Abstract
The embryonic self-renewal factor SALL4 has been implicated in the development of human acute myeloid leukemia (AML). Transgenic mice expressing the human SALL4B allele develop AML, which indicates that this molecule contributes to leukemia development and maintenance. However, the underlying mechanism of SALL4-dependent AML progression is unknown. Using SALL4B transgenic mice, we observed that HoxA9 was significantly upregulated in SALL4B leukemic cells compared with wild-type controls. Downregulation of HoxA9 in SALL4B leukemic cells led to decreased replating capacity in vitro and delayed AML development in recipient mice. In primary human AML cells, downregulation of SALL4 led to decreased HOXA9 expression and enhanced apoptosis. We found that SALL4 bound a specific region of the HOXA9 promoter in leukemic cells. SALL4 overexpression led to enhanced binding of histone activation markers at the HOXA9 promoter region, as well as increased HOXA9 expression in these cells. Furthermore, we observed that SALL4 interacted with mixed-lineage leukemia (MLL) and co-occupied the HOXA9 promoter region with MLL in AML leukemic cells, which suggests that a SALL4/MLL pathway may control HOXA9 expression. In summary, our findings revealed a molecular mechanism for SALL4 function in leukemogenesis and suggest that targeting of the SALL4/MLL/HOXA9 pathway would be an innovative approach in treating AML.
Introduction
SALL4, a member of the zinc finger transcription factor SALL gene family, is the human homolog of the Drosophila homeotic gene spalt.
The role of SALL4 as a transcription factor has been very well established in early embryonic development, as Sall4-null mice die shortly after implantation (1). Our group and others have also shown that in mice, murine Sall4 plays an essential role in regulating the pluripotency and self-renewal properties of ES cells through directly regulating the expression of Oct4 and coordinating with Nanog to control ES cell differentiation (2). Sall4-deficient ES cells differentiate spontaneously and form the trophectoderm in mouse blastocysts, indicating its vital role in maintaining pluripotency (3).
The protein is present in humans in 2 isoforms, SALL4A and SALL4B, as a result of alternative splicing (4). Human SALL4 mutations are associated with Duane-radial ray syndrome (DRRS; also known as Okihiro syndrome) (5–8). This condition is an autosomal-dominant disorder involving radial-sided hand anomalies in association with Duane syndrome (DS), a congenital disorder of eye movement characterized by strabismus.
SALL4 is also involved in normal hematopoiesis and leukemogenesis. During normal hematopoiesis, SALL4 is preferentially expressed in human CD34+ hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) and downregulated in CD34– cells during hematopoietic differentiation (4). Depletion of SALL4 in CD34+ cells impaired their proliferation and self-renewal ability (9). Recently, SALL4 was reported to be a robust stimulator of HSC expansion (10). In addition to its role in normal hematopoiesis, we previously found that SALL4 is aberrantly expressed in myeloid leukemia cell lines and primary acute myeloid leukemia (AML) samples by using immunohistological staining and quantitative real-time RT-PCR (qRT-PCR) analysis (4). Moreover, SALL4 expression correlates with disease progression in human chronic myeloid leukemia (11). Its expression in AML patients correlates with treatment status (12). Further exploration revealed the role of SALL4 in drug resistance, in which SALL4 was involved in the maintenance of side population (SP) cells by regulating ATP-binding cassette drug transport genes (12). Therefore, SALL4 may be used as a marker for diagnosis and prognosis in AML.
Previously, we also demonstrated that constitutive expression of SALL4 contributes to leukemogenesis in adult mice (4). Mice transgenic for SALL4B, one of the SALL4 isoforms we identified, developed preleukemic myelodysplastic syndrome–like (MDS-like) features and subsequent AML, which suggests that SALL4 contributes to the initiation of leukemia (4). In addition, loss-of-function studies have demonstrated that SALL4 is a key regulator in leukemic cell survival, and downregulation of SALL4 leads to significant apoptosis of leukemic cells (13), which suggests that SALL4 is essential for the maintenance of leukemia cells.
HOXA constitutes 1 of the 4 families of HOX genes, which are transcription factors characterized by a homeobox domain. Mutations of the HOX genes have been linked to defects of limb and genital development (14, 15). In addition, HoxA9 is critical to murine granulopoiesis, and dysregulated HOXA9 expression is implicated in more than 70% of human AML. Its expression is enriched in human CD34+CD38– stem cells compared with normal CD34– cells (16). Consistent with its role in leukemogenesis, HoxA9-knockout mice demonstrate the most severe phenotype of all the Hox knockout models, with multilineage hematopoietic differentiation defects (17, 18), as well as defects in HSC repopulation and proliferation (19). Additionally, the ABD HOXA genes, HoxA7–HoxA10, are overexpressed in mice with expression of a partial tandem duplication of the mixed-lineage leukemia (MLL) allele (MLL PTD), which has been described in AML (20–22). This overexpression is also correlated with increased histone acetylation in HOX gene promoters, suggesting a unique role of epigenetic modification in the regulation of HOX genes (20). Downregulation of HOXA9 has been shown by Armstrong’s group to be critical for survival in human leukemia with MLL rearrangement (23).
The HOXA and SALL gene families have been linked during development through protein-protein interactions. Both have been implicated in segmentation across various species. In fact, the SALL family modulates HOX expression in murine limb development (24). We recently showed that SALL4 can regulate HOXA9 during normal human myelopoiesis (9). While both the homeotic gene SALL4 and the homeobox gene HOXA9 play important roles in myeloid leukemogenesis, the connection between them during leukemia development has not yet been investigated. Here, through gene expression profiles of cells from SALL4B-induced leukemic mice (referred to herein as SALL4B leukemic cells), we identified HoxA9 among the upregulated genes. Functional studies showed that downregulation of HoxA9 in SALL4B leukemic cells led to decreased replating capacity in vitro and delayed AML development in recipient mice. Using human AML leukemic cells, we further confirmed that HOXA9 expression was regulated by SALL4, through its interaction and co-occupation of the HOXA9 promoter region with MLL. In summary, we demonstrated a unique SALL4/MLL/HOXA9–mediated process in murine and human AML, a novel pathway to be targeted in AML.
Results
Upregulation of HoxA9 in SALL4B leukemic cells.
To investigate the functional role of SALL4 in leukemogenesis, we generated a transgenic SALL4B mouse model that develops AML and is transplantable (4). To explore the molecular mechanism of SALL4B in the development of leukemia, we used serial transplantations to identify the leukemic initiating cells (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI62891DS1). First, we sorted the Lin–Sca-1+c-kit+ (LSK) and HPC subpopulations, such as common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte/erythroid progenitors (MEPs), from primary leukemic SALL4B transgenic donor mice according to previously described methods (25). The sorted SALL4B leukemic cells were then transplanted into immunodeficient NOD-SCID mice, and leukemia development was monitored in the recipients. Aggressive fatal AML with onset ranging 1–3 months developed in the recipient mice after primary transplantation of SALL4B leukemic cells. The transplanted disease was characterized by immature blasts (c-kit+) in the peripheral blood, BM, and tissues such as the liver, lymph nodes, and spleen (Figure 1, A–H). These were the same phenotypes as reported previously for SALL4B-induced AML donor mice, and SALL4 expression was maintained in the transplanted leukemic cells as well (Figure 1I).
Figure 1. Upregulation of HoxA9 in SALL4B leukemic cells.
(A–H) Leukemia development in recipient mice. Images are from a representative recipient of a primary transplant. Leukocytosis was observed in the blood smears (A). Blasts were present in the peripheral blood (B), BM smear (C), lymph node (D), spleen (E and F), and liver (G and H). Original magnification, ×40 (A, E, and G), ×200 (D, F, and H), ×600 (B and C). (I) SALL4 expression was determined by IHC staining in SALL4B leukemic BM cells. Original magnification, ×600. (J) Validation of HOXA9 upregulation in SALL4B leukemic GMP population. qRT-PCR analysis of gene expression in SALL4B leukemic GMPs compared with control normal GMPs. Measurements were from 3 individual secondary transplanted recipients (n = 3), each performed in triplicate. *P < 0.05. (K) Flow analysis of donor cell subtype after secondary transplantation.
The HPC subpopulations from primary transplanted leukemic recipient mice were sorted again and used for secondary transplantations. All secondary recipients from leukemic GMP donors developed AML within 30 days after transplantation. The GMPs continued to expand in the leukemic recipients, becoming the dominant HPC population after secondary transplantation (Figure 1K). We then isolated the enriched SALL4B leukemic GMP cells from 3 independently transplanted NOD-SCID recipients and compared their gene expression profiles with their counterpart GMP populations from recipient control NOD-SCID as well as SALL4B wild-type littermates. Analysis using dChip revealed an expression signature for SALL4B leukemic cells, consisting of upregulated (83 probe sets) and downregulated (394 probe sets) genes (Supplemental Table 1). HoxA9 was among the top upregulated genes, as determined by the level of change in expression in SALL4B leukemic cells compared with those from controls, and this was verified by qRT-PCR (Figure 1J).
Downregulation of HoxA9 in SALL4B leukemic cells leads to decreased leukemogenic potential.
HoxA9 has been reported to be important for myeloid leukemogenesis and is a key survival factor for human leukemia with MLL rearrangement (23). We tried to reduce HoxA9 expression in SALL4B leukemic cells to test whether this can, at least in part, decrease SALL4B-mediated leukemogenesis. HoxA9 lentiviral shRNA viruses were generated as previously reported by Armstrong’s group (23).
During our serial transplant experiments, we noticed that the leukemic development in the recipient mice became more aggressive, with a shortened leukemia-free period. In addition, fewer unsorted donor leukemic cells were needed for the induction of leukemic phenotype in the recipients. After the second transplant, BM cells from leukemic recipients were isolated by Ficoll-Paque PLUS and labeled with GFP to trace donor cells. Limited numbers of donor cells (10 or 100 cells) were transplanted into recipient mice to test their leukemogenic ability. We found that as few as 10 of these GFP+ cells could give rise to leukemic phenotype in the recipients (Supplemental Figure 2). We then used secondary transplants as an enrichment method for leukemic initiation cells. In the subsequent mouse experiments, Ficoll-Paque–isolated leukemic cells after 2 rounds of transplants from recipients were used. Knockdown of HoxA9 was performed using IF2-HOXA9 shRNA and IF3-HOXA9 shRNA, which have been demonstrated by Armstrong’s group to specifically downregulate HOXA9 expression in both murine and human hematopoietic cells (23). Using qRT-PCR, we found that the 2 pairs of HOXA9 shRNAs had some differences in their ability to downregulate murine HoxA9 expression in SALL4B leukemic cells (Figure 2A). To test whether HoxA9 downregulation in SALL4B leukemic cells has any functional effects, we used a previously described replating assay (23).
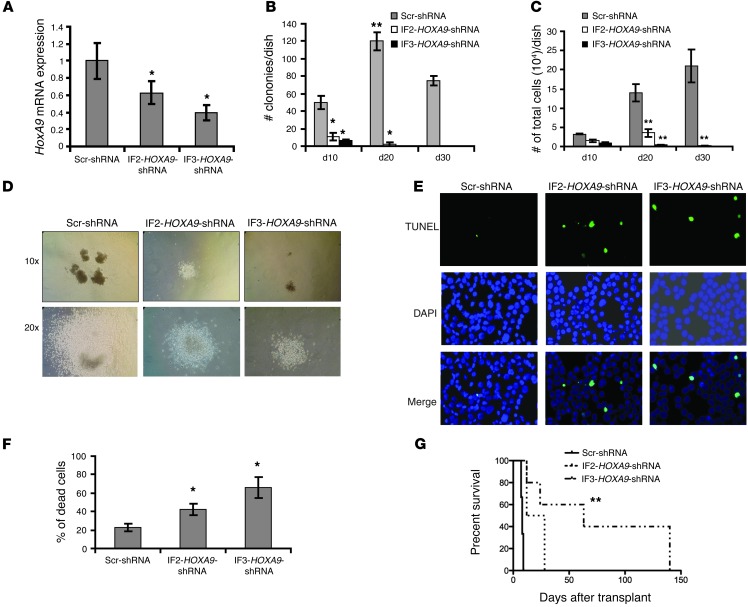
Figure 2. HoxA9 is critical in SALL4-mediated AML.
(A) qRT-PCR analysis of HoxA9 expression in SALL4B leukemic cells after HOXA9 shRNA or control scrambled shRNA (Scr-shRNA) virus infection following puromycin (1 μg/ml) selection for 72 hours on semisolid medium. (B) SALL4B leukemic cell colony formation and replating capacity after HoxA9 suppression. 1,000 HOXA9 shRNA– or scrambled shRNA–treated SALL4B leukemic cells were seeded on semisolid medium with puromycin (1 μg/ml) and replated every 10 days. The number of colonies per dish was recorded after each round of replating. (C) Total number of cells per dish after each round of replating. (D) Representative colony images 10 days after the first round of plating. Original magnification, ×10 (top); ×20 (bottom). (E and F) Increased cell death in HOXA9-knockdown cells was observed by TUNEL assay (E) and Trypan blue stain (F) 3 days after the first round of plating. Original magnification, ×20 (E). (G) Log-rank (Mantel-Cox) survival curve of mice transplanted with SALL4B leukemic cells treated with IF2-HOXA9 shRNA (n = 8), IF3-HOXA9 shRNA (n = 7), or scrambled shRNA (n = 11). Data are mean ± SD from 3 independent experiments. *P < 0.05, **P < 0.001.
Using a semisolid medium, we could replate SALL4B leukemic cells treated with control scrambled shRNA for more than 3 rounds (30 days). However, SALL4B leukemic cells with moderated HoxA9 reduction (i.e., treated with IF2-HOXA9 shRNA) could not be replated for more than 2 rounds (20 days), while cells treated with the more potent IF3-HOXA9 shRNA could only be replated once (Figure 2B). In addition, upon HoxA9 knockdown, the total colony number and total cell number in each plate was significantly decreased compared with the scrambled shRNA–treated group (Figure 2, B and C). Furthermore, the SALL4B leukemic colonies treated with scrambled shRNA viruses were larger than those treated with HOXA9 shRNA viruses (Figure 2D).
The decreased replating capacity of SALL4B leukemic cells after downregulation of HoxA9 could be due to cell death and apoptosis, since HoxA9 has been implicated in maintaining leukemic cell survival (23). To test this possibility, SALL4B leukemic cells were treated with scrambled or HOXA9 shRNA for 3 days with puromycin selection on semisolid medium. The cells were then harvested and cytospun upon coverslips, and apoptotic cells were measured using TUNEL assay. As shown in Figure 2E, there was an increase in cell apoptosis in HOXA9 shRNA–treated groups compared with the scrambled control, which suggests that HoxA9 suppression can decrease SALL4B leukemic cell replating capacity by promoting cell apoptosis. This was further supported by the increased cell death in the HOXA9 shRNA–treated group, as measured by Trypan blue staining (Figure 2F).
To evaluate the effect(s) of HoxA9 downregulation in vivo, 1 × 103 SALL4B leukemic cells after second transplants treated with scrambled or HOXA9 shRNAs were transplanted into NOD-SCID mice. Scrambled shRNA–treated recipient mice developed fatal leukemia within 2 weeks, with a median survival of 8.5 days (n = 11). In contrast, recipient mice treated with the potent IF3-HOXA9 shRNA showed a statistically significant (P = 0.004) delay in leukemic development, with a median survival of 63 days (n = 7). Treatment of the SALL4B leukemic cells with the less-effective IF2-HOXA9 shRNA also prolonged the survival of its recipient mice, with a median survival of 20 days (P = 0.06, n = 8; Figure 2G and Supplemental Table 2). Once the HOXA9 shRNA–treated recipient mice developed leukemia, expression of HoxA9 in the leukemic BM samples was back to its baseline level, as tested by qRT-PCR, which suggests that the leukemic cells could come from the non–HoxA9-knockdown cells.
Taken together, the upregulation of HoxA9 observed in SALL4B leukemic cells and the loss-of-function study of HoxA9 in these cells provide direct evidence that HoxA9 is an important downstream target of SALL4 in the SALL4B leukemic murine model.
SALL4 is essential for proper HOXA9 expression and leukemic cell viability in primary human AML samples.
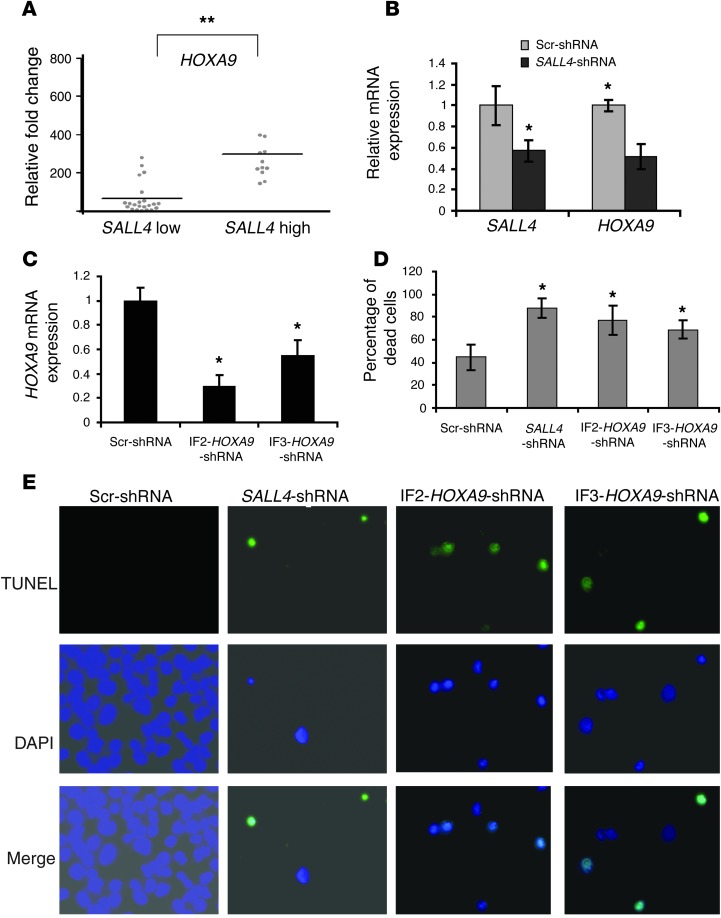
To test whether the SALL4/HOXA9 pathway is also important in human primary AML, we first evaluated the correlation of expression between SALL4 and HOXA9 in primary human AML samples. Using publicly available data sets, we performed analysis on SALL4 and HOXA cluster gene expression correlation, derived from cDNA microarray analysis of 385 previously described AML specimens (see Methods and ref. 26). There was positive correlation of expression between SALL4 and HOXA genes, particularly in AML M4 subgroups (Supplemental Table 3). We then verified the expression correlation of SALL4 and HOXA9 by qRT-PCR. 34 primary AML samples were subjected to qRT-PCR (Supplemental Table 4) and stratified into SALL4hi and SALL4lo groups using the mean intensity of SALL4 as a threshold. Expression of HOXA9 in the primary samples in these 2 groups was then examined. HOXA9 showed positive correlation with SALL4; its expression was significantly higher in the SALL4hi versus the SALL4lo group (P = 0.0001; Figure 3A). Next, we performed SALL4 knockdown experiments using a lentiviral mediated shRNA approach. Treatment of 3 independent human primary AML samples expressing both SALL4 and HOXA9 with this SALL4 shRNA significantly decreased SALL4 expression (Figure 3B).
Figure 3. Downregulation of SALL4 decreases HOXA9 expression in human primary AML and leads to apoptosis.
(A) Expression of SALL4 and HOXA9 was correlated in human primary AML samples. qRT-PCR was performed on 34 primary samples with primers for SALL4 and HOXA9 mRNA. Mean value of SALL4 was used to stratify the samples to SALL4hi or SALL4lo groups. Each dot represents a patient. Data are mean ± SD from 3 independent experiments. **P < 0.001. (B and C) Downregulation of SALL4 or HOXA9 expression in primary AML. Primary AML cells were transduced at a density of 1 × 106 cells/ml with lentiviruses expressing scrambled, SALL4, or HOXA9 shRNA. After 2 days of culturing in media containing cytokines (10 ng/ml IL-3, 25 ng/ml SCF, and 10 ng/ml IL-6), transduced cells were selected with 1 μg/ml puromycin for 3 days on semisolid medium. Expression of SALL4 (B) and HOXA9 (C) in primary AML after SALL4, HOXA9, or scrambled shRNA treatment was measured by qRT-PCR. (D and E) Increased apoptosis after downregulation of SALL4 or HOXA9 using the approach described above in primary AML cells was further evaluated by TUNEL assay (D) and Trypan blue staining (E). Original magnification, ×20 (E). Data are mean ± SD from 3 independent experiments. *P < 0.05.
We then tested whether knocking down SALL4 affected the expression of HOXA9 in primary human AML cells. Using qRT-PCR, we found that HOXA9 expression was reduced 50% compared with that of scrambled shRNA–treated cells (Figure 3B). We next tested whether downregulation of SALL4 and/or HOXA9 could lead to increased apoptosis/cell death in primary AML samples. Transduction of human primary AML cells with IF2-HOXA9 shRNA and IF3-HOXA9 shRNA reduced HOXA9 expression by 70% and 50%, respectively (Figure 3C). Downregulation of SALL4 or HOXA9 in these AML samples resulted in increased apoptosis, as determined by Trypan blue stain and TUNEL assay (Figure 3, D and E).
In summary, the correlation of expression and the loss-of-function studies of SALL4 and HOXA9 in primary human AML and murine leukemic model indicate that the SALL4/HOXA9 pathway is maintained in human and murine leukemia and is critical for leukemic cell survival.
SALL4 binds to a specific region of HOXA9 promoter.
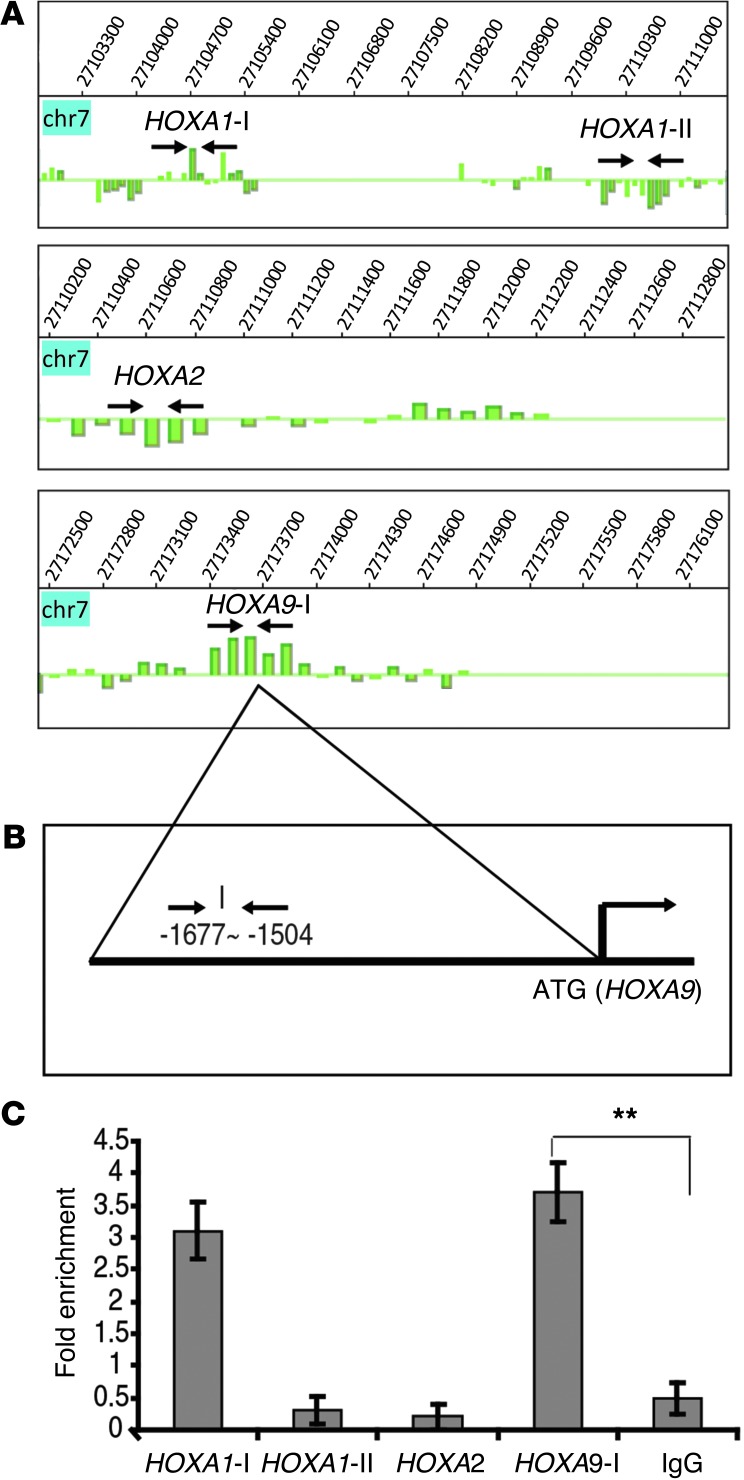
As part of the search for the transcriptional network responsible for the function of SALL4, we conducted a parallel genome-wide analysis of SALL4 target genes in primary CD34+ cells and murine ES cells using ChIP followed by a promoter microarray (ChIP-chip; refs. 9, 27). This analysis demonstrated that SALL4 bound to the promoter regions of several HOXA genes, including HOXA9 (Figure 4A). Comparison of SALL4 leukemic GMP expression signature with global ChIP-chip data sets also revealed that HOXA9 was among the overlapping genes (Supplemental Figure 3). In order to validate SALL4 binding to the HOXA9 promoter, we used ChIP coupled with qPCR assay (ChIP-qPCR). Leukemic CD34+ KG1a cells endogenously expressing both SALL4 and HOXA9 were used for this experiment. Enrichment of DNA fragments pulled down from these cells using our previously validated anti-SALL4 antibody (refs. 9, 27, and Supplemental Figure 4) was compared with input control. Primers were designed around positive binding peaks according to the ChIP-chip data. Additional regions from HOXA1-I, HOXA1-II, and HOXA2 locus served as positive and negative controls (see Supplemental Table 5 for primers). As shown in Figure 4, B and C, SALL4 bound specifically to the HOXA9 promoter region (–1,677 to –1,504 upstream of the ATG site) compared with the negative region control and IgG control.
Figure 4. SALL4 binds to the promoter region of HOXA9 in the AML cell line.
(A) Analysis of ChIP-chip data showing SALL4 binding sites at the promoter regions of HOXA genes. Arrows indicate location of primers for validation by ChIP-qPCR. (B) Human HOXA9-I promoter region. (C) SALL4 bound to the promoter region of HOXA9 in KG1a cells, as evaluated by ChIP-qPCR. All values represent the average of at least 2 separate pulldowns (biological duplicates) and qPCR assays. Standard error bars were calculated from the SD of separate trials (biological repeats). Promoter regions from HOXA1-II and HOXA2 were used as internal negative controls, the region for HOXA1-I was used as a positive control, and rabbit IgG was used as a negative control for antibody specificity. **P < 0.001.
Overexpression of SALL4 results in enrichment of binding of SALL4 and epigenetic activation markers at the HOXA9 promoter region.
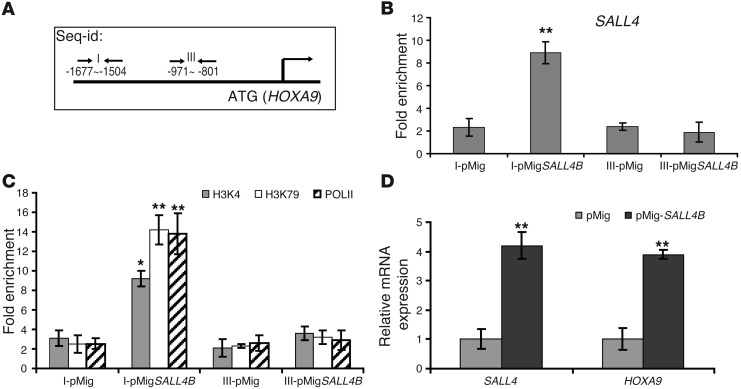
We then tested whether increased SALL4 expression can have an effect on its binding at the HOXA9 promoter region. In addition to the HOXA9-I binding site (–1,677 to –1,504; Figure 4B), we also included another region of HOXA9 promoter, designated HOXA9-III (–971 to –801), for the subsequent ChIP-qPCR studies (Figure 5A). After overexpression of SALL4B in KG1a cells, we evaluated the binding enrichment at these 2 regions of the HOXA9 promoter. Increased SALL4 binding (4-fold) was only observed using primers spanning HOXA9-I (Figure 5B). We next investigated the mechanism by which SALL4 activated HOXA9 expression. We observed that epigenetic activation markers, including H3K4 (methylation of lysine 4 of histone 3) and H3K79 (methylation of lysine 79 of histone 3), were enriched at least 6-fold in the same HOXA9-I region bound by SALL4. After overexpressing SALL4B, we also observed that RNA polymerase II (POLII), a marker for gene transcription, bound to this specific region of HOXA9 promoter (Figure 5C). Along with binding of SALL4 and induction of epigenetic activation markers, increased expression of SALL4B led to increased HOXA9 RNA expression (Figure 5D), consistent with our previous observation of upregulated HoxA9 expression in SALL4B leukemic cells (Figure 1J).
Figure 5. Overexpression of SALL4 induces enrichment of SALL4 binding and epigenetic activation markers at the HOXA9 promoter region and increases HOXA9 expression.
(A) Human HOXA9 promoter region. The locations of the primers used in ChIP-qPCR (arrows) are relative to the translation start ATG. (B) ChIP-qPCR demonstrated that SALL4 binding at HOXA9-I was enriched by overexpression of SALL4B in KG1a cells compared with cells treated with control vector (pMig). In contrast, HOXA9-III showed no enrichment of SALL4 binding. ChIP was performed using SALL4 antibody with KG1a cells transduced with control pMig-GFP or pMig-GFP-SALL4B vectors. GFP+ cells were used in this experiment. All values are averages calculated from 3 separate SALL4 pulldowns. (C) Enrichment of epigenetic markers in the HOXA9 promoter region induced by SALL4B overexpression. ChIP was performed using H3K4, H3K79, or POLII antibody on KG1a cells treated as in B. (D) qRT-PCR analysis of SALL4B and HOXA9 expression in pMig-SALL4B– and control vector–transduced GFP+ KG1a cells. Data in B–D are mean ± SD from 3 independent experiments. *P < 0.05, **P < 0.001.
In summary, our data indicate that SALL4 can promote HOXA9 expression by binding to the HOXA9 promoter region, and this process is associated with enriched binding of epigenetic activation markers.
SALL4 binds to a specific site in the HOXA9 promoter.
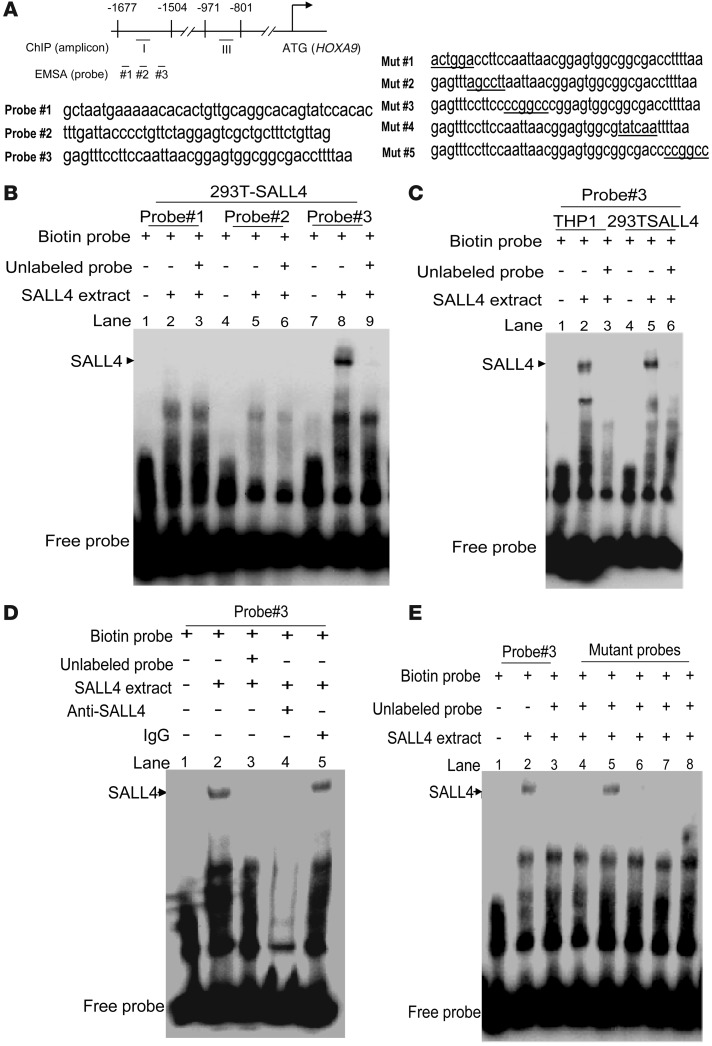
In order to further confirm that SALL4 binds to the HOXA9 promoter region, we designed 3 pairs of oligonucleotide probes and performed EMSAs (Figure 6A). As shown in Figure 6, B and C, probe 3 (–1,575 to –1,535) was markedly bound by SALL4 overexpressed in 293T cells as well as by endogenous SALL4 in THP1 cells. We further demonstrated that this SALL4/probe complex could be abolished by incubation with an anti-SALL4 antibody, but not by control IgG (Figure 6D).To further define the SALL4 binding site in the HOXA9 promoter region, we made a series of 5 mutants of probe 3. Among these, only mutant 2 could not compete with the binding between SALL4 and wild-type probe (Figure 6E), which indicates that the mutated residues were required for DNA sequence–specific binding of SALL4 to the HOXA9 promoter.
Figure 6. Identification of a SALL4 DNA binding site in the promoter region of HOXA9 by EMSA assays.
(A) Human HOXA9 promoter region. Corresponding locations of amplicons for qPCR and oligo probes for EMSA are indicated. Left: 3 probe sequences. Right: Mutant probe sequences of probe 3. (B) EMSAs were performed to identify the SALL4 DNA binding cells to the HOXA9 promoter region using nuclear extracts from 293T cells transfected with a SALL4 expression construct. Of 3 pairs of oligonucleotide sequences, only probe 3 was specifically bound by SALL4 protein (lane 8); however, this shift could be prevented by competition from 200-fold excess of nonlabeled probe (lane 9). (C) Endogenous SALL4 from THP1 nuclear extracts bound probe 3 specifically (lanes 1–3). Lanes 4–6 show nuclear extracts from 293T-SALL4 as a positive control. (D) SALL4 antibody abolished SALL4 and probe 3 binding (lane 4), but not IgG control antibody (lane 5). (E) Among the 5 mutant probes, only mutant probe 2 (lane 5) failed to inhibit wild-type probe 3 binding to SALL4 protein.
Interaction between SALL4 and MLL.
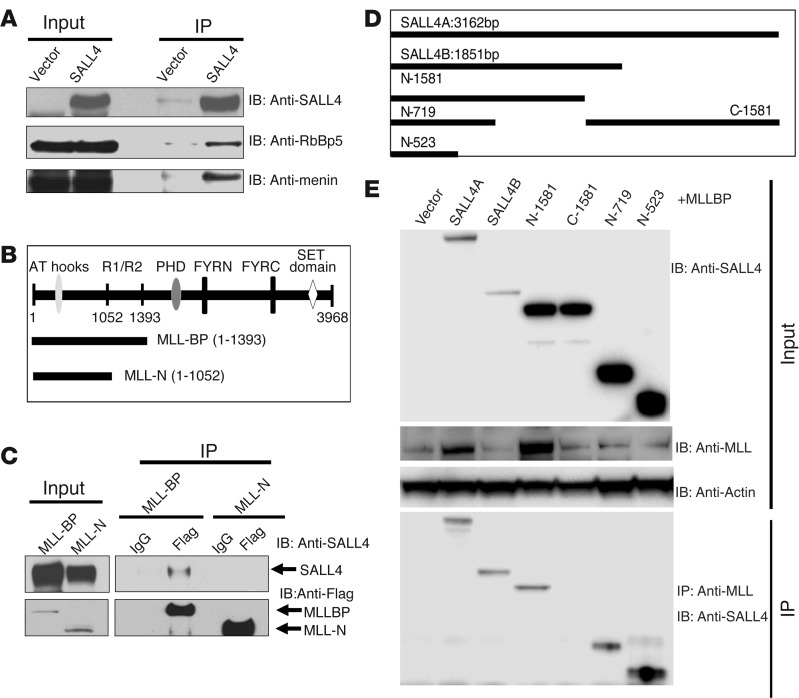
Overexpression of SALL4 induced POLII binding on the HOXA9 promoter. In addition, the MLL complex has been reported to recruit POLII to activate HOXA9 expression (28). One possibility is that there is a direct connection between SALL4 and MLL in regulation of HOXA9 expression. To test this hypothesis, we first carried out co-IP experiments. Extracts from 293T cells overexpressing SALL4 were subjected to IP with the SALL4-specific antibody, followed by Western blotting with antibodies against various components of the MLL complex. Members of the MLL complex, including RbBp5 and Menin, were found in the SALL4 pulldown (Figure 7A). In addition, the N terminus of MLL is known to interact with other proteins (29, 30), raising the possibility that MLL might interact with SALL4 through its N-terminal domain. To test this possibility, 293T cells coexpressing SALL4 with either a longer N-terminal Flag-tagged MLL construct (MLL-BP; 1–1,393 aa), including the protein interacting domain R1/R2, or a shorter N-terminal Flag-tagged MLL construct (MLL-N; 1–1,052 aa) lacking this region (Figure 7B), were subjected to IP with an anti-Flag antibody, followed by Western blot analysis with the SALL4 antibody. Interestingly, only MLL-BP, not MLL-N, could interact with SALL4 (Figure 7C). Notably, the MLL-BP domain was preserved in both wild-type MLL and MLL fusion proteins, such as MLL-AF9. We extended our experiments mapping the interaction between SALL4 and MLL by making several SALL4-truncated proteins. When tested for their abilities to interact with MLL-BP, in addition to full-length SALL4A and SALL4B, the N terminus of SALL4 (N-1581) could interact with MLL-BP, whereas the C terminus of SALL4 (C-1581) failed to show any binding. Additional mapping focused on the N terminus of SALL4 demonstrated that the N-523 fragment (1–523 bp) retained its ability to interact with MLL-BP (Figure 7, D and E).
Figure 7. SALL4 interacts with MLL.
(A) Co-IP with anti-SALL4 antibody was done using 293T cell lysates after SALL4 overexpression. The protein complex from Co-IP was resolved in SDS-PAGE and immunoblotted using antibodies against SALL4, RbBP5, and Menin. (B) MLL full-length and truncated proteins (MLL-BP and MLL-N). (C) 293T cells were cotransfected with SALL4 and Flag-tagged truncated MLL constructs. IP was performed with Flag antibody, followed by Western blot analysis with SALL4 antibody, which demonstrated that SALL4 could only be detected in pulldown with MLL-BP. Flag antibody could pull down both overexpressed MLL-BP and MLL-N proteins from 293T transfected cells. (D) SALL4 truncated vectors used for interaction with MLL-BP. (E) 293T cells were cotransfected with various SALL4 vectors and Flag-tagged truncated MLL-BP constructs. IP was performed with MLL antibody, followed by Western blot analysis with SALL4 antibody. Whereas the C terminus of SALL4 (C-1581) did not interact with MLL-BP, the N terminus (N-523) retained its ability to bind to MLL-BP.
SALL4 and MLL co-occupy the same HOXA9 promoter region in AML cells.
To verify whether the interaction between SALL4 and the MLL complex could occur in a leukemic context, we repeated the co-IP experiments using SALL4-expressing AML cells (Supplemental Figure 5). As shown in Figure 8A, a MLL-specific antibody could efficiently pull down endogenous SALL4 from leukemic KG1 cells. In addition, since the MLL-AF9 fusion protein has been extensively studied for its role in leukemogenesis, we also tested whether SALL4 could interact with the MLL-AF9 fusion protein. THP1, a human acute myelomonocytic leukemia cell line with the MLL-AF9 translocation, was chosen as a cell model. Notably, in this line harboring the MLL rearrangement, SALL4 protein was still detected in the MLL pulldowns. More importantly, when tested in a primary AML patient sample, we observed the interaction between SALL4 and MLL as well (Figure 8A).
Figure 8. Co-occupancy of the HOXA9 promoter region by SALL4 and MLL in primary AML cells and cell lines.
(A) Endogenous SALL4 protein was subjected to IP using MLL antibody in KG1, THP1, and primary AML cells, followed by Western blot analysis with SALL4 antibody. (B) ChIP–Re-CHIP demonstrated co-occupancy of the HOXA9 promoter region by SALL4 and MLL. The first ChIP assay was performed with antibody against MLL, and the second ChIP (Re-ChIP) was done with antibody against SALL4. Controls for the Re-ChIP assay were performed with anti-rabbit and anti-mouse IgG (rIgG/mIgG). Data are representative of at least 2 independent experiments. *P < 0.05, **P < 0.01.
Since both SALL4 and the MLL complex can regulate HOXA9 protein expression by binding to its promoter region, we next tested whether SALL4 and MLL could co-occupy the same region on the HOXA9 promoter. The MLL binding region on the HOXA9 gene has been well studied. Based on published data on the MLL binding site (31), we designed a sequential ChIP-qPCR assay, referred to herein as ChIP–Re-ChIP. As shown in Figure 8B, specific co-occupancy was observed for SALL4 and MLL in KG1, THP1, and primary AML cells using primers spanning the region of HOXA9-I.
The molecular mechanism of MLL and its fusion proteins in AML development occurs, at least in part, through activation of the HOX genes, including HoxA9 (32). The upregulation of HoxA9 in SALL4B leukemic cells and the interaction between SALL4 and MLL prompted us to compare our SALL4B leukemic cell gene expression profiling with that from a MLL-AF9 murine model (17). Gene set enrichment analysis (GSEA) was used for this comparison study. GSEA is an algorithm used to compare 2 sets of expression profiles to determine whether a particular expression signature is enriched or depleted (32–34). There was a statistically significant similarity of the gene expression signatures between SALL4B leukemic GMPs and MLL-AF9 leukemic GMPs (Supplemental Figure 6); the top 50 genes shared by the 2 groups, including HoxA9, are listed in Supplemental Table 6.
In order to identify whether other MLL target genes are modulated by the SALL4/MLL interaction, we overexpressed SALL4 in the KG1a AML cell line. A SALL4 mutant lacking the ability to interact with MLL was used as a control. Using qRT-PCR, we found that SALL4 overexpression significantly upregulated most of a set of well-known MLL target genes (8 of 10 tested; refs. 35–39), such as HOXA cluster genes and MEIS1. In contrast, the SALL4 mutant lacking the MLL interaction domain had no effect on the expression of these MLL target genes (Supplemental Figure 7).
Taken together, our data strongly suggest that SALL4 regulates HOXA9 expression through interacting with MLL, and this novel SALL4/MLL/HOXA9 pathway may play a critical role in human leukemogenesis.
Discussion
While both HOXA9 and SALL4 play important role(s) in myeloid leukemogenesis, the connection between these 2 transcription factors has not been investigated. Further characterization of the underlying mechanism should allow for better understanding of leukemogenesis and provide potential therapeutic targets for AML treatment. Here, through gene expression profiling of our previously characterized SALL4B transgenic mouse model (which develops AML), we observed upregulated HoxA9 expression in the SALL4B leukemic cell population. Downregulation of HoxA9 in SALL4B leukemic cells led to apoptosis and decreased replating ability in vitro as well as decreased AML development in recipient mice in vivo. Parallel human studies demonstrated that expression of SALL4 and HOXA9 was correlated in primary AML patient samples. Furthermore, downregulation of SALL4 in human primary AML cells leads to decreased HOXA9 expression, and loss-of-function studies of each gene in primary human AML samples showed a similar cell apoptosis/death phenotype. This prompted us to examine whether HOXA9 is a direct target of SALL4, and whether SALL4 can regulate HOXA9 in the hematopoietic system.
Additional ChIP-chip studies suggested that HOXA9 is a potential SALL4 target. We confirmed this finding by ChIP-qPCR, which showed that SALL4 bound to the specific promoter region of HOXA9 in leukemic cells at the endogenous level. Furthermore, overexpressing SALL4 enhanced markers of histone activation, such as H3K4 and H3K79 methylation, as well as POLII binding in the same promoter region, which increased HOXA9 expression in these cells.
MLL, which codes for a histone methyltransferase, is a global gene transcription regulator.
The molecular mechanism of MLL and its fusion proteins in AML development occurs, at least in part, through activation of HOX genes, including HoxA9 (32). We then explored whether SALL4 regulates HOXA9 through interaction with MLL. Using both 293T cells and human leukemic cells, we observed that SALL4 not only physically interacted with MLL, but also co-occupied on the HOXA9 promoter region with MLL. Others have shown that both MLL fusions and the normal MLL allele are important for leukemogenesis (40). Our finding that SALL4 interacted with both wild-type MLL and MLL fusion proteins prompted us to compare our SALL4B leukemic cell gene expression profile with that of a MLL-AF9 murine model (17). There was a statistically significant similarity of the gene expression signatures between SALL4B leukemic GMPs and MLL-AF9 leukemic GMPs (Supplemental Figure 6), further supporting the connections among SALL4, HOXA9, and MLL.
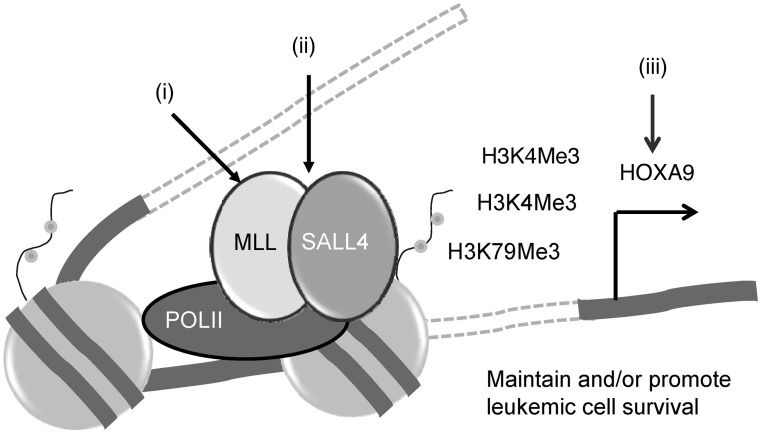
In conclusion, we propose that a novel SALL4/MLL/HOXA9 pathway plays an important role in myeloid leukemogenesis by promoting leukemic survival (Figure 9). Based on this model, we propose the following novel therapeutic approaches in targeting AML: (a) inhibiting the function of MLL or its complex with menin, as recently proposed by others (41); (b) disrupting the SALL4/MLL interaction, which we have recently shown to disrupt the protein interaction between SALL4 and NuRD (42); and (c) targeting HOXA9 directly. As the HOXA and SALL gene families are linked during development through protein-protein interactions (24), it is possible that SALL4 forms a complex with HOXA9 and coregulates common downstream target genes in various cellular biological processes, such as myeloid differentiation, proliferation, and cell survival. Given that SALL4 interacts with both wild-type MLL and a MLL fusion protein, further studies will determine whether modulation of the SALL4/MLL/HOXA9 pathway or interruption of the connection between SALL4 and MLL represent novel approaches in treating AML, including leukemia with MLL rearrangements. As SALL4, MLL, and HOXA9 have all been implicated in the development of leukemia, our findings provide new insights into the pathogenesis of AML.
Figure 9. Targeting the SALL4/MLL/HOXA9 pathway in AML.
SALL4 can upregulate HOXA9 expression by interacting with MLL, which results in H3K4 and H3K79 methylation. The SALL4/MLL/HOXA9 pathway is critical for leukemic cell survival, as disruption of this pathway results in leukemic cell death. Based on this model, we propose the following novel therapeutic approaches in targeting AML: (i) inhibiting the function of MLL and its complex (MLL inhibitor); (ii) disrupting the SALL4/MLL interaction; and (iii) targeting HOXA9 (HOXA9 inhibitor).
Methods
Further information can be found in Supplemental Methods.
Mice and leukemic transplantation experiments.
SALL4B transgenic mice were generated as previously described as well as murine leukemic transplants (4). Whole BM cells, spleen cells, and sorted LSKs, CMPs, GMPs, and MEPs from SALL4B-induced leukemic mice were transplanted into NOD-SCID mice by tail vein injection. The transplantation cell dose range was 1 × 106 for whole BM and spleen; 3.9 × 103 (primary) or 1,334 (secondary) for LSKs; and 8 × 104 (primary) or 2,300 (secondary) for GMPs. 8 × 103 CMPs or 80 × 103 MEPs were used for primary leukemic transplants; there were no leukemic CMPs or MEPs for secondary transplant, since only leukemic GMPs remained. For HoxA9 knockdown experiments, BM cells were harvested from serial transplant leukemic mice, and blast cells were isolated with Ficoll-Paque separation. 1,000 cells were transplanted into NOD-SCID recipient mice by tail vein injection.
Fluorescence-activated cell sorting (FACS) analysis and cell depletion.
HSCs were isolated from mouse BM cells as described previously (25). Briefly, murine lineage-positive cells were depleted with antibodies against CD3, CD4, CD8, Gr-1, CD19, and B220/CD45RA (Invitrogen) using magnetic beads (Dynal Biotech ASA). After depletion, cells were stained for the following markers: Sca-1, CD34, Fcγ receptors II and II (FcgRII/III), and c-kit (BD Biosciences — Pharmingen). Finally, cells were resuspended in 1× PBS with 2% heat-inactivated FBS and 1 μg/ml PI, then analyzed or sorted using a MoFlo cell sorter (Dako). The cell surface marker Sca-1, obtained as a biotin conjugate, was visualized using a streptavidin–APC-Cy7 antibody (BD Biosciences — Pharmingen).
PCR and qRT-PCR.
PCR and qRT-PCR were performed as previously described (4). Briefly, a PCR kit (Qiagen) was used to genotype the SALL4B founder mice and transmission of the transgene. Genomic DNA was purified from mouse tail using a high-quality DNA kit (Gentra Systems). SALL4B primer sequences were as follows: forward, 5′-AGCAGAGCTCGTTTAGTGAACCG-3′; reverse, 5′-CTGTCATTCATGATGAGGACAGG-3′. Total RNA was isolated using a phenol-free and filter-based RNA isolation system (Qiagen) digested with DNase I to remove DNA contamination. Primer sequences for qRT-PCR, designed using Primer Express software (Applied Biosystems), were as follows: GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; GAPDH reverse, 5′-GAAGATGGTGATGGGATTTC-3′; SALL4B forward, 5′-ACATCTCCGCGGTGGATGT-3′; SALL4B reverse, 5′-TGCTCCGACACTTGTGCTTG-3′; Gapdh forward, 5′-ACTCCACTCACGGCAAATTC-3′; Gapdh reverse, 5′-TCTCCATGGTGGTGAAGACA-3′; Hoxa9 forward, 5′-ATGGCATTAAACCTGAACCG-3′; Hoxa9 reverse, 5′-GTCTCCGCCGCTCTCATTC-3′. All reactions were performed in an ABI-7000 sequence detection system using TaqMan PCR core reagents according to the manufacturer’s instructions (Applied Biosystems). For each sample, GAPDH expression was used to normalize the amount of investigated transcript.
Gene expression profiling and data analysis.
Total RNA of various populations (2–5 × 104 cells/population) from BM of wild-type mice, NOD-SCID mice, and NOD-SCID mice with leukemic cell transplants was extracted using TRIzol reagent (Invitrogen). RNAs (50 ng) were reverse transcribed to cDNAs, then amplified by single primer isothermal amplification (SPIA). Finally, amplified products (3.75 μg) were fragmented and biotin labeled by Ovation RNA Amplification System V2 and FL-OvationTM cDNA Biotin Module V2 kits (NuGEN Technologies), according to the manufacturer’s instructions. Labeled cDNA was hybridized to an Affymetrix Mouse Genome 430A 2.0 Array and detected by the Microarray Core Facility at the Dana-Farber Cancer Institute.
SALL4 and HOXA cluster gene expression correlation was analyzed using publicly available data sets, derived from cDNA microarray analysis of 385 AML specimens (accession no. GSE14468; ref. 26). Gene expression data were deposited in the European Bioinformatics Institute ArrayExpress database (accession no. E-MEXP-2072; http://www.ebi.ac.uk/arrayexpress). Data were analyzed by dChip ( http://biosun1.harvard.edu/complab/dchip/) and GSEA 2.0 algorithm ( http://www.broad.mit.edu/gsea/). With dChip, raw expression data were normalized to account for differences in chip intensities and then filtered using 0.4 < SD/SEM < 1,000; presence call percentage in the arrays used ≥20% and expression level of at least 20 in ≥20% samples. Hierarchical clustering was performed among the filtered genes. For comparisons of gene expression between the 2 groups, transcripts were considered to be up- or downregulated when their transcript levels in SALL4B transgenic mice changed 1.5-fold (increase or decrease) compared with control samples, with P < 0.05 and lower confidence bound > 90%. GSEA was carried out as described by Subramanian et al. (33).
ChIP followed by qPCR and ChIP–Re-ChIP.
We previously described this assay when it was used to identify SALL4 targets in ChIP experiments in leukemic NB4 and murine ES cells (13, 27, 43). Briefly, cells were crosslinked with 1% formaldehyde at room temperature for 10 minutes and washed once with ice-cold PBS containing protease inhibitor cocktail (Roche Diagnostics). Sonication conditions were optimized for Omni Sonic Ruptor 400 for 7 sonications. In addition to sonication, the DNA was also sheared via an enzyme shearing kit (Active Motif) according to the manufacturer’s instructions. These ChIP samples were incubated overnight with ChIP-qualified antibodies (anti-H3K4me3, catalog no. MC315; anti-H3K79me2, catalog no. NL59; anti–RNA POLII, catalog no. 17-620; all from Millipore; and our own SALL4 antibody) (13, 27, 43). Samples were subjected to reverse crosslinking with NaCl (final concentration, 200 mM) at 65°C followed by RNase A and proteinase K treatment and phenol-chloroform extraction. Nucleotide sequences were obtained from NimbleGen ChIP-chip design files for positive probes, and real-time PCR primers were designed for the promoter regions to include these positive probes in their product fragments (average size, 160 bp). qPCR was performed on the immunoprecipitated DNA fragments with the BioRad SYBR Green real-time PCR kit. To detect enrichment of SALL4 binding, expression of SALL4 pulldown DNA was compared with that of input (no IP) and IgG controls using Gene Expression Macro software (provided by BioRad) designed to analyze qPCR and qRT-PCR data.
For ChIP–Re-ChIP experiments, protein-DNA complexes after the first ChIP with anti–MLL-N antibodies (catalog no. A300-086A, Bethyl Laboratories) were washed with washing buffer and Tris-EDTA buffer as described above. Complexes were eluted by incubation for 30 minutes at 37°C in 50 μl elution buffer (1× TE containing 2% SDS, 15 mM DTT, and protease inhibitors). After centrifugation, the supernatant was diluted 30 times with dilution buffer supplemented with protease inhibitors. Samples were then subjected to the ChIP procedure with SALL4 antibody (27) and IgG control.
EMSA.
EMSA was carried out as previously described (12). Briefly, nuclear extracts from SALL4-transfected 293T and THP1 cells was prepared with nuclear extract kit (catalog no. 78833, Pierce) according to the manufacturer’s instructions. Synthetic complementary of oligonucleotides were labeled using the biotin 3′-end DNA labeling kit (catalog no. 89818, Pierce). Binding reactions were carried out for 20 minutes at room temperature in the presence of 50 ng/μl poly (dI-dC), 0.05% Nonidet P-40, 10 mm EDTA, and 2.5% glycerol in 1× binding buffer (LightShift chemiluminescent EMSA kit, catalog no. 20148, Pierce), using 20 fmol biotin-end-labeled target DNA and 2 μg nuclear extract. Unlabeled target DNA (4 pmol), 1 μl anti-SALL4 antibody, or normal rabbit IgG (Cell Signaling) was added per 20 μl of binding reaction, as indicated.
Construction of pMIG-SALL4 plasmid, production of retroviral particles, and transduction of KG1a cells.
The human SALL4B coding sequence was inserted into the pMIG retroviral plasmid. The amphotropic packaging cell line Phoenix was transfected using calcium phosphate/chloroquine. Retroviral supernatants were harvested and used for infection of KG1a cells. After addition of 1 ml viral particles (viral titer adjusted to 1 × 106 transducing units/ml) to 1 × 106 cells and supplementation with 8 μg/ml polybrene, spinoculation was performed at 1,044 g for 90 minutes at 37°C. 2 infection cycles were performed, after which cells were placed back in original complete growth medium. After 48 hours, GFP+ cells were sorted and used for ChIP-qPCR or RT-PCR analysis.
Co-IP.
Subconfluent 293T cells were cotransfected with pCXN2-FLAG–MLL-BP (1–1,393 aa) or pCXN2-FLAG–MLL-N (1–1,052 aa) and pCDNA3-SALL4 using Fugene 6 (Roche). Various SALL4 vectors were gifts from Y. Ma (Nevada Cancer Institute, Las Vegas, Nevada, USA). At 48 hours after transfection, cells were lysed in 50 mM Tris (pH 8.0), 150 mM NaCl, 2 mM MgCl2, 10% glycerol, 1% NP-40, 10 mM NaF, and protease inhibitor. Flag antibody–agarose (Sigma-Aldrich) or mouse IgG-protein bead as a control was added into lysate and incubated overnight at 4°C. Next, protein complexes were washed 5 times with 1 ml lysis buffer and resolved in 3%–8% SDS-PAGE, after which Western blots were performed with SALL4 antibody (27). For detecting MLL components, cells were transfected with the SALL4 construct, subjected to IP with anti-FLAG–agarose, and immunoblotted with the antibodies anti-RbBp5 (catalog no. A300-109A, Bethyl Laboratories), anti-Menin (catalog no. A300-105A, Bethyl Laboratories), anti-MLL1 (catalog no. A300-087A, Bethyl Laboratories), and anti-Flag (catalog no. F1804, Sigma-Aldrich). Endogenous MLL and SALL4 co-IP was performed in KG1, THP1, and primary AML cells using the regular co-IP protocol with MLL antibody (catalog no. A300-086A, Bethyl Laboratories). Clean-Blot Detection kit (catalog no. 21230, Thermo Scientific) was used to reduce the background after co-IP.
Lentiviral virus production and transduction.
The SALL4 shRNA construct (Puri-7412 and Puri-7410) and scrambled control vectors were verified by our previous studies (13, 44). IF2-HOXA9 and IF3-HOXA9 shRNA constructs were purchased from Invitrogen; sequences were as follows: SALL4 shRNA, 5′-GCCTTGAAACAAGCCAAGCTA-3′; 1F2-HOXA9 shRNA, 5′-CACGCTTGACACTCACACTTT-3′; 1F3-HOXA9 shRNA, 5′-GTGGTTCTCCTCCAGTTGATA-3′; scrambled shRNA, 5′-CCTAAGGTTAAGTCGCCCTCG-3′.
Lentiviral supernatants were obtained in 293T cells by cotransfection of the shRNA plasmids and packaging plasmids containing VSV-G and pHR8.9. For lentiviral infection of primary murine SALL4B or human AML patient samples, 1 × 105 cells were seeded in 12-well plates (200 μl/well) in the appropriate culture media. Polybrene (hexadimerthrine bromide, Sigma-Aldrich) was added at a final concentration of 8 μg/ml. After addition of 1 ml lentiviral particles (titer of each lentiviral shRNA construct adjusted to 1 × 106 transducing unit/ml, to achieve MOI of 10 transducing units/cell), spinoculation was performed at 669 g for 90 minutes at 37°C. Next, cells were brought back to 500 μl in volume using the appropriate fresh culture media and incubated at 37°C, 5% CO2, until being used for subsequent applications.
Primary AML samples.
For gene expression studies, 34 AML samples were collected from patients. In addition, 3 primary AML leukemic patient samples were obtained from Brigham and Women’s Hospital. Culture conditions were adapted from a previously published protocol (23) that supports 40%–50% viability at 72 hours of post-thaw culturing, based on our experience. Only live cells were used for subsequent experiments.
Serial replating assays.
1,000 leukemic cells were plated in methycellulose (StemCell Technologies) containing a full complement of cytokines (GM-CSF, IL-3, IL-6, stem cell factor, and erythropoietin) in the presence of 1 μg/ml puromycin. Cells were cultured for 10 days, collected, and counted, and 1,000 cells were then placed into methylcell medium for the next round of culture.
Analysis of cell death and apoptosis.
Virus-transduced human primary AML cells or murine leukemic cells were seeded on semisolid medium as described above with 0.5–1 μg/ml puromycin selection for 72 hours. Next, cells were cytocentrifuged onto microscope slides, and apoptotic cells were measured using a TUNEL apoptosis kit (catalog no. G3250, Promega). Additional cells were stained with Trypan blue (Invitrogen).
Statistics.
Results are expressed as mean ± SD from at least 3 independent experiments. Statistical significance between 2 groups was determined by unpaired 2-tailed Student’s t test (GraphPad Prism). The Kaplan-Meier method was used to estimate survival curves for murine leukemic transplant data, and log-rank test was used to evaluate statistical differences. A P value less than 0.05 was considered significant.
Study approval.
All animal work was conducted according to relevant national and international guidelines and in accordance with the recommendations of the Weatherall report under protocol 10-10-1832 at Children’s Hospital Boston animal facility. The 34 primary AML samples were collected from patients with informed consent under protocol 2007/00173 from the Domain Specific Review Board of National Healthcare Group of Singapore (NHG DSRB); use of these samples was also approved by NHG DSRB under protocol 2009/00495. The 3 primary AML leukemic patient samples obtained from Brigham and Women’s Hospital were approved under IRB protocol 2011-P-000096/1.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants PO1 DK080665 and CA66996 to D.G. Tenen and by NIH grants RO1 HL092437 and PO1 HL095489 to L. Chai. This research is supported by the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award (to D.G. Tenen). We thank Wee Joo Chng for assistance in obtaining AML samples and Nicole Tenen for assistance in preparation of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(10):4195–4207. doi:10.1172/JCI62891.
References
- 1.Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc Natl Acad Sci U S A. 2006;103(44):16319–16324. doi: 10.1073/pnas.0607884103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8(10):1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108(8):2726–2735. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Baradie R, et al. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet. 2002;71(5):1195–1199. doi: 10.1086/343821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borozdin W, et al. Novel mutations in the gene SALL4 provide further evidence for acro-renal-ocular and Okihiro syndromes being allelic entities, and extend the phenotypic spectrum. J Med Genet. 2004;41(8):e102. doi: 10.1136/jmg.2004.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlhase J, et al. Mutations at the SALL4 locus on chromosome 20 result in a range of clinically overlapping phenotypes, including Okihiro syndrome, Holt-Oram syndrome, acro-renal-ocular syndrome, and patients previously reported to represent thalidomide embryopathy. J Med Genet. 2003;40(7):473–478. doi: 10.1136/jmg.40.7.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohlhase J, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002;11(23):2979–2987. doi: 10.1093/hmg/11.23.2979. [DOI] [PubMed] [Google Scholar]
- 9.Gao C, et al. SALL4 is a key transcription regulator in normal human hematopoiesis. Transfusion. 2013;53(5):1037–1049. doi: 10.1111/j.1537-2995.2012.03888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguila JR, et al. SALL4 is a robust stimulator for the expansion of hematopoietic stem cells. Blood. 2011;118(3):576–585. doi: 10.1182/blood-2011-01-333641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, et al. Dissecting the role of SALL4, a newly identified stem cell factor, in chronic myelogenous leukemia. Leukemia. 2011;7(7):1211–1213. doi: 10.1038/leu.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong HW, et al. SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes. PLoS One. 2011;6(4):e18372. doi: 10.1371/journal.pone.0018372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Chai L, Gao C, et al. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112(3):805–813. doi: 10.1182/blood-2007-11-126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utsch B, Becker K, Brock D, Lentze MJ, Bidlingmaier F, Ludwig MA. A novel stable polyalanine [poly(A)] expansion in the HOXA13 gene associated with hand-foot-genital syndrome: proper function of poly(A)-harbouring transcription factors depends on a critical repeat length? Hum Genet. 2002;110(5):488–494. doi: 10.1007/s00439-002-0712-8. [DOI] [PubMed] [Google Scholar]
- 15.Nakano K, et al. Novel mutations of the HOXD13 gene in hand and foot malformations. Int Surg. 2007;92(5):287–295. [PubMed] [Google Scholar]
- 16.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13(5):687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 17.Izon DJ, Rozenfeld S, Fong ST, Komuves L, Largman C, Lawrence HJ. Loss of function of the homeobox gene Hoxa-9 perturbs early T-cell development and induces apoptosis in primitive thymocytes. Blood. 1998;92(2):383–393. [PubMed] [Google Scholar]
- 18.Lawrence HJ, et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89(6):1922–1930. [PubMed] [Google Scholar]
- 19.Lawrence HJ, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106(12):3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorrance AM, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116(10):2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schichman SA, et al. ALL-1 partial duplication in acute leukemia. Proc Natl Acad Sci U S A. 1994;91(13):6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caligiuri MA, et al. Molecular rearrangement of the ALL-1 gene in acute myeloid leukemia without cytogenetic evidence of 11q23 chromosomal translocations. Cancer Res. 1994;54(2):370–373. [PubMed] [Google Scholar]
- 23.Faber J, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawakami Y, et al. Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development. 2009;136(4):585–594. doi: 10.1242/dev.027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 26.Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, et al. Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(50):19756–19761. doi: 10.1073/pnas.0809321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102(41):14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeleznik-Le NJ, Harden AM, Rowley JD. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci U S A. 1994;91(22):10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford BD, Hess JL. MLL core components give the green light to histone methylation. ACS Chem Biol. 2006;1(8):495–498. doi: 10.1021/cb600367v. [DOI] [PubMed] [Google Scholar]
- 31.Guenther MG, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102(24):8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirstetter P, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong P, Iwasaki M, Somervaille TCP, So CWE, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21(21):2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang QF, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117(25):6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tim CP, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiel AT, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17(2):148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi A, et al. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood. 2012;120(2):4461–4469. doi: 10.1182/blood-2012-05-429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao C, et al. Targeting transcription factor SALL4 in acute myeloid leukemia by interrupting its interaction with an epigenetic complex. Blood. 2013;121(8):1413–1421. doi: 10.1182/blood-2012-04-424275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS One. 2010;5(5):e10766. doi: 10.1371/journal.pone.0010766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, et al. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci U S A. 2007;104(25):10494–10499. doi: 10.1073/pnas.0704001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.