Abstract
The knowledge that many pathogens rely on cell-to-cell communication mechanisms known as quorum sensing, opens a new disease control strategy: quorum quenching. Here we report on one of the rare examples where Gram-positive bacteria, the ‘Staphylococcus intermedius group’ of zoonotic pathogens, excrete two compounds in millimolar concentrations that suppress the quorum sensing signaling and inhibit the growth of a broad spectrum of Gram-negative beta- and gamma-proteobacteria. These compounds were isolated from Staphylococcus delphini. They represent a new class of quorum quenchers with the chemical formula N-[2-(1H-indol-3-yl)ethyl]-urea and N-(2-phenethyl)-urea, which we named yayurea A and B, respectively. In vitro studies with the N-acyl homoserine lactone (AHL) responding receptor LuxN of V. harveyi indicated that both compounds caused opposite effects on phosphorylation to those caused by AHL. This explains the quorum quenching activity. Staphylococcal strains producing yayurea A and B clearly benefit from an increased competitiveness in a mixed community.
Author Summary
While studying the potential interaction of staphylococci with Gram-negative bacteria, we came across another communication system in a Staphylococcus species group, which consists of closely related coagulase-positive bacterial species that play a role as zoonotic pathogens. We found that these species excrete two small compounds that inhibit both the expression of QS-controlled toxins and other QS-regulated compounds as well as growth in Gram-negative bacteria. The excreted compounds, which we named yayurea A and B, were isolated from S. delphini and structurally characterized. They represent new bacterial products, which quench the QS regulation in a wide spectrum of Gram-negative bacteria by stimulating the LuxN-mediated phosphorylation of LuxU. Furthermore, growth of yayurea A and B producing S. delphini is not suppressed by respiratory toxins when co-cultured with P. aeruginosa. This suggests that the quorum quenchers have a function in self-protection and competitiveness in natural environments shared with Gram-negatives. Here we show one of the rare cases of inter-phylum interference between firmicutes (Gram-positive) and beta-/gammaproteobacteria (Gram-negative).
Introduction
In many bacteria, pathogenicity is controlled and coordinated by an inter-cellular communication process named quorum sensing (QS). QS is based on the synthesis and secretion of small hormone-like molecules termed autoinducers that bind to cognate receptors. The signal-activated receptor controls directly or indirectly expression of target genes. Since the concentration of signaling molecules in liquid culture is proportional to cell density in the culture, gene expression is coordinated in response to the bacterial population density [1], [2]. In V. harveyi, the QS system consists of three autoinducers and three cognate receptors functioning in parallel to channel information into a shared regulatory pathway [2]. Similar to other Gram-negative bacteria, V. harveyi produces an AHL signal termed HAI-1, 3-hydroxy-C4-homoserine lactone [3], which binds to the membrane-bound sensor histidine kinase (LuxN). The second molecule is AI-2, a furanosyl borate diester, which binds to the periplasmic protein LuxP. The LuxP-AI-2 complex interacts with another membrane-bound sensor histidine kinase, LuxQ. The third molecule is termed CAI-1 (for cholera autoinducer-1), a long chain ketone [4], [5], which is recognized by the membrane-bound sensor histidine kinase, CqsS [6]. At low cell density, in the absence of appreciable amounts of autoinducers, the three sensors (LuxN, LuxQ, and CqsS) act as autophosphorylating kinases that subsequently transfer the phosphate to the cytoplasmic protein LuxU, which passes the phosphate to the DNA-binding response regulator protein LuxO [7], [8]. Phosphorylated LuxO represses the master regulator of QS, LuxR, via sigma factor σ54 and regulatory small RNAs [9], [10].
Similar to V. harveyi, P. aeruginosa coordinates the expression of nearly 10% of its genome through three hierarchically arranged QS systems, namely Las, Rhl and Pqs [11]. Each system consists of enzymes involved in autoinducer synthesis and the target regulator: LasI produces 3-oxo-C12-HSL for activation of LasR [12], RhlI produces C4-HSL for the activation of RhlR [13], [14], and the biosynthetic enzymes PqsABCDE and PhnAB produce PQS (2-heptyl-3-hydroxy-4-quinolone) for activation of PqsR [15]–[17]. QS systems are also prevalent in many other Gram-negative bacteria.
QS system is a promising target for anti-virulence therapy [1], [18]. In contrast to classic antibiotics, quorum-quenching compounds are inhibitors of bacterial virulence, rather than of bacterial growth [19]. Since the first studies on QS inhibitors, the halogenated furanones [20], more compounds have been identified [21], [22].
In contrast to Gram-negative bacteria, many Gram-positive bacteria communicate using modified oligopeptides as signals and “two-component”-type membrane-bound sensor histidine kinases as receptors. The well-studied QS system in Staphylococcus is the agr QS system [23]. The excreted signal is a thiolactone- or lactone-based peptide [24] (AIP, autoinducer peptide) that mediates communication with other staphylococci in a cell density dependent way [25], [26].
While studying the potential interaction of staphylococci with Gram-negative bacteria [27], [28], we came across another communication system in a Staphylococcus species group, named ‘intermedius group’. This group consists of closely related mainly coagulase-positive bacterial species including S. delphini, S. intermedius, S. lutrae, S. pseudintermedius, and S. schleiferi. They are all phylogenetically related, are zoonotic pathogens, and only rarely occur in human infections. We found that these species excrete two low molecular compounds that inhibit the expression of QS-controlled toxins and other QS-regulated compounds in Gram-negative bacteria. The excreted compounds, which we named yayurea A and B, were isolated from S. delphini and structurally characterized. Yayurea A and B represent new bacterial products, and were able to quench the QS regulation in a wide spectrum of Gram-negative bacteria. Furthermore, growth of yayurea A and B producing S. delphini is not suppressed by respiratory toxins when co-cultured with P. aeruginosa. This suggests that the quorum quenchers have a function in self-protection and competitiveness in natural environments shared with Gram-negatives. Here we show an example of inter-phylum interference between Firmicutes (Gram-positive) and the Gram-negative beta- and gammaproteobacteria.
Results
Staphylococcus delphini suppresses production of QS-regulated phenotypes in various Gram-negative bacteria
Our aim was to find out if some staphylococcal species are able to suppress the QS controlled phenotypes in Gram-negative bacteria. To investigate this, we tested the ability of several staphylococcal species to inhibit pyocyanin production of P. aeruginosa in a co-cultivation assay, as pyocyanin production is QS controlled. We found that S. delphini DSMZ 20771 completely inhibited pyocyanin production over 24 h co-cultivation with P. aeruginosa PAO1, while Staphylococcus aureus showed no such activity (Fig. 1A). This led us to investigate if S. delphini could also suppress QS-controlled phenotypes in other Gram-negative bacteria such as the QS-regulated prodigiosin production in Serratia marcescens [29]; bioluminescence in Vibrio harveyi [7], [30]; or violacein production in Chromobacterium subtsugae, a pathogen of potato beetles [31]. Indeed, in co-cultivation studies with these Gram-negative bacteria, S. delphini did also suppress prodigiosin and violacein production as well as bioluminescence, while S. aureus did not (Fig. 1BCD). Sterile filtered culture supernatant of a 24 h S. delphini culture had the same QS-inhibiting effect as the co-culture, indicating that the QS-inhibiting compound(s) were excreted. The supernatants of S. aureus and S. epidermidis caused no effect.
Figure 1. Quenching of QS-regulated pigments and bioluminescence by S. delphini.

P. aeruginosa (A), S. marcescens (B), V. harveyi (C) and C. subtsugae (D) were each co-cultivated with S. aureus (1) or S. delphini (2) for 24 h. Pyocyanin, which is excreted by P. aeruginosa, was determined in the supernatant at its absorption maximum A520 nm. Prodigiosin, which is cell wall bound in S. marcescens, was ethanol-extracted from the cell pellet and determined at its absorption maximum A534 nm. Bioluminescence of V. harveyi was intensified by aeration before measuring in a bioluminescence reader. Violacein from C. subtsugae was quantitatively extracted with butanol and determined at its absorption maximum A585 nm.
Structural analysis of the QS-inhibiting compounds from S. delphini
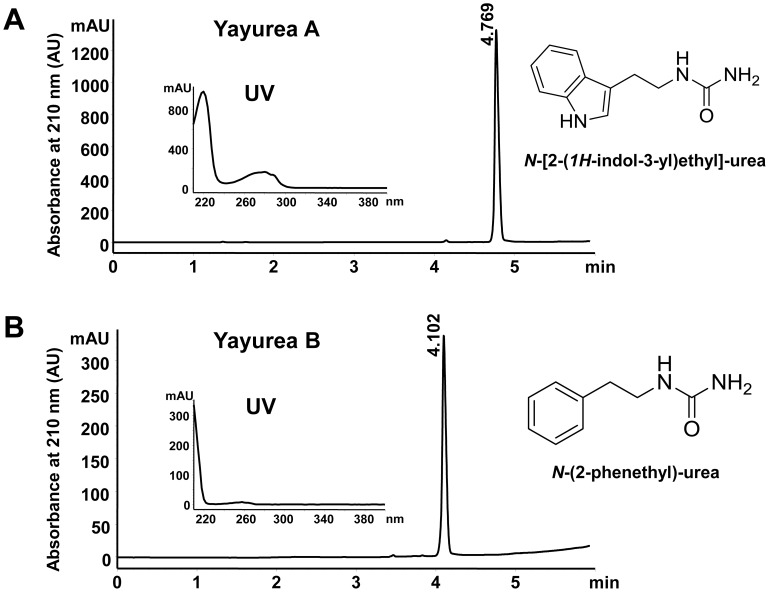
The QS inhibitors were isolated from the supernatant of an overnight culture of S. delphini DSMZ 20771. Further purification revealed that the supernatant contained two compounds with different retention times (Rt) in HPLC and distinct UV spectra. We named the two compounds yayurea A and B.
Yayurea A (indole-ethylurea) was isolated as a brownish solid, revealed an ion peak at m/z = 161 ([M+H]+) in ESI-MS, and showed the molecular formula, C10H13N2, in FT-ICR-MS analysis. The 1H-indole-3-ethylamine moiety was deduced from GC-EI-MS (Rt = 29.0 min). 1H-NMR spectra showed a characteristic singlet at δH = 8.55 ppm. The remaining signals were assigned to 2-(3-indoyl) ethylamine. The 13C-NMR spectrum and additional HSQC experiments displayed signals pointing to a carbonyl group at δ = 170.4 ppm, five methin groups, and two methylene groups to reveal a urea moiety. Additionally, 2D NMR experiments supported the structure of yayurea A as N-[2-(1H-indole-3-yl) ethyl]-urea (Fig. 2A).
Figure 2. RP-HPLC profile, UV-spectrum and structures of the two QS-inhibitors purified from S. delphini.
(A) QS-inhibitor, N-[2-(1H-indol-3-yl)ethyl]-urea (yayurea A). (B) QS-inhibitor, N-(2-phenethyl)-urea (yayurea B). RP-HPLC was carried out on an Agilent 1200 and Waters xBridge C18, 5 mm, 4.6×150 mm column; compounds were eluted with a 15 min linear gradient of 0.1% phosphoric acid to acetonitrile at a flow rate of 1.5 ml/min.
Yayurea B (phenethylurea) was obtained as a colorless solid. A preliminary molecular formula, C8H10N, was deduced from the FT-ICR-MS spectrum which showed an ion at m/z = 121 ([M+H]+). GC-EI-MS provided a signal at Rt = 15.0 min pointing to 2-phenethylamine. The 1H-NMR again showed a set of aromatic protons and a singlet at δH = 8.54 ppm, while the 13C-NMR displayed a signal at δH = 170.4 ppm (C = O) and five aromatic and 4 aliphatic protons. In summary, the structure of N-(2-phenethyl)-urea was assigned for yayurea B (Fig. 2B). UV-Absorption maxima emphasized the phenyl- (λ = 260 nm) and indole chromophores (λ = 225 nm and λ = 280 nm respectively) of the yayureas. Comprehensive physicochemical characteristics and mass spectra of yayurea A and B are shown (Table 1 and Figure S1). In the meantime, yayurea A and B could also be chemically synthesized and their identity confirmed by mass spectral and NMR analyses (Wölfle et al. manuscript in preparation). Both the synthesized and the natural compounds revealed the same chemical properties and the same QS-quenching activity in Gram-negative bacteria (data not shown).
Table 1. Physical data for yayurea A and B.
| N-[2-(1H-indol-3-yl)ethyl]-urea (yayurea A) | N-(2-phenethyl)-urea (yayurea B) | |
| Formula | C11H13O1N3 (203.24 g/mol) | C9H11O1N2 (163.20 g/mol) |
| Melting point | 240.8°C | 214.9°C |
| Rf-values | 0.10 (CHCl3/MeOH 9∶1) 0.63 (MeOH/H2O 7∶3) | 0.28 (CHCl3/MeOH 9∶1) 0.94 (MeOH/H2O 7∶3) |
| 1H NMR | (600 MHz, MeOH-d4) d 3.11 (t, J = 7.0, 7.4 Hz, 2H), 3.22 (t, J = 7.1, 7.3 Hz, 2H), 7.04 (dd, J = 7.4, 7.5 Hz, 1H), 7.13 (dd, J = 7.7, 7.9 Hz, 1H), 7.17 (s, 1H), 7.37 (d, J = 8.2 Hz, 1H), 7.57 (d, J = 7.9 Hz, 1H), 8.55 (s, 1H). | (600 MHz, MeOH-d4) d 2.95 (t, J = 7.4, 8.1 Hz, 2H), 3.16 (t, J = 7.4, 8.1 Hz, 2H), 7.28 (m, 3H), 7.35 (m, 2H), 8.54 (s, 1H). |
| 13C NMR | (150 MHz, MeOH-d4) d 24.9, 41.5, 110.6, 112.7, 119.0, 120.2, 122.9, 124.4, 128.4, 138.5, 170.5. | (150 MHz, MeOH-d4) d 34.9, 42.1, 128.4, 129.9, 130.2, 138.1, 170.2. |
| MS (ESI) | (Positive ions) m/z (%) [M+2H-CONH2]+ 161.11. | (Positive ions) m/z (%) [M+2H-CONH2]+ 122.10. |
Suppression of QS-regulated respiratory toxins protects S. delphini from killing by Pseudomonas aeruginosa
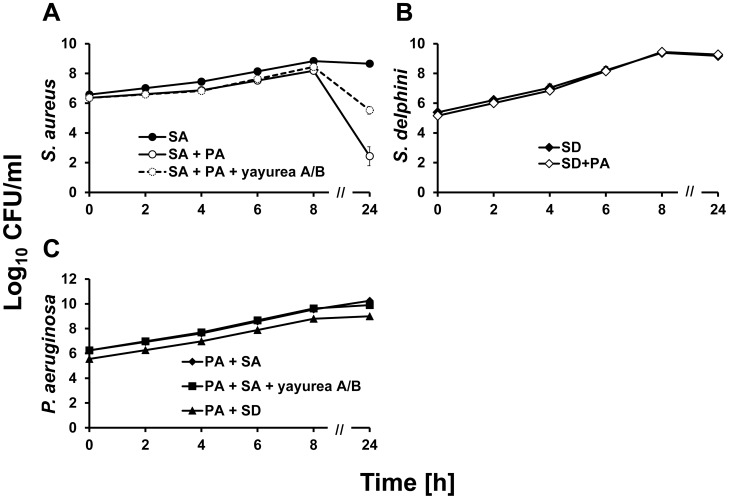
P. aeruginosa produces various QS-controlled respiratory toxins such as pyocyanin and hydrogen cyanide [32], [33], which kill S. aureus [27]. If yayurea A and B repress the production of QS-regulated pyocyanin or hydrogen cyanide, one would expect that S. delphini survives better in a co-culture with P. aeruginosa than, for example, S. aureus. Indeed, co-cultivation studies with S. aureus (non-producer) or S. delphini with P. aeruginosa SH1 revealed that the viability (CFU) of S. aureus significantly decreased in the stationary growth phase, most likely due to the respiratory toxins produced by P. aeruginosa (Fig. 3A), while that of S. delphini was uninfluenced (Fig. 3B). The addition of yayurea A and B to the mixed S. aureus and P. aeruginosa culture protected S. aureus from killing in the stationary phase (Fig. 3A). Furthermore, the CFU of P. aeruginosa SH1 was unaffected while co-cultured with S. delphini or S. aureus, indicating that none of the two staphylococcal species was able to kill P. aeruginosa. Co-cultivation of P. aeruginosa with S. aureus in the presence of yayureas (100 µg/ml yayurea A and 900 µg/ml yayurea B) had also no effect on viability of P. aeruginosa (Fig. 3C). Other tested Pseudomonas strains PAO1 and DSMZ 50071 showed similar results (data not shown). All in all, these results showed that production of yayurea A and B enables staphylococci to coexist with Gram-negative bacteria in a mixed community.
Figure 3. Survival of staphylococcal strains in mixed culture with P. aeruginosa.
(A) CFU of S. aureus alone (SA) and in co-culture with P. aeruginosa (SA+PA). For the protection test, yayurea A (100 µg/ml) and B (900 µg/ml) were added to the mixture of S. aureus and P. aeruginosa (SA+PA+yayurea A/B). (B) CFU of S. delphini alone (SD) and in co-culture with P. aeruginosa (SD+PA). (C) CFU of P. aeruginosa SH1 co-cultured with S. aureus (PA+SA), yayureas (PA+SA+yayurea A/B) or S. delphini (PA+SD). Values represent the means of three independent experiments. Bars indicate mean standard deviation, SD.
Yayurea A and B are mainly produced in stationary growth phase of S. delphini
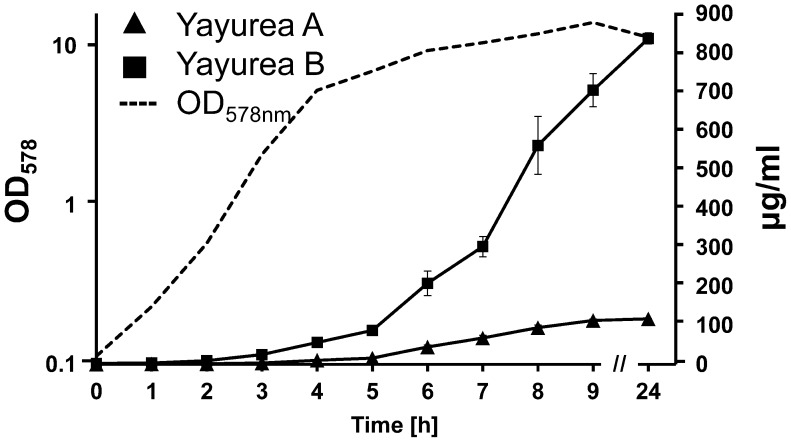
We followed the production of yayurea A and B over 24 h in the supernatant S. delphini grown in TSB. The amount of produced yayurea A and B was determined by HPLC-analysis; peak integration was correlated with standard yayurea A and B. The production of both compounds started at the transition of exponential to stationary growth phase (after approximately 4 h) and increased rapidly for the next 5 h; after 24 h little more was produced (Fig. 4). The production kinetics is reminiscent of a QS controlled expression. Both compounds were produced in amazingly high concentrations: yayurea A reached concentration of 120 µg/ml and yayurea B even 900 µg/ml. This high concentration is entirely sufficient to suppress QS-systems in Gram-negative bacteria as can be seen below.
Figure 4. Quantification of yayurea A and B production in relation to growth.
S. delphini was grown in TSB at 37°C. Supernatant was collected and OD578 was measured hourly for the first 9 h and after 24 h. Amounts of yayurea A and B in supernatants were quantified by triplicate HPLC measurements. Bars indicate mean standard deviation, SD.
Yayurea A is more active than yayurea B in inhibiting various QS-controlled traits in Gram-negative bacteria
Purified yayurea A and B inhibited QS-regulated factors in Gram-negative bacteria in a dose-dependent manner. We tested prodigiosin production in S. marcescens, bioluminescence in V. harveyi, and pyocyanin production in P. aeruginosa (Fig. 5 and 6).
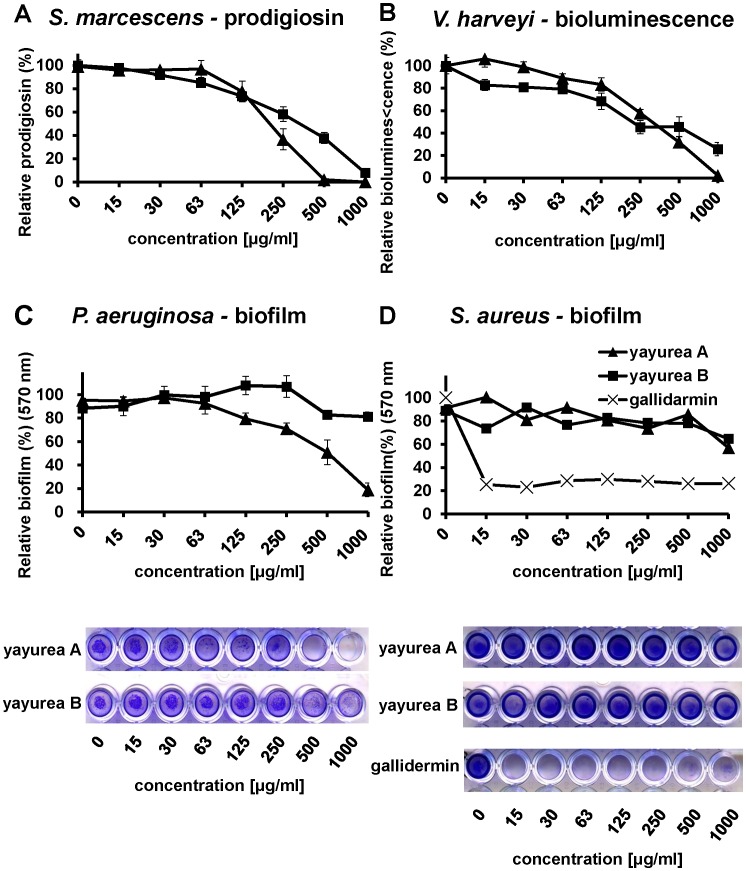
Figure 5. Concentration-dependent inhibition of QS-regulated phenotypes in Gram-negative bacteria.
(A) Prodigiosin production in S. marcescens. Cells were grown in MB medium with serial dilutions of yayurea A or B at 28°C. Relative prodigiosin production was calculated as the ratio between prodigiosin content (absorbance at 534 nm) and cell density (absorbance at 600 nm). (B) Bioluminescence in V. harveyi. Cells were grown in marine broth with serial dilutions of the compounds at 28°C for 24 h. Relative luminescence units were normalized by the cell density. (C) Biofilm formation of P. aeruginosa. Cells were grown in LB with serial dilutions of yayurea A or B at 37°C for 24 h. (D) Biofilm formation of S. aureus. Cells were grown in TSB with serial dilutions of yayurea A, B, or gallidermin (positive control) at 37°C for 24 h. Biofilm cell layer was visualized by crystal violet staining and measured at 590 nm. Microtiter plates presented are representative of at least three independent sets of experiments. Bars indicate standard deviation of the mean, SD.
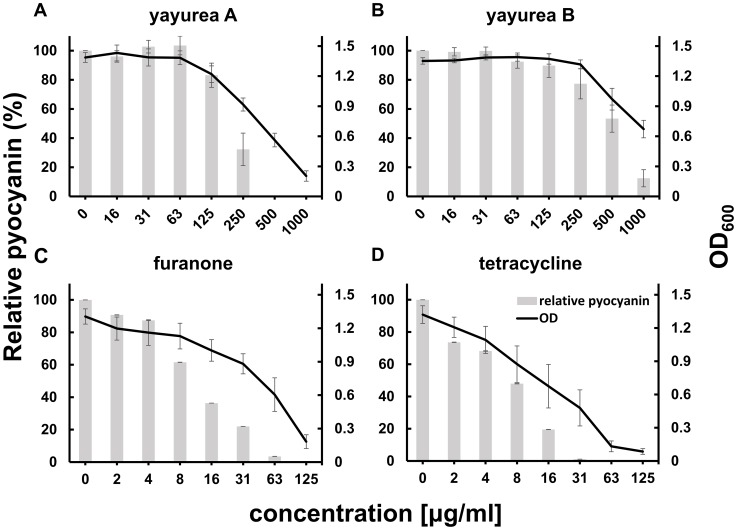
Figure 6. Concentration-dependent inhibition of pyocyanin production and growth of P. aeruginosa.
P. aeruginosa PAO1 was grown in LB at 30°C with serial dilutions of yayurea A (A), yayurea B (B), furanone (C) and tetracycline (D). Relative pyocyanin production was calculated as the ratio between pyocyanin content and cell density (absorbance at 600 nm). Values represent the means of three independent experiments. Bars indicate standard deviation of the mean, SD.
Yayurea A and B inhibited production of prodigiosin in S. marcescens in a dose-dependent way. Inhibition already started at low concentrations (15 µg/ml) and increased with increasing concentrations of yayurea A or B. At 250 µg/ml, prodigiosin production was inhibited at 60% (yayurea A) and 40% (yayurea B). At a concentration of 1000 µg/ml, prodigiosin production was completely inhibited by yayurea A, and to approximately 70% by yayurea B (Fig. 5A). At 500 µg/ml, bioluminescence of V. harveyi was inhibited by yayurea A and B by 99% and 76% respectively (Fig. 5B). We also investigated whether yayurea A and B inhibited biofilm formation in P. aeruginosa and S. aureus. Again yayurea A quite efficiently inhibited biofilm formation in P. aeruginosa, while yayurea B was less effective (Fig. 5C). In contrast to gallidermin, a good biofilm inhibitor in staphylococci [34], both yayureas showed no biofilm-inhibiting effect with S. aureus (Fig. 5D).
We noticed that high dose of yayurea A and B (especially yayurea A) inhibit the growth of Gram-negative bacteria. We used P. aeruginosa as an example to verify the effect of growth and QS inhibition by yayurea A and B, and used the antibiotic tetracycline and the well-known QS-inhibitor furanone [35] as controls (Fig. 6). For QS inhibition, around 50% of the yayurea A and B concentration was necessary compared to that needed for growth inhibition (Fig. 6 A, B). Furanone revealed a similar correlation of growth and QS inhibition; the concentration (31 µg/ml) that inhibited 40% of growth inhibited 80% of QS (Fig. 6C). In contrast to yayurea and furanone, tetracycline inhibited growth and QS almost linearly (Fig. 6D). Besides P. aeruginosa, yayureas affect also growth of S. marcescens, V. harveyi, and V. cholerae (Figure S2); however, growth of E. coli was not affected. All staphylococcal species tested, such as S. aureus, S. carnosus, S. delphini or S. schleiferi, are resistant to yayurea A and B, independently whether they are producing these compounds or not.
Yayurea A and B are perceived by the AHL-receptor LuxN of Vibrio harveyi
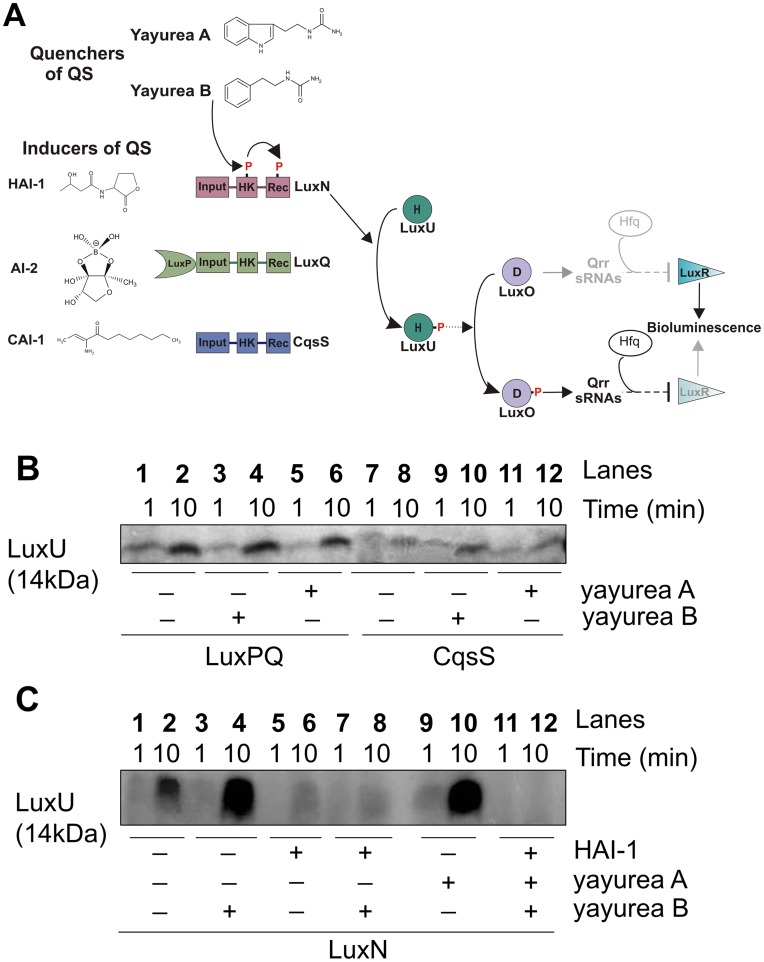
To gain insight into the molecular mechanism behind why yayurea A and B cause a decrease in bioluminescence of V. harveyi, we performed in vitro phosphorylation assays of the corresponding signaling proteins. The full-length hybrid kinases LuxN, LuxQ, CqsS (tagged with 6 histidine residues) were heterologously expressed in an E. coli strain that lacks the F1/Fo-ATPase (to prevent ATP degradation during phosphorylation experiments), and inverted membrane vesicles prepared from this strain were directly used for the phosphorylation experiments. Then we tested the effect of yayurea A and B on the LuxN, LuxQ [in interplay with LuxP (LuxPQ)], and CqsS-mediated time-dependent phosphorylation of the HPt protein LuxU (Fig. 7A). Yayurea A and B had no effect on LuxPQ or CqsS-mediated phosphorylation of LuxU (Fig. 7B). However, they significantly stimulated the LuxN-mediated phosphorylation of LuxU in comparison to the control (Fig. 7C, compare lane 2 with lanes 4 and 10). LuxN is the sensor for the AHL autoinducer N-(3-hydroxybutyryl)-homoserine lactone (HAI-1) (Fig. 7A). It is known [36] that the presence of HAI-1 inhibits the autophosphorylation activity of LuxN, thus decreasing the level of phospho-LuxU (Fig. 7C, compare lanes 2 and 6).
Figure 7. Yayurea A and B are perceived by V. harveyi LuxN receptor.
(A) Schematic representation of the QS phosphorelay in V. harveyi. In the absence of autoinducers (HAI-1, AI-2 and CAI-1) at low cell density, each of the three receptors, LuxN, LuxQ and CqsS, respectively, autophosphorylates at a conserved histidine of their histidine kinase domain (HK). The phosphoryl group is first transferred to the receiver domain (Rec) of the receptor kinase and then to the HPt protein LuxU. LuxP is a periplasmic binding protein. P denotes phosphorylation sites. Upon perception of the autoinducers at high cell density, autophosphorylation of the receptors and the subsequent phosphosphrylation cascade is inhibited. Yayurea A and B stimulated the phosphorylation of the cascade via LuxN. (B) LuxPQ and CqsS mediated phosphorylation of LuxU in the presence of yayurea A and B. (C) LuxN mediated phosphorylation of LuxU in the presence of yayurea A and B and HAI-1. LuxQ, CqsS, LuxN-bearing membrane vesicles and LuxU, were incubated with 100 µM [γ-32P] ATP. The effect of yayurea A, B and HAI-1 (C) on the initial rate of LuxU phosphorylation was evaluated. Each reaction was sampled and stopped at two different time points: after 1 and 10 minutes. Final concentrations were 20 µM for HAI-1, 1.1 mM for yayurea A and 1.3 mM yayurea B, which reflects the in vivo situation. Absence of HAI-1 or yayurea A or B is indicated by “−” and presence by “+”.
Yayurea A and B caused the opposite effect, leading to an increase in the level of phospho-LuxU, which explains the decrease in bioluminescence when V. harveyi is exposed to these compounds. Under our test conditions, autoinducer HAI-1 was dominant in relation to yayurea A or B (Fig. 7C, compare lane 4 with lanes 8 and 12). Taken together these data show that the receptor kinase LuxN of V. harveyi specifically recognizes yayurea A and B. It is still unclear whether yayurea A and B also bind to the HAI-1 binding site or whether LuxN contains an independent binding site for these new compounds.
Phylogenetic position of yayurea A and B producing species in the Staphylococcus taxa
We tested a number of staphylococcal species representatives (listed in Table 2) by co-cultivation with P. aeruginosa for suppression of pyocyanin production. Only five species exerted such an activity: S. delphini, S. intermedius, S. lutrae, S. pseudintermedius, and S. schleiferi, and they all produced yayurea A and B as determined by HPLC analysis. Based on 16S rRNA- and multilocus sequence typing (MLST) the species are phylogenetically related and summarized in the ‘intermedius group’ [37] (Figure S3). Typically, the species are coagulase positive (with exception of S. schleiferi subsp. schleiferi), and oxidase negative. Interestingly, they all colonize various animals, and many represent zoonotic pathogens [38], [39]. None of the other listed staphylococcal species (even not the next related S. hyicus, S. chromogenes or S. muscae) inhibited QS-system in Gram-negative bacteria or produced yayurea A and B.
Table 2. Strains tested for Q-quenching activity and yayurea A and B production.
| Strains | QS-inhibition |
| S. aureus RN4220 | − |
| S. aureus SA113 | − |
| S. aureus 8325-4 | − |
| S. aureus RN1 | − |
| S. aureus HG001 | − |
| S. aureus HG002 | − |
| S. aureus HG003 | − |
| S. aureus Newman | − |
| S. arlettae DSMZ 20672T | − |
| S. carnosusTM300 DSMZ 20501 | − |
| S. capitis subsp. capitis LK499 ATCC 27840 | − |
| S. caprae DSMZ 20608T | − |
| S. chromogenes DSMZ 20454T | − |
| S. cohnii subsp. cohnii DSMZ20260 | − |
| S. condiment DSMZ 11674T | − |
| S. delphini DSMZ 20771 | +++ |
| S. epidermis ATCC14990 | − |
| S. equorum subsp. equorum DSMZ 20674T | − |
| S. gallinarum DSMZ 20610T | − |
| S. muscae DSMZ 7068T | − |
| S. haemolyticus CCM2737 | − |
| S. hominis DSMZ 20328 | − |
| S. hyicus NCTC 10350 | − |
| S. intermedius CCM 5739 | + |
| S. lentus DSMZ 20352T | − |
| S. lugdunesis ATCC 43809 | − |
| S. lutrae DSMZ10244T | + |
| S. pasteuri ATCC 51129 | − |
| S. pseudintermedius ED99 | +++ |
| S. saprophyticus subsp. saprophyticus DSMZ 200229 | − |
| S. schleiferi subsp. coagulans ATCC49545 | ++ |
| S. schleiferi subsp. schleiferi DSMZ 4807 | +++ |
| S. simulans MK148 ATCC 27848 | − |
| S. warneri DSMZ 20316T | − |
| S. xylosus DSMZ 20266 | − |
(+), Species that produce yayurea A and B.
Discussion
In this study, we found that some staphylococcal species excrete two novel compounds, yayurea A and B that interfere with the QS system of diverse Gram-negative bacteria. We tested 24 staphylococcal species with respect to production of yayurea A and B by co-cultivation with Gram-negative bacteria. The results showed that only five species (S. delphini [40], S. intermedius [41], S. pseudintermedius [42], S. lutrae [43], and S. schleiferi [44]) produced these compounds and were able to inhibit QS-regulated markers of Gram-negative test bacteria. All five species are naturally associated with various animals, where they act as zoonotic pathogens. They are only rarely associated with human infections. It is remarkable that these species are clustered in various phylogenetic trees, based on either 16S rRNA [45], [46], on thermonuclease (nuc) sequences [47], on major autolysin (atl) sequences [48], or on multilocus sequence typing (MLST) [37]. The five species form a phylogenetic cluster, termed “intermedius group” (Figure S3).
To gain insight into the mode of QS quenching by yayurea A and B, we performed in vitro phosphorylation assays with the autoinducer receptors LuxN, LuxQ, CqsS (Fig. 7A). Yayurea A and B had no effect on LuxPQ or CqsS-mediated phosphorylation of LuxU. However, they significantly stimulated the LuxN-mediated phosphorylation of LuxU (Fig. 7B–C), suggesting that they interact with LuxN and cause LuxU activation. While the LuxN autoinducer HAI-1, an AHL, inhibits the autophosphorylation of LuxN, consequently leading to a decrease in LuxU-phosphorylation, yayurea A and B caused the opposite effect by increasing LuxU phosphorylation. Thus, it is suggested that yayurea A and B keep the V. harveyi in a phenotypic state of low cell density, although the cells have grown to high density.
When both AI-1 and yayurea A or B were applied, HAI-1 overruled the effect of yayurea A and B in vitro. Bioluminescence of the wild type strain in vivo significantly decreased after exposure to yayurea A and B, although the effect of the latter compound was smaller. Obviously, HAI-1 did not overrule the effect of yayurea A and B in vivo. This difference can be explained by the fact that bioluminescence was measured in stationary phase grown cells at a time when the impact of HAI-1 on QS induction is low [49]. Furthermore, the contribution of the other receptors (LuxQ and CqsS) and their cognate autoinducers (AI-2 and CAI-1) needs to be considered in vivo [49]. The ratio of the three receptors and the impact of their kinase and phosphatase activities on the output of the phosphorylation cascade is not yet fully understood. This could be another explanation for the strong effect of the quorum quenchers in vivo. Membrane-topology analysis predicts that LuxN is bound to the bacterial inner-membrane by nine transmembrane (TM) spanning domains [50], and periplasmic loop 3 might be the HAI-1 binding site [21]. We believe that yayurea A and B interfere with the AHL quorum sensing response of Gram-negative bacteria; since the tested bacteria on which the effects were observed have at least one AHL-based quorum sensing system (Pseudomonas, Chromobacterium, Vibrio, and Serratia).
Most natural environments harbor a stunningly diverse collection of microbial species. One example, is the marine bacterium Halobacillus salinus, which produces N-(2′-phenylethyl)-isobutyramide and 2,3-methyl-N-(2′-phenylethyl)-butyramide [51]. These compounds are unrelated to yayurea A (N-[2-(1H-indol-3-yl)ethyl]-urea) and B (N-(2-phenethyl)-urea). Within these communities, bacteria compete with their neighbors for space and resources [52]. Zoonotic commensals and pathogens, including yayurea producing staphylococcal species, use animals as a habitat. Whether these animals are also colonized by Gram-negative bacteria, other than in the gut, has barely been investigated. However, one can assume that animals bathe in puddles, lakes, and rivers, suggesting that their mucus, skin, fur, or feathers may easily encounter Gram-negative bacteria, being transient to permanent colonizers. One can also assume, that the ‘intermedius group’ share the habitat animal with Gram-negative bacteria. S. delphini was isolated from dolphin. Since both V. harveyi and S. delphini are marine bacteria and animal pathogens, it is conceivable that both bacteria share the same habitat and moreover, the same host surface. Our results demonstrated suppression of the quorum sensing regulated bioluminescence of V. harveyi during co-culture with S. delphini (Fig. 1), indicating that yayurea A and B are effective quorum quenchers.
The in vitro phosphorylation assay gave first mechanistic insights on how yayurea A and B affect quorum sensing of V. harveyi. We detected a significant and specific effect of yayurea A and B on the AHL-receptor LuxN. The concentrations used for these in vitro assays might not be physiological, especially when the influence of both molecules (HAI-1 and yayurea) was studied. Furthermore, it should be noted that HAI-1 is not constantly produced [49]. Therefore, in the natural habitat there might be times, when yayurea from S. delphini can fully interact with LuxN from V. harveyi in the absence of any competition with HAI-1. It therefore makes sense that the zoonotic staphylococci impair the QS-system of Gram-negative bacteria for competitive reasons. The growth of these staphylococci is not impaired by P. aeruginosa because the excreted yayureas suppress the production of the QS-controlled toxins and are thereby protected from being killed by the toxins (Fig. 3). While biofilm formation appears to be modulated by many regulators and environmental conditions in P. aeruginosa, pyocyanin and cyanide are controlled by the Las-QS system [53], implying that yayurea A and B might compete with 3-oxo-C12-HSL for LasR interaction.
When yayurea A (125 µg/ml) was added, the growth of P. aeruginosa was not affected (Fig. 6A), but quorum sensing and biofilm formation (Figs. 6A and 5C) were inhibited by 20%, which indicates that biofilm inhibition takes place prior to the onset of growth inhibition. In addition to their quorum quenching activity in Gram-negative bacteria, yayureas also inhibited their growth at higher dose (Fig. 2), which is a further advantage in the race for space and resources. For this advantage the zoonotic staphylococci are apparently prepared to pay a certain price, namely the production and excretion of comparatively high amounts of yayurea A and B. However, the benefit in competitiveness appears to prevail the cost disadvantages. As QS controls not only virulence factors but also many metabolic functions important for fitness, we don't know whether the inhibition of growth is a consequence of QS-inhibition or vice versa [54]–[56]. For some antibiotics (azithromycin, ceftazidime, and ciprofloxacin), it has been shown that they decrease the expression of QS-regulated virulence and many other genes [57].
The yayureas are potential candidates for use as anti-infectives. The only disadvantage might be the rather high concentration needed to completely quench QS; on the other hand, preliminary results suggest that they hardly have cytotoxic activities.
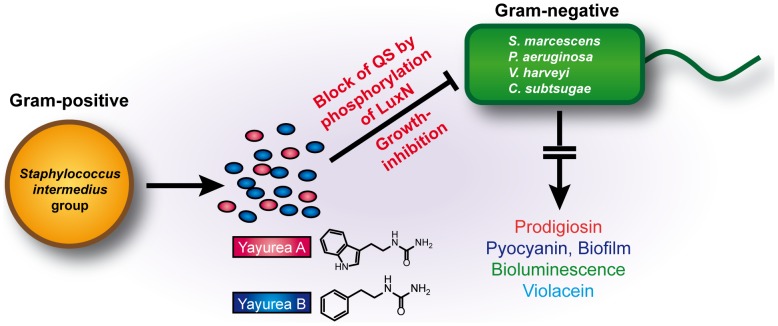
An interesting question is why just the ‘intermedius group’ is producing yayureas. The most likely answer is that this group shares its habitat with Gram-negative bacteria. There must be a benefit to producing yayurea A and B because they are excreted in such high amounts certainly costing energy. In addition, the yayureas show growth inhibition only at quantities that exceed the physiological concentrations. It is a clever arrangement, that S. delphini produces just enough yayurea A and B to almost completely silence the expression of the studied QS-regulated compounds or biofilm formation in diverse Gram-negative bacteria. In mixed cultures, the yayurea-producing staphylococci arrest Gram-negative bacteria in a pheno- and genotypic state of low cell density, although the cells have grown to high density. The advantage for the staphylococci is twofold, on one hand, they are protected from QS-controlled toxins and on the other hand, yayurea A and B affect stationary growth of particularly those Gram-negative bacteria with prominent QS-control systems. This is one of the rare cases of inter-phylum interference between firmicutes (Gram-positive) and beta-/gammaproteobacteria (Gram-negative). A schematic presentation of the interference is shown in Fig. 8.
Figure 8. Schematic presentation of the interference between zoonotic staphylococcal species and Gram-negative bacteria.
Animal associated (zoonotic) Staphylococcus species excrete novel quorum-quenching compounds, yayurea A and B, which block quorum sensing system in various Gram-negative bacteria by activating LuxN phosphorylation and have also growth inhibiting activity. The benefit for the staphylococcal species is better survival and increased competitiveness in a joint ecosystem.
Materials and Methods
Bacterial strains and growth conditions
Staphylococcus strains were grown in Tryptic Soy Broth (TSB, Sigma) medium at 37°C, Vibrio cholerae SP27459 (O1 El Tor), Serratia marcescens, Chromobacterium subtsugae DSMZ 17043 and Pseudomonas aeruginosa were grown in lysogenic broth (LB) medium, and Vibrio harveyi DSMZ6904 in marine broth (MB; 5 g peptone, 3 g yeast extract and 75% sea water per liter of deionized water) at 28°C. The Staphylococcus strains tested in this work are listed in Table 2.
Mixed cultivation of Staphylococcus sp. with P. aeruginosa
In co-culture experiments, we inoculated TSB with S. aureus (OD578 0.005) and P. aeruginosa SH1 (OD578 0.005). For the protection test, yayurea A (100 µg/ml), yayurea B (900 µg/ml), or water (control) were added to the mixed culture of S. aureus and P. aeruginosa after 3 h incubation. For co-cultivation of S. delphini with P. aeruginosa, each of the strains was inoculated with OD578 0.001, because doubling time of S. delphini was shorter than that of S. aureus. Co-cultures were aerobically grown at 37°C. For CFU determination, samples were diluted and plated on Chapman agar (selective medium for staphylococci, while growth of P. aeruginosa is retarded) and BM agar (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) on which P. aeruginosa grows better than the staphylococci. Colonies were counted after incubating for 24 h at 37°C; staphylococcal and pseudomonas colonies could be easily distinguished by colony shape and pigmentation.
Purification and structural analysis of the QS-inhibiting compounds from S. delphini
The QS inhibitors were isolated from aerobically-grown overnight culture of S. delphini (vigorous shaking at 37°C, 20 h). Cells were centrifuged and the supernatant was applied to an Amberilite XAD 16 resin (Rhom& Haas, Germany). The column was first washed with water, then with 40% and 60% methanol and finally eluted with solvent A (80% methanol containing 5% acetic acid). Yayureas were significantly enriched in the last fraction. In the second purification step, the eluate was evaporated in vacuo, suspended in water, and further separated on an Amberilite IRC 50 cation exchange resin (Serva, Heidelberg, Germany) after the pH was adjusted to 7.0 with 1 M NaOH. The column was washed with water, then 70% ethanol and eluted with solvent A. The eluate was concentrated in vacuo, diluted, and adjusted to a final concentration of 50 mM sodium phosphate buffer with a pH of 4.2. In the third purification step, the eluate was separated on a SP Sepharose cation exchange column (GE Healthcare, Germany) with a linear 1 M NaCl gradient in 50 mM sodium phosphate buffer on a Äkta FPLC (GE Healthcare, Germany). The two active compounds were eluted separately at 120 mM and 280 mM NaCl concentration respectively. The final purification and desalting of each peak was carried out by reversed phase preparative HPLC (RP-HPLC) (Bischoff, Leonberg, Germany) on a nucleosil 100 C-18, 8×250 mm column (Machery Nagel, Düren, Germany) with a linear water acetonitrile (containing 0.1% TFA) gradient of 0% to 60% in 25 min. Purified compounds were lyophilized and stored at −20°C. Qualitative analysis was carried out on an Agilent 1200 HPLC system (Agilent technologies, Waldbronn, Germany) and a RP-HPLC Waters xBridge C18, 5 mm, 4.6×150 mm column. Compounds were eluted with a 15 min linear gradient of 0.1% phosphoric acid to acetonitrile at a flow rate of 1.5 ml/min. For structure elucidation, mass spectrometry was carried out on GC-MS and FT-ICR MS (Bruker, ApexII). NMR spectra were measured in d4-methanol and recorded on a Bruker AMX600 spectrometer (600 MHz for 1H, 150 MHz for 13C), solvent was used as internal standard (δH/C 3.31/49.15 for MeOH-d4).
Assessing QS-regulated compounds in S. marcescens, P. aeruginosa, C. subtsugae, and bioluminescence in V. harveyi in co-cultivation with Staphylococcus sp.
Overnight cultures of S. aureus and S. delphini cells were diluted in LB medium containing 0.3% glucose to an OD578 value of 1.0 (we also tried 0.1, 0.5, but OD 1.0 results were most pronounced), incubated for 4 h at 37°C, then co-cultivated at 30°C with S. marcescens or P. aeruginosa; each strain was inoculated with a starting OD600 of 0.01. After 24 h of cultivation, prodigiosin of S. marcescens was extracted from the cell pellets by ethanol acidified with 4% of 1 M hydrochloric acid and then quantified by A534 nm determination. Pyocyanin in the supernatant was determined by its absorption maximum at 520 nm [58]. For bioluminescence tests, overnight-cultured V. harveyi was diluted to an OD578 of 1.0 and co-cultivated with an equal amount of S. aureus or S. delphini cells in LB-MB (LB medium containing 0.1% glucose mixed with equal volumes of MB) for 18 h. Overnight cultures of S. aureus and S. delphini cells were diluted to an OD578 of 0.1, incubated for 4 h at 37°C, then co-cultivated at 30°C with C. subtsugae, which was diluted to a final OD600 value of 2; this high OD was necessary to pronounce violacein production, whose expression is dependent on AHL at higher cell density. After 24 h of incubation, cell pellets were collected and resuspended in water. Cells were lysed by 10% sodium dodecyl sulfate and incubated for 5 min at room temperature. Violacein was quantitatively extracted from the cell by adding water-saturated butanol. The butanol phase containing the violacein was collected and determined at its absorption maximum at 585 nm.
Production of yayurea A and B in S. delphini and their effect on bacterial growth
S. delphini was inoculated to a starting OD578 of 0.1 and incubated at 37°C. During the 24 h incubation, cell density was followed by OD578 and the active compounds in the supernatants were determined by RP-HPLC and phosphoric acid – acetonitrile gradient as described above. The Gram-negative representatives, E. coli, P. aeruginosa, S. marcescens, V. cholerae and V. harveyi were inoculated to OD600 of 0.1 and grown in LB for 2 h. After that time, 1 mg/ml yayurea A or B (as solution in H2O) or an equal volume of water as negative control was added. Cells were either incubated at 37°C (S. aureus or S. delphini) or 30°C (E. coli, S. marcescens, V. harveyi, V. cholerae and P. aeruginosa). Cell density was followed for 24 h.
Activity assays for QS-regulated compounds, bioluminescence, and biofilm formation in Gram-negative bacteria
Pyocyanin production
An overnight culture of P. aeruginosa PAO1 was diluted with LB medium to an OD600 of 0.1. The (Z-)-4-bromo-5-(bromomethylene)-2(5H)-furanone (Sigma), yayureas and tetracycline were dissolved in 10% DMSO. 190 µl diluted PAO1 was cultured in 96-well plates with 10 µl different concentrations of pure yayurea A, B, furanone, or 10% DMSO (negative control) at 30°C. After 24 h of incubation, cell density was measured at 600 nm and pyocyanin was isolated as described previously [58]. To measure pyocyanin production, the supernatants were subjected to HPLC separation on a Nucleosil 100, C-18 column and a 0–100% Water-ACN Gradient (water containing 0.1% phosphoric acid) in 15 min at a flow rate of 1.5 ml/min. The content of pyocyanin was determined at its absorption maximum at 520 nm [58]. The relative pyocyanin production was calculated as the ratio of the amount of pyocyanin content from the HPLC measurements to cell density.
Prodigiosin production
S. marcescens cells were inoculated from an overnight culture to a starting OD600 of 0.1. Then 100 µl of the diluted culture was mixed with 100 µl of different amounts of the purified compounds or water in wells of 96-well microtiter plates and incubated for 24 h at 28°C. Cell density was measured at 600 nm. Prodigiosin was extracted as described above and the relative prodigiosin production was calculated as the ratio between the amount of prodigiosin at its absorption maximum at 534 nm and cell density values at 600 nm. The prodigiosin suppressing effect of yayurea A and B was determined (H2O was used as control).
Biofilm formation assay
Biofilm formation assays were performed as described previously [34]. A full-grown culture of P. aeruginosa PAO1 or S. aureus was diluted with LB medium containing 0.3% (w/v) glucose to an OD600 of 0.1. 100 µl of the dilution was mixed with 100 µl of the yayurea solutions with the indicated concentrations, and incubated in 96-wellplates at 37°C for 24 h. To determine the biofilm formation, planktonic cells were discarded and the plates gently washed with PBS and air-dried for 30 min. The wells were stained with 200 µl of a 0.1% crystal violet solution at room temperature for 30 min. The stained biofilm was rinsed with distilled water followed by the addition of 200 µl DMSO. Absorbance of crystal violet as indicator for biofilm-forming bacteria was measured at 590 nm. The effects of the compounds were evaluated for which the sample with water was set to 100%.
Bioluminescence
An overnight culture of V. harveyi was diluted to an OD600 of 0.1 and grown with yayurea A, B, or water for 24 h at 28°C. Bioluminescence was determined in a Tecan infinite M200 plate reader (Tecan, Groedig, Austria). Relative luminescence was normalized by the OD600 values. The effects of the compounds were calculated by the relative luminescence, for which the sample with water was set to 100%.
Investigation of the QS-target(s) of yayureas in vitro
Production of QS-receptors and QS-molecules
Inverted membrane vesicles were prepared using E. coli strainTKR2000 expressing plasmids pNKN, pNKQ, and pNKS encoding wild type LuxN, LuxQ, and CqsS (each with a C-terminal His-tag), respectively. Inverted membrane vesicles were prepared as described by [36]. LuxP was produced in, and purified from, E. coli MDAI-2 transformed with the plasmid pGEX_LuxP as described before [59]. LuxU was produced and purified as described before, using E. coli JM109 transformed with plasmid pQE30LuxU-6His [36]. Synthetic autoinducer, HAI-1, (from the University of Nottingham) was dissolved in a minimal volume of acetonitrile [10% (v/v)], diluted with water to a concentration of 100 mM and stored at −20°C.
Phosphorylation assays
Each QS-receptor kinase was tested as full-length membrane integrated proteins in inverted membrane vesicles. The buffer used for phosphorylation reactions was as follows: 50 mM Tris/HCl pH 8.0, 10% (v/v) glycerol, 500 mM KCl, 2 mM DTT. Each phosphorylation reaction contained equimoloar receptor concentrations [the total protein concentration were accordingly adapted: 3.2 mg/ml (LuxN), 5.5 mg/ml (LuxQ), and 5.2 mg/ml (CqsS) membrane proteins], and purified LuxU at 0.1 mg/ml. For phosphorylation of LuxQ, LuxP was added at a concentration of 0.4 mg/ml. LuxP integration into LuxQ-bearing membrane vesicles was triggered by several cycles of thawing and freezing. When indicated, yayurea A or B were added to the reaction mix at a final concentration of 220 µg/ml, and HAI-1 at 20 µM. The reaction was started by adding radio labeled Mg2+-ATP, typically 100 µM [γ-32P]ATP (specific radioactivity of 0.94 Ci/mmol; Perkin Elmer) and 110 µM MgCl2. The reaction was stopped after 1 and 10 minutes by adding SDS Laemmli-loading buffer followed by separation of the proteins on SDS-polyacrylamide gels. Gels were dried at 80°C on filter paper, exposed to a phosphoscreen for at least 24 h, and subsequently scanned using a PhosphorImager SI (GE Healthcare). The gels presented are representative of at least three independent sets of experiments.
Supporting Information
Mass spectra of Yayurea A and B. Mass spectrometry was carried out on GC-MS and FT-ICR MS (Bruker, ApexII).
(TIF)
Influence of yayurea A and B on growth. Growth curve of S. marcescens, P. aeruginosa, V. harveyi and V. cholerae in BM with 1000 µg/ml yayurea A or B or equal volume of water. Arrow indicates time point (after 2 h) of the addition of compounds to the growing culture. All the measurements were made in triplicate. Bars indicate standard deviation, SD.
(TIF)
Phylogenetic tree among Staphylococcus species. The tree is based on 16S rRNA relationships according to [45]. The phylogenetic position of the ‘intermedius group’ composed of S. intermedius, S. pseudintermedius, and S. delphini is marked in bold. Based on a combinatin of 16S rRNA and multilocus data, the group was recently complemented by the next related species S. lutrae, S. schleiferi subsp. schleiferi and S. schleiferi subsp. coagulans [37]. All members of this group produce yayurea A and B.
(TIF)
Acknowledgments
We thank Karl-Heinz Schleifer for critical review of the manuscript, Ross Fitzgerald for offering S. pseudintermedius strain and Regine Stemmler, Vera Augsburger, and Sabine Scheu for technical assistance.
Funding Statement
This study was partially funded by grants from the Landesgraduiertenförderung Baden Württemberg “Antimicrobial compounds”, the Deutsche Forschungsgemeinschaft SFB 766, TR-SFB 34, Exc114-1 and SPP1617 and Open Access Publishing Fund of Tuebingen University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Atkinson S, Williams P (2009) Quorum sensing and social networking in the microbial world. J R Soc Interface 6: 959–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346. [DOI] [PubMed] [Google Scholar]
- 3. Hanzelka BL, Parsek MR, Val DL, Dunlap PV, Cronan JE Jr, et al. (1999) Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol 181: 5766–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, et al. (2007) The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450: 883–886. [DOI] [PubMed] [Google Scholar]
- 5. Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, et al. (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415: 545–549. [DOI] [PubMed] [Google Scholar]
- 6. Henke JM, Bassler BL (2004) Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi . J Bacteriol 186: 6902–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freeman JA, Bassler BL (1999) A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi . Mol Microbiol 31: 665–677. [DOI] [PubMed] [Google Scholar]
- 8. Freeman JA, Bassler BL (1999) Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi . J Bacteriol 181: 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, et al. (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82. [DOI] [PubMed] [Google Scholar]
- 10. Lilley BN, Bassler BL (2000) Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol 36: 940–954. [DOI] [PubMed] [Google Scholar]
- 11. Williams P, Camara M (2009) Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12: 182–191. [DOI] [PubMed] [Google Scholar]
- 12. Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, et al. (1994) Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A 91: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pearson JP, Passador L, Iglewski BH, Greenberg EP (1995) A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa . Proc Natl Acad Sci U S A 92: 1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, et al. (1995) Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol 17: 333–343. [DOI] [PubMed] [Google Scholar]
- 15. Deziel E, Lepine F, Milot S, He J, Mindrinos MN, et al. (2004) Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101: 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C (2002) Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa . J Bacteriol 184: 6472–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGrath S, Wade DS, Pesci EC (2004) Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol Lett 230: 27–34. [DOI] [PubMed] [Google Scholar]
- 18. Njoroge J, Sperandio V (2009) Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol Med 1: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ (2008) The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manefield M, Welch M, Givskov M, Salmond GP, Kjelleberg S (2001) Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora . FEMS Microbiol Lett 205: 131–138. [DOI] [PubMed] [Google Scholar]
- 21. Swem LR, Swem DL, Wingreen NS, Bassler BL (2008) Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi . Cell 134: 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wermuth CG (2006) Selective optimization of side activities: the SOSA approach. Drug Discov Today 11: 160–164. [DOI] [PubMed] [Google Scholar]
- 23. Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, et al. (1995) The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus . Mol Gen Genet 248: 446–458. [DOI] [PubMed] [Google Scholar]
- 24. Otto M, Süssmuth R, Jung G, Götz F (1998) Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett 424: 89–94. [DOI] [PubMed] [Google Scholar]
- 25. Ji G, Beavis R, Novick RP (1997) Bacterial interference caused by autoinducing peptide variants. Science 276: 2027–2030. [DOI] [PubMed] [Google Scholar]
- 26. Lyon GJ, Novick RP (2004) Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25: 1389–1403. [DOI] [PubMed] [Google Scholar]
- 27. Biswas L, Biswas R, Schlag M, Bertram R, Götz F (2009) Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa . Appl Environ Microbiol 75: 6910–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, et al. (2006) Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas . J Bacteriol 188: 8079–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomson NR, Crow MA, McGowan SJ, Cox A, Salmond GP (2000) Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol Microbiol 36: 539–556. [DOI] [PubMed] [Google Scholar]
- 30. Anetzberger C, Pirch T, Jung K (2009) Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi . Mol Microbiol 73: 267–277. [DOI] [PubMed] [Google Scholar]
- 31. Martin PA, Gundersen-Rindal D, Blackburn M, Buyer J (2007) Chromobacterium subtsugae sp. nov., a betaproteobacterium toxic to Colorado potato beetle and other insect pests. Int J Syst Evol Microbiol 57: 993–999. [DOI] [PubMed] [Google Scholar]
- 32. Castric PA (1975) Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa . Can J Microbiol 21: 613–618. [DOI] [PubMed] [Google Scholar]
- 33. Hassan HM, Fridovich I (1980) Mechanism of the antibiotic action pyocyanine. J Bacteriol 141: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saising J, Dube L, Ziebandt AK, Voravuthikunchai SP, Nega M, et al. (2012) Activity of Gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 56: 5804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, et al. (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22: 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Timmen M, Bassler BL, Jung K (2006) AI-1 influences the kinase activity but not the phosphatase activity of LuxN of Vibrio harveyi. J Biol Chem 281: 24398–24404. [DOI] [PubMed] [Google Scholar]
- 37. Lamers RP, Muthukrishnan G, Castoe TA, Tafur S, Cole AM, et al. (2012) Phylogenetic relationships among Staphylococcus species and refinement of cluster groups based on multilocus data. BMC Evol Biol 12: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bannoehr J, Ben Zakour NL, Waller AS, Guardabassi L, Thoday KL, et al. (2007) Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J Bacteriol 189: 8685–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ben Zakour NL, Beatson SA, van den Broek AH, Thoday KL, Fitzgerald JR (2012) Comparative Genomics of the Staphylococcus intermedius Group of Animal Pathogens. Front Cell Infect Microbiol 2: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Varaldo PE, Kilpper-Bälz R, Biavasco F, Satta G, Schleifer KH (1988) Staphylococcus delphini sp. nov., a coagulase-positive species isolated from dolphins. Int J Syst Bacteriol 38: 436–439. [Google Scholar]
- 41. Hájek V (1976) Staphylococcus intermedius, a new species isolated from animals. Int J Syst Bacteriol 26: 401–408. [Google Scholar]
- 42. van Duijkeren E, Kamphuis M, van der Mije IC, Laarhoven LM, Duim B, et al. (2011) Transmission of methicillin-resistant Staphylococcus pseudintermedius between infected dogs and cats and contact pets, humans and the environment in households and veterinary clinics. Vet Microbiol 150: 338–343. [DOI] [PubMed] [Google Scholar]
- 43. Foster G, Ross HM, Hutson RA, Collins MD (1997) Staphylococcus lutrae sp. nov., a new coagulase-positive species isolated from otters. Int J Syst Bacteriol 47: 724–726. [DOI] [PubMed] [Google Scholar]
- 44. Bes M, Guerin-Faublee V, Freney J, Etienne J (2002) Isolation of Staphylococcus schleiferi subspecies coagulans from two cases of canine pyoderma. Vet Rec 150: 487–488. [DOI] [PubMed] [Google Scholar]
- 45.Götz F, Bannerman T, Schleifer KH (2006) The Genera Staphylococcus and Macrococcus. In: Dworkin M, editor. Procaryotes. New York: Springer. pp. 5–75.
- 46. Takahashi T, Satoh I, Kikuchi N (1999) Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol 49 Pt 2: 725–728. [DOI] [PubMed] [Google Scholar]
- 47. Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, et al. (2010) Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol 48: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Albrecht T, Raue S, Rosenstein R, Nieselt K, Götz F (2012) Phylogeny of the staphylococcal major autolysin and its use in genus and species typing. J Bacteriol 194: 2630–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anetzberger C, Reiger M, Fekete A, Schell U, Stambrau N, et al. (2012) Autoinducers act as biological timers in Vibrio harveyi . PLoS One 7: e48310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jung K, Odenbach T, Timmen M (2007) The quorum-sensing hybrid histidine kinase LuxN of Vibrio harveyi contains a periplasmically located N terminus. J Bacteriol 189: 2945–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Teasdale ME, Liu J, Wallace J, Akhlaghi F, Rowley DC (2009) Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl Environ Microbiol 75: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, et al. (2007) RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa . Mol Microbiol 66: 1557–1565. [DOI] [PubMed] [Google Scholar]
- 54. Dandekar AA, Chugani S, Greenberg EP (2012) Bacterial quorum sensing and metabolic incentives to cooperate. Science 338: 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heurlier K, Denervaud V, Haas D (2006) Impact of quorum sensing on fitness of Pseudomonas aeruginosa . Int J Med Microbiol 296: 93–102. [DOI] [PubMed] [Google Scholar]
- 56. van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL (2013) Individual and Combined Roles of the Master Regulators AphA and LuxR in Control of the Vibrio harveyi Quorum-Sensing Regulon. J Bacteriol 195: 436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, et al. (2008) Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52: 3648–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cox CD (1986) Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun 52: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM (2005) Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell 18: 507–518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectra of Yayurea A and B. Mass spectrometry was carried out on GC-MS and FT-ICR MS (Bruker, ApexII).
(TIF)
Influence of yayurea A and B on growth. Growth curve of S. marcescens, P. aeruginosa, V. harveyi and V. cholerae in BM with 1000 µg/ml yayurea A or B or equal volume of water. Arrow indicates time point (after 2 h) of the addition of compounds to the growing culture. All the measurements were made in triplicate. Bars indicate standard deviation, SD.
(TIF)
Phylogenetic tree among Staphylococcus species. The tree is based on 16S rRNA relationships according to [45]. The phylogenetic position of the ‘intermedius group’ composed of S. intermedius, S. pseudintermedius, and S. delphini is marked in bold. Based on a combinatin of 16S rRNA and multilocus data, the group was recently complemented by the next related species S. lutrae, S. schleiferi subsp. schleiferi and S. schleiferi subsp. coagulans [37]. All members of this group produce yayurea A and B.
(TIF)