Abstract
Myeloid-derived suppressor cells (MDSCs) comprise immature myeloid populations produced in diverse pathologies, including neoplasia. Because MDSCs can impair antitumor immunity, these cells have emerged as a significant barrier to cancer therapy. Although much research has focused on how MDSCs promote tumor progression, it remains unclear how MDSCs develop and why the MDSC response is heavily granulocytic. Given that MDSCs are a manifestation of aberrant myelopoiesis, we hypothesized that MDSCs arise from perturbations in the regulation of interferon regulatory factor–8 (IRF-8), an integral transcriptional component of myeloid differentiation and lineage commitment. Overall, we demonstrated that (a) Irf8-deficient mice generated myeloid populations highly homologous to tumor-induced MDSCs with respect to phenotype, function, and gene expression profiles; (b) IRF-8 overexpression in mice attenuated MDSC accumulation and enhanced immunotherapeutic efficacy; (c) the MDSC-inducing factors G-CSF and GM-CSF facilitated IRF-8 downregulation via STAT3- and STAT5-dependent pathways; and (d) IRF-8 levels in MDSCs of breast cancer patients declined with increasing MDSC frequency, implicating IRF-8 as a negative regulator in human MDSC biology. Together, our results reveal a previously unrecognized role for IRF-8 expression in MDSC subset development, which may provide new avenues to target MDSCs in neoplasia.
Introduction
A major barrier to effective cancer immunotherapy is immune suppression, and the accumulation of myeloid-derived suppressor cells (MDSCs) has recently been recognized as a major mechanism to promote immune suppression (1, 2). MDSCs comprise a mixture of myeloid cells reflecting various stages of differentiation, and in mouse models, these cells are typically distinguished from other inhibitory myeloid populations based on their unique coexpression of macrophage (CD11b) and granulocyte (Gr-1) markers (1).
Tumor-induced MDSCs are further dichotomized into monocytic and granulocytic subsets based on the differential expression of the Ly6G and Ly6C epitopes (3, 4). Intriguingly, granulocytic MDSCs outnumber monocytic MDSCs in numerous mouse tumor models (3, 5), although the basis for this subset dichotomy remains unclear. The phenotypes in humans are more complex and vary with tumor type. However, there is general agreement that a common lineage-negative MDSC subset observed among a range of human cancers bears the core phenotype CD33+HLA-DR– (6–11). Interestingly, this subset resembles promyelocytes, a granulocytic population reflecting an early stage of differentiation (6, 7).
Although many studies have been dedicated to the phenotypic characterization of MDSCs and unraveling mechanisms by which these cells mediate tumor progression, a large gap remains in our understanding of the mechanisms that initiate their development. It is known, however, that MDSC subsets emerge in response to tumor-derived factors (TDFs) and the signaling pathways these molecules engage. As a number of TDFs engage the STAT3 or STAT5 signaling pathway, STAT3 or STAT5 activation has been associated with various stages in MDSC biology (1, 12–19).
Thus, in this study, we sought to further expand our understanding of the transcriptional control of tumor-induced MDSC development. To this end, we tested the central hypothesis that tumor-mediated downregulation of interferon regulatory factor–8 (IRF-8) facilitates MDSC development and that the loss of IRF-8 is key to the disproportionate balance of the two major subsets. IRF-8, unlike other IRF family members, is intimately involved in diverse stages of myelopoiesis, namely the differentiation and lineage commitment of monocytes/macrophages versus granulocytes (20–25). IRF-8 deficiency in mice leads to myeloproliferative disorders, largely characterized by the robust expansion of granulocytic and, to a lesser extent, monocytic cell types, reflecting various stages of differentiation (20, 21). Based on the knowledge that myeloid differentiation is tightly controlled by IRF-8 expression levels (20–23, 25) and that MDSCs arise as a consequence of impaired myeloid differentiation (1, 2), we reasoned that tumor-mediated events facilitate IRF-8 downregulation, ultimately leading to the accumulation of MDSCs.
Additional support for this hypothesis comes from our previous study showing that IRF-8 expression is strongly diminished in tumor-induced MDSCs and that overexpressing IRF-8 in mice ameliorated some of the M2-like features of MDSCs (26). Notably, however, these earlier studies utilized an IRF-8–Tg model driven by a ubiquitous CMV promoter in which the outcome may have been influenced through either intrinsic or extrinsic mechanisms. Furthermore, it remained unclear whether the modulation of the MDSC response through IRF-8 had any significant bearing on tumor growth. Therefore, to directly examine the impact of IRF-8 expression on CD11b+Gr-1+ MDSC generation and tumor growth, we produced a new IRF-8–Tg mouse model based on tissue-specific CD11b expression. Conversely, we also used a loss-of-function genetic approach to determine whether IRF-8 deficiency produces CD11b+Gr-1+ myeloid cells that phenotypically, functionally, and molecularly resembled MDSCs of tumor-bearing mice.
In this study, we addressed the following unanswered questions: (a) Does the expression of IRF-8 provide a molecular basis for the disproportionate frequencies of the two major MDSC subsets? (b) Does the modulation of IRF-8 levels in MDSCs affect tumor growth, suggesting that IRF-8 enhancement has therapeutic implications? (c) What is the mechanism of IRF-8 regulation, with emphasis on the roles of STAT3 or STAT5, as a number of MDSC-inducing factors have been shown to act through these signaling pathways? (d) Does IRF-8 expression have clinical value in human MDSC biology? Together, our results revealed a previously unrecognized role for IRF-8 in MDSC subset development, which offers a novel explanation for the robust accumulation of the granulocytic MDSC population. Furthermore, we identified IRF-8 as a novel target of STAT3 or STAT5 signaling, thereby filling a fundamental gap in MDSC transcriptional biology. Moreover, we showed that IRF-8 expression in the MDSCs of breast cancer patients (but not of healthy donors) is inversely correlated with the frequency of these cells, implicating IRF-8 as an important component in human MDSC tumor biology.
Results
Irf8 expression is reduced in two murine MDSC subsets.
Recently, we demonstrated a significant reduction in IRF-8 expression in splenic-derived CD11b+Gr-1+ MDSCs of tumor-bearing mice compared with phenotypically matched populations of non-tumor-bearing control mice (26). We used two well-established orthotopic mammary tumor models, 4T1 and AT-3, for MDSC induction. Although both models were derived from mammary carcinomas, they were selected largely for their capacity to generate MDSC responses (27, 28). Here, we determined whether the loss of IRF-8 expression was MDSC subset specific. Monocytic (CD11b+Ly6Chi/+Ly6G–) or granulocytic (CD11b+Ly6CloLy6G+) populations were purified through flow cytometry of the splenocytes from tumor-bearing mice (of the two different models) or non-tumor-bearing control mice and then analyzed for IRF-8 expression (Figure 1, A and B, and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI68189DS1).
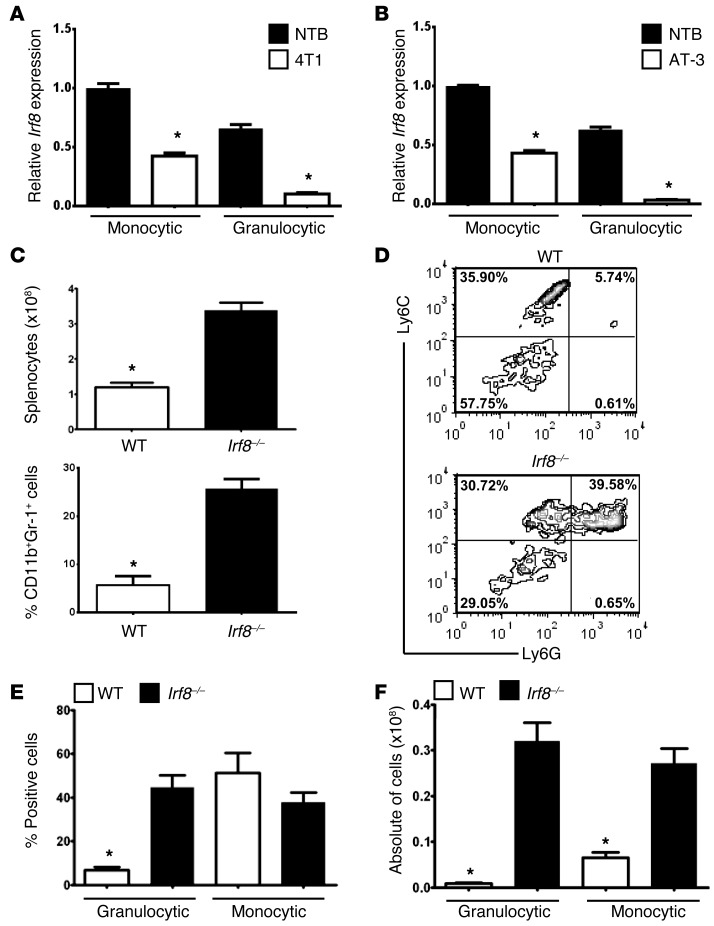
Figure 1. Irf8 expression is downregulated in tumor-induced MDSC subsets.
Irf8 mRNA levels in purified myeloid subsets from spleen of non-tumor-bearing or 4T1 (A) or AT-3 (B) tumor–bearing mice (≥1,000 mm3). The gating strategy for MDSC subset purification is shown in Supplemental Figure 1. Irf8 mRNA levels were measured through qPCR (n = 3 separate mice/group, *P < 0.01) and are representative of two separate experiments. For both panels, the data are expressed as the mean ± SEM of the fold changes relative to the first bar set at 1. (C) Upper panel: quantification of total splenocytes from Irf8–/– or WT littermate controls (8–10 weeks of age) (n = 5 each, *P < 0.01). Lower panel: CD11b and Gr-1 coexpression on splenocytes from Irf8–/– mice versus age-matched WT controls (n = 3 mice each, *P < 0.002). (D) Representative contour plot of Ly6C and Ly6G expression in the gated CD11b+ cells from WT and Irf8–/– mice in C. (E) Quantification of the percentages of granulocytic and monocytic subsets (*P < 0.003). (F) Total numbers of monocytic and granulocytic cells (*P < 0.001).
First, we confirmed that under non-tumor-bearing conditions, granulocytic cells expressed lower levels of Irf8 compared with monocytic cells (29). Second, we observed that monocytic and granulocytic subsets from 4T1 tumor–bearing mice displayed significantly lower levels of Irf8 compared with phenotypically matched cells from control mice. Interestingly, the loss of Irf8 expression was greater in the granulocytic fraction compared with the monocytic fraction (Figure 1A). Similar results were observed using a second mammary tumor model (AT-3) in a different MHC haplotype (H-2b) (Figure 1B). Collectively, these data demonstrate that Irf8 levels are depressed in both murine MDSC subsets under tumor-bearing conditions.
IRF-8 deficiency results in the accumulation of MDSC-like populations.
To determine a causal link between IRF-8 expression and MDSC development, we first employed a loss-of-function approach using an Irf8-null mouse model (Irf8–/–). Irf8–/– mice develop a hematopoietic disorder characterized by a robust accumulation of myeloid populations, particularly those of the granulocytic lineage (20). An analysis of H&E-stained whole spleen sections from Irf8–/– mice (~6 months of age) revealed increased extramedullary hematopoiesis (EMH) compared with matched WT mice (Supplemental Figure 2A).
We hypothesized that if MDSCs are produced as a result of tumor-induced IRF-8 downregulation, then IRF-8 loss caused by other means should elicit myeloid populations that resemble MDSCs. As expected, Irf8–/– mice displayed significant splenomegaly compared with matched WT controls (Figure 1C), primarily reflecting the expansion of CD11b+Gr-1+ myeloid cells (Figure 1C). Further characterization of the CD11b+Gr-1+ myeloid response in Irf8–/– mice revealed an approximately 7-fold expansion of the granulocytic subset compared with the WT controls. In contrast, the percentages of cells within the monocytic fraction were similar between the two genotypes (Figure 1, D and E). Although converting these data to absolute counts showed significant increases in the total numbers of each myeloid subset compared with the WT controls, the rise in the granulocytic subset (~30-fold) still far exceeded the rise in the monocytic subset (~4-fold) (Figure 1F).
Next, we adopted several functional and molecular assays to determine whether CD11b+Gr-1+ cells from Irf8–/– mice displayed MDSC characteristics. Purified CD11b+Gr-1+ cells from Irf8–/– mice were examined for their ability to inhibit polyclonal (anti-CD3 mAb)– or allogeneic-induced T cell proliferation (Figure 2). The data indicated that CD11b+Gr-1+ cells from Irf8–/– mice strongly inhibited T cell proliferation, compared with T cells mixed with CD11b+Gr-1+ cells from WT mice or T cells stimulated with anti-CD3 mAb in the absence of CD11b+Gr-1+ cells (average cpm, 127,151) (Figure 2A). These data are also consistent with the finding that myeloid cells from control mice, comprising the bulk population or individual subsets, are not suppressive (1). Because neither subset is suppressive, we concluded that the use of the bulk population in this particular context was appropriate. Similarly, CD11b+Gr-1+ cells from Irf8–/– mice also suppressed alloreactive T cell proliferation relative to the controls incubated with CD11b+Gr-1+ cells from WT mice (Figure 2B). Thus, the pattern/magnitude of suppression observed with CD11b+Gr-1+ cells from Irf8–/– mice (Figure 2, A and B) versus those from tumor-bearing mice (see additional data below) was similar.
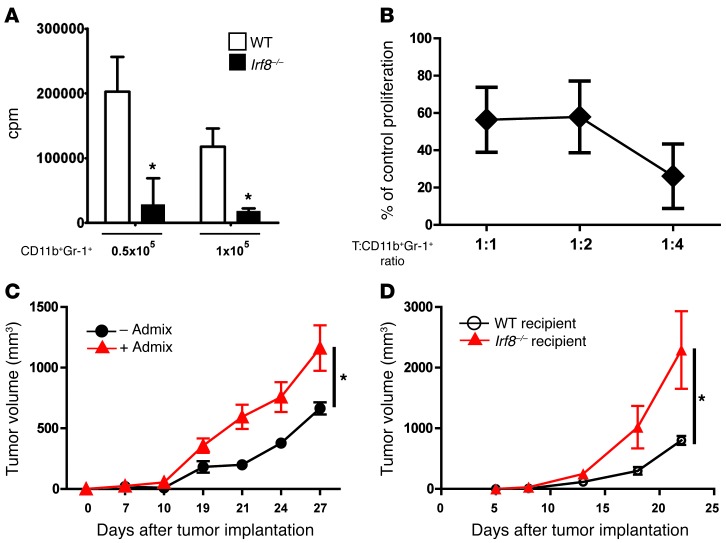
Figure 2. Irf8-deficient mice harbor MDSC-like cells.
(A) Ability of CD11b+Gr-1+ cells isolated from WT or Irf8–/– mice to inhibit anti-CD3–stimulated T cells at the indicated cell densities (n = 3 determinations, *P < 0.02; T cells plus anti-CD3 mAb control, average cpm, 127,151). (B) Ability of purified CD11b+Gr-1+ cells from Irf8–/– mice to suppress alloreactive (H-2b anti–H-2d) T cell proliferation relative to the control, incubated with CD11b+Gr-1+ cells from WT mice. The CD11b+Gr-1+ cells matched the haplotype of the responder T cells and were tested at the indicated lymphocyte to myeloid cell ratios. Proliferation in A and B was measured through 3H-thymidine uptake and is representative of 2 separate experiments. (C) AT-3 tumor cells were co-mixed with or without CD11b+Gr-1+ cells derived from Irf8–/– mice, and tumor growth was recorded. The results represent the mean ± SEM (n = 8 recipient mice/group pooled from 2 separate experiments; *P = 0.002). (D) AT-3 tumor growth rate in WT versus Irf8–/– mice (n = 5 recipient mice/group; P = 0.02).
CD11b+Gr-1+ cells from Irf8–/– mice were examined for their ability to enhance tumor growth using a myeloid-tumor adoptive transfer approach. This proof-of-concept strategy provides a direct method to determine the impact of the myeloid population on tumor growth within the tumor microenvironment (13, 26, 28). Here, CD11b+Gr-1+ cells from Irf8–/– mice were mixed with AT-3 tumor cells (reflecting the haplotype of the knockout mouse), followed by the orthotopic implantation of the co-mixture into syngeneic B6 recipients. We observed that CD11b+Gr-1+ cells from Irf8–/– mice significantly increased tumor growth (Figure 2C) in a manner similar to that observed with cells from tumor-bearing mice (ref. 26 and see additional data below), further supporting the hypothesis for a causal link between IRF-8 expression and MDSC biology. Notably, tumor growth was faster in Irf8–/– mice compared with WT mice (Figure 2D), consistent with the notion that IRF-8 deficiency supports a pro-tumor environment.
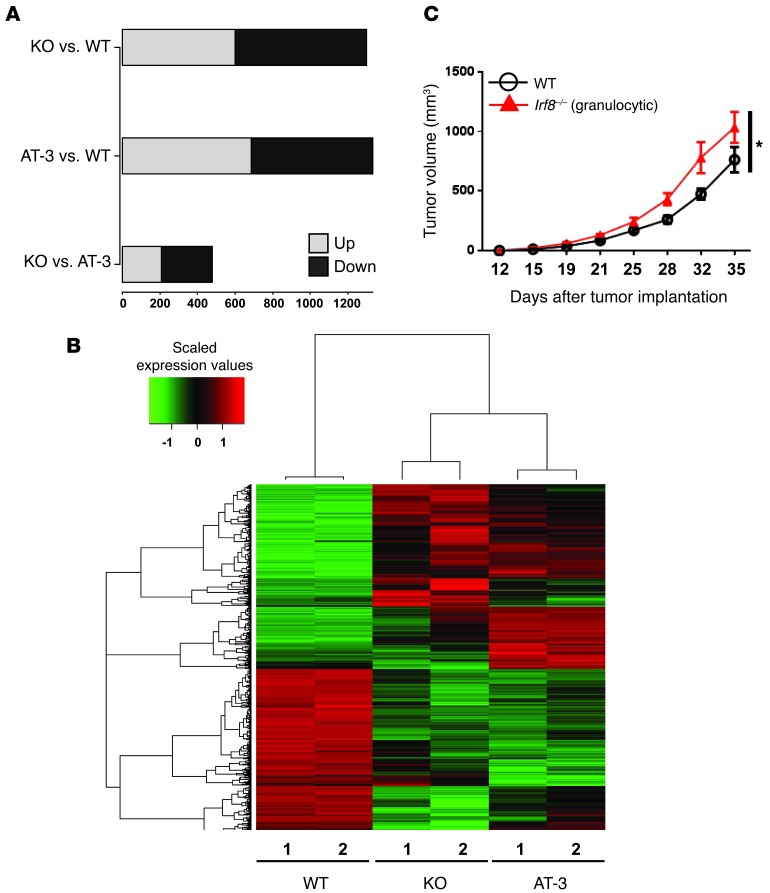
Global gene expression studies were performed to determine whether the CD11b+Gr-1+ cells from Irf8–/– mice resembled those from tumor-bearing mice more than those from the non-tumor-bearing control mice (i.e., WT) at a molecular level. To this end, we focused on the granulocytic population, as these cells represented the predominant subset. The granulocytic phenotype is defined as CD11b+Ly6G+Ly6Clo or CD11b+Gr-1hi. Here, splenic CD11b+Gr-1hi cell populations from WT, AT-3 tumor–bearing, or Irf8–/– mice were purified in two independent experiments through flow cytometry (>97% purity; Supplemental Figure 2B) and subjected to whole genome expression profiling. The post-sort contour plots showed a high degree of purity and phenotypic similarity between the CD11b+Gr-1hi cells recovered from WT and Irf8–/– mice. Differential expression analysis of the microarray data revealed that the gene expression patterns of cells from Irf8–/– mice resembled those of tumor-bearing hosts more than those of the WT group (Figure 3). Specifically, we identified 1,332 genes showing significant expression changes (i.e., >2-fold change, P < 0.01) in tumor-bearing versus WT mice. Similarly, we identified 1,229 significantly different genes between and Irf8–/– and WT mice. In contrast, only 479 genes were differentially expressed between tumor-bearing and Irf8–/– mice using the same significance criteria (Figure 3A).
Figure 3. The gene expression profile of CD11b+Gr-1hi cells from Irf8–/– mice resembles that of tumor-bearing mice.
(A) Mouse whole-genome microarray analysis using purified splenic CD11b+Gr-1hi cells from WT, Irf8–/–, and AT-3 tumor–bearing mice. Three sets of comparisons were made to obtain differentially expressed genes (>2-fold change; P < 0.01): Irf8–/– (KO) versus WT; AT-3 versus WT; and Irf8–/– versus AT-3. (B) Hierarchical clustering of genes down- (green) or upregulated (red) from each sample (>2-fold change; P < 0.01). Each group was set up in biological duplicates. (C) As in Figure 2C, except that AT-3 tumor cells were co-mixed with the purified granulocytic fraction from Irf8–/– mice. Purified CD11b+Gr-1+ cells from WT mice were included as a control. The results represent the mean ± SEM (n = 8 WT; n = 7 Irf8–/–; *P = 0.005).
Unbiased pathway gene set enrichment analysis (GSEA) revealed a similar pattern. Among the 268 curated pathways analyzed, we identified 57 pathways different between WT and tumor-bearing mice, 42 pathways different between WT and Irf8–/– mice and 18 pathways different between tumor bearing and Irf8–/– mice (Supplemental Figure 2C). The identities of these pathways, in order of statistical significance, are illustrated in Supplemental Tables 1–3. Hierarchical clustering based on the identified differentially expressed genes showed that Irf8–/– and tumor-bearing mice were clustered into a group separate from that of the WT samples (Figure 3B). Taken together, the results of the microarray gene expression profiling analysis showed that CD11b+Gr-1hi cells from Irf8–/– mice resembled those of tumor-bearing mice considerably more than those from the non-tumor-bearing control mice. Myeloid-tumor adoptive transfer experiments confirmed that this purified granulocytic fraction possessed pro-tumor activity, as these cells enhanced the tumor growth rate relative to AT-3 cells co-transferred with myeloid cells from WT mice (Figure 3C).
IRF-8 overexpression reduces MDSC accumulation.
We used a gain-of-function approach to further explore the causal relationship between IRF-8 expression and MDSC development. To this end, we generated an IRF-8–Tg mouse driven by the murine CD11b promoter. We first compared basal Irf8 mRNA levels in purified CD11b+ myeloid cells among several founder mice against those of an Irf8-Tg mouse model driven by the CMV promoter (CMV-IRF8 mice) (26). Two founder lines (lines 370, 371) exhibited substantially higher Irf8 levels compared with several other founder lines, and they were comparable to those of CMV-IRF8 mice (Supplemental Figure 3A). Further characterization of these two founder lines revealed that the Irf8 transgene did not alter steady-state hematological parameters (Supplemental Figure 3, B–D, and Supplemental Table 4).
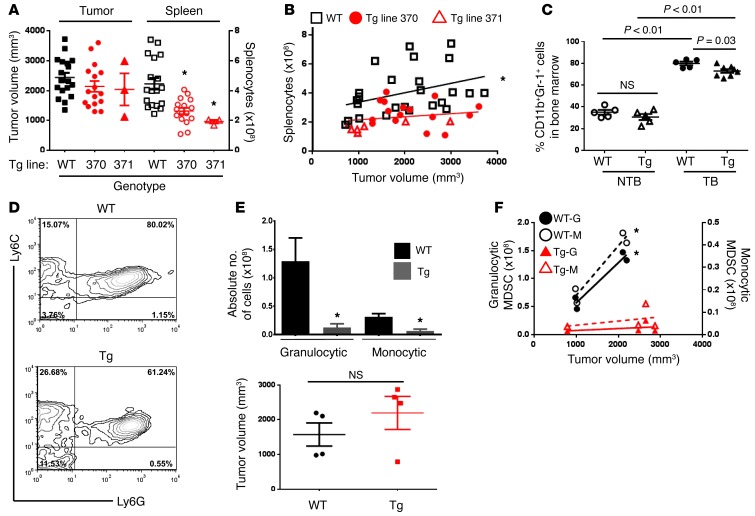
To determine whether IRF-8 modulation influenced MDSC development, we first quantified the frequencies of MDSCs in the spleens of Tg and WT cohorts harboring similar AT-3 tumor loads (Figure 4, A and B). All Tg progeny were produced in a founder line–specific manner, and the data represent the results from separate founder lines (i.e., either 370 or 371 distinguished with different red symbols). We observed that Irf8 overexpression significantly attenuated splenomegaly in either founder line (Figure 4A and Supplemental Figure 4A). We then evaluated changes in splenocyte number as a function of tumor load (Figure 4B). In WT mice, increasing tumor volume was accompanied by a significant increase in the number of splenocytes. In contrast, the correlation between tumor size and splenocyte number observed in WT mice was greatly reduced in Irf8-Tg mice (in either founder line), indicating that IRF-8 expression modulated hematopoiesis under tumor-bearing conditions.
Figure 4. MDSC accumulation is diminished in Irf8-Tg mice.
(A) Splenocyte counts (on the right y axis) from AT-3 tumor–bearing WT mice or Irf8-Tg mice of the indicated founder line. The data are shown for mice with larger tumor volumes (>1,200 mm3) to illustrate the impact of Irf8 under the most advanced tumor conditions. *P < 0.003. (B) Relationship between tumor size and splenocyte number for all mice, including those with smaller tumor volumes (<1,200 mm3) to generate a broader curve (Spearman r value = 0.59; *P = 0.008 for WT; not significant for either Tg line). Each symbol represents a single mouse. (C) Quantification of CD11b+Gr-1+ cells from bone marrow of WT or Irf8-Tg mice with (TB) or without (NTB) tumor growth (>1,200 mm3). (D) Representative contour plots of Ly6C and Ly6G expression on the CD11b-gated splenic fraction. (E) Upper panel: quantification of splenic granulocytic and monocytic subsets using a gating strategy similar to that described in Supplemental Figure 1 (n = 4 mice each; in the case the Tg group, 3 from line 370 and 1 from line 371). Lower panel: tumor sizes in individual mice. (F) Granulocytic (left y axis) and monocytic (right y axis) MDSC subsets plotted in relation to their respective tumor sizes from E. *P < 0.04 for WT subsets; not significant for Tg subsets. The data are reported as the mean ± SEM for the number of mice or experiments shown.
Further analysis revealed significant reductions in the percentage of CD11b+Gr-1+ cells from Irf8-Tg mice compared with the WT controls, as measured in the spleen (Supplemental Figure 4, B and C), bone marrow (albeit to a lesser extent) (Figure 4C), and tumor site (see Figure 5). In tumor-draining lymph nodes, we observed that the frequency of CD11b+Gr-1+ cells was low (<2%), and there was no observed difference from non-tumor-bearing control mice (data not shown). Although it remains unclear why MDSCs did not significantly accumulate in the draining lymph nodes, our data are consistent with previous studies (30, 31). Detailed MDSC subset analysis performed in the spleen showed a significant decrease in the percentage of granulocytic cells and a slight increase in monocytic cells in Irf8-Tg mice compared with tumor sized–matched WT controls (Figure 4, D and E, and Supplemental Figure 4D). Although converting these data to absolute numbers revealed significant decreases in the total numbers of each subset compared with the WT controls, the largest decline was observed with the granulocytic subset (~10-fold) compared with the monocytic subset (~4.5-fold) (Figure 4E). Furthermore, as observed with total splenocytes (Figure 4B), the relationship between tumor size and MDSC subset frequency was significantly reduced in Irf8-Tg mice compared with WT mice (Figure 4F; red versus black data points), indicating that IRF-8 indeed modulated MDSC subset accumulation under tumor-bearing conditions.
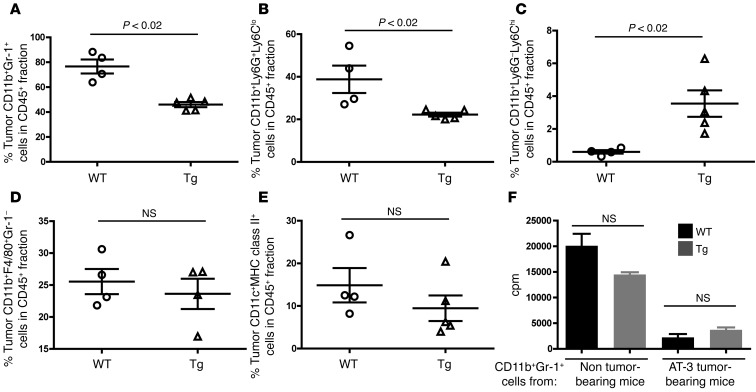
Figure 5. Intratumoral MDSC accumulation is also diminished in Irf8-Tg mice.
Flow analysis of the indicated myeloid populations from primary tumor tissue of WT or Irf8-Tg mice at endpoint, as shown in Figure 4, A or C. Total live cells were first gated on the CD45+ leukocyte fraction. The gated CD45+ fraction was then plotted in relation to the myeloid markers shown in a manner similar to that in Supplemental Figure 1. The data are illustrated for total MDSCs (A), MDSC subsets (B and C), macrophages (D), and total DCs (E). (F) Ability of CD11b+Gr-1+ cells from AT-3 tumor–bearing WT or AT-3 tumor–bearing Irf8-Tg mice to suppress T cell proliferation in response to immobilized anti-CD3 mAb. T cells (1 × 105/well) and CD11b+Gr-1+ cells (5 × 104/well) (n = 3 determinations; *P < 0.01; T cells + anti-CD3 mAb without any CD11b+Gr-1+ cells, 23,212 ± 1,865).
In primary tumor tissue, CD11b+Gr-1+ cells represented a substantial portion of the overall CD45+ leukocyte population. Importantly, the extent of CD11b+Gr-1+ cell accumulation was significantly reduced in Irf8-Tg mice (Figure 5A; gating strategy shown in Supplemental Figure 5), similar to what we observed systemically in the spleen and, to a lesser extent, the bone marrow (Figure 4C). And as with the spleen, the decrease in MDSC load of tumor-bearing Irf8-Tg mice was observed in the granulocytic subset (Figure 5B). Interestingly, a small yet significant increase in the percentage of the monocytic subset was observed (Figure 5C), similar to the pattern in the spleen (Figure 4D). Because the tumor volumes at the time of these analyses were comparable, the increase in the percentage of the tumor-infiltrating monocytic subset is consistent with a concomitant increase in the relative cell number. We also evaluated the impact of transgenic Irf8 expression on the frequencies of two other major tumor-infiltrating myeloid populations, macrophages and DCs. In contrast to the MDSC analysis, no significant differences were noted in the percentages of intratumoral macrophages (CD11b+F4/80+Gr-1–) or total DCs [(CD11c+MHC class II+ (I-Ab)] (Figure 5, D and E). Further analyses of both MDSC subsets revealed a lack of MHC class II expression (i.e., average I-Ab MFI values for each MDSC subset were less than 150, which was similar to the unstained samples). Together, these data suggest that under these tumor-bearing conditions, IRF-8 expression selectively altered the frequency of MDSCs compared with several other major myeloid subsets.
Interestingly, the tumor-induced CD11b+Gr-1+ cells from Irf8-Tg mice still blocked T cell proliferation (Figure 5F), despite their ability to maintain high Irf8 levels (data not shown, but see Supplemental Figure 4E for expression patterns in single- versus double-Tg MTAG mice). Nonetheless, these data demonstrated that IRF-8 expression can significantly mitigate MDSC load systemically and in the tumor microenvironment, but not necessarily the immunosuppressive behavior of the residual population.
Combinatorial strategies enhance antitumor activity.
Thus far, our data demonstrate an inverse causal relationship between IRF-8 expression and the accumulation of MDSCs. However, no significant impact was observed in tumor growth rate (Figure 6A). These data suggested the following two hypotheses: (a) the remaining MDSCs were still sufficient to impede host immunosurveillance; and/or (b) alterations in MDSCs alone were insufficient to enable direct antitumor effects but might predispose the host to increased susceptibility to immunotherapy. To test and distinguish between these two possibilities, we took advantage of two distinct biological strategies: cell depletion and immune stimulation.
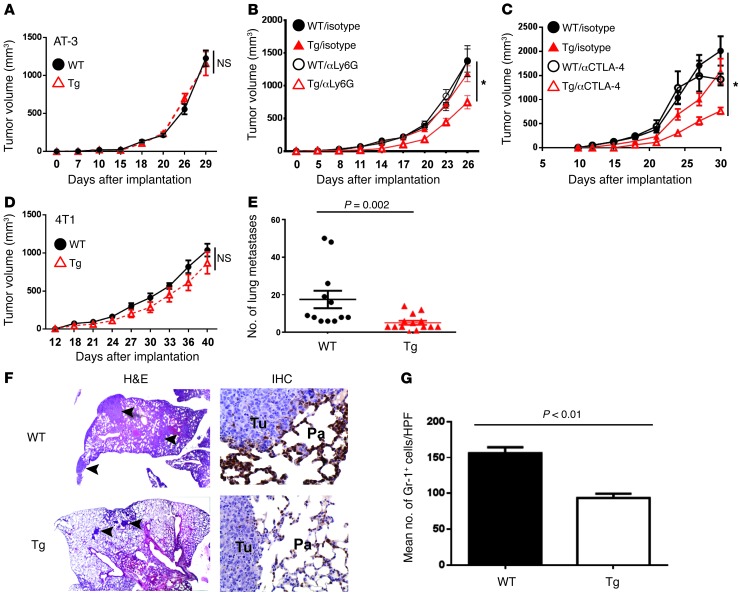
Figure 6. Influence of Irf8 enhancement on tumor growth during immunosurveillance or immunotherapy.
(A) Tumor growth rate of AT-3 cells in WT versus Irf8-Tg mice (n = 9 for WT and n = 10 for Tg line 370). (B) WT or Tg mice (line 370) were treated with either anti-Ly6G mAb or an isotype control after the tumors became palpable. The data are representative of 2 experiments (n = 10 mice/group). *P = 0.009 for differences in tumor growth rate between anti-Ly6G–treated Irf8-Tg mice versus all other cohorts. (C) As in B, except that the mice were treated with either anti–CTLA-4 mAb or an isotype control. The data are representative of 2 separate experiments (n = 5 mice/group). *P < 0.04 for differences in tumor sizes between anti–CTLA-4–treated and vehicle-treated Irf8-Tg mice at days 18, 21, 27, and 30; P < 0.05 at day 15. (D) Tumor growth rate of 4T1 cells in WT versus Irf8-Tg mice (Tg line 370). (E) Spontaneous lung metastasis was quantified at endpoint tumor volumes for mice in D (n = 12 WT; n = 14 Tg). (E) H&E-stained lung tissues were used to quantify metastasis. (F) Representative H&E-stained lung images (left; original magnification, ×20) and representative images of anti–Gr-1 staining (1 of 3 mice tested), analyzed through IHC (right; original magnification, ×400). The arrows indicate examples of discrete foci; Tu, tumor; Pa, parenchyma. H&E analysis comfirmed myeloid morphology. (G) Quantification of IHC data in F, based on the average number of stained cells per high-power field (×400) from 5 random sections of each slide. The data represent the mean ± SEM of 3 mice per group.
With regard to cell depletion, we employed a mAb-based approach to deplete Ly6G+ MDSCs after AT-3 tumor implantation (32), as this subset constituted a larger proportion of the remaining MDSCs (Figure 4, D and E, and Supplemental Figure 4D). We postulated that anti-Ly6G mAb in concert with IRF-8 enhancement would have a greater antitumor effect in Irf8-Tg mice compared with WT mice, as fewer MDSCs would have to be depleted to relieve suppression. Our results indicated that treatment of Irf8-Tg mice with anti-Ly6G mAb significantly reduced the tumor growth rate compared with treatment with the isotype control (Figure 6B). Under these experimental conditions, we observed no appreciable antitumor effect in WT mice treated with the anti-Ly6G mAb. With regard to the second approach (i.e., immune stimulation), we tested anti–CTLA-4 mAb as an immunotherapy agent, in part because of the well-documented ability of this antibody to boost antitumor responses under certain preclinical and clinical settings (33). Here, we observed that CTLA-4 blockade significantly inhibited tumor growth at multiple time points in Irf8-Tg mice, but not in the WT controls.
As in the AT-3 model, the growth rate of the 4T1 primary tumor was unaltered in Irf8-Tg mice (Figure 6, A versus D). However, in contrast to AT-3, 4T1 tumors metastasize to the lungs, even when the primary tumor remains intact. Therefore, we examined whether transgenic Irf8 expression influenced the extent of lung metastasis at the experimental endpoint tumor measurement. The lungs were removed, and the degree of metastatic burden was histologically quantified in a blinded fashion. Our findings demonstrated that the extent of metastatic disease was significantly lower in Irf8-Tg mice compared with the WT controls, as determined after counting total lung nodules (Figure 6, E and F) or estimating the percentage of lung tissue occupied by cancer (WT: 8.4% ± 3%; Tg: 2.9% ± 0.5%; P = 0.016). Moreover, the reduction in lung metastasis was accompanied by a significant reduction in the presence of Gr-1–expressing myeloid populations, as determined through immunohistochemistry (IHC) (Figure 6, F and G). Thus, our data suggest that enhancement of IRF-8 is bioactive and effective against tumor growth under certain circumstances of immune surveillance (i.e., metastasis) or immunotherapy (i.e., combination strategies that further reduce MDSCs or activate host immune responses).
IRF-8 enhancement reduces autochthonous tumor growth rate.
Next, we examined the impact of IRF-8 enhancement in a mouse tumor model of autochthonous tumor development and progression, termed MTAG. MTAG mice develop multifocal mammary tumors as a result of tissue-specific transgene expression of the polyomavirus middle T antigen (34). Moreover, the vast majority of MTAG mice eventually develop lung metastasis (26, 34). We previously showed that MTAG mice develop MDSCs and that MDSC burden is proportional to tumor load (26). To determine the impact of IRF-8 expression on MDSC accumulation and tumor growth, we bred MTAG mice with CD11b-IRF8 mice to generate single-Tg mice (MTAG) and double-Tg mice (MTAG expressing the Irf8 transgene). In these studies, we randomly chose founder line 370 for breeding. Interestingly, the tumor growth rate under autochthonous conditions was significantly reduced in double-Tg compared with single-Tg mice. This pattern was observed after recording the growth of the largest single tumor or monitoring its “time to progression” to approximately 1,000 mm3, a surrogate endpoint reflecting approximately 50% of the maximal limit (Figure 7, A and B). We observed that double-Tg mice took approximately 2 weeks longer to achieve this burden compared with single-Tg mice, suggesting a delay in the onset of tumor growth. Similar patterns were observed after summing the growth of all measurable tumors at each time point (data not shown).
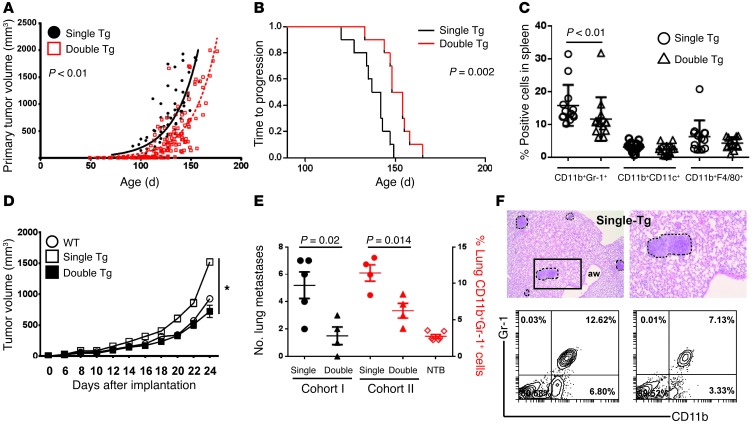
Figure 7. Irf8 enhancement slows autochthonous tumor growth.
(A) Tumor growth rate in double-Tg versus single-Tg mice, as determined after tracking the single largest tumor. Each symbol denotes a tumor measurement in a single mouse over time (n = 13 mice each; all Irf8-Tg mice from line 370). (B) Kaplan-Meier plot of the data in A, based on time to progression to approximately 50% of maximal tumor growth as a surrogate endpoint. (C) Each data point represents flow analysis of the indicated myeloid subset from single-Tg or double-Tg mice at endpoint. (D) Myeloid/tumor admixture experiments, as in Figure 2. AT-3 cells were mixed with splenic CD11b+Gr-1+ cells recovered from single-Tg, double-Tg, or non-tumor-bearing mice (WT), and tumor growth was recorded. The cells from single-Tg mice, but not those of double-Tg mice, showed a significant (*P < 0.001) increase in the AT-3 tumor growth rate relative to cells from WT mice (n = 5 mice/group; one of 2 separate experiments). The data are expressed as the mean ± SEM for the indicated number of mice. (E) Quantification of lung metastasis using histology (termed cohort 1) or CD11b+Gr-1+ cell frequency using flow cytometry analysis of total lung digests (termed cohort 2) from single-Tg or double-Tg mice. NTB, non-tumor-bearing WT mice. Both cohorts were matched based on age and primary tumor burden, as indicated in Results. (F) Representative H&E-stained images of metastatic foci in MTAG mice (upper left, original magnification, ×40; upper right, original magnification, ×100) and flow plots of CD11b+Gr-1+ cell populations (bottom panel).
At endpoint, we evaluated three additional parameters of IRF-8 bioactivity: (a) impact on the frequencies of the CD11b+Gr-1+ cells compared with two other CD11b+ myeloid subsets, myeloid DCs and macrophages, as in Figure 5; (b) impact on MDSC function; and (c) impact on metastasis. Indeed, we observed that double-Tg mice harbored a significantly lower frequency of CD11b+Gr-1+ cells compared with single-Tg mice (Figure 7C). In contrast, no significant differences in the percentages of myeloid DCs or macrophages were observed (Figure 7C and Supplemental Figure 4F), similar to what we observed in tumor-infiltrating myeloid populations of the AT-3 model (Figure 5). These data suggest that under these autochthonous tumor-bearing conditions, transgenic Irf8 expression selectively altered the frequency of MDSCs compared with two other major myeloid populations. These relationships are further explored in Figure 8.
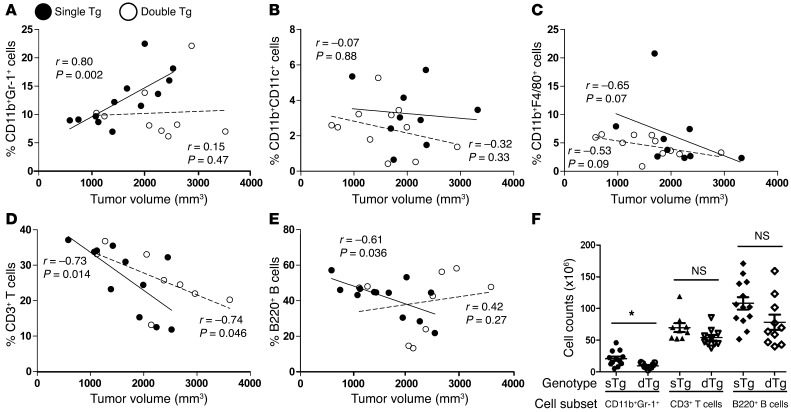
Figure 8. IRF-8 selectively modulates CD11b+Gr-1+ MDSC frequency under autochthonous tumor growth conditions.
(A–E) Spearman correlation r values and P values for each genotype, reflecting the indicated comparisons. Each symbol represents a single mouse based on a new series of experiments. The percentages signify the fraction of each subset relative to the total splenocyte population, whereas the tumor volume denotes the summation of all tumors from an individual single- or double-Tg mouse at endpoint. (F) The endpoint data points in A, D, and E were converted to absolute cell counts (*P < 0.02; NS, not significant).
With regard to MDSC function, we performed in vivo admix experiments instead of suppression assays, as our earlier data revealed that IRF-8 modulation did not affect the immune suppressive behavior of MDSCs. Interestingly, we observed that while CD11b+Gr-1+ cells from single-Tg mice enhanced tumor growth rate, the cells from double-Tg mice showed no effect (Figure 7D). Together, these results indicate that under autochthonous neoplastic conditions, myeloid IRF-8 levels can modulate primary tumor growth rate, accompanied by alterations in MDSC numbers and/or certain MDSC functional properties. These observations also strengthen the notion that autochthonous tumor growth operates under a host-tumor dynamic somewhat different from that under implantable conditions (“chronic” versus “acute”), likely impacting how the immune response is ultimately shaped. Notably, CD11b+Gr-1+ cells from double-Tg mice maintained high Irf8 levels compared with the non-tumor-bearing WT or Irf8-Tg controls (Supplemental Figure 4E). In contrast, Irf8 levels were significantly reduced in single-Tg mice compared with the WT controls.
Because MTAG mice can develop lung metastasis, we examined whether IRF-8 expression also influenced the extent of disease in the lungs at endpoint tumor measurements. Furthermore, we extended this analysis to characterize the frequency of lung-derived CD11b+Gr-1+ cells using flow cytometry analysis of total lung digests. However, as we wanted to use the entire lung tissue for each read-out, we established two groups of mice: one group for histological assessment of lung metastasis (i.e., cohort 1) and a second group (i.e., cohort 2) for CD11b+Gr-1+ cell quantification (Figure 7, E and F). Both groups were matched based on age (range 132–138 days) and primary tumor burden (range 1,580–2,300 mm3) at endpoint. Similar to what we observed in the 4T1 model, double-Tg mice displayed a significant decrease in the number of lung metastasis compared with single-Tg mice (cohort 1; left axis of Figure 7E). Moreover, the decline in metastasis was accompanied by a significant reduction in the frequency of CD11b+Gr-1+ cells within the lung tumor microenvironment (cohort 2; right axis of Figure 7E), approaching the levels observed in the non-tumor-bearing controls. Figure 7F shows representative images of metastatic foci in the MTAG model and contour plots of the CD11b+Gr-1+ cell populations.
To further test the hypothesis that IRF-8 selectively impacts MDSC accumulation, we evaluated changes in the frequencies of splenic CD11b+Gr-1+ cells, myeloid DCs, macrophages, T cells, and B cells as functions of tumor load (Figure 8). As the slope of the line shows, increasing tumor volume in single-Tg mice was accompanied by a significant increase in the percentage of CD11b+Gr-1+ cells (Figure 8A). In contrast, in double-Tg mice, the slope of the line remained virtually unchanged, despite increases in tumor load, indicating that IRF-8 was able to modulate the accumulation of CD11b+Gr-1+ cells under tumor-bearing conditions. Different patterns, however, were observed with myeloid DCs, macrophages, total CD3+ T cells (Figure 8, B–D), and NK cells (data not shown). Here, the slope of the lines for both genotypes paralleled each other and generally declined with increasing tumor burden. An exception was observed with B cells, although the increase in double-Tg mice was not significantly associated with increasing tumor load (Figure 8E). In addition, there were no significant differences in the percentages of these leukocyte subsets between non-tumor-bearing WT and Irf8-Tg mice (Supplemental Figure 3D). Thus, the impact of IRF-8 expression on tumor-induced myelopoiesis selectively targeted the CD11b+Gr-1+ population. The decline in other leukocyte subsets largely reflected the disproportionate increase in the frequency of CD11b+Gr-1+ cells, a finding common in the MDSC field (1, 2). Analysis of total cell counts confirmed significant differences between genotypes for CD11b+Gr-1+ cells but not, for example, T and B cells (Figure 8F).
Myelopoietic growth factors downregulate Irf8 expression.
After establishing that IRF-8 plays an integral role in MDSC development, we next investigated the potential mechanisms underlying IRF-8 regulation. Because MDSC development can be initiated by various STAT3- or STAT5-activating TDFs, namely G-CSF and GM-CSF (1, 12–18, 28), in vitro or in vivo, we hypothesized that these signaling events converge at the level of Irf8 transcription. To test this hypothesis, we first analyzed the effects of recombinant G-CSF treatment on Irf8 levels in vitro and in vivo. Exposure of bone marrow–derived CD11b+Gr-1+ cells to G-CSF in vitro significantly reduced Irf8 levels as early as 2 hours after treatment relative to the untreated (media) control (Figure 9A). Although Irf8 levels in the controls declined somewhat over time, the Irf8 levels in the G-CSF–treated samples remained significantly lower compared with the time point–matched controls.
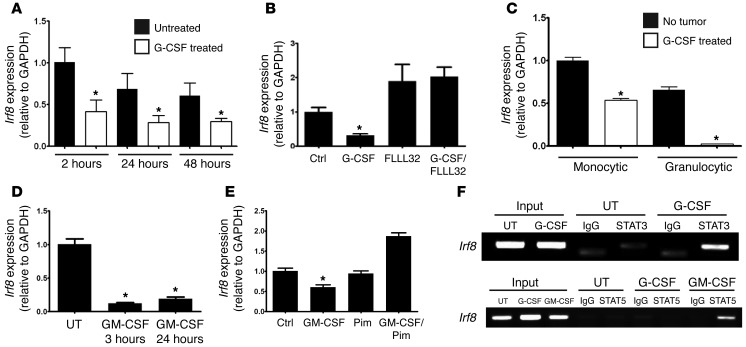
Figure 9. MDSC-associated myelopoietic growth factors inhibit Irf8 expression through STAT-dependent mechanisms.
(A) Irf8 mRNA levels of purified bone marrow–derived CD11b+Gr-1+ cells after treatment with G-CSF (50 ng/ml) (*P < 0.03; the data are representative of 3 separate experiments). As in B, except that the cells were incubated in the absence or presence of FLLL32 (2 μM for 24 hours) (C) Similar to A, except that Irf8 mRNA levels of purified subsets recovered after treatment of BALB/c mice with G-CSF (10 μg/day) or vehicle for 5 consecutive days (n = 3 mice each, *P < 0.001). (D) As in A, except that Irf8 mRNA levels of cells after treatment with GM-CSF (50 ng/ml) (*P < 0.05). (E) As in D, except that the cells were incubated in the absence or presence of pimozide (1 μM for 24 hours). (F) ChIP analysis of the Irf8 promoter in J774.2 cells in the absence (UT) or presence of G-CSF (upper and lower gels) or GM-CSF (lower gel) for 60 minutes to identify a “G-CSF/STAT3/IRF-8 axis” or a “GM-CSF/STAT5/IRF-8 axis” (1 of 3 separate experiments for each).
To demonstrate a role for STAT3 in G-CSF–induced IRF-8 downregulation, we repeated the experiments in the absence or presence FLLL32, a recently characterized JAK2/STAT3 inhibitor (35). Indeed, FLLL32 blocked the ability of G-CSF to alter Irf8 expression, thereby implicating IRF-8 as a downstream target of the STAT3 pathway (Figure 9B). Interestingly, FLLL32 treatment alone increased Irf8 expression, implicating a basal level of STAT3 activity. Prior control experiments in a myeloid cell line model (KG1a; a constitutively active STAT3 variant of KG1) revealed the ability of FLLL32 to inhibit STAT3 phosphorylation in a dose-dependent manner relative to total STAT3 or β-actin (Supplemental Figure 6A). We then evaluated Irf8 levels in mice after G-CSF treatment in vivo to complement our recent study showing that purified CD11b+Gr-1+ myeloid cells from G-CSF–treated mice strongly resemble tumor-induced MDSCs at phenotypic, functional, and gene expression levels (28). Here, we observed that the Irf8 levels in both monocytic and granulocytic subsets were significantly diminished in G-CSF–treated mice compared with the vehicle-treated controls (Figure 9C), a pattern similar to that observed in tumor-bearing mice (Figure 1). Thus, the loss of Irf8 expression in CD11b+Gr-1+ cells of both tumor-bearing and G-CSF–treated mice occurred in both myeloid subsets, implicating STAT3-activating cytokines, such as G-CSF, as relevant players in IRF-8 downregulation.
We expanded this analysis to GM-CSF treatment in vitro, and as with G-CSF, we observed a significant decrease in Irf8 expression (Figure 9D). To determine whether the GM-CSF–induced Irf8 downregulation was STAT5 dependent, we repeated these experiments in the absence or presence pimozide, a newly identified STAT5 inhibitor (36). We observed that pimozide treatment prevented Irf8 downregulation in response to GM-CSF exposure, implicating STAT5 as a potential negative regulator of IRF-8 expression (Figure 9E). Prior control experiments in K562 cells, a STAT5 constitutively active cell line model, indicated the ability of pimozide to block STAT5 phosphorylation (Supplemental Figure 6B). Similar to what we found, Esashi et al. (37) reported that activation of STAT5 by GM-CSF represses Irf8 transcription in a mouse dendritic cell system.
Because G-CSF and GM-CSF preferentially activate STAT3 and STAT5, respectively (38, 39), we next examined whether these STATs directly bind to elements of the Irf8 promoter using the macrophage cell line model J774.2, treated with or without cytokines (Figure 9F). Based on an in silico analysis of the murine Irf8 promoter (40), the PCR step of the ChIP assay was performed using primer pairs encompassing putative STAT3 or STAT5 binding elements (ref. 41 and see Supplemental Data for details regarding the location of these sites). In untreated cells, a weak but specific Irf8 signal was detected following IP with the STAT3 Ab, suggesting a low level of basal STAT3 activity. Importantly, a strong Irf8 band was detected in G-CSF–treated cells following IP with ChIP-certified STAT3 Ab, but not with the STAT5 Ab (Figure 9F), indicating that STAT3 is the preferential mediator between G-CSF signaling and Irf8 promoter engagement. Furthermore, IP with the isotype-matched Ab revealed only a weak, nonspecific signal evident at the incorrect product size (Figure 9F). Similarly, in GM-CSF–treated cells, we detected a strong Irf8 band following IP with the STAT5 Ab, but not the isotype-matched Ab (Figure 9F). Collectively, these data suggest that myelopoietic growth factors associated with MDSC development, such as G-CSF or GM-CSF, downregulate Irf8 transcription via previously undefined STAT3- or STAT5-dependent mechanisms.
IRF-8 levels are depressed in MDSCs of breast cancer patients.
Although monocytic (CD33+CD14+CD15–HLA-DRlo) and granulocytic (CD33+CD14–CD15+HLA-DR–) MDSC subsets have been described in humans, it has been suggested that both subsets originate from a common lineage-negative progenitor population, termed CD33+CD14–CD15–HLA-DR– (6–11). Because this MDSC-like progenitor precedes bifurcation into lineage-defined subsets and is commonly observed in the peripheral blood of patients with breast cancer and other solid tumor types, we investigated the relationship between IRF-8 status and MDSC frequency in this population. First, we retrospectively analyzed the banked peripheral blood leukocytes of breast cancer patients with advanced disease (stage III/IV) for the presence of this common MDSC subset (6–8, 10, 11) relative to healthy controls. We reasoned that this comparison, based on sampling sources of cells from contrasting ends of the health spectrum, was appropriate to determine the impact of disease on the frequency of MDSCs and their IRF-8 status. Patient characteristics are shown in Supplemental Table 5.
Because patient materials were limiting, we developed a multicolor, flow-based assay to quantify intracellular IRF-8 levels in the gated population. To optimize the IRF-8 staining step of the assay, however, we initially used THP-1 cells, a myeloid cell line model that constitutively expresses high levels of IRF-8. Furthermore, in these pilot experiments, IRF-8 expression was analyzed using Image Stream instrumentation, which distinguishes Ab localization in the nucleus versus the cytoplasm. To validate specificity, we modified the assay to impede Ab binding to its target using peptide competition. In this case, we preincubated THP-1 cells with the IRF-8 peptide sequence, reflecting the immunogen used to generate the original Ab. Our data indicated that IRF-8 strongly stained THP-1 cells and staining was strongly localized to the nucleus (Supplemental Figure 6C), determined according to the similarity score, a measure of the coincidence of IRF-8 localization with a nuclear dye (DAPI). Specificity of staining was validated through peptide competition, which greatly attenuated the similarity score.
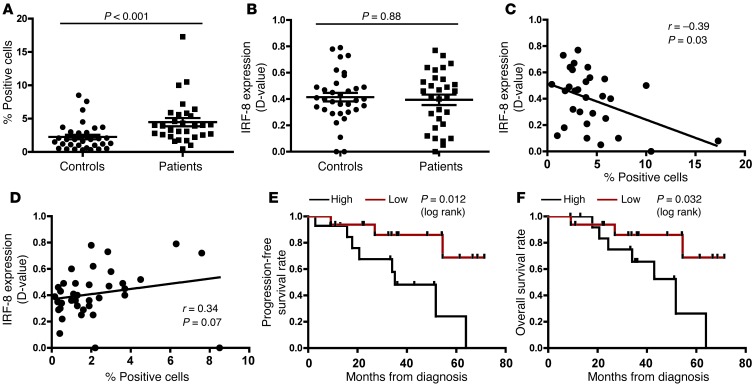
We subsequently analyzed the patient samples, which were batch tested using flow cytometry (see gating strategy in Supplemental Figure 7). Consistent with previous reports (6–10), we observed a significant increase in the percentage of this MDSC subset compared with female age- and race-matched controls (Figure 10A). Surprisingly, no significant difference in IRF-8 expression was observed between healthy donors and patients (Figure 10B). However, when the patient IRF-8 data were plotted versus the corresponding cell percentages, we observed a strong negative correlation between these two parameters (with or without the highest MDSC value). In contrast, no inverse correlation was observed with the controls (Figure 10, C and D). Indeed, these latter data showed a trend toward a positive correlation, suggesting that the inverse relationship between MDSC frequency and IRF-8 status is disease dependent. To determine whether MDSC presence conveyed clinical significance, we stratified the patient MDSC population into high and low fractions in a manner similar to that employed for intratumoral macrophage or T cell quantification (42, 43). These data were plotted in relation to progression-free or overall survival (Figure 10, E and F). Interestingly, we observed statistically significant inverse correlations between MDSC levels and progression-free or overall survival, reinforcing the notion that MDSC levels have prognostic merit. Collectively, these data support the hypothesis that MDSC frequency is IRF-8 dependent and that higher MDSC frequencies coincide with poorer patient outcomes.
Figure 10. Relationship between MDSC frequency and IRF-8 expression.
(A) The percentage of CD33+HLA-DR– cells in the peripheral blood of stage III/IV breast cancer patients and matched healthy controls was determined using multicolor flow cytometry. (B) Quantification of the IRF-8 levels in A, based on the methods described in Methods and Supplemental Figure 6. Correlation between the frequencies of the cells in A versus the IRF-8 levels in B for the controls (C) or patients (D). In all panels, each data point represents an individual specimen. Kaplan-Meier plots for progression-free survival (E) or overall survival (F) of all patients stratified into 2 MDSC cohorts based on the MDSC load. The median was used to define high (n = 14) versus low (n = 16) patient subgroups.
Discussion
Here, we provide the first evidence of a critical role for IRF-8 expression in the transcriptional control of MDSC subset development. We also observed an inverse relationship between IRF-8 levels and MDSC frequency in patients with breast cancer, but not healthy controls, and that elevated MDSC frequencies coincided with a poorer prognosis. While these results reflect a limited number of patients, the findings are consistent with our mouse studies. These data also strengthen the hypothesis that IRF-8 downregulation can occur through STAT3- or STAT5-dependent mechanisms. Taken together, these findings provide new insights into potential therapeutic options that target MDSC-driven transcriptional pathways.
Studies have long revealed an essential role for IRF-8 in regulating myelopoiesis (20–25). This seminal finding was made in Irf8–/– mice, which exhibit a myeloproliferative phenotype consisting predominantly of granulocytic cells and, to a lesser extent, macrophages, reflecting diverse stages of myelopoiesis (20, 21). The observation that MDSC accumulation is a manifestation of aberrant myelopoiesis, coupled with the observation that IRF-8 deficiency profoundly impairs myeloid differentiation, established a rationale to test the hypothesis that MDSCs arise as a consequence of tumor-induced IRF-8 downregulation. Loss-of-function studies using Irf8–/– mice revealed that resident myeloid populations resembled tumor-induced MDSCs at phenotypic, functional, and molecular levels. These data also provided evidence for a previously undescribed consequence of IRF-8 deficiency. For the molecular analyses, we focused on the granulocytic fraction, as these cells represented the predominant subset present in both Irf8–/– and tumor-bearing mice. While the gene expression profiles were similar between both groups of cells, they were not identical, consistent with the notion that MDSCs arise via IRF-8–dependent and –independent mechanisms. Whether the monocytic fraction of Irf8–/– mice is similar to monocytic MDSCs remains to be explored in detail.
Conversely, we showed that IRF-8 overexpression attenuated tumor-induced MDSC development in both implantable and autochthonous mouse tumor models. An analysis of the MDSC subsets revealed that transgenic IRF-8 expression reduced the numbers of both granulocytic and monocytic subsets. However, the magnitude of the contraction was greater with the former than with the latter subset, consistent with the observation that IRF-8 deficiency in null mice conversely favors granulocytic expansion (20). In addition to the effects on cell numbers, we showed that MDSCs derived from tumor-bearing IRF-8–Tg mice were less pro-tumorigenic in vivo compared with those from tumor-bearing WT mice.
Unexpectedly, IRF-8 enhancement did not lessen MDSC-mediated immunosuppression, whereas IRF-8 deficiency effectively elicited immunosuppressive MDSC-like populations. Although it remains unclear why IRF-8 enhancement did not alter the immune suppressive behavior of the remaining MDSC fraction, it is possible that the magnitude of IRF-8 enhancement in this transgenic model remained below the functional threshold required to ameliorate all MDSC characteristics. In contrast, IRF-8 expression in Irf8–/– mice is completely compromised, likely rendering a more profound impact on myeloid function. Nonetheless, the decrease in MDSC numbers was accompanied by improved tumor growth control under autochthonous conditions, spontaneous metastasis, or implantable conditions when combined with an additional anti-MDSC strategy or an immunotherapy. These data are consistent with other studies showing that immune-based therapies mediate stronger antitumor effects when combined with strategies that eliminate MDSCs or inhibit their pro-tumor behavior (44–47).
Notably, IRF-8 overexpression was targeted to CD11b+ cells, which reduced the likelihood of “off-target” cellular effects compared with the use of a more ubiquitous CMV promoter. We used the CD11b promoter to drive IRF-8 expression for at least two major reasons: (a) CD11b expression is common to the broad MDSC population (1); and (b) to our knowledge, no MDSC-specific marker has been reported for use as a promoter. Despite the additional advantages of the CD11b promoter over the CMV promoter, the CD11b marker can also be expressed in macrophages, NK cells, and myeloid DCs. However, our data showed that IRF-8 did not alter the frequency of other leukocyte subsets, either systemically or intratumorally, supporting the hypothesis that IRF-8 selectively modulates the dynamics of the CD11b+Gr-1+ response. Although IRF-8 did not modulate the numbers of macrophages or various DC subsets, these data do not necessarily preclude the possibility that IRF-8 levels influence the functionality of these cells. Thus, this hypothesis requires further detailed investigation.
Moreover, we sought to identify the mechanisms underlying IRF-8 downregulation. Based on the knowledge that G-CSF and GM-CSF are relevant MDSC-inducing TDFs (1, 12–18, 28) that preferentially signal through STAT3 or STAT5 (38, 39), we hypothesized that IRF-8 is a downstream target (direct or indirect) of STAT-based mechanisms. Indeed, our findings indicated that treatment of bone marrow–derived CD11b+Gr-1+ cells with G-CSF or GM-CSF in vitro led to a rapid decline in IRF-8 levels. Additional experiments showed that G-CSF treatment in vivo decreased IRF-8 levels in both granulocytic and monocytic subsets. The results of pharmacological studies supported a causal role for STAT3 or STAT5 signaling in cytokine-induced IRF-8 downregulation, while ChIP assays illustrated direct binding interactions between STAT3 or STAT5 protein and elements of the IRF-8 promoter. In addition to IRF-8, the S100A9 protein (48) or the CCAAT/enhancer-binding protein-β (C/EBPβ) (49) have recently been identified as downstream targets of STAT3 signaling, although their relationship with each other or with IRF-8 has not been studied in detail. Nonetheless, it is unlikely that C/EBPβ is a direct repressor of IRF-8, as IRF-8 transcription/expression in C/EBPβ-knockout mice remains unimpaired (50). One possibility, however, is that these proteins (as well as other yet unidentified transcription factors) are regulated independently through a common STAT3-based mechanism, and that each protein, in turn, might act at discrete stages in monocytic and/or granulocytic MDSC differentiation.
Taken together, the results of the present study have provided new insights into the transcriptional control of MDSC biology, namely (a) the identification of IRF-8 as a transcriptional regulator of MDSC subset development, which offers a novel explanation for the predominance of the granulocytic over the monocytic subset; (b) the conceptual value of IRF-8 enhancement to ameliorate granulocytic MDSC accumulation, which has potential therapeutic implications; and (c) MDSC-inducing factors that act through STAT3 or STAT5 pathways that converge at the level of IRF-8 transcription, thereby providing a mechanistic underpinning for the importance of such STATs in MDSC biology. Future studies, however, are warranted to identify events downstream of IRF-8 that positively or negatively regulate myeloid differentiation. Indeed, our microarray data reveal a significant downregulation of activators of myeloid differentiation (e.g., Egr1, Egr2, and RARα), and the upregulation of repressors of myeloid differentiation (e.g., NAB2), common to cells of both Irf8–/– and tumor-bearing hosts. While further studies are required to examine these events, this study revealed a previously undescribed role for IRF-8 in myeloid-tumor biology.
Methods
Mice.
Female BALB/c and C57BL/6 (B6) mice, 6–8 weeks of age, were obtained from the NCI-Frederick Cancer Research Animal Facility. The procedures for developing and genotyping CD11b-Irf8-Tg and MTAG-Irf8-Tg (double-Tg) mice are detailed in the Supplemental Data. Irf8–/– mice on a B6 background (20) were a gift of K. Ozato (NIH, Bethesda, Maryland, USA). The MTAG (MMTV-PyMT) Tg mouse (34) on a B6 background was a gift of S. Gendler (Mayo Clinic, Scottsdale, Arizona, USA). WT littermates were used as controls.
Patient samples.
Clinical data and samples were obtained from breast cancer patients (all with invasive ductal carcinoma, stage III/IV; see Supplemental Table 5) at diagnosis, and all patients provided informed consent to the Data Bank and Biorepository (DBBR) at RPCI, as previously described (51), via a de-identified IRB-approved protocol. The blood samples were processed within 1 hour to collect buffy coats, which were then stored in liquid nitrogen. A total of 30 breast cancer patients were analyzed compared with female age- and race-matched healthy donor samples stored under the same conditions.
Tumor growth studies.
The AT-3 tumor cell line was established from a primary mammary gland carcinoma of MTAG mice (26). The J774.2 macrophage cell line and 4T1 mammary carcinoma cells (52) were obtained from Sigma-Aldrich and ATCC, respectively. AT-3 and 4T1 tumor cells were implanted orthotopically (5 × 105 and 5 × 104 cells, respectively) into a single mammary gland. For the myeloid/tumor admix experiments, 2.5 × 105 splenic CD11b+Gr-1+ cells were mixed with 5 × 105 AT-3 cells and implanted orthotopically as previously described (26). Tumor growth was measured 2–3 times weekly, and the volumes were calculated using the formula (w2 × l)/2. Although female MTAG mice can develop multiple tumors, the mice were euthanized when any single tumor mass approached 2 cm in diameter or when signs of morbidity were observed. The volume of the largest tumor was calculated over time, and the total tumor burden reflecting the sum of all tumors was determined. In the anti-Ly6G studies, when AT-3 tumors became palpable (~5–10 days after implantation), the mice were injected i.p. with either 250 μg of anti-Ly6G mAb (clone IA8) or rat IgG2a (clone 2A3), as previously described (32). The injections were continued biweekly until differences in tumor growth between groups showed significant separation. In the anti–CTLA-4 studies, the mice were injected i.p. with either 100 μg of anti–CTLA-4 (clone 9H10) or hamster IgG (polyclonal isotype control) when AT-3 tumors first became palpable. A total of 3 injections were administered at 3-day intervals, as previously described (26). All Abs used in vivo were purchased from BioXcell.
Isolation of myeloid subsets.
CD11b+ myeloid cells were isolated from lymphoid tissues using double-column magnetic bead technology (AutoMACS; Miltenyi Biotec) as previously described (28) or through flow cytometry, as outlined below. In the case of the former approach, greater than 90% of the CD11b+ cells coexpressed Gr-1 (26).
Flow analyses.
Cells were analyzed using a FACSCalibur flow cytometer (BD). The data were analyzed using FCS Express software (De Novo Software). Directly conjugated Abs (and isotype controls) included those reactive with CD11b (BioLegend), Gr-1, Ly6C, Ly6G, F4/80, CD11c, CD3, CD4, CD8, B220 (CD45R), CD19, MHC class II (I-Ab), or CD45.2 (BD Biosciences). All preparations were preincubated with CD16/32 antibody (BD Biosciences) to reduce Fc receptor binding. The FACSAria cell sorter was used for aseptic sorting, and the data were analyzed using FACSDiva software. The purified cells (>97%) were used for the indicated experiments. Colocalization studies were performed using Image Stream Instrumentation (Amnis/EMD Millipore).
Human blood leukocyte samples were analyzed for CD33, HLA-DR (BD Biosciences), and IRF-8 expression. The samples were first surface stained for CD33 and HLA-DR, followed by intracellular staining for IRF-8 (C-19; Santa Cruz Biotechnology Inc.), as previously described (53). To control for nonspecific IRF-8 staining, a matched sample was incubated with an IRF-8–blocking peptide (Santa Cruz Biotechnology Inc.) for 2 hours prior to staining. Specific IRF-8 reactivity was determined after correcting for the difference between unblocked and blocked samples. Kolmogorov-Smirnov (KS) statistics, a nonparametric test to measure the difference between two sample distributions, generated D-value scores ranging from 0 (i.e., no difference) to 1 (i.e., maximum difference). All human samples were collected using an LSR Fortessa (BD Biosciences) and statistically analyzed using WinList (Verity Software).
Phenotypic analyses of tumor-infiltrating myeloid populations.
Tumor masses were removed at the indicated time points and fragmented into smaller pieces, followed by enzymatic digestion for 4–6 hours at room temperature using a sterile mixture consisting of collagenase type IA (1 mg/ml) and hyaluronidase type V (0.1 mg/ml) (both from Sigma-Aldrich) to form single-cell suspensions. After digestion, the cell suspensions were collected, washed, and prepared for flow cytometry, as described above.
Immune suppression assays.
T cell populations (Thy1.2+) were isolated from the spleens of naive mice using an AutoMACS system. Before culture, anti-CD3 mAb (1 μg/well) was added to 96-well, flat-bottomed plates for overnight incubation at 4°C. T cells (5 × 104/well) and syngeneic CD11b+Gr-1+ splenocytes were added at the indicated ratios from the different groups. 3H-thymidine (1 μCi/well) was dispensed during the final 18–24 hours of a 72-hour culture period (28).
Molecular analyses.
RNA was isolated using an RNeasy Mini kit (QIAGEN). The cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). The cDNA was then used for all gene expression studies, as described in the Supplemental Data. All data were deposited in a MIAME-compliant manner and are publically available in the GEO database under accession #GSE39228.
ChIP assay.
J774.2 cells (5 × 106/sample) were treated with vehicle, recombinant G-CSF, or GM-CSF (each at 50 ng/ml; Peprotech) for 1 hour at 37°C. The cytokine concentrations were consistent with those used in other studies concerning myeloid cell biology (39, 54). Chromatin was prepared using the EZ ChIP kit (Millipore) according to the manufacturer’s instructions. Protein-bound chromatin was immunoprecipitated using either rabbit IgG (Cell Signaling Technology), ChIP-certified STAT3 Ab (C-20X; Santa Cruz Biotechnology Inc.), or phospho-STAT5 Ab (Tyr694, Cell Signaling Technology). PCR analysis of the purified chromatin was conducted as described in the Supplemental Data.
Histology.
Four-micrometer sections were cut from formalin-fixed, paraffin-embedded lung tissues of tumor-bearing WT or IRF-8–Tg mice, as indicated in Results. The specimens were stained with H&E for histology or prepared for immunohistochemistry using purified rat anti–mouse Gr-1 Ab (clone RB6-8C5; 1:100 dilution, BioLegend). The sections were first deparaffinized and rehydrated. No pretreatment for antigen retrieval was required. Endogenous peroxidase was quenched with aqueous 3% hydrogen peroxide. The specimens were incubated for 60 minutes with the primary Ab (or rat IgG2b isotype control; Invitrogen), rinsed, and stained with an appropriate secondary Ab (biotinylated goat anti-rat IgG, 1:500 dilution; BD Biosciences — Pharmingen). Elite ABC kits were used for the blocking, secondary Ab, and immunoperoxidase steps according to the manufacturer’s instructions (Vector Laboratories). Color was developed after incubation with 3′3-diaminobenzidine, followed by counterstaining with hematoxylin. The positive cells were quantified in a blinded manner by averaging the number of cells counted in 5 random fields, reflecting regions of similar cell densities under high power (×40).
Statistics.
Pairwise comparisons of the mean measurements between groups were analyzed using 2-sided unpaired t tests or exact 2-sided Wilcoxon rank-sum tests. Linear mixed model methods were used to assess the effect of IRF-8 expression (i.e., transgenic IRF-8 versus non-transgenic IRF-8 expression) on tumor growth rate. The tumor size measurements were modeled as a function of a random mouse effect, and fixed effects for an IRF-8 exposure indicator, mouse age at the time of measurement, and the interaction. The age and tumor size measurements were log-transformed to satisfy linearity and distributional assumptions. The statistical significance of the IRF-8 effect on the tumor growth rate was determined by the P value of the interaction term. The survival outcomes were determined using log-rank tests. Modeling was performed using SAS version 9.3. In all figures where error bars are shown, the data are recorded as the mean ± SEM of the indicated number of mice or biological replicates. P values less than 0.05 were considered statistically significant.
Study approval.
All studies performed on mice were approved by the Institutional Animal Care and Use Committee (under protocol 1117M) of the Roswell Park Cancer Institute, and all studies performed on archived human specimens were approved through our IRB (under protocol NHR 008510) in accordance with NIH and Public Health Service policies. Informed consent was not required for the experiments performed in this study, as this research fulfilled the criteria as de-identified, non-human subject research.
Supplementary Material
Acknowledgments
The authors thank Keiko Ozato (NIH) for the Irf8–/– mice, Sandra Gendler (Mayo Clinic) for the MTAG mice, and Lionel Feigenbaum (NIH) for assistance with the development of the IRF-8–Tg mice. The authors also thank Pui-Kai Li and Greg Lesinski (The Ohio State University, Columbus, Ohio, USA) for the gift of FLLL32. All RPCI core resources, including Flow and Image Cytometry, DBBR, Clinical Data Network, and Genomics, were supported through NCI Cancer Center Support Grant R01 CA016056. This work was also supported through National Cancer Institute/NIH grants R01 CA140622 (to S.I. Abrams) and R01 CA133085 and R01 CA168512-DRP1 (to K. Liu), an Alliance Developmental Award from the Roswell Park Alliance Foundation (to S.I. Abrams), and NIH training grant T32CA085183 (to J.D. Waight).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(10):4464–4478. doi:10.1172/JCI68189.
References
- 1.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol. 2012;22(4):275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181(8):5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawanobori Y, et al. Chemokine-mediated rapid turnover of myeloid-derived suppressor cells in tumor-bearing mice. Blood. 2008;111(12):5457–5466. doi: 10.1182/blood-2008-01-136895. [DOI] [PubMed] [Google Scholar]
- 5.Sinha P, Chornoguz O, Clements VK, Artemenko KA, Zubarev RA, Ostrand-Rosenberg S. Myeloid-derived suppressor cells express the death receptor Fas and apoptose in response to T cell-expressed FasL. Blood. 2011;117(20):5381–5390. doi: 10.1182/blood-2010-11-321752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58(1):49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandruzzato S, et al. IL4Ralpha+ myeloid-derived suppressor cell expansion in cancer patients. J Immunol. 2009;182(10):6562–6568. doi: 10.4049/jimmunol.0803831. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez PC, et al. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69(4):1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60(10):1419–1430. doi: 10.1007/s00262-011-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raychaudhuri B, et al. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro Oncol. 2011;13(6):591–599. doi: 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. 2013;190(7):3783–3797. doi: 10.4049/jimmunol.1201449. [DOI] [PubMed] [Google Scholar]
- 12.Tu S, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14(5):408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kujawski M, Kortylewski M, Lee H, Herrmann A, Kay H, Yu H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118(10):3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko JS, et al. Direct and differential suppression of myeloid-derived suppressor cell subsets by sunitinib is compartmentally constrained. Cancer Res. 2010;70(9):3526–3536. doi: 10.1158/0008-5472.CAN-09-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalmin F, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120(2):457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayne LJ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21(6):822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, et al. TNF signaling drives myeloid-derived suppressor cell accumulation. J Clin Invest. 2012;122(11):4094–4104. doi: 10.1172/JCI64115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasquez-Dunddel D, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123(4):1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87(2):307–317. doi: 10.1016/S0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 21.Gabriele L, et al. Regulation of apoptosis in myeloid cells by interferon consensus sequence-binding protein. J Exp Med. 1999;190(3):411–421. doi: 10.1084/jem.190.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13(2):155–165. doi: 10.1016/S1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 23.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 24.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker AM, Michael DG, Satpathy AT, Sciammas R, Singh H, Bhattacharya D. IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood. 2012;119(9):2003–2012. doi: 10.1182/blood-2011-06-364976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart TJ, Liewehr DJ, Steinberg SM, Greeneltch KM, Abrams SI. Modulating the expression of IFN regulatory factor 8 alters the protumorigenic behavior of CD11b+Gr-1+ myeloid cells. . J Immunol. 2009;183(1):117–128. doi: 10.4049/jimmunol.0804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha P, Parker KH, Horn L, Ostrand-Rosenberg S. Tumor-induced myeloid-derived suppressor cell function is independent of IFNγ IL-4Rα. Eur J Immunol. 2012;42(8):2052–2059. doi: 10.1002/eji.201142230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waight JD, Hu Q, Miller A, Liu S, Abrams SI. Tumor-derived G-CSF facilitates neoplastic growth through a granulocytic myeloid-derived suppressor cell-dependent mechanism. PLoS One. 2011;6(11):e27690. doi: 10.1371/journal.pone.0027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsujimura H, Nagamura-Inoue T, Tamura T, Ozato K. IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J Immunol. 2002;169(3):1261–1269. doi: 10.4049/jimmunol.169.3.1261. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, et al. Gr-1+CD11b+ cells are responsible for tumor promoting effect of TGF-β in breast cancer progression. Int J Cancer. 2012;131(11):2584–2595. doi: 10.1002/ijc.27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer C, et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc Natl Acad Sci U S A. 2011;108(41):17111–17116. doi: 10.1073/pnas.1108121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fridlender ZG, et al. Polarization of tumor-associated neutrophil phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell. 2009;16(3):183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bill MA, et al. Structurally modified curcumin analogs inhibit STAT3 phosphorylation and promote apoptosis of human renal cell carcinoma and melanoma cell lines. PLoS One. 2012;7(8):e40724. doi: 10.1371/journal.pone.0040724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson EA, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117(12):3421–3429. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28(4):509–520. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehtonen A, Matikainen S, Miettinen M, Julkunen I. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J Leukoc Biol. 2002;71(3):511–519. [PubMed] [Google Scholar]
- 39.Panopoulos AD, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108(12):3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanno Y, et al. Ozato K. The genomic structure of the murine ICSBP gene reveals the presence of the gamma interferon-responsive element, to which an ISGF3 alpha subunit (or similar) molecule binds. Mol Cell Biol. 1993;13(7):3951–3963. doi: 10.1128/mcb.13.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci U S A. 1995;92(7):3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin Cancer Res. 2005;11(18):6713–6721. doi: 10.1158/1078-0432.CCR-05-0883. [DOI] [PubMed] [Google Scholar]
- 45.Shojaei F, et al. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. . Nat Biotechnol. 2007;25(8):911–920. doi: 10.1038/nbt1323. [DOI] [PubMed] [Google Scholar]
- 46.Ozao-Choy J, et al. The novel role of tyrosine kinase inhibitor in the reversal of immune suppression and modulation of tumor microenvironment for immune-based cancer therapies. Cancer Res. 2009;69(6):2514–2522. doi: 10.1158/0008-5472.CAN-08-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagaraj S, et al. Anti-inflammatory triterpenoid blocks immune suppressive function of MDSCs and improves immune response in cancer. Clin Cancer Res. 2010;16(6):1812–1823. doi: 10.1158/1078-0432.CCR-09-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205(10):2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marigo I, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPβ transcription factor. Immunity. 2010;32(6):790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. Critical role for transcription factor C/EBP-β in regulating the expression of death-associated protein kinase 1. Mol Cell Biol. 2008;28(8):2528–2548. doi: 10.1128/MCB.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ambrosone CB, Nesline MK, Davis W. Establishing a cancer center data bank and biorepository for multidisciplinary research. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1575–1577. doi: 10.1158/1055-9965.EPI-06-0628. [DOI] [PubMed] [Google Scholar]
- 52.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–1405. [PubMed] [Google Scholar]
- 53.Yamamoto M, et al. Shared and distinct functions of the transcription factors IRF4 and IRF8 in myeloid cell development. PLoS One. 2011;6(10):e25812. doi: 10.1371/journal.pone.0025812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bharadwaj U, Li M, Zhang R, Chen C, Yao Q. Elevated interleukin-6 and G-CSF in human pancreatic cancer cell conditioned medium suppress dendritic cell differentiation and activation. Cancer Res. 2007;67(11):5479–5488. doi: 10.1158/0008-5472.CAN-06-3963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.