We carried out large-scale, quantitative analyses of the relative abundance of thousands of peptides and phosphopeptides from maize leaf proteins at four developmental stages. Our data, which comprise an important community resource, highlight the role of posttranslational regulation in developmental transitions and provide insights into proteins with cell wall– and hormone-related functions.
Abstract
We performed large-scale, quantitative analyses of the maize (Zea mays) leaf proteome and phosphoproteome at four developmental stages. Exploiting the developmental gradient of maize leaves, we analyzed protein and phosphoprotein abundance as maize leaves transition from proliferative cell division to differentiation to cell expansion and compared these developing zones to one another and the mature leaf blade. Comparison of the proteomes and phosphoproteomes suggests a key role for posttranslational regulation in developmental transitions. Analysis of proteins with cell wall– and hormone-related functions illustrates the utility of the data set and provides further insight into maize leaf development. We compare phosphorylation sites identified here to those previously identified in Arabidopsis thaliana. We also discuss instances where comparison of phosphorylated and unmodified peptides from a particular protein indicates tissue-specific phosphorylation. For example, comparison of unmodified and phosphorylated forms of PINFORMED1 (PIN1) suggests a tissue-specific difference in phosphorylation, which correlates with changes in PIN1 polarization in epidermal cells during development. Together, our data provide insights into regulatory processes underlying maize leaf development and provide a community resource cataloging the abundance and phosphorylation status of thousands of maize proteins at four leaf developmental stages.
INTRODUCTION
Maize (Zea mays) has long been an important food crop and a classical genetic system. The recent completion of the maize genome (Schnable et al., 2009) has facilitated the development of tools and resources to study maize. A useful phenomenon of maize biology is that leaves mature from the tip to the base, such that growing leaves display a developmental gradient, making the maize leaf an attractive model system for developmental studies (Sylvester et al., 1990). In unexpanded leaves, proliferative, symmetric cell division is confined to the leaf base. Distal to this basal division zone, symmetric divisions give way to fate-specifying asymmetric divisions as cells differentiate and begin to expand. Further distal still, cells are no longer dividing but continue to expand and mature. While the roles of a few proteins and phytohormones in these important developmental events have been studied, a broad understanding of the underlying processes has yet to be achieved. The developmental gradient of the maize leaf has been exploited by researchers characterizing C4 photosynthesis to create large data resources: Complementary RNaseq (Li et al., 2010) and proteomics (Majeran et al., 2010) studies analyzed the mRNA and protein content at successive stages of photosynthetic development. Another study measured hormone levels in a series of early maize leaf development stages and demonstrated that gibberellic acid is important for the transition from cell division to growth (Nelissen et al., 2012). While an atlas of maize transcript expression using microarrays has characterized gene expression in many maize tissues, including the leaf base (encompassing all phases of cell division and growth) and leaf tip (mature photosynthetic tissue) (Sekhon et al., 2011), there are no publicly available large-scale data comparing global gene or protein expression as maize leaf cells transition from division to differentiation to expansion.
It has long been acknowledged that protein abundance and transcript abundance correlate poorly (Gygi et al., 1999), which is largely explained by differing translational rates (Piques et al., 2009; Schwanhäusser et al., 2011). Hence, obtaining a complete systems-level analysis of molecular events requires direct measurements of proteins. Indeed, while the genotype specifies the potential of an organism, proteins that carry out cellular processes, and the interactions between these proteins and the environment, dictate phenotype. Thus, knowledge of the proteotype, or protein complement, is required to fully understand the biology of an organism and its constituent parts. An important component of the proteotype includes posttranslational modifications that may affect protein function, such as protein phosphorylation. Using proteomics, quantitative measurements of protein abundance and phosphorylation status are possible. Data resources for plant proteomics and phosphoproteomics include the pep2pro (Baerenfaller et al., 2008) and PhosPhat resources for Arabidopsis thaliana (Heazlewood et al., 2008), the Plant Proteome Database for Arabidopsis and maize (Sun et al., 2009), and the Medicago Phosphoprotein Database for Medicago truncatula (Rose et al., 2012). (For a recent review of large-scale plant proteomics, see Nakagami et al. [2012].) Most of these proteomic analyses identify peptides, or phosphopeptides and their corresponding phosphorylation sites, but few quantify the relative abundance of these peptides in different tissues, especially in the case of phosphopeptides. Moreover, none of these resources provide simultaneous quantification of unphosphorylated and phosphorylated forms of a protein. Such parallel comparisons are essential to distinguish changes in the phosphorylation state of a particular protein from changes in the overall abundance of that protein, as illustrated by studies in plants (Reiland et al., 2011; Zhang et al., 2013) and other systems (Rigbolt et al., 2011). Until very recently, comprehensive coverage of the proteome was not possible; therefore, the likelihood of identifying the same protein in both a phosphoproteomic and proteomic analysis was low, making such parallel comparisons difficult. However, as proteomic technology has developed, the number of proteins that can be identified in a complex protein sample has increased greatly, and spectral counting for comparing relative abundance of proteins has gained popularity (Liu et al., 2004; Huttlin et al., 2010).
Here, we present parallel proteomic and phosphoproteomic analyses of developing maize leaves, focusing on early stages of growth and differentiation. Using a label-free proteomics method, we quantified peptides and phosphopeptides from four developmental zones of the leaf. In total, we identified more than 81,000 peptides from over 12,000 proteins and over 11,000 phosphorylated peptides from more than 3500 proteins, providing both quantitative and qualitative information about the distribution of maize proteins and their phosphorylation status through successive stages of maize leaf development. Using examples from cell wall and hormone biology, we demonstrate how parallel analyses of the proteome and phosphoproteome fuel hypotheses regarding protein function.
RESULTS
Maize Leaves Were Divided into Regions Containing Dividing, Differentiating, Expanding, or Mature Cells
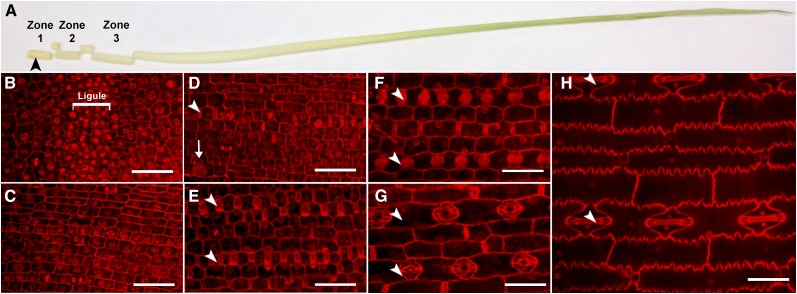
For analysis of proteotypes, maize leaf tissue was isolated at a series of developmental stages. Leaf tissue was harvested from ∼4-week-old maize plants when leaf 8 was at least 50 cm long and leaf 10 was just emerging from the whorl (see Supplemental Figure 1A online). Several fully expanded leaves were removed to expose developing tissue at the bases of remaining leaves; three basal leaf zones were excised for analysis based on the developmental stages they represented (Figure 1A). Zone 1, 0 to 1.25 cm from the leaf base, contains cells that are dividing, primarily isodiametric in shape, and either undifferentiated or undergoing early stages of differentiation (Figures 1B and 1C; see Supplemental Figure 1B online). Zone 2, 1.5 to 2.75 cm from the leaf base, contains cells of varying size and shape, indicating cellular differentiation (Figures 1D and 1E; see Supplemental Figure 1C online). Some cells in zone 2 are still dividing, but these divisions are predominantly asymmetric, giving rise to stomata and other specialized cell types. Zone 3, 3.5 to 5.5 cm from the leaf base, contains postmitotic, expanding cells that have undergone extensive differentiation (Figures 1F and 1G; see Supplemental Figure 1D online). A mature leaf sample comprised of blade tissue (excluding the midrib and sheath) from leaf 8 was also analyzed (Figure 1H). Four to six biological replicates, each composed of tissue from 11 to 24 plants, were analyzed by HPLC–tandem mass spectrometry (MS/MS).
Figure 1.
Leaf Tissues Used for Proteomic Analyses.
(A) To isolate zones 1 to 3, outer leaves of 30-d-old plants were removed to expose a leaf whose developing ligule (black arrowhead) was within 0.5 cm of the leaf base, and the remaining leaves were cut into sections as illustrated.
(B) to (H) Confocal images of propidium iodide–stained, formaldehyde/alcohol/acetic acid–fixed epidermis of zone 1 ([B] and [C]), zone 2 ([D] and [E]), zone 3 ([F] and [G]), and mature leaves (H). Bars = 50 μm.
(B) and (C) The base of zone 1 contains the preligular band (labeled as ligule). Symmetrically dividing cells can be seen at the base (B) and top (C) of zone 1.
(D) At the base of zone 2, some cell files have started to form stomata (arrowhead) and hair primordia (arrow).
(E) At the top of zone 2, stomatal rows are completing their divisions (arrowheads).
(F) and (G) At both the base (F) and top (G) of zone 3, cells have completed division and are expanding. Stomatal rows are indicated by arrowheads.
(H) Mature leaves are fully expanded with a variety of cell types, including stomatal rows (arrowheads).
[See online article for color version of this figure.]
HPLC-MS/MS Identified 12,093 Proteins and 3557 Phosphoproteins in the Maize Leaf
Proteins extracted from each tissue sample were separated and analyzed by HPLC-MS/MS as described in Methods. The numbers of spectra collected and matched are listed in Supplemental Table 1 online. Across all four leaf regions, 81,051 peptides from a database composed of maize 5a working set proteins, and 73 peptides were identified from the decoy database. These peptides were matched to a maximum possible number of 28,504 proteins (see Supplemental Table 2 online). In many cases, peptides map to more than one possible protein, all of which are included in this set of 28,504 proteins. This number includes not only closely related proteins, but also different splice isoforms of the same locus. Therefore, this is likely an overestimate of the true number of proteins identified. To obtain a more conservative estimate, proteins to which such shared peptides mapped were grouped together. A total of 12,032 protein groups were identified. Group leaders, which either have the highest number of peptide identifications or correspond to the longest protein in the case of ties, were assigned for each group. Throughout the remainder of the article, “proteins” are synonymous with “group leaders.” Supplemental Data Set 1 online lists the peptide sequences of all protein models, and Supplemental Data Set 2 online lists the group leaders, as well as all additional members of the group.
Although 12,032 proteins are assigned within our false discovery rate and, therefore, likely present within our tissues, we further filtered the data set to 8005 proteins for quantitative analysis using the steps summarized in Supplemental Table 2 online. To estimate the relative abundance of each protein, spectral counts (SPCs) were normalized to obtain a normalized spectral abundance factor (NSAF) (Florens et al., 2006). The list of all proteins identified is given in Supplemental Data Set 1 online, and the list of proteins in the filtered, normalized data set is shown in Supplemental Data Set 2 online. Supplemental Figure 2 online shows a validation of our quantitation methodology demonstrating that the NSAFs of several proteins correlate well with protein abundance assessed by protein gel blotting across a large range of NSAF values.
Majeran et al. (2010) recently completed a comprehensive, quantitative proteomics study on maize leaves to investigate the developmental progression of photosynthesis and metabolism. They used partially expanded juvenile leaves (leaf 3) rather than expanded leaf 8 (or mature sample) or unexpanded leaves 10 or older (zones 1, 2, and 3), which was used here. Juvenile and adult leaves have several morphological differences, such as the presence of epicuticular wax in juvenile leaves only and the presence of specialized hairs on adult leaves (Sylvester et al., 1990). Additionally, leaf 3 is smaller than leaf 10, resulting in compressed zones in leaf 3 relative to leaf 10. Therefore “3 cm” is not necessarily equivalent in both studies; this can be seen qualitatively as the leaf they used appears to be green as early as 3 cm, while ours is still white in zone 3. Additionally, Majeran et al. (2010) excluded the developing ligule, which was included in this study in zone 1. Nonetheless, since both studies identified proteins in developing leaves, considerable overlap is expected and is shown in the Venn diagram in Supplemental Figure 3A online. The correlation coefficient of our mature leaf sample, which correlated poorly with our younger zones (Figure 2A), correlates much better with their oldest leaf sample (see Supplemental Figures 3B and 3C online). Comparison of the normalized spectral counts (nSPCs) of several proteins identified in both studies shows good agreement, validating both studies (see Supplemental Figure 3D online).
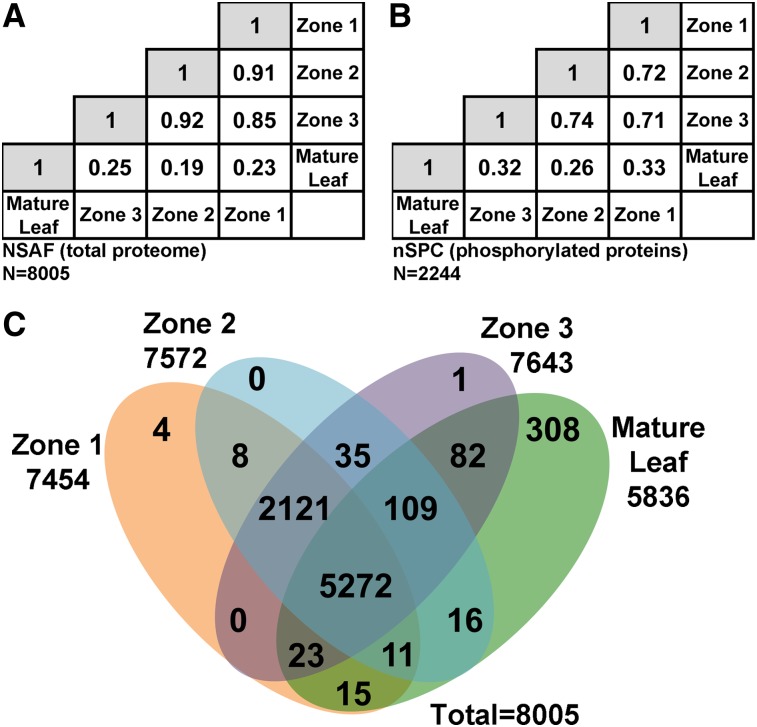
Figure 2.
Comparison of Proteins and Phosphorylated Proteins Identified in Different Regions of the Maize Leaf.
(A) and (B) Matrix of correlation coefficients of the NSAF values calculated from the total proteome (A) and nSPC calculated from phosphorylated proteins (B). The correlation coefficient between two tissues is at the intersection of the row and column.
(C) Venn diagram of proteins showing the unmodified proteomes of the four leaf tissue segments. “Present” is defined as having at least one SPC in one replicate.
[See online article for color version of this figure.]
To identify phosphorylated proteins, phosphopeptides were enriched using cerium oxide prior to analysis by HPLC-MS/MS. A total of 13,925 phosphorylated amino acids were identified in 11,429 peptides (see Supplemental Table 3 online). In some cases, at least one phosphorylated residue could be definitively localized (7200 residues in 6278 phosphopeptides). In other cases, at least one phosphorylation site is present, but the specific Ser, Thr, or Tyr could not confidently be assigned and is therefore considered “nonlocalized” (additional 6725 residues in 5151 phosphopeptides) (see Supplemental Table 3 online). The 11,429 identified phosphopeptides are from a maximum of 7126 proteins corresponding to 3471 protein groups defined as discussed earlier (see Supplemental Table 4 online). As for the unmodified proteome, additional filtering steps were applied to the phosphoprotein data set to define a set of 2244 proteins used for quantitative analyses (see Supplemental Table 4 online). Two data sets were used for analyses of phosphorylation. The first is an unfiltered data set composed of the 11,429 phosphopeptides that were identified but not derived from the decoy database (see Supplemental Data Set 3 online). To gain statistical power, a second data set was analyzed, which combines SPCs from all phosphopeptides from a given protein (see Supplemental Data Set 4 online). This data set was filtered as indicated in Supplemental Table 4 online. For both the phosphoprotein and phosphopeptide data sets, the relative abundance is indicated by the nSPCs.
Protein Enrichments in Distinct Developmental Zones Reflect Processes Occurring in Those Zones
To analyze the proteotypes associated with proliferative division, differentiation, and cell expansion in growing maize leaves, we compared the proteins identified in the three zones of growing leaves to one another and to those of mature maize leaves (Figure 2). Pairwise Pearson’s correlation coefficients of NSAFs calculated from the proteome (Figure 2A) and nSPCs calculated from phosphoproteome (Figure 2B) are shown in correlation matrices. Strikingly, correlations among zones 1, 2, and 3 are much higher than correlations between the mature leaf and any of the three younger zones, for both proteins and phosphoproteins. This phenomenon is further illustrated in the Venn diagram shown in Figure 2C classifying all proteins present in each zone (irrespective of their quantitative values). Although the majority (5272 of 8005) of proteins identified in our study were present in all regions of the leaf, many (2121) were exclusively found in zones 1, 2, and 3 but not mature leaves. Also striking is that while none of the three developing zones had many unique proteins, 308 proteins were exclusively found in the mature leaf. Thus, differences between mature leaves and the three young zones are largely attributable to proteins absent from the mature leaves, and vice versa, rather than differences in expression values in these tissues.
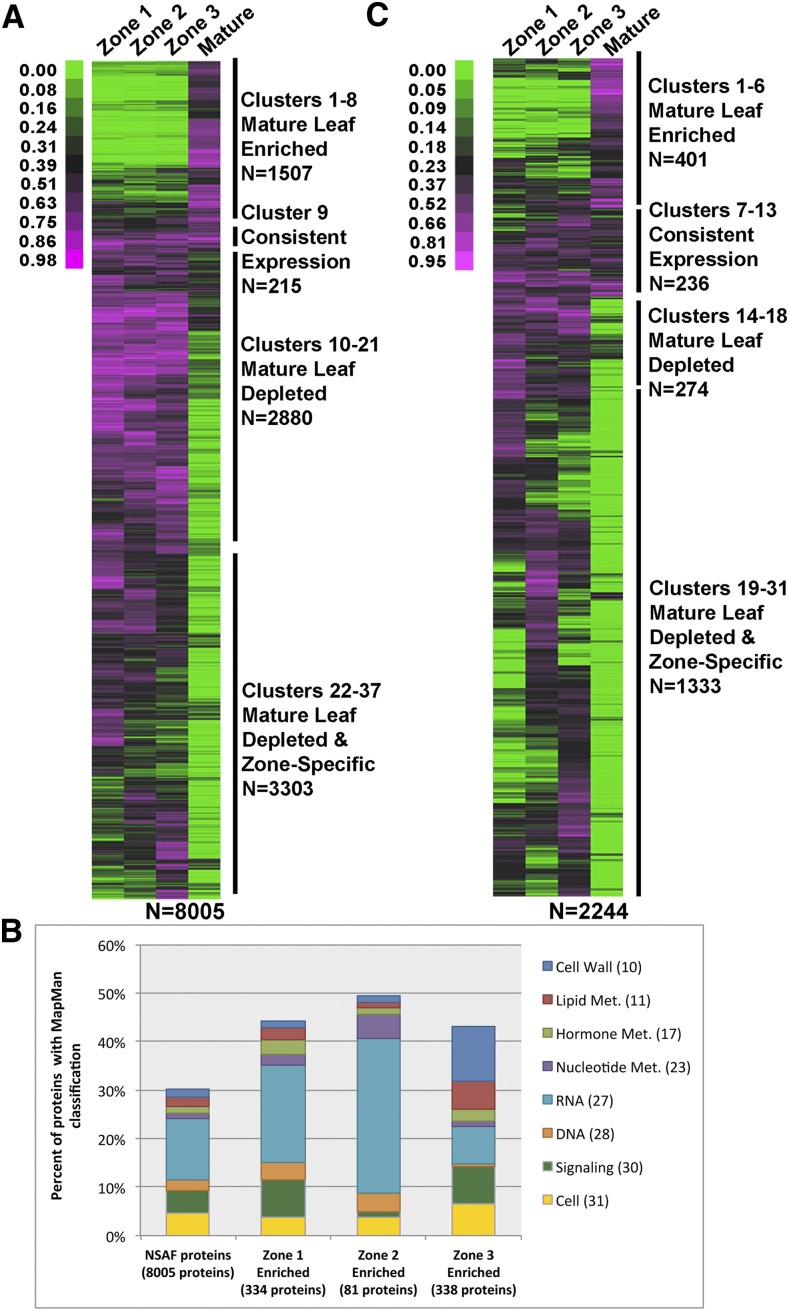
To characterize the global protein expression patterns and to identify proteins enriched in one or more of the four leaf zones, we performed a hierarchical clustering analysis of NSAF scores scaled from 0 to 1 (Figure 3A). The clusters fall into four major groups: (1) clusters 1 to 8, containing proteins enriched in mature leaf tissue; (2) cluster 9, containing proteins with NSAF scores unchanged across all four leaf zones; (3) clusters 10 to 21, containing proteins depleted in mature leaf tissue but with similar expression in the three young zones; and (4) clusters 22 to 37, containing proteins depleted in mature leaves and enriched in one or more developing zones. To determine whether certain functional classes of proteins were overrepresented within the four identified cluster groups, each protein was assigned to a MapMan bin (Thimm et al., 2004) and each cluster group was analyzed for overrepresented bins (see Supplemental Table 5 online). Predictably, cluster 9, containing proteins with unchanged expression, contains an overrepresentation of housekeeping proteins, such as those involved in glycolysis and the tricarboxylic acid cycle. The mature leaf–enriched cluster group (clusters 1 to 8) shows an overrepresentation of proteins involved processes such as photosynthesis, carbohydrate metabolism, and secondary metabolism, and proteins in the last two cluster groups (10 to 21 and 22 to 37), which are depleted in the mature leaf relative to the three younger zones, show an enrichment of proteins assigned to the RNA, DNA, and signaling bins.
Figure 3.
Enrichment of Proteins and Phosphoproteins in Distinct Zones of Developing Maize Leaves.
(A) Heat map of unmodified proteome NSAF values from Supplemental Data Set 2 online scaled from 0 to 1 and ordered by hierarchical clustering. Four main groups of clusters were identified, as marked. Each protein was assigned to a MapMan bin, and classes of proteins that were overrepresented in each cluster were calculated (see text and Supplemental Table 5 online).
(B) Percentage of proteins assigned to selected MapMan bins differs between proteins enriched specifically in zones 1, 2, or 3. A complete list of all bins in all four leaf zones is in Supplemental Table 6 online.
(C) Heat map of phosphoprotein nSPC values from Supplemental Data Set 4 online values, scaled from 0 to 1, and ordered by hierarchical clustering.
The last cluster group, clusters 22 to 37, is of greatest interest from a developmental standpoint because this group distinguishes between the three developing zones of the leaf. Figure 3A shows that while many of these proteins are enriched in two of the three zones, surprisingly few are enriched specifically in a single zone and these are not easily identifiable from the cluster analysis. To identify such proteins, an enrichment factor (EF) was calculated (see Methods). Each protein has four EFs: EFZone1, EFZone2, EFZone3, and EFMature, which are listed in Supplemental Data Set 2 online. Proteins with an EF > 2 with a q-value of <0.05 are considered “enriched” and were also analyzed to identify overrepresented MapMan bins (see Supplemental Table 6 online). More than 1000 proteins had an EFMature > 2; overrepresented bins among these proteins were similar to those in clusters 1 to 8. EFs identified 332, 81, and 338 proteins uniquely enriched within zones 1, 2, and 3, respectively. Analysis of overrepresented MapMan bins point to unique processes within these three zones (Figure 3B). Proteins enriched in zone 1 are overrepresented within MapMan Bins of signaling, RNA, and hormone, which is consistent with cells in this zone undergoing active division. Zone 2, where cells are undergoing asymmetric division and cellular differentiation, has surprisingly few enriched proteins. Within this zone, RNA and nucleotide metabolism bins are overrepresented, suggesting active mRNA synthesis during differentiation. Within the RNA bin, subclasses of enriched proteins differed between zone 1 and zone 2; for example, members of the Argonaute family and the Squamosa transcription factor family are enriched in zone 1, while members of the Alfin-like, TEOSINTE/CYCLOIDEA/PCP, and GROWTH REGULATING FACTOR transcription factor families are overrepresented in zone 2. Finally, zone 3 contained 338 proteins with an EFZone3 > 2. This includes an overrepresentation of proteins involved in cell wall biosynthesis and lipid biosynthesis, which is likely related to the high rates of cell expansion occurring in this zone.
A High Proportion of Phosphoproteins Show Zone-Specific Enrichment
A comparison of the distribution of MapMan Bin assignments for the 2244 proteins in the phosphoprotein data set to the 8005 proteins in the protein data set reveals that the types of proteins identified in the phosphoproteome differ from the unmodified proteome (see Supplemental Table 7 online). This suggests that particular functional classes of proteins are preferentially phosphorylated. Within the phosphoprotein data set there is an overrepresentation of proteins that fall within the RNA, DNA, signaling, and cell bins and a corresponding decrease in several bins associated with different types of metabolism (see Supplemental Table 7 online). This is consistent with the idea that proteins that play a regulatory role, such as transcription factors and kinases, are more often subject to posttranslational regulation via phosphorylation than are metabolic enzymes.
To observe patterns of protein phosphorylation across developing leaves, a clustering analysis like that performed for the proteome was performed on the phosphorylated protein data set. Clustering of scaled phosphoprotein nSPC values resulted in the same four major groups of clusters seen previously (mature leaf-enriched, unchanging phosphorylation across all zones, mature leaf-depleted, and mature-leaf depleted with zone-specific phosphorylation; Figure 3C). However, relative to the unmodified proteome, far more phosphoproteins were specifically enriched in one of the three young leaf zones (cf. Figures 3A to 3C). MapMan bin overrepresentation analysis was also extended to the four phosphoprotein cluster groups. Generally, the overrepresented bins were similar between the phosphoproteome and the proteome for three of the four cluster groups. The exception was the cluster of proteins with unchanging phosphorylation. Rather than housekeeping proteins, phosphoproteins within the RNA bin were overrepresented in this cluster relative to the phosphoproteome, especially those involved in RNA processing as opposed to those involved in regulation of transcription (see Supplemental Table 7 online).
This comparison of the phosphoproteome to the unmodified proteome highlights the importance of phosphorylation during cellular differentiation. While zones 1, 2, and 3 all have a larger proportion of enriched phosphoproteins than proteins, this is especially notable for zone 2, which has very few enriched proteins. For example, within the NSAF protein data set, there were a total of 104 proteins (1.3% of 8005 identified proteins) with an EFZone2 > 2, and 81 of these (1.0%) had a q-value of <0.05. Comparatively, within the nSPC phosphoprotein data set, there were 364 proteins (16.2% of 2244 phosphoproteins) with an EFZone2 > 2, and 53 of these (2.4%) had a q-value of <0.05. This result is significant for several reasons. First, it suggests that the low number of zone 2–enriched proteins found is not merely due to overlap of developmental processes, or lack of resolution, between zone 2 and its two flanking zones. Second, it suggests that the differentiation processes taking place in this zone are regulated more extensively by protein phosphorylation than by upregulation/downregulation of protein levels. Third, it emphasizes the importance of posttranslational modifications in defining the proteotype and the corresponding developmental phenotypes.
Motif Analysis of Phosphorylated Proteins Suggests Transition in Kinase Activities as the Leaf Ages
Given that protein phosphorylation appears to be an important feature of the proteotype, motifs associated with localized phosphorylation sites were identified using Motif-X (Schwartz and Gygi, 2005). Ser, Thr, and Tyr phosphorylation comprised 85.7, 13.5, and 0.8% of all localized phosphorylation sites observed, respectively. In total, 55 phosphoserine motifs, eight phosphothreonine motifs, and a single phosphotyrosine motif were identified (see Supplemental Data Set 5 online). This phosphotyrosine motif has been previously shown in mammalian cells to be a site of autophosphorylation resulting in activation in glycogen synthase kinase 3 and dual specificity Tyr kinase (Lochhead et al., 2005, 2006). Since Ser phosphorylation sites comprise the majority of the observed phosphorylation sites, further analysis focused on these motifs only. Most of the phosphoserine motifs could be grouped into three major classes: Pro-directed, basic, and acidic (see Supplemental Data Set 5 online). Although only 10 of the 55 phosphoserine motifs fall into the Pro-directed class, over 40% of the identified Ser phosphopeptides possess this motif (Table 1). Acidic motifs account for 29 of the 55 identified phosphoserine motifs and nearly 40% of Ser phosphopeptides possess this motif. Finally, basic phospho-motifs were represented by seven motifs and were represented in nearly 8% of the Ser phosphopeptides.
Table 1. Three Classes of Phosphoserine Motifs Identified and Their Distributions across Different Tissues.
| Motif Class | Motif Pattern | Total No. of Motifs in Class | Proportion of Phosphopeptides with Motif |
||||
|---|---|---|---|---|---|---|---|
| All | Zone 1 Enriched | Zone 2 Enriched | Zone 3 Enriched | Mature Leaf Enriched | |||
| Pro-directed | sP | 10 | 43.2% | 50.2% (1.2E-5) | 47.3% (9.0E-6) | 53.2% (5.2E-11) | 33.8% (6.4E-11) |
| Acidic | [D/E]s, s[D/E],sX[D/E], sXX[D/E] | 29 | 39.6% | 35.9% (0.003) | 39.0% (0.020) | 32.2% (2.6E-7) | 45.1% (6.2E-5) |
| Basic | RXXs | 7 | 7.9% | 6.5% (0.022) | 6.6% (0.003) | 14.2% (0.053) | 15.5% (0.028) |
The 55 Ser phospho-motifs identified by Motif-X were classified, and the three most common motifs and their class name, motif pattern, and total number of motifs are listed in the first three columns. The motifs were mapped back to all localized Ser phosphopeptides, and the percentage of either all of these peptides or peptides with an EF > 2 in a particular motif is shown. The P value from a hypergeometric test is shown in parentheses below the percentage. A complete list of all motifs is in Supplemental Data Set 5 online.
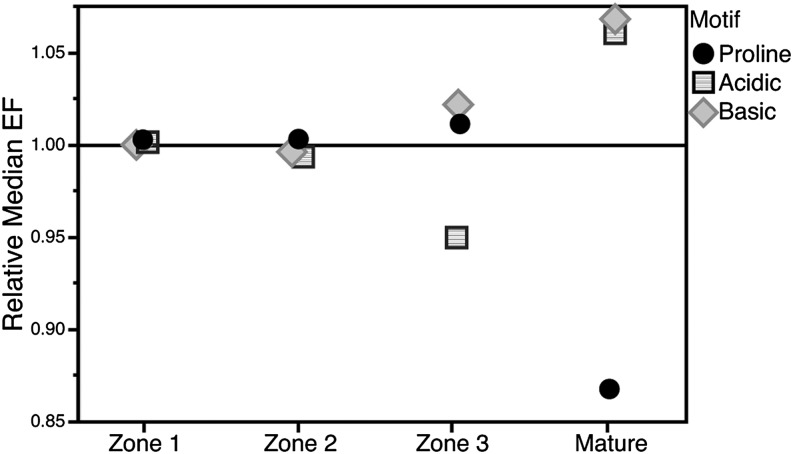
Since clustering analysis suggested that phosphorylation differences define distinct zones of developing leaves, we asked whether there are differences in the distribution of phospho-motifs in each zone. This was done in two ways: a tissue-based frequency measure, which compares the percentage of peptides with a given motif that are enriched in a particular tissue (Table 1), or a motif-based quantitative measure, which calculates the median EFs of peptides possessing a particular motif (Figure 4). Both analyses reveal that mature leaves differ from the other three tissues, with fewer Pro-directed motifs and more acidic and basic motifs. Conversely, zone 3 has fewer acidic motifs in favor of more Pro-directed and basic motifs. Both analyses suggest a gradual increase in the basic motifs as the leaves age, with many fewer Pro-directed motifs in mature leaves. This implies that different kinase families are active in regulating the processes occurring in leaves as they age.
Figure 4.
Relative Median EFs in Four Leaf Tissues Reveals Different Distribution of Motifs in the Leaf.
All phosphopeptides with either a Pro-directed, acidic, or basic motif were identified. For each motif, the median EF was calculated for each tissue. The median EF was then divided by the median EF calculated for all localized phosphopeptides identified to get the relative median EF. Relative median EFs >1 suggest there are more motifs of a particular type in this tissue relative to all tissues and <1 suggest fewer motifs.
Cell Wall–Related Proteins Show Zone-Specific Enrichment or Phosphorylation and Previously Unidentified Phosphorylation Sites
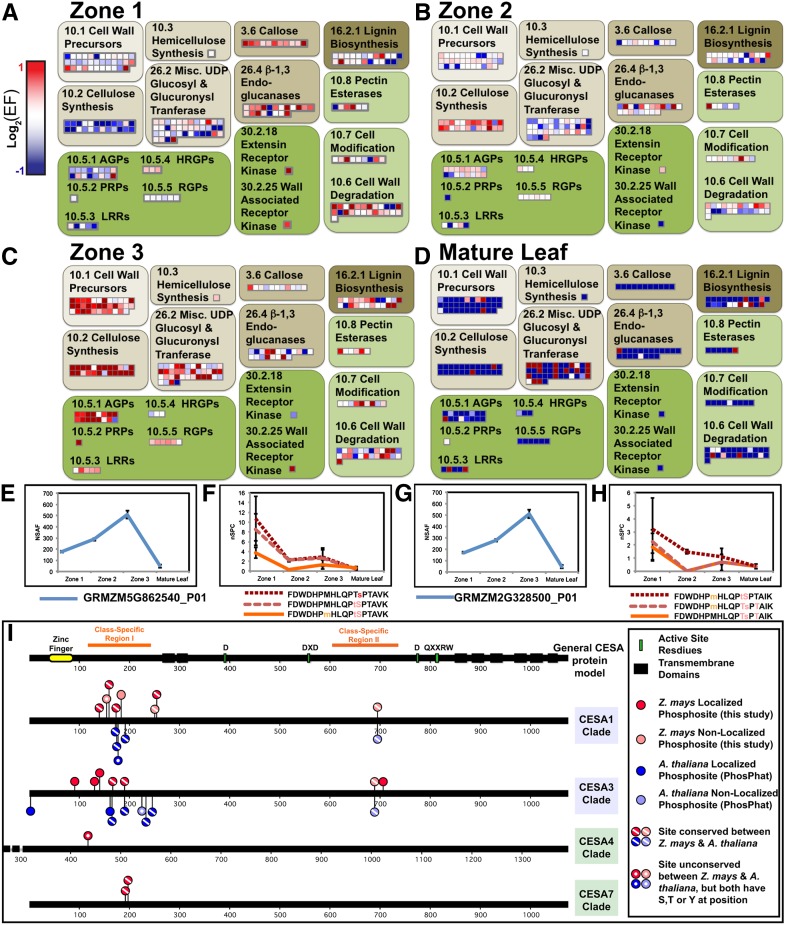
To gain further insight into maize leaf growth and to illustrate how the data sets presented here can be exploited, we performed a focused analysis of cell wall biosynthesis, which is crucial for cell expansion. Using MapMan, heat maps were created to illustrate the relative enrichment or depletion of a particular protein (Figure 5). Each square represents a protein; red squares represent enriched proteins, while blue squares represent depleted proteins. Similar to the EF analysis presented earlier (Figure 3B), this analysis shows that zone 3 (expansion zone) shows the highest relative enrichment of most cell wall–related proteins, consistent with the need for active biosynthesis of wall carbohydrates to sustain rapid cell expansion in zone 3 (compare Figure 5C with 5A, 5B, and 5D). However, there are two notable exceptions. First, most enzymes involved in callose synthesis and degradation (bins 3.6 and 26.4) are enriched in zone 1 (compare Figure 5A to 5B to 5D). Since zone 1 contains a high frequency of dividing cells, enrichment of these callose-synthesizing and -degrading enzymes likely reflects synthesis of callose in newly formed cell plates and its subsequent removal as cell plates mature. The other notable exception is bin 26.2 (miscellaneous UDP-glucosyl and -glucuronosyl transferases), containing many proteins that are enriched in mature leaf tissue (Figure 5D). UDP-glucosyl and -glucuronosyl transferases either produce cell wall polymers, such as pectin and hemicellulose, or add a glycoside to other compounds, such as hormones or secondary metabolites. Many of the closest Arabidopsis homologs of the maize proteins in this bin with EFMature > 2 are predicted to add sugar molecules to compounds such as indole-3-acetic acid, zeatin, or secondary metabolites. Therefore, comparative expression analysis of glycosyltransferases can aid in subclassifying these enzymes and coupled with homology data help predict what types of molecules these enzymes add sugars to.
Figure 5.
Analysis of Relative Abundance and Phosphorylation Status of Cell Wall–Related Proteins in Growing Leaves.
(A) to (D) The log2 of EFs for all identified cell wall–related proteins heat-mapped using MapMan in zone 1 (A), zone 2 (B), zone 3 (C), and mature leaves (D). Each square represents a single protein. Enriched proteins with an EF >2 are red squares, unchanged proteins are white, and depleted proteins are blue squares.
(E) to (H) Examples of two UDPGDH isoforms enriched in zone 3 but showing higher phosphorylation in zone 1. The protein NSAF and phosphopeptide nSPCs for the first isoform are shown in (E) and (F), respectively, while the NSAF and nSPCs for the second isoform are shown in (G) and (H).
(I) Phosphorylation of CESA proteins. Localized (dark red and dark blue) and nonlocalized (light red and light blue) phosphorylation sites mapped to proteins alignments of maize and Arabidopsis CESA proteins. Maize phosphorylation sites from this study are shown in red, above the line. Arabidopsis sites are shown in blue, below the line. Arabidopsis sites are from the PhosPhat 4.0 database (http://phosphat.mpimp-golm.mpg.de/) and are current as of February, 2013. The error bars indicate standard errors across biological replicates in Figures 5 to 7.
Because our global analysis indicated that phosphorylation is important in defining proteotype, we identified cell wall proteins whose phosphorylation patterns differed from protein expression patterns. Examples of this include two proteins belonging to the cell wall precursors bin 10.1 that encode isoforms of UDP-Glc dehydrogenase (UDPGDH), the key enzyme that converts Glc to glucuronic acid, which in subsequent enzymatic steps can be converted to Xyl, apiose, and/or Ara (Carpita and McCann, 2000). These two protein models each have uniquely identifying peptides as well as share peptides with one another but do not share peptides with any other protein in our database (see Supplemental Data Set 6A online). These proteins are enriched in zone 3 (Figures 5E and 5G) but their phosphopeptides are enriched in zone 1 (Figures 5F and 5H). Thus, proportionally fewer of these proteins are phosphorylated as cells transition from proliferative cell division to differentiation and expansion. Our findings suggest that phosphoregulation of precursor biosynthesis is important for the transition from proliferative cell division to cellular differentiation.
To further characterize cell wall proteins, phosphorylation sites on cellulose synthase (CESA) family proteins were compared with those previously identified in Arabidopsis. Phosphorylation of CESA has been shown to regulate its velocity in the membrane and thus is important for the regulation of cell wall biogenesis (Chen et al., 2010; Bischoff et al., 2011). CESA isoforms are associated with either primary or secondary wall biosynthesis. A phylogenetic tree of all CESA genes in Arabidopsis and maize was created (see Supplemental Figure 4A online), and consensus sequences were constructed. Phosphorylation sites identified in this study (shown in red) as well as all phosphorylation sites on the Arabidopsis CESAs listed in the PhosPhat database (Heazlewood et al., 2008; Durek et al., 2010) are marked on these consensus sequences in Figure 5I. This study identified 15 phosphorylation sites in the primary cell wall enzymes CESA1 and CESA3 compared with 13 identified in the PhosPhat database, which are derived from nine prior studies. All peptides mapping to CESA1 and CESA3 isoforms are in Supplemental Data Set 6 online. Phosphorylated sites include some that were observed in both Arabidopsis and maize; some that were observed in only one species, but potentially could be conserved between maize and Arabidopsis (i.e., there is a corresponding Ser, Thr, or Tyr in the orthologous protein); and some that are species specific because there is no orthologous site in the other species (Figure 5I). In addition to identifying previously undescribed phosphorylation sites in CESA1 and CESA3, we also identified three phosphorylation sites absent from the PhosPhat data set that are in two secondary cell wall CESA proteins (CESA4 and CESA7). Identification of potential sites of regulation unique to secondary cell wall biosynthesis could be of potential use in engineering the rate of secondary wall biosynthesis. Comparison of CESA phosphorylation sites identified in our study to those previously identified demonstrates the broad coverage of phosphorylation sites in our data set and the ability to use the data to identify phosphorylation sites that are potentially conserved in other plants but have not been identified in those plants.
Auxin-Related Proteins Are Enriched in Zone 1 and Show Complex Phosphorylation Patterns
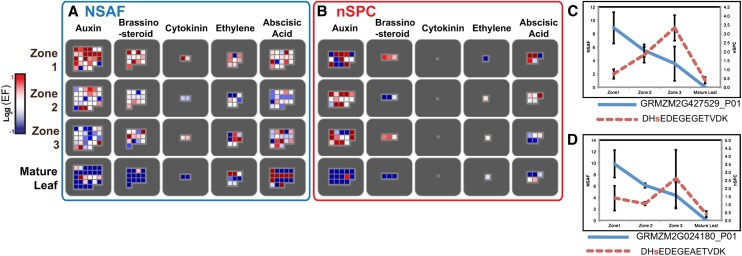
Because hormones are integral to plant growth and development, we analyzed the abundance and phosphorylation status of proteins involved in hormone biosynthesis, degradation, and response (Figure 6A). As described earlier for cell wall–related proteins, heat maps were created illustrating the relative abundance of proteins (Figure 6A) and phosphoproteins (Figure 6B). As shown in Figure 6A, auxin-related proteins were the most numerous within our data set, followed by proteins associated with brassinosteroids. These proteins, and the smaller number with cytokinin- and ethylene-related functions that we identified, show the highest relative abundance in zone 1, gradually decrease through zones 2 and 3, and are absent or expressed at the lowest levels in mature leaves. By contrast, several proteins associated with abscisic acid, especially abscisic acid biosynthesis, show mature leaf enrichment.
Figure 6.
Enrichment of Hormone-Related Proteins and Phosphoproteins.
(A) and (B) Heat maps of log2(EF) for protein NSAFs (A) and phosphoprotein nSPCs (B) for all identified proteins annotated as hormone-related in MapMan. See Figure 5 legend for explanation of colored squares.
(C) and (D) Relative NSAF (blue lines) and phosphopeptide nSPC values (red lines) for two putative F-box proteins. For each graph, the identified phosophopeptide is written in the legend with the phosphorylated Ser in red lowercase letters.
We also identified several examples of hormone-related proteins whose phosphopeptides are more abundant in zone 2 or 3 despite having higher protein abundance in zone 1. All peptides for these proteins are listed in Supplemental Data Set 6 online, and these peptides do not map to other proteins encoded in the maize genome. Two of these proteins share 60% identity with Arabidopsis Auxin F-Box (AFB) 4 and 5. AFBs are auxin receptors that, in the presence of auxin, no longer facilitate the degradation of Aux/indole-3-acetic acid transcription factors, thereby permitting transcription of genes involved in auxin response (Parry and Estelle, 2006). The relative abundance of these two putative maize F-box proteins gradually decreases through the transition from zones 1 to 3, and they are not detected in mature leaves (Figures 6C and 6D). Reciprocally, the abundance of a phosphopeptide present in both proteins shows the opposite pattern increases from zone 1 to zone 3 (Figures 6C and 6D). This negative correlation suggests that the abundance of these two F-box proteins may be negatively regulated by phosphorylation at this site.
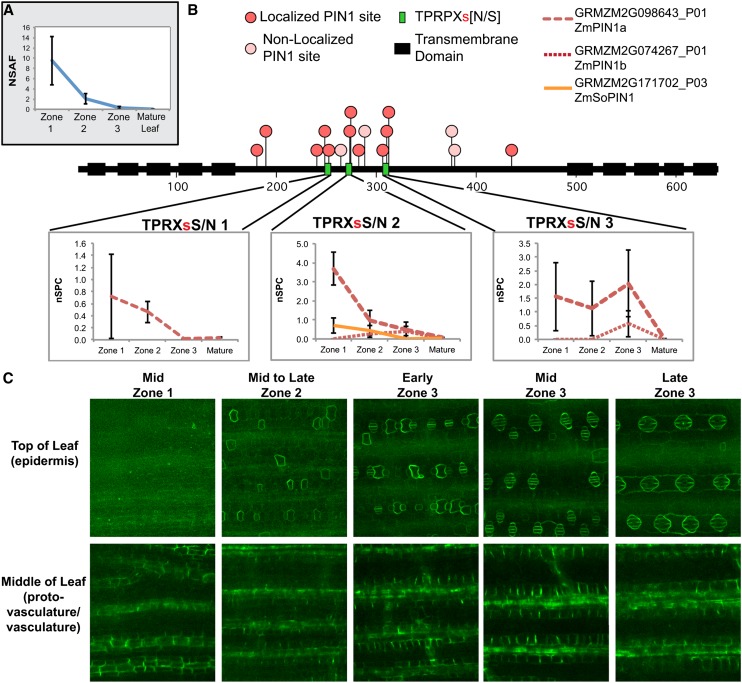
Auxin transport is a fundamental and highly studied aspect of auxin biology, and recent findings indicate that phosphorylation of PINFORMED (PIN) auxin transporters is crucial for their correct intracellular localization (Huang et al., 2010; Zhang et al., 2010; Dhonukshe et al., 2010; Ganguly et al., 2012). Therefore, we examined the relative abundance of peptides and phosphopeptides mapping to PIN1 auxin transporters. Maize contains three PIN1 orthologs (PIN1a, PIN1b, and PIN1c; Carraro et al., 2006) as well as a gene in an adjacent clade, not found in Arabidopsis, named SISTER OF PIN1 (SoPIN1; D. O’Connor, personal communication). Due to their high sequence identity, the three maize PIN1s are assigned to a single protein group with PIN1a as the group leader. PIN1s are most abundant in zone 1 (Figure 7A). Sixteen phosphorylation sites we identified in PIN1a, b, c, and/or SoPIN1 that are also conserved in AtPIN1 are marked on the schematic in Figure 7B. Not marked are two sites identified only in SoPIN1 that are not conserved in any of the other isoforms of PIN1. The conserved phosphorylated residues we identified include Ser residues within three TPRXsS/N motifs in the hydrophilic loop (HL-PINS); phosphorylation of these motifs by PID/WAG kinases is important for intracellular localization of PIN1 (Dhonukshe et al., 2010; Huang et al., 2010; Zhang et al., 2010; Ganguly et al., 2012). The abundance of peptides phosphorylated at the first two TPRXxS/N motifs (Figure 7B) and PIN1 peptides phosphorylated outside the TPRXxS/N motifs (see Supplemental Data Set 4 online) correlates well with total PIN1 protein abundance. We noticed that several peptides mapping to the third TPRXsS/N motif are slightly enriched in zone 3. Since a phosphosite may be represented in multiple peptides due to variations, such as additional phosphosites in the peptide, missed cleavages, or oxidized Met residues, we summed together the nSPC of all peptides containing the phosphorylated Ser from each of the TPRXsS/N sites (Figure 7B). These peptides are listed in Supplemental Data Set 6 online.
Figure 7.
PIN1 Expression Is Highest in Zone 1, but a Ser Residue Is Differentially Phosphorylated in Zone 3.
(A) NSAF scores of PIN1 indicate highest expression is in zone 1. The NSAF score shown is for the group leader GRMZM2G098643_P01 but may also include peptides from PIN1b, and PIN1c.
(B) PIN phosphorylation sites marked by red dots in a consensus sequence of PIN1a, PIN1b, PIN1c, and SoPIN1. All phosphopeptides containing the phosphorylated Ser highlighted in red from the TPRXsS/N were summed together for each of the four PIN proteins and are graphed below their respective sites. The peptides and their SPCs are individually listed in Supplemental Data Set 6 online.
(C) Expression of PIN1a-YFP is strongest in the vasculature in all three zones (bottom panels) but also appears in the epidermis in zone 3 where it changes cell-type expression and localization (top panels).
The foregoing analysis suggests differential phosphorylation of PIN1 in zone 3, predicting possible differences in PIN1 intracellular localization in this zone relative to others. Little is known about the behavior of PIN1 in developing maize leaves, so using plants expressing PIN1a-YFP (for yellow fluorescent protein) driven by the native promoter (Gallavotti et al., 2008), we analyzed PIN1 expression and localization in the three growing zones of the leaf. In all three zones, the most conspicuous expression was in the developing vasculature, and it is likely the majority of the PIN1 expression comes from vasculature expression (Figure 7C, bottom panels). However, PIN1a-YFP was also observed in the epidermis and showed dynamic, zone-specific expression and localization patterns (Figure 7C, top panels). In zone 1 and the basal portion of zone 2, PIN1a-YFP was not observed in the epidermis. Toward the end of zone 2, PIN1a-YFP started to appear in the epidermis, in cells separating guard mother cells (interstomatal cells). In zone 3, expression shifted from interstomatal cells to guard mother cells, persisting in guard cells formed by division of guard mother cells. In addition, uniform PIN1 was observed in newly formed subsidiary cells (flanking the guard cells) and then conspicuously polarized away from adjacent guard cells. Thus, multiple changes in both epidermal cell-type specificity and subcellular localization of PIN1a occurs as cells transit through zone 3, correlating with an increase in phosphorylation of the third TPRxSS/N site. Thus, phosphorylation of the third TPRxSS/N site might underlie some of these changes in PIN1 expression or localization.
DISCUSSION
The work presented in this study had dual goals: (1) analysis of changes in the maize leaf proteome accompanying the transition from proliferative division to cellular differentiation to expansion during maize leaf development aimed at obtaining insights into processes regulating these developmental transitions; and (2) creation of a community resource that can be mined by others to address additional questions about the maize proteome and leaf development. Proteins and their phosphorylation sites were indexed and quantified in parallel; when coupled, these two data types become especially powerful, permitting changes in phosphorylation state to be distinguished from changes in protein abundance and identifying potential sites of phosphoregulation on a genome-wide scale. We identified protein classes, specific proteins, and phosphorylation sites enriched in different developmental zones of the leaf, focusing primarily on proteins involved in cell wall biosynthesis and hormone biology. This analysis revealed both expected and unexpected features of the proteotypes of each zone. In presenting our analysis, we provided several different examples of how the data can be mined to answer additional questions in the future.
Protein Phosphorylation Status Better Distinguishes Developmental Zones of the Leaf Than Protein Abundance
In this analysis, the growing maize leaf was divided into three zones dominated by sequential events in leaf development: proliferative cell division (zone 1), differentiation (zone 2), and postmitotic cell expansion (zone 3). Comparisons of the proteomes of these three developing leaf zones as well as mature leaves identified proteins enriched in each region; however, the number of enriched proteins, especially in zone 2, was relatively low. Importantly, comparison of the phosphoproteomes of these four leaf regions revealed many differences among them, including zone 2. Therefore, the proteotype of zone 2 can be distinguished from that of zones 1 and 3 primarily by differences in phosphorylation status (and presumably activity) of proteins, rather than the protein complement itself. A parallel proteomic and phosphoproteomic analysis of differentiating human embryonic stem cells similarly found more dynamic changes in the phosphoproteome than in the proteome (Rigbolt et al., 2011). This suggests a common mechanism among differentiating cells wherein regulation of protein activity is more important than the presence or absence of a protein. This may reflect the precise orchestration of timing required for differentiation and development. Having the required proteins present within the cell in a broader window than required and then carefully regulating each player through posttranslational modifications that affect their interactions or activity may allow cells to precisely and efficiently execute differentiation processes.
It is notable that while we found enrichment of many phosphorylation sites within zone 2, there was not an overrepresentation of signaling proteins in this zone. There are examples of specific kinases that showed protein enrichment in this zone, but they were not overrepresented as a group. Indeed, while there was significant enrichment of signaling proteins within zones 1 and 3, the opposite is true of zone 2. This seemingly paradoxical finding might be explained by an increase in kinase activity in zone 2 rather than kinase abundance. We observed a transition from zone 2 to zone 3 in the prevalence of different types of phosphomotifs, which may reflect a transition of kinase activities as the leaf ages. Further studies correlating specific kinase activities with the abundance of kinases and potential targets may help in explaining the phenomena described above. Some inferences can still be drawn, however. For example, kinases targeting Pro-directed motifs include both cyclin-directed and mitogen-activated kinases (Lu et al., 2002); therefore, it is not surprising that motifs of this class are common in zones 1 and 2, since these zones both contain actively dividing cells. The enrichment of signaling proteins in zone 3 may reflect synthesis of new kinases, as it correlates with a transition from Pro to acidic phosphomotif classes.
Parallel Analyses of the Unmodified and Phosphoproteomes Identify Potential Regulatory Sites Important for Maize Leaf Development
Using examples of proteins important for the growth-related processes of cell expansion and hormone biology, we provide specific examples of proteins that are uniquely enriched or differentially phosphorylated in distinct developmental zones of the maize leaf. We further illustrate how comparison of the relative abundance of unmodified peptides versus phosphopeptides from a given protein reveals changes in phosphorylation status that can drive hypotheses regarding function. For example, consistent with the rapid expansion of cells in zone 3, we found that most proteins in our data set with cell wall–related functions were enriched in zone 3 relative to other zones. We also provide clear evidence of a change in phosphorylation status of two UDPGDH isoforms, suggesting that phosphoregulation of these enzymes may mediate a change in the availability of glucuronic acid associated with the transition from zone 1 to zone 2. In maize, mutations in these genes reduce the amount of Ara and Xyl in cell walls without altering cellulose content (Kärkönen et al., 2005), but the impact of the identified phosphorylation sites on enzymatic activity is unclear. UDPGDH mutants of Arabidopsis have a notable change in pectic cell wall polymers accompanied by several developmental defects (Reboul et al., 2011), illustrating the importance of this enzyme in cell wall assembly. Although phosphoregulation of this protein in plants is uncharacterized, phosphorylation (on a different residue to that observed here) of a similar protein in bacteria is required for activity (Mijakovic et al., 2003). Given the importance in UDPGDH in cell wall biosynthesis, modulation of phosphorylation of this protein may be an effective way of engineering plant cell walls. We also compare phosphorylation of CESA proteins in maize and Arabidopsis and identify several phosphorylation sites that are conserved between maize and Arabidopsis.
Analysis of proteins associated with hormone biosynthesis, degradation, and perception reveals that the majority of these proteins are enriched in zone 1. This is consistent with a previously published report indicating that levels of indole-3-acetic acid (auxin), castasterone (BR), and zeatin (cytokinin) are highest in the maize leaf base and gradually decrease in successively older regions of the leaf (Nelissen et al., 2012). Moreover, we identified a phosphosite in a pair of F-box proteins whose abundance is anticorrelated with protein abundance. Given that F-box protein stability is regulated by the proteasome (Stuttmann et al., 2009), phosphorylation at this site may direct these proteins to the proteasome.
We identified 16 phosphorylation sites in PIN1, all of which are conserved between Arabidopsis and maize. The relative abundance of peptides containing most of these phosphosites correlates closely with the overall abundance of PIN1, suggesting no differential phosphorylation across the different leaf zones. However, phosphorylation of a Ser within the third TPRXsS/N motif did not follow this pattern and appears to occur predominantly in zone 3. The zone-specific enrichment of this phosphosite correlated with dynamic changes in epidermal expression and localization of Zm-PIN1. Mutational analysis of At-PIN3 has shown that subcellular polarization of At-PIN3 in root hairs is dependent on the first TPRXsS/N in conjunction with several phosphosites preceding it, but not the second or third TPRXsS/N motif (Ganguly et al., 2012). Given the diversity of PIN1 localization in a broad array of cell types, differential phosphorylation of the three TPRXsS/N sites, in addition to other phosphorylation sites, may be an important aspect of PIN1 regulation. Confirmation of the observed difference in phosphorylation using directed proteomics such as AQUA peptides or selective reaction monitoring (both of which use heavy isotope-labeled synthetic peptides as internal controls for quantification) (Gerber et al., 2003; Lange et al., 2008) coupled with directed mutagenesis will surely reveal the importance of different phosphosites in different biological contexts. Identification of candidate sites using parallel, quantitative analyses of protein and phosphorylation site abundance in different tissues and/or cell types, such as those provided here, can greatly expedite identification of relevant phosphorylation sites.
METHODS
Plant Material
Maize (Zea mays; B73) plants were grown in the greenhouse without supplemental lighting in 8-inch pots for 30 d, when leaf 8 was at least 50 cm long and leaf 10 was emerging from the whorl. Leaf 8 was used for mature leaf analysis. Blade tissue from the ligule to leaf tip, excluding the midrib, was harvested. For zones 1, 2, and 3, the older, outer leaves were removed until the ligule was within 0.5 cm of the base, typically leaf 10. These young, remaining leaves were sectioned into zone 1 (0 to 1.25 cm), zone 2 (1.5 to 2.75 cm), and zone 3 (3.5 to 5.5 cm) and immediately frozen in liquid nitrogen.
Protein Extraction and Preparation
Approximately 2 g of tissue was ground in liquid nitrogen and precipitated using acetone. The protein pellets were then extracted in buffer containing 0.1% SDS, 1 mM EDTA, and 50 mM HEPES buffer, pH 7. Cys residues were reduced and alkylated using 1 mM Tris (2-carboxyethyl)phosphine) (Fisher; AC36383) at 95°C for 5 min followed by 2.5 mM iodoacetamide (Fisher; AC12227) at 37°C in the dark for 15 min. Proteins were digested with trypsin, and digested peptides were purified on a Waters Oasis MCX cartridge. Peptides were eluted with 50% isopropyl alcohol and 400 mM NH4HCO3, pH 9.5, dried in a vacuum concentrator, and resuspended in 1% formic acid (nonmodified peptides) or 3% trifluoroacetic acid (phosphopeptides).
Phosphopeptide enrichment was performed by adding 1% colloidal CeO2 (Sigma-Aldrich; 289744) to the acidified peptide solution at 1:10 (w/w). After brief vortexing, CeO2 with captured phosphopeptides was pelleted and washed with 1% trifluoroacetic acid. Phosphopeptides were eluted using 200 mM (NH4)2HPO4, 2 M NH3.H2O, and 10 mM EDTA, pH 9.5. CeO2 was precipitated by adding 10% formic acid with 100 mM citric acid centrifuging. The supernatant containing phosphopeptides was removed and used for mass spectrometry analysis.
Mass Spectrometry and Peptide Identification
An Agilent 1100 HPLC system (Agilent Technologies) delivered a flow rate of 600 nL min−1 to a three-phase capillary chromatography column through a splitter. Using a custom pressure cell, 5 µm Zorbax SB-C18 (Agilent) was packed into fused silica capillary tubing (250-µm i.d., 360-µm o.d., and 30 cm long) to form the first-dimension reverse phase column (RP1). A 5-cm-long strong cation exchange (SCX) column packed with 5 µm polysulfoethyl was connected to RP1 using a zero dead volume 1-µm filter (Upchurch; M548) attached to the exit of the RP1 column. A fused silica capillary (200-µm i.d., 360-µm o.d., and 20 cm long) packed with 5 µm Zorbax SB-C18 (Agilent) was connected to SCX as the analytical column (RP2). The electrospray tip of the fused silica tubing was pulled to an inner diameter smaller than 1 µm using a laser puller (Sutter P-2000). The peptide mixtures were loaded onto the RP1 column using the custom pressure cell. A new set of columns was used for each HPLC-MS/MS analysis. Peptides were first eluted from the RP1 column to the SCX column using a 0 to 80% acetonitrile gradient for 150 min. For total proteome profiling experiments, peptides were fractionated by the SCX column using a series of 27 step salt gradients (10 mM, 15 mM, 20 mM, 22.5 mM, 25 mM, 27.5 mM, 30 mM, 32.5 mM, 35 mM, 37.5 mM, 40 mM, 42.5 mM, 45 mM, 47.5 mM, 50 mM, 52.5 mM, 55 mM, 57.5 mM, 60 mM, 65 mM, 70 mM, 75 mM, 80 mM, 85 mM, 90 mM, 150 mM, and 1 M ammonium acetate for 20 min each), followed by high-resolution reverse-phase separation using an acetonitrile gradient of 0 to 80% for 120 min. For phosphoproteome profiling experiments, peptides were fractionated by the SCX column using a series of 18 step salt gradients (5 mM, 6 mM, 7 mM, 8 mM, 9 mM, 10 mM, 12 mM, 15 mM, 20 mM, 30 mM, 40 mM, 50 mM, 60 mM, 70 mM, 80 mM, 90 mM, 100 mM, and 1 M ammonium acetate). It took 3 d to finish one full proteome analysis and 2 d to finish one phosphoproteome analysis.
Spectra were acquired on an LTQ Velos linear ion trap tandem mass spectrometer (Thermo Electron) employing automated, data-dependent acquisition. The mass spectrometer was operated in positive ion mode with a source temperature of 250°C. As a final fractionation step, gas phase separation in the ion trap was employed to separate the peptides into three mass classes prior to scanning; the full mass spectrometry scan range was divided into three smaller scan ranges (300 to 800, 800 to 1100, and 1100 to 2000 D) to improve dynamic range. The three smaller scan ranges were done in a single run. Each mass spectrometry scan was followed by vie MS/MS scans of the most intense ions from the parent mass spectrometry scan. A dynamic exclusion of 1 min was used to improve the duty cycle. The LTQ Velos scan rate is ∼5.5 spectra per second. The LTQ Velos used in this study does not have an OrbiTrap analyzer; thus, all data were low mass resolution. A maximum charge state of 3 was used in the Spectrum Mill database search. The charge states of majority MS/MS spectra (>90%) could not be determined. Thus, each spectrum was searched three times as singly, doubly, and triply charged precursors. No high-resolution precursor scan was used.
Database Search
The raw data were extracted and searched using Spectrum Mill v3.03 (Agilent Technologies). MS/MS spectra with a sequence tag length of 1 or less were considered to be poor spectra and were discarded. The sequence tag length is a Spectrum Mill quality filter and is the longest path of amino acids that is represented in the MS/MS spectrum. A minimum sequence tag length of 2 means there are at least three fragment ions with the two mass differences matching two amino acids. The remaining MS/MS spectra were searched against maize B73 RefGen_v2 5a Working Gene Set downloaded from www.maizesequence.org. The enzyme parameter was limited to full tryptic peptides with a maximum miscleavage of 1. All other search parameters were set to Spectrum Mill’s default settings (carbamidomethylation of Cys residues, ±2.5 D for precursor ions, ±0.7 D for fragment ions, and a minimum matched peak intensity of 50%). Ox-Met, n-term pyro-Gln, and phosphorylation on Ser, Thr, or Tyr defined as variable modifications for phosphoproteome data. A maximum of two modifications per peptide was used. A 1:1 concatenated forward-reverse database was constructed to calculate the false discovery rate. The tryptic peptides in the reverse, or decoy, database were compared with the forward database and were shuffled if they matched to any tryptic peptides from the forward database. Peptide cutoff scores were dynamically assigned to each data set to maintain the false discovery rate < 0.1% at the peptide level for unmodified peptides and <1% for the phosphopeptides. Phosphorylation sites were localized to a particular amino acid within a peptide using the variable modification localization score in Agilent’s Spectrum Mill software (Chalkley and Clauser, 2012). Proteins that share common peptides were grouped using principles of parsimony to address the protein database redundancy issue. Thus, proteins within the same group shared the same set or subset of peptides. The database search and group assignment was performed across all replicates simultaneously, ensuring the same group (and therefore group leader) is consistent across all tissues.
Normalization and Filtering
For both the proteome and phosphoproteome, multiple mass spectrometry runs of a single biological replicate were summed together. For proteome analysis, proteins with only one peptide identification or <25 SPCs across all replicates were excluded, as indicated in Supplemental Table 2 online. Any SPC = 0 was substituted for SPC = 0.01 prior to normalization. Biological replicates were normalized using NSAF normalization by dividing by the total SPCs in addition to the protein length (Florens et al., 2006). Since the resulting score is much smaller than 1, all NSAF values were multiplied by a constant (1,000,000) to obtain numbers similar to the observed SPC. For phosphopeptide and phosphoprotein analysis, phosphorylation levels were quantified via spectral counting (Liu et al., 2004; Huttlin et al., 2010; Qiao et al., 2012) and the resulting SPC were multiplied by 10,000. No data filters were applied to phosphopeptide data set, and the phosphoproteins were filtered as indicated in the text and in Supplemental Table 4 online.
Bioinformatics
Correlation coefficients and hierarchical clustering was performed in JMP (SAS). Prior to clustering, NSAF or nSPC were scaled between 0 and 1 for each protein by dividing the NSAF for each biological replicate by the maximum NSAF across all biological replicates that protein. The scaled scores were then averaged and used for hierarchical clustering in JMP using the Ward method (without normalization).
MapMan bins were assigned using the mapping file Zm_B73_5b_FGS_cds_2011, which was downloaded from the MapMan Store (http://mapman.gabipd.org/web/guest/mapmanstore). In cases were more than one bin applied to a single protein, a bin was randomly chosen to minimize bias. Only 128 of 8005 proteins had multiple annotations, most of which were in the same parent bin or in functionally related bins; hence, discarding these bin assignments results in minimal information loss while avoiding double counting of proteins in closely related or identical parent bins. Since this mapping file only contained proteins from the filtered gene set of the maize 5b genome release, those proteins that we identified that were exclusively in the working genome set were assigned a bin number of 99.
Functional overrepresentation of MapMan bins in cluster groups and EF groups was tested using the hypogeometric test in Excel, where the distribution of MapMan bins from either the 8005 identified NSAF proteins or the 2244 phosphoproteins was used as a reference.
To determine statistical significance of the null hypothesis that all NSAF (or nSPC) values for a given protein are the same across all leaf tissues, the linear regression formula Y= βMature + X1βZone1 + X2βZone2 + X3βZone3 was used. The log2 of the NSAF or nSPC was used for the Y variables. The P values derived from the resulting F-statistic were corrected using the Benjamini and Hochberg multiple testing correction using a threshold of 0.05 to get a q-value.
EFs for each protein were calculated by taking the average NSAFs for one leaf zone and dividing by the median of the average NSAFs for that protein. EFs for phosphopeptides and phosphoproteins were similarly calculated.
Graphs represent measured NSAFs or NSPCs mean values across biological replicates, and the error bars are standard errors.
Motif Analysis
Overrepresented motifs from localized phosphopeptides were identified using Motif-X (Schwartz and Gygi, 2005). Localized phosphopeptides were mapped back to the maize proteome to identify the surrounding amino acids. Phosphopeptides with two phosphorylation sites were broken into two separate entries. Ser, Thr, and Tyr phosphorylation sites were analyzed separately. Motif-X default settings of width = 13, occurrence = 20, and significance = 0.000001 were used for Ser and Thr. For Tyr, less stringent settings of width = 13, occurrence = 3, and significance = 0.0005 were used since only 49 peptides contained localized phosphotyrosine motifs. A background file of the protein sequences from all uniquely identified group leaders from the unmodified proteome and phosphoproteome (13,303 total) was created and used, since the entire maize proteome exceeds the 10-MB limit.
Phylogenetic Analysis
Alignment and phylogenetic analysis of CESA proteins was performed using MEGA (Tamura et al., 2011). Protein sequences were aligned with MUSCLE and then hand-edited. The following default settings were used for the alignment: open gap penalty 10, extend gap penalty 0.2, and hydrophobicity multiplier 1.2. The alignment is available in Supplemental Table 8 online as a fasta protein document. The phylogeny was built using the maximum parsimony method, resulting in a bootstrap consensus tree inferred from 1000 iterations, and the bootstrap values are shown next to the branches. The Subtree-Pruning-Regenerating method was used. Sites with less than 95% coverage were eliminated from the analysis.
Protein Gel Blotting
Proteins were extracted for protein gel blotting by grinding in liquid nitrogen followed by homogenization in extraction buffer (100 mM HEPES-KOH, pH 7.5, 5% glycerol, 330 mM Suc, 0.5% PVP, 15 mM EGTA, 5 mM EDTA, 50 mM Na4P2O7-10H2O, 25 mM NaF, 3 mM DTT, 1 mM PMSF, 10 μM leupeptin A hydrochloride, and 1 nM calyculin A). Insoluble debris was removed by centrifugation at 10,000g, and the soluble extracts were measured using Bio-Rad protein assay. For SDS-PAGE, 30 μg of protein was boiled in SDS loading buffer with 100 mM DTT for each lane and separated on AnykD TGX polyacrylamide gels (Bio-Rad). Proteins were transferred to Immobilon-P (Millipore) membranes for immunodetection. Protein detection using primary antibodies was as follows: mouse anti-α-tubulin was used at 1/4000 dilution (Sigma-Aldrich T6074); rabbit anti-PAN1 was used at 8 μg/mL (Cartwright et al., 2009); rabbit anti-PAN2 was used at 2 μg/mL (Zhang et al., 2012); rabbit antivacuolar ATPase was used at 1/800 (Agrisera ASO7 213); the maize homolog of Nap1 was a custom rabbit antibody raised and affinity purified against the peptide CRMAKSGRTKQEADLE and was used at 12 μg/mL; and anti-ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit was used at 1/12,000 (Agrisera AS01 017). Antibody detection was done using either anti-rabbit AP-conjugated (Promega), anti-mouse AP-conjugated (Promega), or anti-hen horseradish peroxidase–conjugated (Agrisera) secondary antibodies, as appropriate.
Microscopy
All microscope images were taken using a custom-assembled spinning disk confocal microscope system described previously (Walker et al., 2007). Microscope images were processed using ImageJ and Adobe Photoshop, applying only linear adjustments to pixel values. In the case of PIN1-YFP images, images from different regions were processed identically. For propidium iodide staining, leaf segments from each of the four regions described above were harvested and fixed in formaldehyde/alcohol/acetic acid. After fixation, leaves were stained for 10 min in 1 mg/mL propidium iodide, rinsed, mounted in water, and imaged. PIN1-YFP plants were a gift from Andrea Galvotti (Gallavotti et al., 2008).
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL libraries under the following accession numbers: for maize CESA1 clade proteins, GRMZM2G104092, GRMZM2G039454, GRMZM2G112336, and GRMZM2G027723; CESA3 clade proteins, GRMZM2G150404, GRMZM2G111642, GRMZM2G424832, and GRMZM2G018241; CESA4 clade proteins, GRMZM2G445905; CESA8 clade proteins, GRMZM2G055795 and GRMZM2G037413; UDPGH, GRMZM5G862540 and GRMZM2G328500; PIN1a, GRMZM2G098643; PIN1b, GRMZM2G074267; PIN1c, GRMZM2G149184; SoPIN1, GRMZM171702. For Arabidopsis: CESA1, AT4G32410; CESA2, AT4g39350; CESA3, AT5g05170; CESA4, AT5g44030; CESA5, AT5g09870; CESA6, AT5g64740; CESA7, AT5g17420; CESA8, AT4g18780; CESA9, AT2g21770; and CESA10, At2g25540.
Supplemental Data
The following materials are available in the online version of this article. All supplemental data for this article are deposited in the DRYAD repository: http://dx.doi.org/10.5061/dryad.9qv2k. The spectral files are available for FTP download from ftp://maizeproteome.ucsd.edu/leaf/.
Supplemental Figure 1. Maize Leaf Tissues Used for Proteomic Analyses.
Supplemental Figure 2. Relative Protein Abundance Measured by NSAF Correlates with Relative Protein Abundance Measured by Protein Gel Blotting.
Supplemental Figure 3. Comparison of Proteins Identified by Majeran et al. (2010) to Proteins Identified in This Study.
Supplemental Figure 4. Phylogenetic Tree and Expression of Primary and Secondary Cell Wall Cellulose Synthases.
Supplemental Table 1. Total Number of Spectra Collected.
Supplemental Table 2. Number of Unmodified Proteins Identified in Total and in Each Leaf Tissue before and after Filtering for Unmodified Proteome Analysis.
Supplemental Table 3. Number of Phosphopeptides with Localized and Nonlocalized Phosphorylation Sites Identified in Total and in Each Tissue.
Supplemental Table 4. Number of Identified Phosphoproteins in Total and in Each Tissue before and after Filtering.
Supplemental Table 5. Overrepresentation of MapMan Bins among Proteins Belonging to the Unmodified Proteome Cluster Groups Illustrated in Figure 3A.
Supplemental Table 6. Overrepresentation of MapMan Bins among Proteins with an EF > 2.
Supplemental Table 7. Overrepresentation of MapMan Bins among Proteins Belonging to the Phosphoproteome Cluster Groups Illustrated in Figure 3C.
Supplemental Table 8. MUSCLE Alignment in Fasta Text Format of the 29 CESA Proteins from Maize and Arabidopsis Shown in Supplemental Figure 4 Online.
Supplemental Data Set 1. Unfiltered, Unnormalized Data for Protein Identifications.
Supplemental Data Set 2. 8005 Proteins and Corresponding NSAF Values.
Supplemental Data Set 3. Normalized Phosphopeptide nSPCs.
Supplemental Data Set 4. Phosphoprotein Normalized Spectral Counts.
Supplemental Data Set 5. Motifs Identified via Motif-X Surrounding Localized Phospho-Ser, -Thr, and -Tyr Residues.
Supplemental Data Set 6. Individual Peptides and Phosphopeptides Used in This Study.
Acknowledgments
This work was supported by National Science Foundation Grant DBI-0924023. We thank Andrea Galvotti for the PIN1a-YFP transgenic lines, Ah Young Lee for help in tissue harvesting, Jesse Coull for advice on statistical analysis, Josh Osborn for loading mass spectrometry samples, and Carolyn Rasmussen, Justin Walley, and Eric Bennett for helpful comments.
AUTHOR CONTRIBUTIONS
M.R.F., Z.S., S.P.B., and L.G.S. designed the research. M.R.F. and Z.S. performed research. F.R.B. helped with analyzing different tools for localizing phosphorylation sites. M.R.F. and Z.S. analyzed data. M.R.F. wrote the article with input from the other authors.
Glossary
- MS/MS
tandem mass spectrometry
- NSAF
normalized spectral abundance factor
- SPC
spectral count
- nSPC
normalized spectral count
- EF
enrichment factor
- UDPGDH
UDP-Glc dehydrogenase
- SCX
strong cation exchange
References
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Bischoff V., Desprez T., Mouille G., Vernhettes S., Gonneau M., Höfte H. (2011). Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr. Biol. 21: 1822–1827 [DOI] [PubMed] [Google Scholar]
- Carpita, N., and McCann, M. (2000). The Cell Wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Waldorf, MD: American Society of Plant Physiologists), pp. 52–109. [Google Scholar]
- Carraro N., Forestan C., Canova S., Traas J., Varotto S. (2006). ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol. 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright H.N., Humphries J.A., Smith L.G. (2009). PAN1: A receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323: 649–651 [DOI] [PubMed] [Google Scholar]
- Chalkley R.J., Clauser K.R. (2012). Modification site localization scoring: Strategies and performance. Mol. Cell. Proteomics 11: 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D.W., Somerville C.R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Durek P., Schmidt R., Heazlewood J.L., Jones A., MacLean D., Nagel A., Kersten B., Schulze W.X. (2010). PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38 (Database issue): D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L., Carozza M.J., Swanson S.K., Fournier M., Coleman M.K., Workman J.L., Washburn M.P. (2006). Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Yang Y., Schmidt R.J., Jackson D. (2008). The relationship between auxin transport and maize branching. Plant Physiol. 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Lee S.-H., Cho H.-T. (2012). Functional identification of the phosphorylation sites of Arabidopsis PIN-FORMED3 for its subcellular localization and biological role. Plant J. 71: 810–823 [DOI] [PubMed] [Google Scholar]
- Gerber S.A., Rush J., Stemman O., Kirschner M.W., Gygi S.P. (2003). Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA 100: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi S.P., Rochon Y., Franza B.R., Aebersold R. (1999). Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W.X. (2008). PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36 (Database issue): D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkönen A., Murigneux A., Martinant J.P., Pepey E., Tatout C., Dudley B.J., Fry S.C. (2005). UDP-glucose dehydrogenases of maize: A role in cell wall pentose biosynthesis. Biochem. J. 391: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V., Picotti P., Domon B., Aebersold R. (2008). Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 4: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liu H., Sadygov R.G., Yates J.R., III (2004). A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Lochhead P.A., Kinstrie R., Sibbet G., Rawjee T., Morrice N., Cleghon V. (2006). A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Mol. Cell 24: 627–633 [DOI] [PubMed] [Google Scholar]
- Lochhead P.A., Sibbet G., Morrice N., Cleghon V. (2005). Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121: 925–936 [DOI] [PubMed] [Google Scholar]
- Lu K.P., Liou Y.-C., Zhou X.Z. (2002). Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 12: 164–172 [DOI] [PubMed] [Google Scholar]
- Majeran W., Friso G., Ponnala L., Connolly B., Huang M., Reidel E., Zhang C., Asakura Y., Bhuiyan N.H., Sun Q., Turgeon R., van Wijk K.J. (2010). Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I., Poncet S., Boël G., Mazé A., Gillet S., Jamet E., Decottignies P., Grangeasse C., Doublet P., Le Maréchal P., Deutscher J. (2003). Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 22: 4709–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Sugiyama N., Ishihama Y., Shirasu K. (2012). Shotguns in the front line: Phosphoproteomics in plants. Plant Cell Physiol. 53: 118–124 [DOI] [PubMed] [Google Scholar]
- Nelissen H., Rymen B., Jikumaru Y., Demuynck K., Van Lijsebettens M., Kamiya Y., Inzé D., Beemster G.T. (2012). A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 22: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Parry G., Estelle M. (2006). Auxin receptors: A new role for F-box proteins. Curr. Opin. Cell Biol. 18: 152–156 [DOI] [PubMed] [Google Scholar]
- Piques M., Schulze W.X., Höhne M., Usadel B., Gibon Y., Rohwer J., Stitt M. (2009). Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.S., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul R., Geserick C., Pabst M., Frey B., Wittmann D., Lütz-Meindl U., Léonard R., Tenhaken R. (2011). Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J. Biol. Chem. 286: 39982–39992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S., Finazzi G., Endler A., Willig A., Baerenfaller K., Grossmann J., Gerrits B., Rutishauser D., Gruissem W., Rochaix J.D., Baginsky S. (2011). Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt K.T.G., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B. (2011). System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4: rs3. [DOI] [PubMed] [Google Scholar]
- Rose, C.M., Venkateshwaran, M., Grimsrud, P.A., Westphall, M.S., Sussman, M.R., and Ané, J.-M. (2012). Medicago PhosphoProtein Database: A repository for Medicago truncatula phosphoprotein data. Front. Plant Sci. 3: 122. [DOI] [PMC free article] [PubMed]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Schwartz D., Gygi S.P. (2005). An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23: 1391–1398 [DOI] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Stuttmann J., Lechner E., Guérois R., Parker J.E., Nussaume L., Genschik P., Noël L.D. (2009). COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J. Biol. Chem. 284: 7920–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37 (Database issue): D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester A.W., Cande W.Z., Freeling M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Walker K.L., Müller S., Moss D., Ehrhardt D.W., Smith L.G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhou H., Berke L., Heck A.J., Mohammed S., Scheres B., Menke F.L. (2013). Quantitative phosphoproteomics after auxin-stimulated lateral root induction identifies an SNX1 protein phosphorylation site required for growth. Mol. Cell. Proteomics 12: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Facette M., Humphries J.A., Shen Z., Park Y., Sutimantanapi D., Sylvester A.W., Briggs S.P., Smith L.G. (2012). Identification of PAN2 by quantitative proteomics as a leucine-rich repeat-receptor-like kinase acting upstream of PAN1 to polarize cell division in maize. Plant Cell 24: 4577–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]