Abstract
Background
O6-methylguanine-DNA methyltransferase is one of the few proteins to directly remove alkylating agents in the human DNA direct reversal repair pathway. A large number of case-control studies have been conducted to explore the association between MGMT Leu84Phe polymorphism and cancer risk. However, the results were not consistent.
Methods
We carried out a meta-analysis of 44 case-control studies to clarify the association between the Leu84Phe polymorphism and cancer risk.
Results
Overall, significant association of the T allele with cancer susceptibility was verified with meta-analysis under a recessive genetic model (P<0.001, OR=1.30, 95%CI 1.24-1.50) and TT versus CC comparison (P=0.001, OR=1.29, 95% CI 1.12-1.50). In subgroup analysis, a significant increased risk was found for lung cancer (TT versus CC, P=0.027, OR=1.67, 95% CI 1.06-2.63; recessive genetic model, P=0.32, OR=1.64, 95% CI 1.04-2.58), whereas risk of colorectal cancer was significantly low under a dominant genetic model (P=0.019, OR=0.84, 95% CI 0.72-0.97). Additionally, a significant association between TT genetic model and total cancer risk was found in the Caucasian population (TT versus CC, P=0.014, OR=1.29, 95% CI 1.05-1.59; recessive genetic model, P=0.009, OR=1.31, 95% CI 1.07-1.61), but not in the Asian population. An increased risk for lung cancer was also verified in the Caucasian population (TT versus CC, P=0.035, OR=1.62, 95% CI 1.04-2.53; recessive genetic model, P=0.048, OR=1.57, 95% CI 1.01-2.45).
Conclusions
These results suggest that MGMT Leu84Phe polymorphism might contribute to the susceptibility of certain cancers.
Introduction
Over the past decades, there has been an increasing understanding of the disease process in human carcinoma. It is now well established that carcinoma can be initiated by DNA damage from UV exposure, ionizing radiation, environmental chemical agents, and byproducts of cell metabolism. Normally, when DNA damage occurs, DNA repair systems recognize the DNA lesions, excise them, and restore the DNA to maintain genome stability and integrity [1]. However, if genetic alterations occur in genes encoding DNA repair proteins, the DNA repair process may be impaired, potentially contributing to an increased risk for developing cancers.
The O6-methylguanine-DNA methyltransferase (MGMT) is one of the most important proteins in the DNA repair process. It is a 207 amino acid zinc-bound protein which is encoded by MGMT gene located on chromosome 10 at 10q26 and spans approximately 300kb [2]. It has been shown that MGMT has basic methyl-transferring activity [3] and plays a central role in the cellular defense against alkylating agents within the human DNA direct reversal repair pathway.
Also known as O6-alkylguanine–DNA alkyltransferase (ATase, AGT, or AGAT), MGMT protein can directly remove alkyl or methyl adducts from the O 6position of guanine to an internal cysteine residue at codon 145 of the protein [4]. By which, it protects cells against potential DNA alkylation damage from endogenous and exogenous alkylating species such as cigarette consumption, environmental contaminants, and diet [5]. Additionally, it seems that MGMT lacks the ability to dealkylate itself. MGMT therefore can take part only in a single reaction, in which it is irreversibly inactivated [6]. Hence, the reaction should be stoichiometric rather than catalytic. The MGMT expression shows significant variation not only among different body tissues [7], but also among individuals in the same specific tissue [8]. Though the causes of the inter-individual differences in MGMT protein expression levels remain unclear to date, functional polymorphisms in the MGMT gene may have the potential to affect DNA repair capacity. Because of its important role in human DNA direct reversal repair pathway, MGMT has attracted significant attention as a candidate susceptibility gene for cancer.
A large number of molecular epidemiology studies have been carried out to assess the roles of the MGMT polymorphisms in various types of cancer, including lung cancer, head and neck cancer, and colorectal cancer [9,10,11,12,13,14,15,16,17,18,19,20,21]. The MGMTLeu84Phe substitution is the most widely studied polymorphism in MGMT due to a (C->T) transition at nt.262 (MGMT Leu84Phe, rs12917). However, numerous studies on the association of the MGMT Leu84Phe polymorphism with cancer risk have yielded inconsistent results and even partially contradictory conclusions. Several factors may contribute to the discrepancies among different studies. The differences of tumor sites, ethnicities or sample size may all cause the bias of the result of each individual study.
Since single studies may have been underpowered in clarifying the associations of MGMT polymorphisms with cancer susceptibility, to address the controversy among literatures, in the present study we conducted an evidence-based quantitative meta-analysis of the association between the MGMT Leu84Phe polymorphism and susceptibility to cancer.
Materials and Methods
Identification and eligibility of relevant studies
To identify all studies that explored the association of MGMT Leu84Phe polymorphism with cancer risk, we carried out a computerized literature search of the PubMed database (up to July 20, 2012), using the following key words: ‘MGMT,’ ‘polymorphism,’ and ‘cancer,’ without any restriction on language or publication year. The searched papers were read and assessed for their appropriateness of including. All references cited in the articles were also read to identify relevant publications. Eligible studies should meet two criteria: (1) case-control studies; and (2) genotype frequencies in both cancer cases and controls were available. Exclusion criteria were as follows: (a) not relevant to MGMT Leu84Phe polymorphism; (b) not case-control study; (c) control population included malignant tumor cases; and (d) article was a review or duplication of previous publication.
Data extraction
The data was extracted by two investigators (Jun Liu and Fei Chen) from each article independently. Discrepancies were not solved until consensus was reached on every item. From each study, the following data were collected: author’s name, year of publication, country of origin, racial descent, cancer type, source of the control population, genotyping methods, matched factors as well as adjusted factors, number of cases and controls, genotype frequencies for cases and controls, characteristics of cancer cases, and controls. If data of subpopulation from different ethnicities was available in one paper, we took each subpopulation as an individual study.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) for each study was assessed using goodness-of-fit test (x2 of Fisher’s exact test) only in control groups [22]. Crude odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to evaluate the strength of association between MGMTLeu84Phe polymorphism and cancer susceptibility. In the overall and subgroup meta-analysis, we evaluated the associations of genetic variants with cancer risk in homozygous genetic contrast (TT vs. CC), dominant geneticmodel (CT+TT vs. CC), recessive genetic model (TT vs. CT+CC) and T allele vs C allele. The significance of the pooled OR was assessed by the Z-test (P<0.05 shows a significant association). In addition to overall meta-analysis, stratified analysis on ethnicity (Asians, Caucasians, and the other ethnicities group) and tumor site was also performed A x2-based Q-test was carried out to assess the heterogeneity of the ORs [23]. If the result of heterogeneity test was P>0.1, ORs were pooled according to the fixed-effects model (Mantel-Haenszel model). Otherwise, the random-effects model (DerSimonian and Laird model) was applied [24]. The Egger regression test and Begg-Mazumdar test were utilized to measure the potential publication bias [25]. All statistical tests were conducted with the software STATA v.10.0 (Stata Corporation, College Station, TX, USA) using two-side P values.
Results
Characteristics of studies
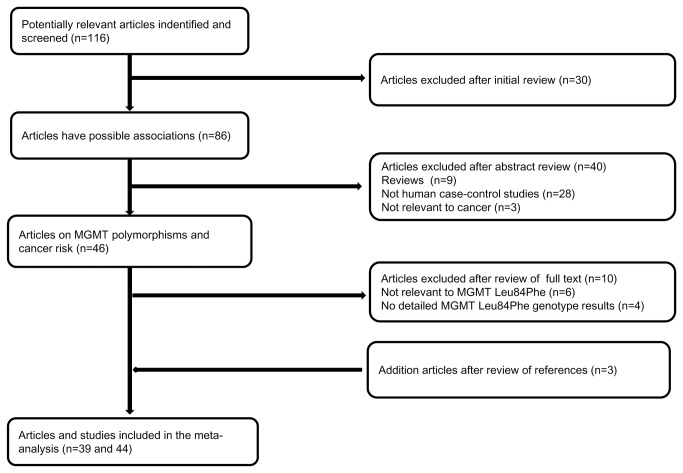
The preliminary literature search yielded 46 articles that explored the association of MGMT polymorphisms with the susceptibility to different cancers. However, six articles [26,27,28,29,30,31] irrelevant to MGMT Leu84Phe polymorphism and four articles [32,33,34,35] without detailed MGMT Leu84Phe genotypes data were excluded. In addition, three articles [10,36,37] were included by literature reading and manual searching. Therefore, 39 articles [9-21, 36-61] were identified and included in the final meta-analysis (Figure 1 ). Five papers [14], [18] [56], [59], and [61] presented data including more than one racial populations and each subgroup in these studies was taken as a separate study. Therefore, a total of 44 studies from 39 papers (18938 cancer patients and 28796 controls) were included. All of the cases were confirmed by histological or pathological examination. A classic polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay was adopted only in 7 of 44 studies and some other genotyping methods were also used widely, such as Taqman, sequencing and Illumina SNP genotyping BeadLab platform. All the genotyping methods are valid for the present meta-analysis. All studies stated that the gender status and the age range were matched between case and control population. The characteristics of included studies are listed in Table 1 . All studies were case-control studies or nested case-control studies within prospective cohort studies, including 9 upper aerodigestive tract squamous cell carcinoma (UADT SCC) studies, 7 colorectal cancer studies, 5 lung cancer studies, 4 brain cancer studies, 3 prostate studies and 13 studies on “other cancers”. There were 15 studies of Caucasian ethnicity, 13 studies of Asian ethnicity, and 16 studies of “mixed ethnicities” (including studies of American, Australian, Black and unspecified population, which cannot be categorized as a unique group since it is mixed). The detailed MGMT Leu84Phe genotype distributions and allele frequencies for cancer cases and controls were presented in Table 2 . The equilibrium of genotypes in the controls was consistent with HWE in all but five studies [9,10,17,21,45] (P=0.01, P=0.06, P=0.02, P<0.01, P=0.04, respectively) (Table 2 ).
Figure 1. Studies identified with criteria for inclusion and exclusion.
Table 1. Characteristics of studies included in the meta-analysis.
| First author and published year | Country | Cancer | Racial descent | Source of controls | No. of cases/controls | Matching |
|---|---|---|---|---|---|---|
| Inoue (2003) | Japan | Brain tumors | Asian | Population | 73/224 | Age |
| Krzensniak (2004) | Poland | Lung cancer | Caucasian | Population | 96/96 | Age,Sex,Smoking |
| Bigler (2005) | America | Colorectal cancer | American | Hospital | 517/615 | None |
| Huang (2005) | Poland | Gastric cancer | Caucasian | Population | 280/387 | Age,Sex |
| Huang (2005) 1 | America | Head and neck SCC | Caucasian | Population/hospital | 400/665 | Age,Sex, Race |
| Huang (2005) 2 | America | Head and neck SCC | Non-white American | Population/hospital | 114/89 | Age,Sex, Race |
| Li (2005) | China | Bladder cancer | Asian | Population | 167/204 | Age,Sex, Smoking |
| Ritchey (2005) | China | Prostate cancer | Asian | Population | 161/246 | Age |
| Shen (2005) | America | Breast cancer | American | Population | 1064/1107 | Age |
| Chae (2006) | Korea | Lung cancer | Asian | Hospital | 432/432 | Age,Sex |
| Han (2006) | America | Endometrial cancer | Caucasian | Population | 434/1085 | Age |
| Han (2006) | America | Breast cancer | Caucasian | Population | 1276/1714 | Age |
| Jiao (2006) | America | Pancreatic cancer | American | Hospital | 370/340 | Age,Sex, Race |
| Kietthubthew (2006) | Thailand | Oral SCC | Asian | Population | 106/164 | Age,Sex |
| Moreno (2006) | Spain | Colorectal cancer | Caucasian | Hospital | 272/299 | None |
| Tranah (2006) 1 | America | Colorectal cancer | American (PHS)c | Hospital | 186/2137 | Age,Smoking |
| Tranah (2006) 2 | America | Colorectal cancer | American (NHS)d | Hospital | 257/429 | Age |
| Wang (2006) | America | Lung cancer | Caucasian | Hospital | 1121/1163 | Age,Sex, Race,Smoking, |
| Zienolddiny (2006) | Norway | Lung cancer | Caucasian | Population | 304/363 | Age,Smoking |
| Felini (2007) | America | Gliomas | American | Population | 379/459 | Age,Sex, Race |
| Hall (2007) | Europea | UADT SCC | Caucasian | Hospital | 803/1062 | Age,Sex, Residence |
| Hu (2007) | China | Lung cancer | Asian | Hospital | 500/517 | Age,Sex, residence |
| Huang (2007) | China | Cervical cancer | Asian | Hospital | 539/800 | Age,Residence |
| Shen (2007) | Australia | Non-Hodgkin’s lymphoma | Australian | Population | 555/495 | Age,Sex, Residence |
| Stern (2007) | Singapore | Colorectal cancer | Asian | Population | 292/1166 | None |
| Doecke (2008) | Australia | Esophageal adenocarcinoma | Australian | Population | 566/1337 | Age,Residence |
| Zhang (2008) | China | Biliary tract cancer | Asian | Population | 406/782 | None |
| Hazra (2008) | America | Colorectal cancer | American | Population | 358/357 | Age |
| kbari (2009) | Iran | Esophageal SCC | Asian | Hospital | 196/250 | None |
| Gu (2009) | America | Melanoma | American | Population | 214/212 | Age, Race |
| Khatami (2009) | Iran | Colorectal cancer | Asian | Hospital | 200/201 | Age,Sex |
| Liu (2009) | America | Glioma | American | Population | 369/363 | Age,Sex, Race |
| McKean-Cowdin (2009) | America | Glioblastoma | Caucasian | Population/hospital | 998/1968 | Age,Sex, Race |
| Yang (2009) | China | Non-Hodgkin’s lymphoma | Asian | Hospital | 48/352 | None |
| Agalliu (2010) 1 | America | Prostate cancer | Caucasian | Population | 1250/1237 | Age |
| Agalliu (2010) 2 | America | Prostate cancer | African-American | Population | 147/81 | Age |
| Huang (2010) | America | Oral SCC | Asian | Hospital | 176/110 | None |
| Palli (2010) | China | Gastric cancer | Caucasian | Population | 291/537 | None |
| Zhang (2010) | Italy | Head and neck SCC | Caucasian | Hospital | 721/1234 | Age,Sex |
| Bye (2011) 1 | America | Esophageal SCC | Black | Population | 346/469 | Age,Sex, Race |
| Bye (2011) 2 | South Africa | Esophageal SCC | Mixed ethnicities | Population | 196/423 | Age,Sex, Race |
| Loh (2011) | South Africa | Cancers | Caucasian | Population | 188/1120 | None |
| O’Mara (2011) 1 | UKb | Endometrial cancer | Australian | Population | 1173/1099 | Age,Residence |
| O’Mara (2011) 2 | Australia | Endometrial cancer | Caucasian | Population | 397/406 | Age |
SCC- squamous cell carcinoma;UADT SCC - Upper Aerodigestive Tract Squamous Cell Carcinoma
a: Include 5 central and eastern European countries
b: Indlude Norfolk, East Anglia and United Kingdom
c: PHS- Physicians’ Health Study d: NHS-Nurses’ Health Study
Table 2. Distribution of MGMT Leu84Phe genotypes and allelic frequency.
| Study (year) | Distribution of MGMT Leu85Phe genotypes |
Frequency of MGMT Leu85Phe alleles |
HWE P value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case (n) |
Control (n) |
Case (n) |
Control (n) |
|||||||||||
| CC | CT | TT | CC | CT | TT | C | T | C | T | |||||
| Inoue (2003) | 55 | 18 | 0 | 160 | 55 | 9 | 128 | 18 | 375 | 73 | 0.13 | |||
| Krzensniak (2004) | 67 | 23 | 6 | 74 | 17 | 5 | 157 | 35 | 165 | 27 | 0.01 | |||
| Bigler (2005) | 403 | 108 | 6 | 466 | 136 | 13 | 914 | 120 | 1068 | 162 | 0.41 | |||
| Huang (2005) | 190 | 82 | 8 | 279 | 99 | 9 | 462 | 98 | 657 | 117 | 0.95 | |||
| Huang (2005) a | 315 | 80 | 5 | 468 | 179 | 18 | 710 | 90 | 1115 | 215 | 0.86 | |||
| Huang (2005) b | 71 | 37 | 6 | 61 | 25 | 3 | 179 | 49 | 147 | 31 | 0.82 | |||
| Li (2005) | 132 | 34 | 1 | 173 | 28 | 3 | 298 | 36 | 374 | 34 | 0.15 | |||
| Ritchey (2005) | 123 | 36 | 2 | 213 | 32 | 1 | 282 | 40 | 458 | 34 | 0.86 | |||
| Shen (2005) | 778 | 265 | 21 | 824 | 263 | 20 | 1821 | 307 | 1911 | 303 | 0.85 | |||
| Chae (2006) | 344 | 84 | 4 | 341 | 81 | 10 | 772 | 92 | 763 | 101 | 0.06 | |||
| Han (2006) | 344 | 82 | 8 | 822 | 242 | 21 | 770 | 98 | 1886 | 284 | 0.52 | |||
| Han (2006) | 964 | 279 | 33 | 1306 | 382 | 26 | 2207 | 345 | 2994 | 434 | 0.75 | |||
| Jiao (2006) | 264 | 101 | 5 | 257 | 82 | 1 | 629 | 111 | 596 | 84 | 0.04 | |||
| Kietthubthew (2006) | 84 | 21 | 1 | 130 | 33 | 1 | 189 | 23 | 293 | 35 | 0.48 | |||
| Moreno (2006) | 213 | 47 | 12 | 225 | 63 | 11 | 473 | 71 | 513 | 85 | 0.02 | |||
| Tranah (2006) a | 147 | 33 | 6 | 1634 | 471 | 32 | 327 | 45 | 3739 | 535 | 0.77 | |||
| Tranah (2006) b | 204 | 47 | 6 | 330 | 93 | 6 | 455 | 59 | 753 | 105 | 0.85 | |||
| Wang (2006) | 832 | 259 | 30 | 872 | 272 | 19 | 1923 | 319 | 2016 | 310 | 0.67 | |||
| Zienolddiny (2006) | 189 | 102 | 13 | 247 | 106 | 10 | 480 | 128 | 600 | 126 | 0.73 | |||
| Felini (2007) | 289 | 84 | 6 | 369 | 84 | 6 | 662 | 96 | 822 | 96 | 0.63 | |||
| Hall (2007) | 574 | 198 | 31 | 764 | 277 | 21 | 1346 | 260 | 1805 | 319 | 0.48 | |||
| Hu (2007) | 418 | 77 | 5 | 421 | 93 | 3 | 913 | 87 | 935 | 99 | 0.38 | |||
| Huang (2007) | 372 | 156 | 11 | 592 | 198 | 10 | 900 | 178 | 1382 | 218 | 0.15 | |||
| Shen (2007) | 432 | 112 | 11 | 373 | 110 | 12 | 976 | 134 | 856 | 134 | 0.26 | |||
| Stern (2007) | 251 | 40 | 1 | 959 | 194 | 13 | 542 | 42 | 2112 | 220 | 0.37 | |||
| Doecke (2008) | 416 | 136 | 14 | 1029 | 281 | 27 | 968 | 164 | 2339 | 335 | 0.13 | |||
| Zhang (2008) | 352 | 53 | 1 | 631 | 144 | 7 | 757 | 55 | 1406 | 158 | 0.70 | |||
| Hazra (2008) | 271 | 72 | 15 | 254 | 97 | 6 | 614 | 102 | 605 | 109 | 0.34 | |||
| Akbari (2009) | 142 | 53 | 1 | 185 | 63 | 2 | 337 | 55 | 433 | 67 | 0.17 | |||
| Gu (2009) | 152 | 60 | 2 | 168 | 43 | 1 | 364 | 64 | 379 | 45 | 0.32 | |||
| Khatami (2009) | 40 | 160 | 0 | 61 | 140 | 0 | 240 | 160 | 262 | 140 | 0.00 | |||
| Liu (2009) | 299 | 62 | 8 | 267 | 89 | 7 | 660 | 78 | 623 | 103 | 0.89 | |||
| McKean-Cowdin (2009) | 774 | 204 | 20 | 1480 | 453 | 35 | 1752 | 244 | 3413 | 523 | 0.96 | |||
| Yang (2009) | 33 | 14 | 1 | 289 | 58 | 5 | 80 | 16 | 636 | 68 | 0.29 | |||
| Agalliu (2010) a | 949 | 269 | 32 | 916 | 298 | 23 | 2167 | 333 | 2130 | 344 | 0.83 | |||
| Agalliu (2010) b | 106 | 35 | 6 | 60 | 20 | 1 | 247 | 47 | 140 | 22 | 0.64 | |||
| Huang (2010) | 151 | 25 | 0 | 89 | 21 | 0 | 327 | 25 | 199 | 21 | 0.27 | |||
| Palli (2010) | 210 | 77 | 4 | 395 | 131 | 11 | 497 | 85 | 921 | 153 | 0.97 | |||
| Zhang (2010) | 563 | 151 | 7 | 933 | 284 | 17 | 1277 | 165 | 2150 | 318 | 0.38 | |||
| Bye (2011) a | 225 | 111 | 10 | 300 | 155 | 14 | 561 | 131 | 755 | 183 | 0.26 | |||
| Bye (2011) b | 120 | 65 | 11 | 294 | 116 | 13 | 305 | 87 | 704 | 142 | 0.71 | |||
| Loh (2011) | 146 | 37 | 5 | 894 | 212 | 14 | 329 | 47 | 2000 | 240 | 0.72 | |||
| O’Mara (2011) a | 889 | 261 | 23 | 810 | 270 | 19 | 2039 | 307 | 1890 | 308 | 0.52 | |||
| O’Mara (2011) b | 278 | 108 | 11 | 296 | 103 | 7 | 664 | 130 | 695 | 117 | 0.57 | |||
Bold indicates statistically significant P value.
HWE Hardy–Weinberg equilibrium
Quantitative synthesis
In overall analysis, significant associations between the T allele and cancer risk were found under the recessive genetic model (P=0.001, OR=1.28, 95%CI 1.11-1.47) and TT versus CC comparison (P=0.001, OR=1.28, 95% CI 1.11-1.47). And, after we excluded those studies whose genotype equilibrium was not consistent with HWE, significant associations between the T allele and cancer susceptibility was also uncovered under the recessive genetic model (P<0.001, OR=1.30, 95%CI 1.24-1.50) and TT versus CC comparison (P=0.001, OR=1.29, 95% CI 1.12-1.50). However, no significant association was found in the dominant genetic model (TT+TC versus CC) and T versus C comparison. These results were summarized in Table 3 .
Table 3. Summary ORs (95% CI) for MGMT Leu84Phe variant under different genetic models and tumor site.
| MGMT Leu85Phe | N# | TT versus CC |
CT+TTversus CC |
TT versus CT+CC |
T versus C |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
(dominant genetic model) |
(recessive genetic model) |
|
|||||||||
| Tumor site | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | ||||
| Total | 44 | 1.28 (1.11-1.47) | 0.001 | 1.01 (0.94-1.08)b | 0.808 | 1.28 (1.11-1.47) | 0.001 | 1.01 (0.96-1.08)b | 0.504 | |||
| Total in HWE | 39 | 1.29 (1.12-1.50) | 0.001 | 1.00 (0.93-1.07)b | 0.890 | 1.30 (1.24-1.50) | 0.000 | 1.01 (0.95-1.08)b | 0.692 | |||
| UADT SCC | 9 | 1.24 (0.89-1.73) | 0.197 | 0.96 (0.82-1.13)b | 0.626 | 1.25 (0.90-1.73) | 0.189 | 0.98 (0.84-1.15)b | 0.820 | |||
| Colorectal cancer | 7 | 1.29 (0.85-1.95) | 0.234 | 0.89 (0.78-1.02) | 0.091 | 1.35 (0.90-2.04) | 0.152 | 0.94 (0.84-1.05) | 0.267 | |||
| Colorectal cancer in HWE | 5 | 1.25 (0.62-2.50)b | 0.536 | 0.84 (0.72-0.97) | 0.019 | 1.30 (0.64-2.66)b | 0.470 | 0.88 (0.77-1.01) | 0.073 | |||
| Lung cancer | 5 | 1.38 (0.92-2.06) | 0.119 | 1.05 (0.92-1.19) | 0.485 | 1.34 (0.90-2.00) | 0.147 | 1.06 (0.95-1.20) | 0.298 | |||
| Lung cancer in HWE | 3 | 1.67 (1.06-2.63) | 0.027 | 1.05 (0.91-1.21) | 0.526 | 1.64 (1.04-2.58) | 0.032 | 1.08 (0.95-1.23) | 0.232 | |||
| Brain cancer | 4 | 1.11 (0.71-1.73) | 0.664 | 0.89 (0.68-1.16)b | 0.390 | 1.42 (0.73-1.79) | 0.562 | 0.90 (0.72-1.13)b | 0.375 | |||
| Prostate cancer | 3 | 1.48 (0.88-2.48) | 0.136 | 1.22 (0.74-2.00)b | 0.445 | 1.51 (0.91-2.53) | 0.113 | 1.25 (0.81-1.94)b | 0.321 | |||
| Endomtrial cancer | 3 | 1.14 (0.74-1.77) | 0.560 | 0.92 (0.80-1.06) | 0.240 | 1.64 (0.75-1.80) | 0.495 | 0.95 (0.84-1.07) | 0.394 | |||
| Other cancers | 13 | 1.17 (0.88-1.54) | 0.281 | 1.10 (0.97-1.26)b | 0.147 | 1.14 (0.87-1.51) | 0.350 | 1.09 (0.97-1.23)b | 0.152 | |||
| Other cancers in HWE | 12 | 1.14 (0.86-1.51) | 0.368 | 1.09 (0.95-1.26)b | 0.216 | 1.12 (0.84-1.47) | 0.446 | 1.08 (0.95-1.22)b | 0.236 | |||
Bold indicates statistically significant P value
All summary ORs were calculated using fixed-effects models, unless stated otherwise
# Number of studies
b Random-effect models
HWE − Hardy Weinberg Equilibrium
When the subgroup analyses were carried out according to tumor site, the MGMT T allele was associated with a significant increase in risk of lung cancer (TT Versus CC, P=0.027, OR =1.67, 95% CI 1.06-2.63; recessive genetic model, P=0.32, OR=1.64, 95% CI 1.04-2.58). By contrast, a significant protective effect was found for colorectal cancer under the dominant genetic model (P=0.019, OR=0.84, 95% CI 0.72-0.97). However, no significant association was found in other tumor sites subgroups under all genetic models. These results are also listed in Table 3 .
In most of the available studies, there was no difference of MGMT Leu84Phe genotype/allele distribution among different ethnicities. We also performed stratified analysis by ethnicity (Caucasians, Asians, and mixed ethnicities), and by ethnicity and tumor site together (Table 4 ). In subgroup meta-analysis by ethnicity, significant associations between TT and recessive genetic model and total cancer risk were found in the Caucasian population (TT versus CC, P=0.004, OR =1.32, 95% CI 1.10-1.61; recessive genetic model, P=0.002, OR=1.34, 95% CI 1.11-1.62) and in the mixed ethnicities population (TT versus CC, P=0.041, OR =1.27, 95% CI 1.01-1.60; recessive genetic model, P=0.037, OR=1.28, 95% CI 1.02-1.61). And, when those studies without consistency with HWE were excluded, a significant association was still found for the Caucasian population (TT versus CC, P=0.014, OR =1.29, 95% CI 1.05-1.59; recessive genetic model, P=0.009, OR=1.31, 95% CI 1.07-1.61). However, in the Asian subgroup and the mixed ethnicities subgroup, no significant association was observed for any genetic model. In the analysis stratified by ethnicity and tumor site (Table 4 ), we found an increased risk only in the Caucasian subgroup for lung cancer (TT versus CC, P=0.035, OR =1.62, 95% CI 1.04-2.53; recessive genetic model, P=0.048, OR=1.57, 95% CI 1.01-2.45).
Table 4. Summary ORs (95% CI) for MGMT Leu84Phe variant categorized by ethnicity and ethnicity / tumor site under different genetic models.
| MGMT Leu85Phe | N# | TT versus CC |
TT+TC versus CC |
TT versus TC + CC |
T versus C |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
(dominant genetic model) |
(recessive genetic model) |
|
|||||||||
| Ethnicity | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | ||||
| Caucasian | 15 | 1.32 (1.10-1.61) | 0.004 | 0.98 (0.90-1.06)b | 0.560 | 1.34 (1.11-1.62) | 0.002 | 1.00 (0.93-1.09)b | 0.923 | |||
| Caucasian in HWE | 13 | 1.29 (1.05-1.59) | 0.014 | 0.96 (0.88-1.06)b | 0.407 | 1.31 (1.07-1.61) | 0.009 | 0.99 (0.91-1.08)b | 0.827 | |||
| Asian | 13 | 0.97 (0.58-1.61) | 0.898 | 1.07 (0.88-1.31)b | 0.485 | 0.94 (0.57-1.56) | 0.805 | 1.03 (0.86-1.22)b | 0.779 | |||
| Asian in HWE | 11 | 1.19 (0.68-2.09) | 0.546 | 1.04 (0.83-1.30)b | 0.724 | 1.15 (0.65-2.01) | 0.633 | 1.02 (0.83-1.26)b | 0.861 | |||
| Mixed ethnicities | 16 | 1.27 (1.01-1.60) | 0.041 | 1.01 (0.91-1.13)b | 0.813 | 1.28 (1.02-1.61) | 0.037 | 1.04 (0.95-1.13)b | 0.457 | |||
| Mixed ethnicities in HWE | 15 | 1.25 (0.99-1.58) | 0.057 | 1.00 (0.90-1.12)b | 0.997 | 1.26 (1.00-1.58) | 0.052 | 1.08 (0.95-1.22)b | 0.236 | |||
| Caucasian | ||||||||||||
| Lung cancer | 3 | 1.62 (1.04-2.53) | 0.035 | 1.12 (0.96-1.31) | 0.159 | 1.57 (1.01-2.45) | 0.048 | 1.14 (0.99-1.31) | 0.061 | |||
| UADT SCC | 3 | 0.88 (0.33-2.33)b | 0.794 | 0.85 (0.66-1.08)b | 0.182 | 0.92 (0.36-2.35)b | 0.865 | 0.87 (0.66-1.14)b | 0.312 | |||
| Asian | ||||||||||||
| UADT SCC | 3 | 0.94(01.15-5.84) | 0.950 | 0.96(0.7101.30) | 0.800 | 0.93 (0.15-5.76) | 0.939 | 0.97 (0.73-1.28) | 0.802 | |||
| Mixed ethnicities | ||||||||||||
| Colorectal cancer | 4 | 1.46 (0.89-2.38) | 0.134 | 0.85 (0.72-1.01) | 0.059 | 1.53 (0.94-2.50) | 0.088 | 0.91 (0.79-1.06) | 0.220 | |||
Bold indicates statistically significant P value
All summary ORs were calculated using fixed-effects models, unless stated otherwise
# Number of studies
b Random-effect models
UADT SCC − Upper Aerodigestive Tract Squamous Cell CarcinomaHWE − Hardy Weinberg Equilibrium
As shown in Table 3 and Table 4 , heterogeneity widely existed in the present meta-analysis under the dominant genetic mode and T versus C comparison but not under the homozygous comparison and recessive genetic model.
Publication bias
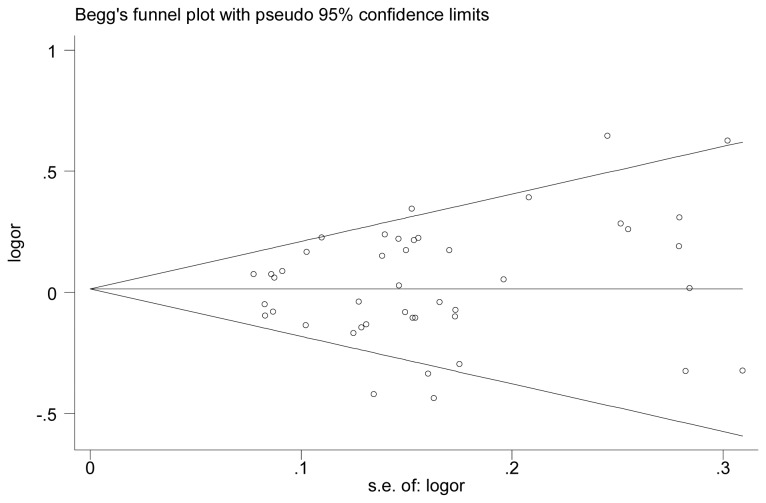
Begg’s funnel plot and Egger’s test were utilized to evaluate the publication bias of the literature. As shown in Figure 2 , the contour-enhanced funnel plot for publication bias did not reveal any evidence of obvious asymmetry in allele contrast (T allele versus C allele), and, as expected, the Egger’s test did not provide any obvious evidence for bias (t=0.12, P=0.902).
Figure 2. Begg’s funnel plot analysis to detect publication bias (MGMT : Leu84Phe T allele versus C allele).
Each point represents a separate study for the indicated association. Logor represents natural logarithm of OR. Horizontal line represents the mean effects size.
Discussion
This meta-analysis including a total of 18938 cancer patients and 28796 controls from 44 independent genetic studies implies that MGMT Leu84Phe polymorphism might contribute to the susceptibility of certain cancers
Although the global analysis indicated that the T variant allele might increase the risk of cancer, the subgroup meta-analysis showed significant association at only two tumor sites (colorectal cancer and lung cancer) and two ethnicity subgroups (Caucasian subgroup and mixed ethnicities subgroup). This phenomenon suggests that the MGMT Leu84Phe polymorphism may play differing roles in cancerogenesis at different sites or in different ethnicities because of variability in genetic backgrounds [62].
Since cancer is a complex disease, it is highly possible that any single genetic factor has only weak effects on an individual’s phenotype. It has been reported that the interaction of different combinations of polymorphisms in the same gene or between and among different genes might together have a pronounced effect on cancer risk [63,64,65]. Studies by Li et al. [66,67] have shown that MGMT is a transcriptional suppressor of ER-dependent signaling upon repair of the O6-methylguanine lesion and that the Lue84 and Ile143 residues lie in close proximity to three conserved leucines of the LXXLL ER-interacting helix. Therefore, it is possible that the ER-dependent signalling could be differentially mediated by the variant 84Phe and 143Val residues. Some studies [9,10,13,40,42,48,49,54] have tried to investigate the combined effects of Lue84Phe, Ile143Val, and other polymorphisms in MGMT on cancer risk. Because the available data were not compatible, we could not evaluate the combined effects of MGMT Leu84Phe and Ile143Val on cancer susceptibility in our meta-analysis.
It is well established that genetic factors may play an important role in the development of tumors. However, there is no doubt that environmental factors such as alcohol consumption, cigarette use, and aging also participate in tumorigenesis. Several studies [11,39,42] reported that heavy cigarette smoking could aggravate the effects of MGMT variants on cancer risk. However, Chae et al. [10] did not find the same results. Li et al. [40] found that both drinking and smoking enhance genetic variants’ effects on bladder cancer risk. It should be noted that alcohol consumption and cigarette use may play different roles at different tumor sites because of the different levels of alkylating agents and different tissue exposure concentrations. Unfortunately, owing to a lack of studies restricted to populations only exposed to alkylating agents, we could not obtain enough original data to further estimate the effects of the gene-environment interactions on cancer susceptibility.
We note several limitations in the present study. First, there was wide heterogeneity due to the nature of our meta-analysis, and the results should be interpreted with caution. Second, our results were based on unadjusted information, and the lack of original data limited estimation of the effect of confounding factors on cancer risk. Notably, confounding factors such as sex, age, alcohol drinking, smoking, and socioeconomic status may alter the association of genetic variants with cancer susceptibility. Third, the number of eligible studies in the subgroup analysis was limited. Subsequently, some subgroup meta-analysis might not have enough statistical power to accurately evaluate the association between the MGMT Leu84Phe polymorphism and cancer risk. More importantly, haplotype analysis has been regarded as a much better approach in genetic association research. However, since more detailed individual information on genotypes of the other polymorphisms of MGMT was unavailable, we were not able to conduct linkage disequilibrium and haplotype analysis in this study.
In conclusion, we observed several significant associations of the MGMT Leu84Phe polymorphism with cancer susceptibility. MGMT Leu84Phe variants may increase lung cancer risk, especially in Caucasians, but reduce colorectal cancer risk, indicating some differences among different tumor sites. In addition, MGMT Leu84Phe variants may increase cancer risk in Caucasians and in the mixed ethnicities group, which suggests an appreciable difference among different ethnic populations. Further well-designed study with greater sample size will be helpful in clarifying the haplotypes, gene–gene and gene–environment interactions on MGMT polymorphisms and tissue-specific cancer risk in ethnicity specific populations, and further mechanistic studies are warranted to elucidate the exact functional roles of MGMT variants.
Supporting Information
PRISMA Checklist.
(DOC)
Funding Statement
This work was supported by Science and Technical Development Foundation of Shandong Province (2011YD18014), China, Doctoral Program of Shandong Province (2007BS03009), China, and Science and Technical Development Foundation of Yantai (2008142-21), China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39-85. doi:10.1146/annurev.biochem.73.011303.073723. PubMed: 15189136. [DOI] [PubMed] [Google Scholar]
- 2. Natarajan AT, Vermeulen S, Darroudi F, Valentine MB, Brent TP et al. (1992) Chromosomal localization of human O6-methylguanine-DNA methyltransferase (MGMT) gene by in situ hybridization. Mutagenesis 7: 83-85. doi:10.1093/mutage/7.1.83. PubMed: 1635460. [DOI] [PubMed] [Google Scholar]
- 3. Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND et al. (2004) DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol 11: 714-720. doi:10.1038/nsmb791. PubMed: 15221026. [DOI] [PubMed] [Google Scholar]
- 4. Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y (1988) Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem 57: 133-157. doi:10.1146/annurev.bi.57.070188.001025. PubMed: 3052269. [DOI] [PubMed] [Google Scholar]
- 5. Bartsch H, Montesano R (1984) Relevance of nitrosamines to human cancer. Carcinogenesis 5: 1381-1393. doi:10.1093/carcin/5.11.1381. PubMed: 6386215. [DOI] [PubMed] [Google Scholar]
- 6. Samson L (1992) The suicidal DNA repair methyltransferases of microbes. Mol Microbiol 6: 825-831. doi:10.1111/j.1365-2958.1992.tb01533.x. PubMed: 1602962. [DOI] [PubMed] [Google Scholar]
- 7. Citron M, Decker R, Chen S, Schneider S, Graver M et al. (1991) O6-methylguanine-DNA methyltransferase in human normal and tumor tissue from brain, lung, and ovary. Cancer Res 51: 4131-4134. PubMed: 1868433. [PubMed] [Google Scholar]
- 8. Margison GP, Povey AC, Kaina B, Santibáñez Koref MF (2003) Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis 24: 625-635. doi:10.1093/carcin/bgg005. PubMed: 12727789. [DOI] [PubMed] [Google Scholar]
- 9. Krześniak M, Butkiewicz D, Samojedny A, Chorazy M, Rusin M (2004) Polymorphisms in TDG and MGMT genes - epidemiological and functional study in lung cancer patients from Poland. Ann Hum Genet 68: 300-312. doi:10.1046/j.1529-8817.2004.00079.x. PubMed: 15225156. [DOI] [PubMed] [Google Scholar]
- 10. Chae MH, Jang JS, Kang HG, Park JH, Park JM et al. (2006) O6-alkylguanine-DNA alkyltransferase gene polymorphisms and the risk of primary lung cancer. Mol Carcinog 45: 239-249. doi:10.1002/mc.20171. PubMed: 16385589. [DOI] [PubMed] [Google Scholar]
- 11. Wang L, Liu H, Zhang Z, Spitz MR, Wei Q (2006) Association of genetic variants of O6-methylguanine-DNA methyltransferase with risk of lung cancer in non-Hispanic Whites. Cancer Epidemiol Biomarkers Prev 15: 2364-2369. doi:10.1158/1055-9965.EPI-06-0437. PubMed: 17164358. [DOI] [PubMed] [Google Scholar]
- 12. Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V et al. (2006) Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27: 560-567. doi:10.1093/carcin/bgi232. PubMed: 16195237. [DOI] [PubMed] [Google Scholar]
- 13. Hu Z, Wang H, Shao M, Jin G, Sun W et al. (2007) Genetic variants in MGMT and risk of lung cancer in Southeastern Chinese: a haplotype-based analysis. Hum Mutat 28: 431-440. doi:10.1002/humu.20462. PubMed: 17285603. [DOI] [PubMed] [Google Scholar]
- 14. Huang WY, Olshan AF, Schwartz SM, Berndt SI, Chen C et al. (2005) Selected genetic polymorphisms in MGMT, XRCC1, XPD, and XRCC3 and risk of head and neck cancer: a pooled analysis. Cancer Epidemiol Biomarkers Prev 14: 1747-1753. doi:10.1158/1055-9965.EPI-05-0162. PubMed: 16030112. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z, Wang L, Wei S, Liu Z, Wang LE et al. (2010) Polymorphisms of the DNA repair gene MGMT and risk and progression of head and neck cancer. DNA Repair (Amst) 9: 558-566. doi:10.1016/j.dnarep.2010.02.006. PubMed: 20206583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bigler J, Ulrich CM, Kawashima T, Whitton J, Potter JD (2005) DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps. Cancer Epidemiol Biomarkers Prev 14: 2501-2508. doi:10.1158/1055-9965.EPI-05-0270. PubMed: 16284370. [DOI] [PubMed] [Google Scholar]
- 17. Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A et al. (2006) Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 12: 2101-2108. doi:10.1158/1078-0432.CCR-05-1363. PubMed: 16609022. [DOI] [PubMed] [Google Scholar]
- 18. Tranah GJ, Bugni J, Giovannucci E, Ma J, Fuchs C et al. (2006) O6-methylguanine-DNA methyltransferase Leu84Phe and Ile143Val polymorphisms and risk of colorectal cancer in the Nurses’ Health Study and Physicians’ Health Study (United States). Cancer Causes Control 17: 721-731. doi:10.1007/s10552-006-0005-y. PubMed: 16633920. [DOI] [PubMed] [Google Scholar]
- 19. Stern MC, Conti DV, Siegmund KD, Corral R, Yuan JM et al. (2007) DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev 16: 2363-2372. doi:10.1158/1055-9965.EPI-07-0268. PubMed: 18006925. [DOI] [PubMed] [Google Scholar]
- 20. Hazra A, Chanock S, Giovannucci E, Cox DG, Niu T et al. (2008) Large-scale evaluation of genetic variants in candidate genes for colorectal cancer risk in the Nurses’ Health Study and the Health Professionals’ Follow-up Study. Cancer Epidemiol Biomarkers Prev 17: 311-319. doi:10.1158/1055-9965.EPI-07-0195. PubMed: 18268114. [DOI] [PubMed] [Google Scholar]
- 21. Khatami F, Noorinayer B, Mohebi SR, Ghiasi S, Mohebi R et al. (2009) Effects of amino acid substitution polymorphisms of two DNA methyltransferases on susceptibility to sporadic colorectal cancer. Asian Pac J Cancer Prev 10: 1183-1188. PubMed: 20192566. [PubMed] [Google Scholar]
- 22. Zintzaras E, Lau J (2008) Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61: 634-645. doi:10.1016/j.jclinepi.2007.12.011. PubMed: 18538260. [DOI] [PubMed] [Google Scholar]
- 23. Zintzaras E, Ioannidis JP (2005) Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol 28: 123-137. doi:10.1002/gepi.20048. PubMed: 15593093. [DOI] [PubMed] [Google Scholar]
- 24. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177-188. doi:10.1016/0197-2456(86)90046-2. PubMed: 3802833. [DOI] [PubMed] [Google Scholar]
- 25. Macaskill P, Walter SD, Irwig L (2001) A comparison of methods to detect publication bias in meta-analysis. Stat Med 20: 641-654. doi:10.1002/sim.698. PubMed: 11223905. [DOI] [PubMed] [Google Scholar]
- 26. Imai Y, Oda H, Nakatsuru Y, Ishikawa T (1995) A polymorphism at codon 160 of human O6-methylguanine-DNA methyltransferase gene in young patients with adult type cancers and functional assay. Carcinogenesis 16: 2441-2445. doi:10.1093/carcin/16.10.2441. PubMed: 7586149. [DOI] [PubMed] [Google Scholar]
- 27. Yang M, Coles BF, Caporaso NE, Choi Y, Lang NP et al. (2004) Lack of association between Caucasian lung cancer risk and O6-methylguanine-DNA methyltransferase-codon 178 genetic polymorphism. Lung Cancer 44: 281-286. doi:10.1016/j.lungcan.2003.12.003. PubMed: 15140540. [DOI] [PubMed] [Google Scholar]
- 28. Crosbie PA, McGown G, Thorncroft MR, O’Donnell PN, Barber PV et al. (2008) Association between lung cancer risk and single nucleotide polymorphisms in the first intron and codon 178 of the DNA repair gene, O6-alkylguanine-DNA alkyltransferase. Int J Cancer 122: 791-795. doi:10.1002/ijc.23059. PubMed: 17957803. [DOI] [PubMed] [Google Scholar]
- 29. Loh YH, Mitrou PN, Bowman R, Wood A, Jeffery H et al. (2010) MGMT Ile143Val polymorphism, dietary factors and the risk of breast, colorectal and prostate cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk study. DNA Repair (Amst) 9: 421-428. doi:10.1016/j.dnarep.2010.01.002. PubMed: 20096652. [DOI] [PubMed] [Google Scholar]
- 30. Ma WJ, Lv GD, Zheng ST, Huang CG, Liu Q et al. (2010) DNA polymorphism and risk of esophageal squamous cell carcinoma in a population of North Xinjiang, China. World J Gastroenterol 16: 641-647. doi:10.3748/wjg.v16.i5.641. PubMed: 20128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong X, Li Y, Chang P, Hess KR, Abbruzzese JL et al. (2012) DNA mismatch repair network gene polymorphism as a susceptibility factor for pancreatic cancer. Mol Carcinog 51: 491-499. doi:10.1002/mc.20817. PubMed: 21681824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Egyhazi S, Ma S, Smoczynski K, Hansson J, Platz A et al. (2002) Novel O6-methylguanine-DNA methyltransferase; SNPs a frequency comparison of patients with familial melanoma and healthy individuals in Sweden. Hum Mutat 20: 408-409. doi:10.1002/humu.9077. [DOI] [PubMed] [Google Scholar]
- 33. Hung RJ, Baragatti M, Thomas D, McKay J, Szeszenia-Dabrowska N et al. (2007) Inherited predisposition of lung cancer: a hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol Biomarkers Prev 16: 2736-2744. doi:10.1158/1055-9965.EPI-07-0494. PubMed: 18086781. [DOI] [PubMed] [Google Scholar]
- 34. Zawlik I, Vaccarella S, Kita D, Mittelbronn M, Franceschi S et al. (2009) Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: a population-based study. Neuroepidemiology 32: 21-29. doi:10.1159/000170088. PubMed: 18997474. [DOI] [PubMed] [Google Scholar]
- 35. Ricceri F, Guarrera S, Sacerdote C, Polidoro S, Allione A et al. (2010) ERCC1 haplotypes modify bladder cancer risk: a case-control study. DNA Repair (Amst) 9: 191-200. doi:10.1016/j.dnarep.2009.12.002. PubMed: 20061190. [DOI] [PubMed] [Google Scholar]
- 36. Hall J, Hashibe M, Boffetta P, Gaborieau V, Moullan N et al. (2007) The association of sequence variants in DNA repair and cell cycle genes with cancers of the upper aerodigestive tract. Carcinogenesis 28: 665-671. PubMed: 17040931. [DOI] [PubMed] [Google Scholar]
- 37. McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M et al. (2009) Associations between polymorphisms in DNA repair genes and glioblastoma. Cancer Epidemiol Biomarkers Prev 18: 1118-1126. doi:10.1158/1055-9965.EPI-08-1078. PubMed: 19318434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inoue R, Isono M, Abe M, Abe T, Kobayashi H (2003) A genotype of the polymorphic DNA repair gene MGMT is associated with de novo glioblastoma. Neurol Res 25: 875-879. doi:10.1179/016164103771954005. PubMed: 14669534. [DOI] [PubMed] [Google Scholar]
- 39. Huang WY, Chow WH, Rothman N, Lissowska J, Llaca V et al. (2005) Selected DNA repair polymorphisms and gastric cancer in Poland. Carcinogenesis 26: 1354-1359. doi:10.1093/carcin/bgi084. PubMed: 15802298. [DOI] [PubMed] [Google Scholar]
- 40. Li C, Liu J, Li A, Qian L, Wang X et al. (2005) Exon 3 polymorphisms and haplotypes of O6-methylguanine-DNA methyltransferase and risk of bladder cancer in southern China: a case-control analysis. Cancer Lett 227: 49-57. doi:10.1016/j.canlet.2005.03.043. PubMed: 15885889. [DOI] [PubMed] [Google Scholar]
- 41. Ritchey JD, Huang WY, Chokkalingam AP, Gao YT, Deng J et al. (2005) Genetic variants of DNA repair genes and prostate cancer: a population-based study. Cancer Epidemiol Biomarkers Prev 14: 1703-1709. doi:10.1158/1055-9965.EPI-04-0809. PubMed: 16030105. [DOI] [PubMed] [Google Scholar]
- 42. Shen J, Terry MB, Gammon MD, Gaudet MM, Teitelbaum SL et al. (2005) MGMT genotype modulates the associations between cigarette smoking, dietary antioxidants and breast cancer risk. Carcinogenesis 26: 2131-2137. doi:10.1093/carcin/bgi179. PubMed: 16014702. [DOI] [PubMed] [Google Scholar]
- 43. Han J, Hankinson SE, De Vivo I (2006) Polymorphisms in O6-methylguanine DNA methyltransferase and endometrial cancer risk. Carcinogenesis 27: 2281-2285. doi:10.1093/carcin/bgl099. PubMed: 16777993. [DOI] [PubMed] [Google Scholar]
- 44. Han J, Tranah GJ, Hankinson SE, Samson LD, Hunter DJ (2006) Polymorphisms in O6-methylguanine DNA methyltransferase and breast cancer risk. Pharmacogenet Genomics 16: 469-474. doi:10.1097/01.fpc.0000215065.21718.4c. PubMed: 16788379. [DOI] [PubMed] [Google Scholar]
- 45. Jiao L, Bondy ML, Hassan MM, Wolff RA, Evans DB et al. (2006) Selected polymorphisms of DNA repair genes and risk of pancreatic cancer. Cancer Detect Prev 30: 284-291. doi:10.1016/j.cdp.2006.05.002. PubMed: 16844323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kietthubthew S, Sriplung H, Au WW, Ishida T (2006) Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health 209: 21-29. doi:10.1016/j.ijheh.2005.06.002. PubMed: 16373199. [DOI] [PubMed] [Google Scholar]
- 47. Felini MJ, Olshan AF, Schroeder JC, North KE, Carozza SE et al. (2007) DNA repair polymorphisms XRCC1 and MGMT and risk of adult gliomas. Neuroepidemiology 29: 55-58. doi:10.1159/000108919. PubMed: 17898525. [DOI] [PubMed] [Google Scholar]
- 48. Huang J, Ye F, Chen H, Lu W, Xie X (2007) Amino acid substitution polymorphisms of the DNA repair gene MGMT and the susceptibility to cervical carcinoma. Carcinogenesis 28: 1314-1322. doi:10.1093/carcin/bgm003. PubMed: 17234722. [DOI] [PubMed] [Google Scholar]
- 49. Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE et al. (2007) Polymorphisms in DNA repair genes and risk of non-Hodgkin’s lymphoma in New South Wales, Australia. Haematologica 92: 1180-1185. doi:10.3324/haematol.11324. PubMed: 17666372. [DOI] [PubMed] [Google Scholar]
- 50. Doecke J, Zhao ZZ, Pandeya N, Sadeghi S, Stark M et al. (2008) Polymorphisms in MGMT and DNA repair genes and the risk of esophageal adenocarcinoma. Int J Cancer 123: 174-180. doi:10.1002/ijc.23410. PubMed: 18386788. [DOI] [PubMed] [Google Scholar]
- 51. Zhang M, Huang WY, Andreotti G, Gao YT, Rashid A et al. (2008) Variants of DNA repair genes and the risk of biliary tract cancers and stones: a population-based study in China. Cancer Epidemiol Biomarkers Prev 17: 2123-2127. doi:10.1158/1055-9965.EPI-07-2735. PubMed: 18708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Akbari MR, Malekzadeh R, Shakeri R, Nasrollahzadeh D, Foumani M et al. (2009) Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res 69: 7994-8000. doi:10.1158/0008-5472.CAN-09-1149. PubMed: 19826048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gu F, Qureshi AA, Kraft P, Guo Q, Hunter DJ et al. (2009) Polymorphisms in genes involved in DNA repair, cell growth, oxidative stress and inflammatory response, and melanoma risk. Br J Dermatol 161: 209-212. doi:10.1111/j.1365-2133.2009.09219.x. PubMed: 19438866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA et al. (2009) Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev 18: 204-214. doi:10.1158/1055-9965.EPI-08-0632. PubMed: 19124499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang F, Shi JY, Xu L, Ren LJ, Zhang QH et al. (2009) [Genetic susceptibility of single nucleotide polymorphism in MGMT to non-Hodgkin lymphoma]. Zhonghua Xue Ye Xue Za Zhi 30: 622-625. PubMed: 19954624. [PubMed] [Google Scholar]
- 56. Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA et al. (2010) Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control 21: 289-300. doi:10.1007/s10552-009-9461-5. PubMed: 19902366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang SH, Chang PY, Liu CJ, Lin MW, Hsia KT (2010) O6-methylguanine-DNA methyltransferase gene coding region polymorphisms and oral cancer risk. J Oral Pathol Med 39: 645-650. doi:10.1111/j.1600-0714.2009.00880.x. PubMed: 20412404. [DOI] [PubMed] [Google Scholar]
- 58. Palli D, Polidoro S, D’Errico M, Saieva C, Guarrera S et al. (2010) Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis 25: 569-575. doi:10.1093/mutage/geq042. PubMed: 20817763. [DOI] [PubMed] [Google Scholar]
- 59. Bye H, Prescott NJ, Matejcic M, Rose E, Lewis CM et al. (2011) Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis 32: 1855-1861. doi:10.1093/carcin/bgr211. PubMed: 21926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loh YH, Mitrou PN, Wood A, Luben RN, McTaggart A et al. (2011) SMAD7 and MGMT genotype variants and cancer incidence in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Cancer Epidemiol 35: 369-374. doi:10.1016/j.canep.2010.09.011. PubMed: 21075068. [DOI] [PubMed] [Google Scholar]
- 61. O’Mara TA, Ferguson K, Fahey P, Marquart L, Yang HP et al. (2011) CHEK2, MGMT, SULT1E1 and SULT1A1 polymorphisms and endometrial cancer risk. Twin Res Hum Genet 14: 328-332. doi:10.1375/twin.14.4.328. PubMed: 21787115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4: 45-61. doi:10.1097/00125817-200203000-00002. PubMed: 11882781. [DOI] [PubMed] [Google Scholar]
- 63. Goodman JE, Mechanic LE, Luke BT, Ambs S, Chanock S et al. (2006. ) Exploring SNP-SNP interactions and colon cancer risk using polymorphism interaction analysis. Int J Cancer 118: 1790-1797. PubMed: 16217767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schwender H, Selinski S, Blaszkewicz M, Marchan R, Ickstadt K et al. (2012) Distinct SNP combinations confer susceptibility to urinary bladder cancer in smokers and non-smokers. PLOS ONE 7: e51880. doi:10.1371/journal.pone.0051880. PubMed: 23284801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chuang LY, Chang HW, Lin MC, Yang CH (2012) Chaotic particle swarm optimization for detecting SNP-SNP interactions for CXCL12-related genes in breastcancer prevention. Eur J Cancer Prev 21: 336-342. PubMed: 22127289. [DOI] [PubMed] [Google Scholar]
- 66. Teo AK, Oh HK, Ali RB, Li BF (2001) The modified human DNA repair enzyme O(6)-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage. Mol Cell Biol 21: 7105-7114. doi:10.1128/MCB.21.20.7105-7114.2001. PubMed: 11564893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oh HK, Teo AK, Ali RB, Lim A, Ayi TC et al. (1996) Conformational change in human DNA repair enzyme O6-methylguanine-DNA methyltransferase upon alkylation of its active site by SN1 (indirect-acting) and SN2 (direct-acting) alkylating agents: breaking a "salt-link". Biochemistry 35: 12259-12266. doi:10.1021/bi9603635. PubMed: 8823159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)