2007.06.005: Neuron | ScienceDirect.com. Neuron. 2007 doi: 10.1016/j.neuron.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Pandey U, Nie Z, Batlevi Y, McCray B. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS : Abstract : Nature. Nature. 2007 doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 28.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 29.Yew EHJ, Cheung NS, Choy MS, Qi RZ, Lee AYW, Peng ZF, et al. Proteasome inhibition by lactacystin in primary neuronal cells induces both potentially neuroprotective and pro-apoptotic transcriptional responses: a microarray analysis. Journal of Neurochemistry. 2005;94:943–956. doi: 10.1111/j.1471-4159.2005.03220.x. [DOI] [PubMed] [Google Scholar]
- 30.Choy MS, Chen MJ, Manikandan J, Peng ZF, Jenner AM, Melendez AJ, et al. Up-regulation of endoplasmic reticulum stress-related genes during the early phase of treatment of cultured cortical neurons by the proteasomal inhibitor lactacystin. J Cell Physiol. 2011;226:494–510. doi: 10.1002/jcp.22359. [DOI] [PubMed] [Google Scholar]
- 31.Adams FS, La Rosa FG, Kumar S, Edwards-Prasad J, Kentroti S, Vernadakis A, et al. Characterization and transplantation of two neuronal cell lines with dopaminergic properties. Neurochem Res. 1996;21:619–627. doi: 10.1007/BF02527762. [DOI] [PubMed] [Google Scholar]
- 32.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods in Enzymology. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 33.Zafar KS, Inayat-Hussain SH, Ross D. A comparative study of proteasomal inhibition and apoptosis induced in N27 mesencephalic cells by dopamine and MG132. Journal of Neurochemistry. 2007;102:913–921. doi: 10.1111/j.1471-4159.2007.04637.x. [DOI] [PubMed] [Google Scholar]
- 34.LOWRY OH, ROSEBROUGH NJ, FARR AL, RANDALL RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 35.Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc Natl Acad Sci USa. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Figueiredo-Pereira ME, Berg KA, Wilk S. A New Inhibitor of the Chymotrypsin-Like Activity of the Multicatalytic Proteinase Complex (20S Proteasome) Induces Accumulation of Ubiquitin-Protein Conjugates in a Neuronal Cell. Journal of Neurochemistry. 2002;63:1578–1581. doi: 10.1046/j.1471-4159.1994.63041578.x. [DOI] [PubMed] [Google Scholar]
- 37.Hoozemans JJM, van Haastert ES, Eikelenboom P, de Vos RAI, Rozemuller JM, Scheper W. Activation of the unfolded protein response in Parkinson’s disease. Biochemical and Biophysical Research Communications. 2007;354:707–711. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 38.Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 40.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods in Enzymology. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 41.Olzmann JA, Li L, Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem. 2008;15:47–60. doi: 10.2174/092986708783330692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNaught KSP, Shashidharan P, Perl DP, Jenner P, Olanow CW. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci. 2002;16:2136–2148. doi: 10.1046/j.1460-9568.2002.02301.x. [DOI] [PubMed] [Google Scholar]
- 43.Johnston JA, Ward CL, Kopito RR. Aggresomes: A Cellular Response to Misfolded Proteins. The Journal of Cell Biology. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Outeiro TF, Klucken J, Strathearn KE, Liu F, Nguyen P, Rochet JC, et al. Small heat shock proteins protect against alpha-synuclein-induced toxicity and aggregation. Biochemical and Biophysical Research Communications. 2006;351:631–638. doi: 10.1016/j.bbrc.2006.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu EJE, Harding HPH, Angelastro JMJ, Vitolo OVO, Ron DD, Greene LAL. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson’s disease. CORD Conference Proceedings. 2002;22:10690–10698. doi: 10.1523/JNEUROSCI.22-24-10690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fribley A. Focused Review: ER Stress and Cancer Proteasome Inhibitor Induces Apoptosis through Induction of Endoplasmic Reticulum Stress. Cancer Biol Ther. 2006 doi: 10.4161/cbt.5.7.2971. [DOI] [PubMed] [Google Scholar]
- 47.Jimbo A, Fujita E, Kouroku Y, Ohnishi J, Inohara N, Kuida K, et al. ER stress induces caspase-8 activation, stimulating cytochrome c release and caspase-9 activation. Exp Cell Res. 2003;283:156–166. doi: 10.1016/s0014-4827(02)00033-2. [DOI] [PubMed] [Google Scholar]
- 48.Wang K. Calpain and caspase: can you tell the difference?, by kevin K.W. Wang Vol. 23, pp. 20–26. Trends Neurosci. 2000;23:59–59. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- 49.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu K, Dunner K, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2009;29:451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and Dynamics of Aggresome Formation by a Cytosolic Gfp-Chimera. The Journal of Cell Biology. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Qian SB. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22:3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fortun J, Dunn WA, Joy S, Li J, Notterpek L. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J Neurosci. 2003;23:10672–10680. doi: 10.1523/JNEUROSCI.23-33-10672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor JP. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Human Molecular Genetics. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 55.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USa. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. Embo J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Molecular Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]