Proteolysis is an essential and irreversible protein modification that influences the function, interaction, and turnover of many proteins. By means of positional proteomics, a comprehensive catalog was generated of the physiological substrates of metacaspase 9 from Arabidopsis seedlings, providing a detailed view into the array of biological processes controlled by this protease.
Abstract
Metacaspases are distant relatives of the metazoan caspases, found in plants, fungi, and protists. However, in contrast with caspases, information about the physiological substrates of metacaspases is still scarce. By means of N-terminal combined fractional diagonal chromatography, the physiological substrates of METACASPASE9 (MC9; AT5G04200) were identified in young seedlings of Arabidopsis thaliana on the proteome-wide level, providing additional insight into MC9 cleavage specificity and revealing a previously unknown preference for acidic residues at the substrate prime site position P1′. The functionalities of the identified MC9 substrates hinted at metacaspase functions other than those related to cell death. These results allowed us to resolve the substrate specificity of MC9 in more detail and indicated that the activity of phosphoenolpyruvate carboxykinase 1 (AT4G37870), a key enzyme in gluconeogenesis, is enhanced upon MC9-dependent proteolysis.
INTRODUCTION
Proteolysis is an irreversible, protease-catalyzed protein modification with diverse effects on protein function and cellular or organismal fate. Proteases mediate protein stability, turnover, and activity and assist in the correct targeting and import of proteins to different organelles. Early development, xylem formation, seed maturation, nutrient supply mobilization, responses to biotic and abiotic stresses, and programmed cell death are facilitated by proteases (Avci et al., 2008; van der Hoorn, 2008; Coll et al., 2011; Hara-Nishimura and Hatsugai, 2011; Vartapetian et al., 2011; Pesquet, 2012). Plant genomes code for numerous proteases (over 700 annotated in Arabidopsis thaliana), but their substrates remain poorly identified (Tsiatsiani et al., 2012). Nevertheless, in the near future, this knowledge gap is expected to be filled by the use of emerging technologies that allow the proteome-wide identification of protease substrates (Huesgen and Overall, 2012).
Mammalian caspases are cysteinyl proteases that, besides functioning in apoptosis, mediate inflammation, cell proliferation, and differentiation by acting on a large number of different cellular substrates (Crawford et al., 2012). Based on their structural similarity to the caspase catalytic domain, metacaspases have been identified in fungi, protozoa, and plants, whereas paracaspases have been found in metazoans and in the slime mold Dictyostelium discoideum (Aravind et al., 1999; Uren et al., 2000; Vercammen et al., 2007). Like caspases, metacaspases and paracaspases carry the hallmarks of the Cys proteases of the CD clan, including the catalytic dyad His/Cys and the hemoglobinase fold (Rawlings and Barrett, 1993; Aravind and Koonin, 2002).
In view of the crucial role of caspases in apoptosis, initial efforts assessed mainly whether metacaspases were responsible for the caspase-like activities detected in plants during cell death events (Bonneau et al., 2008). However, biochemical studies using recombinant metacaspases or protein extracts from loss-of-function or gain-of-function mutants clearly demonstrated that metacaspases, in contrast with the Asp-specific caspases, cleave synthetic peptide substrates after Arg and Lys residues (Vercammen et al., 2004, 2006; Watanabe and Lam, 2005; Bozhkov et al., 2005; González et al., 2007; Moss et al., 2007; Ojha et al., 2010). Therefore, metacaspases are probably not directly responsible for the caspase-like activities in plants, but rather act upstream of the Asp-specific proteases (Meslin et al., 2011).
Besides the multifunctionality of metacaspases, which is apparent mostly in organisms encoding single metacaspase genes, such as Saccharomyces cerevisiae (yeast) and Leishmania major, redundant and antagonistic functions have been demonstrated in organisms encoding multiple metacaspases. The yeast metacaspase, termed Yeast Caspase1 (YCA1), is required for various stress-induced cell death events, including oxidative, osmotic, toxic, and virus-induced stresses and aging (Madeo et al., 2002; Herker et al., 2004; Ivanovska and Hardwick, 2005; Chahomchuen et al., 2009). YCA1 is also involved in cell cycle regulation (Lee et al., 2008) and clearance of insoluble protein aggregates (Lee et al., 2010). The L. major metacaspase MCA is involved in cell cycle regulation and in oxidative stress–induced cell death (Ambit et al., 2008; Zalila et al., 2011). Three of the five Trypanosoma brucei metacaspases are essential for viability of the bloodstream form of the parasite (Helms et al., 2006). Redundant functions in the regulation of senescence-associated death were also shown for two Podospora anserina metacaspases (Hamann et al., 2007) and in the resistance to endoplasmic reticulum stress for two Aspergillus fumigatus metacaspases (Richie et al., 2007). Interestingly, antagonistic functions for homologous metacaspases were found in T. brucei and Arabidopsis in the regulation of cell death events or cell proliferation and differentiation (Coll et al., 2010; Laverrière et al., 2012).
Metacaspases are posttranslationally regulated by multiple mechanisms, and their activity depends on specific cellular conditions. The proteolytic activity of Arabidopsis MC9 is regulated by S-nitrosylation, by a Ser protease inhibitor (Serpin 1), and by autocatalytic processing (Vercammen et al., 2004, 2006; Belenghi et al., 2007). Furthermore, the activities of the Norway spruce (Picea abies) metacaspase (mcII-Pa) and the Arabidopsis MC1, MC4, and MC5 are calcium dependent (Bozhkov et al., 2005; Watanabe and Lam, 2005, 2011a). Recently, a T. brucei metacaspase (MCA4) without detectable proteolytic activity, but necessary for parasitic proliferation and virulence, was found to be processed by another metacaspase, MCA3 (Proto et al., 2011). Despite these results, only a handful of substrates have been identified so far (Tsiatsiani et al., 2011). As knowledge on the corresponding substrates of proteases with unknown function can provide insights into their potential function or role in a specific process, new tools that facilitate substrate identification should assist in expanding our understanding of plant protease functions.
Here, we assessed the degradome (Overall et al., 2004) of the Arabidopsis METACASPASE9 (MC9; AT5G04200) by N-terminal combined fractional diagonal chromatography (COFRADIC). The physiological substrates of this plant protease were identified on the proteome-wide level, and the concrete substrate cleavage sites were characterized by means of positional proteomics. Thanks to these data, the substrate specificity of MC9 could be resolved in more detail, and the activity of phosphoenolpyruvate carboxykinase 1 (PEPCK1; AT4G37870), a key enzyme in gluconeogenesis, was found to be enhanced upon MC9-dependent proteolysis.
RESULTS
Expression Characteristics of the Arabidopsis MC9 Gene
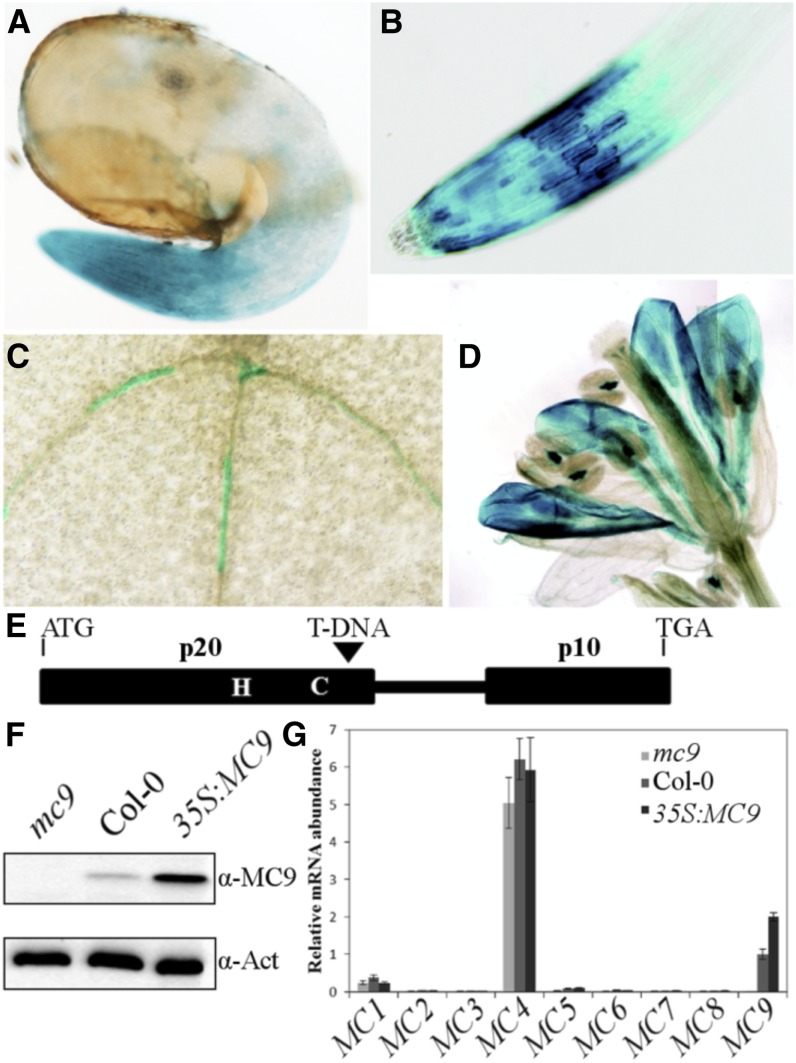
The spatiotemporal expression of Arabidopsis MC9 was studied in ProMC9:GUS reporter lines by means of β-glucuronidase (GUS) histochemical analysis (for a detailed description, see Supplemental Methods 1 online). Several independent homozygous alleles were analyzed with similar results (see representative GUS staining patterns in Figures 1A to 1D). Interestingly, the MC9 promoter activity was already obvious in root caps of young 2-d-old seedlings (corresponding to the developmental stage 0.5 to 0.7, including a 3-d stratification period) (Boyes et al., 2001) (Figure 1A), remained the same in 1-week-old seedlings, and was observed also in epidermal cells (Figure 1B). In hypocotyls and cotyledons, but not in rosette leaves, discrete differentiating primary and secondary tracheary elements were also stained (Figure 1C). In petals, a strong signal appeared just prior to abscission (Figure 1D). Anther connectives also displayed strong MC9 promoter activity (Figure 1D).
Figure 1.
Expression Characteristics of the MC9 Gene and Molecular Characterization of the MC9 Transgenic Lines Used in This Study.
(A) to (D) GUS staining of root cap of 2-d-old seedlings (A), root cap of 1-week-old seedling (B), tracheary elements in expanding cotyledon (C), and petals prior to abscission (D).
(E) Schematic representation of the T-DNA insertion of the GK-540H0 allele. The catalytic His and Cys, the p20 and p10 MC9 protease subunits, and the linker region between the two catalytic subunits are depicted.
(F) Immunoblot analysis of Arabidopsis seedlings' proteins with a MC9 polyclonal antibody. Full-length 35.5-kD MC9 is detected in the wild type (Col-0) and 35S:MC9 overexpressor (Vercammen et al., 2006), but not in the mc9 knockout line (GK-540H0 allele). An actin antibody directed against a common epitope of all actin isoforms (Immuno, clone C4) was used as gel loading control.
(G) Transcript levels of all Arabidopsis metacaspase genes (MC1 to MC9) in 2-d-old seedlings. Values represent the mean of three measurements ± sd relative to the MC9 gene transcript level in the wild type (Col-0).
By quantitative RT-PCR, we assessed the expression of all nine Arabidopsis metacaspases in 2-d-old seedlings of wild-type Columbia-0 (Col-0) and in the T-DNA insertion knockout (mc9) and MC9 overexpression (35S:MC9) (Vercammen et al., 2006) transgenic lines (for a detailed description, see Supplemental Methods 2 online) (Figures 1E to 1G). Both MC9 gain-of-function or loss-of-function plants did not display any obvious aberrant phenotypes under normal vegetative or reproductive growth conditions. MC4 expression was the highest (Figure 1G), consistent with a previous report that it is the most expressed metacaspase at various growth stages (Watanabe and Lam, 2011a). In young seedlings, corresponding to a developmentally active stage with a dynamic proteome, the MC9 expression was the second highest; therefore, this stage was chosen for analysis of the MC9 degradome (Figure 1G).
Identification of MC9 Protein Processing Events by N-Terminal COFRADIC
To explore the function of MC9 through identification of its physiological protein substrates, we used the N-terminal COFRADIC technique (Gevaert et al., 2003; Staes et al., 2011). In contrast with most proteomic approaches, N-terminal COFRADIC is a positional proteomics technology that uses sequential peptide chromatography to enrich for those peptides that contain the protein N termini out of the whole peptide mixture obtained by, for instance, full trypsin digestion of proteomes. Here, this so-called N-terminome consisted of all peptides that possessed the mature protein N termini and the neo-N-termini that had been generated upon proteolysis by MC9. Neo-N-termini that emerged at MC9 processing sites directly conveyed the MC9 proteolytic events and the exact site where the protein cleavage occurred (Van Damme et al., 2005). We analyzed the in vivo N-terminome of 2-d-old seedlings of wild-type and MC9 gain-of-function and loss-of-function transgenic plants (Figures 1E and 1F). First, the N-terminome of the T-DNA insertion knockout (mc9) seedlings was compared with that of wild-type seedlings and, second, to that of seedlings overexpressing MC9 (35S:MC9). Additionally, a mc9 seedling proteome to which Arabidopsis recombinant MC9 (rMC9) (Vercammen et al., 2004) had been added in vitro was studied versus an untreated mc9 control. To compare the N-terminomes, N-hydroxysuccinimide esters of light and heavy isotopic variants of butyric acid were used to mass tag the (neo)-N-terminal peptides (see Supplemental Figure 1 online). This differential labeling strategy was possible because MC9 introduced novel, primary α-amino groups that were uniquely tagged with either a light or heavy variant of butyric acid. After this mass tagging, equal amounts of the proteome preparations were mixed and digested with trypsin, and (neo)-N-terminal peptides were isolated with N-terminal COFRADIC (see Supplemental Figure 1 online). Mass tagging of neo-N-termini enabled us to determine the proteome origin of each labeled peptide. The respective ion signal intensities were further used to identify the MC9 processed sites.
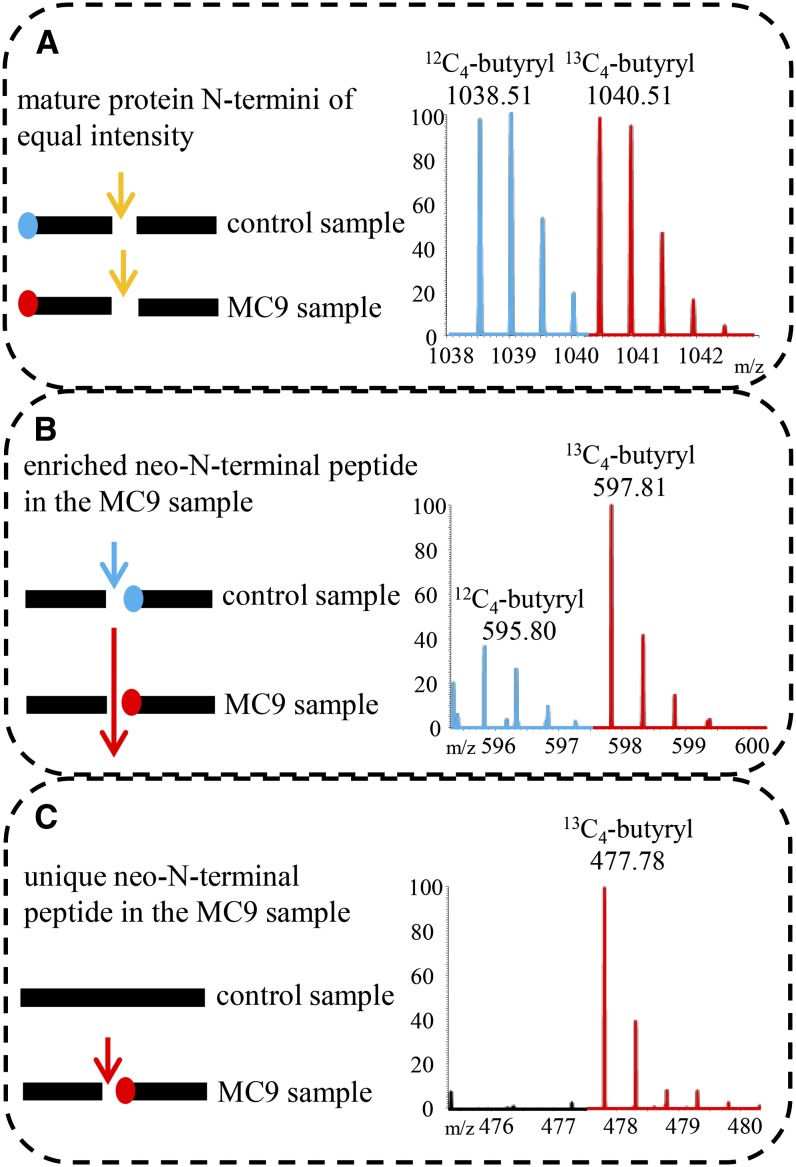
The following MS readouts were expected: (1) Some (neo)-N-terminal peptides would be found at equal intensity in both samples. Such peptides might carry the mature protein N terminus and would not be indicative of MC9 proteolysis (Figure 2A). Furthermore, such N-terminal peptides would imply that the particular protein (form) was not up- or downregulated in the tested proteomes. (2) Some neo-N-terminal peptides would occur in both samples, but with a significantly higher intensity in the sample with either the active MC9 or the supplemented rMC9. This outcome would mean that the corresponding proteins were also cleaved in the control sample (lacking MC9 activity), although to a lesser extent, by another protease, hinting at a redundant protease activity (Figure 2B). Such events could be explained by a scenario in which the MC9 perturbation activated other proteases (i.e., metacaspases). (3) Some neo-N-terminal peptides (so-called singletons) would be uniquely found in the proteome either with the active MC9 (Col-0 or 35S:MC9 proteome) or supplemented with rMC9, implying that the corresponding proteins were cleaved by MC9 (Figure 2C).
Figure 2.
Illustration of the Different Categories of Isolated (Neo)-N-Terminal Peptides.
Mass tagging allowed the differential comparison of N-terminomes derived from samples in which MC9 was active (Col-0 or 35S:MC9 or added rMC9) or absent (control sample mc9). MS spectra of N-terminal peptides were, in most cases, identified as doublet ions of light (in blue) and heavy (in red) isotopes. m/z, mass-to-charge ratio.
(A) Protein cleaved in both samples to the same extent (i.e., by trypsin; yellow arrow) and fragments bearing the mature protein N terminus. As the amounts of these peptides were approximately equal, no MC9-driven proteolysis could be deduced.
(B) Protein cleaved in both samples, but at a higher frequency in the MC9 sample (large red arrow). The neo-N-terminus fragment was generated in both samples, but in significantly higher amounts in the MC9 sample. Proteolysis of the target in the control sample might have occurred by a redundant protease activity (blue arrow). The fragment is identified as a doublet with the highest intensity ion derived from the MC9 sample.
(C) Protein solely cleaved by MC9 (red arrow) in the samples of active MC9 proteases. Thereby, a fragment with a neo-N-terminus was generated, later identified as unique in the MC9 N-terminome and, thus, as a singleton ion.
Peptides were analyzed with an LTQ Orbitrap XL mass spectrometer. The obtained tandem mass spectrometry (MS/MS) spectra were searched against The Arabidopsis Information Resource database (release 8) with the Mascot algorithm and subsequently mapped to the TAIR10 database with DBToolkit (Martens et al., 2005) (for a detailed description, see Supplemental Methods 3 online). In total, 75,310 MS/MS spectra were generated in the mc9/Col-0 analysis, 69,388 in the mc9/35S:MC9 analysis, and 75,042 in the in vitro analysis, identifying 3781, 2879, and 3050 peptides and converging to 1705, 1407, and 1138 proteins, respectively. The ratios of light/heavy peptide signals for all identified peptides were calculated with the Mascot Distiller software and were individually checked for possible neo-N-terminal peptides pointing to MC9 processing. We focused on two classes of peptides: the significantly enriched neo-N-terminal peptides (Z-score ≤ −2.33 or Z-score ≥ 2.33, depending on the labeling used to calculate the light/heavy intensity ratios) and the unique peptides (singletons) in the Col-0, 35S:MC9, or rMC9-treated seedling proteomes in comparison to mc9 (Figures 2B and 2C). As the former might also appear because of differential protein levels in the input proteomes, we scanned the data for the corresponding precursor protein N termini or other neo-N-terminal peptides from these precursor proteins, which were not cleaved by MC9. Comparison of these peptide levels led us to conclude that no large proteome changes occurred between the different proteomes assayed. For an overview of all cleavage sites, their corresponding substrates, and their precursor protein N termini identified in the three analyses, see Supplemental Data Sets 1A to 1C online.
After filtering the data with the known MC9 specificity for Arg and Lys, 72% of all processed sites matched the expected MC9 specificity (28 and 44% cleavage C-terminally to Arg and Lys, respectively), without significant enrichment for a particular amino acid in the other processing events. In total, 551 cleavage events after Arg or Lys (116 in mc9/Col-0, 103 in mc9/35S:MC9, and 332 in vitro) (see Supplemental Figure 2A online) were identified in 392 proteins (97 in mc9/Col-0, 84 in mc9/35S:MC9, and 211 in vitro) (see Supplemental Figure 2B online), from which approximately one-third (160 sites) were reported as singleton peptides in either Col-0, 35S:MC9, or rMC9-treated proteomes (16 in mc9/Col-0, 11 in mc9/35S:MC9, and 133 in vitro) and 391 as significantly enriched peptides in the same proteomes (100 in mc9/Col-0, 92 in mc9/35S:MC9, and 199 in vitro) (see Supplemental Data Sets 1A to 1C online)
Positional Amino Acid Cleavage Specificity of MC9
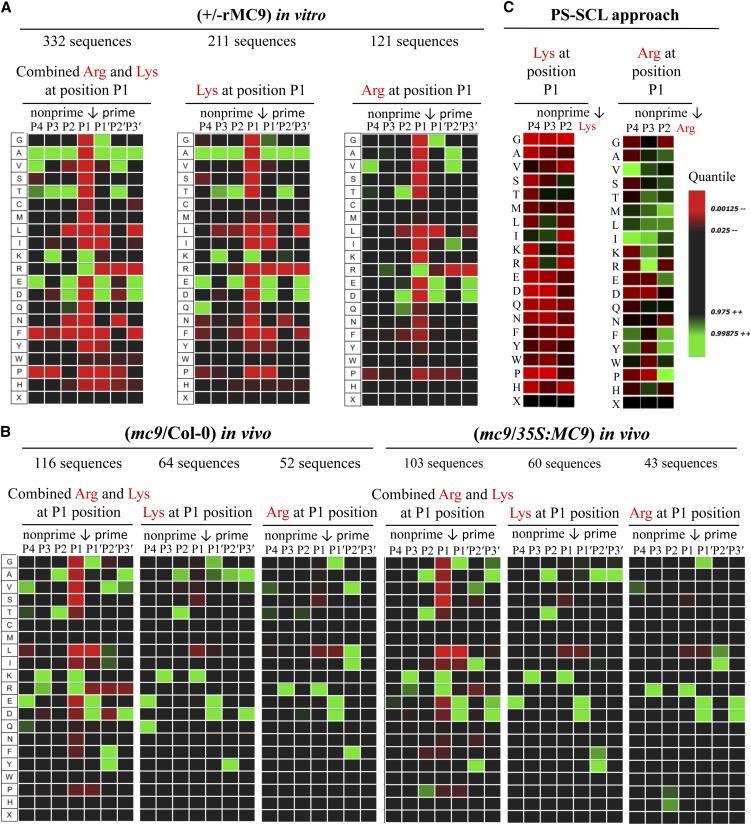
Using iceLogo (Colaert et al., 2009), we inspected the amino acid sequences at the prime and nonprime substrate positions of the Arg and Lys P1-specific cleavage sites (Schechter and Berger, 1967) and analyzed the frequencies of specific residues after statistical correction by means of the natural occurrence of amino acids in Arabidopsis proteins. The specificity information derived from the three data sets was very similar. The large in vitro COFRADIC data set showed that the acidic residues Asp and Glu were often found at neighboring positions to the basic (Arg or Lys) cleavage site. For instance, in 46 and 47% of the Lys-cleaved and Arg-cleaved sequences, respectively, the P1′ position (C-terminally to the cleavage P1 position) was occupied by Asp or Glu and, likewise, the P2 position in 23 and 22% of the Lys-cleaved and Arg-cleaved sequences, respectively. However, only in 5 and 7% of the Lys-cleaved and Arg-cleaved sequences, respectively, both P2 and P1′ were acidic (Figure 3A; see Supplemental Figures 3 and 4A online). At the prime site, Asp or Glu was also observed in 27% of the cases at P3′. At P2′, the small amino acids Thr, Ala, and Val occurred in 38% of all cases, whereas Ala was tolerated at every position. Analogous to the in vivo COFRADIC-derived specificity (Figure 3B), Val and Glu were preferred at P4 and Lys and Arg were frequently found at P3. Ala and Thr were the sole preferred residues at P2 in the in vivo–cleaved substrates, but in vitro–cleaved proteins additionally (23% of the cleaved substrates) tolerated Asp and Glu at this position (Figures 3A and 3B; see Supplemental Figures 3 and 4 online). Regarding the prime site substrate specificity of MC9, the in vivo and in vitro data were largely similar, except for the amino acid tolerance at P2′: In the latter, small residues were present at this position, whereas aromatic residues, such as Phe and Tyr and the aliphatic Ile and Leu, were found also in the former.
Figure 3.
MC9 Specificity Monitored via Its Proteome-Wide Substrate Profiling.
Analysis was performed with iceLogo (Colaert et al., 2009) after multiple sequence alignment and statistical correction with the natural occurrence of amino acids in Arabidopsis proteins. Heat maps illustrating the amino acid frequencies at the P4-P3′ positions in the MC9 substrates as observed in the in vivo mc9/Col-0 and mc9/35S:MC9 COFRADIC studies (A), the in vitro COFRADIC (B), and the PS-SCL approach (C). R software was used to build the PS-SCL heat maps with the published values (Vercammen et al., 2006). Arrows mark the cleaved scissile bonds.
In conclusion, the MC9 specificity for basic residues at the P1 site was accompanied by a strong preference for acidic amino acids at the P1′ site (see Supplemental Figure 4 online). This particular trend of acidic-basic amino acid combinations was also seen at other positions at the nonprime site (P4-P3), whereas Ala was generally tolerated at every position. A relative preference occurred for Lys over Arg at the P1 site and in vivo, often (27% of the cases) a second Arg or Lys at P3. For instance, when Lys was present at P1, 24% of these substrates contained a second Lys at P3.
Previously, in a positional scanning of a synthetic combinatorial library (PS-SCL) of 7-amino-4-methylcoumarin (AMC)–labeled synthetic tetrapeptides spanning the positions P4-P1, VRPR was found as the most preferred rMC9 tetrapeptide substrate, whereas IISK, the best cleaved peptide with Lys at P1, was fivefold less efficiently cleaved than VRPR (for Val-Arg-Pro-Val) (Vercammen et al., 2006). Our current results somewhat contrast with the stronger preference for Arg over Lys at P1, but they are in agreement with the preferred presence of a dibasic amino acid motif (such as RxR) as in VRPR. When the two in vitro approaches (in vitro COFRADIC and PS-SCL) were compared (Figure 3C), Val was the most prevalent residue at P4 in both data sets of Arg-cleaved substrates. However, a major difference was observed at the P2 site: The COFRADIC data indicated that Thr and Asp were preferred and the PS-SCL data showed a preference for hydrophobic residues or Pro. Based on the in vivo and in vitro amino acid frequencies at the P4-P3′ substrate positions derived from the COFRADIC analyses (Figure 3; see Supplemental Figures 3 and 4 online), two revised consensus sequences of the MC9 substrate specificity are proposed for either cleavage after Arg and Lys, namely, [V]-[R]-[T/Q]-[R]-↓-[G/D/E]-[V/F/L/I]-[D/E] and [E/Q]-[K/A]-[T/E]-[K]-↓-[D/E]-[Y/T]-[D/E] to detect the MC9 activity in biochemical assays. By means of GenomeNet (http://www.genome.jp) and Motif Search, we screened the Arabidopsis proteome for the presence of these sequences. The number of protein hits varied depending on the length of the motif. In fact, the P4-P1’ sequences [V]-[R]-[T/Q]-[R]-↓-[G/D/E] and [E/Q]-[K/A]-[T/E]-[K]-↓-[D/E] were found in more than 20 and 380 protein hits, respectively (see Supplemental Data Set 1D online). Nevertheless, these observations are based only on the sequences of the queried proteins and do not take into account whether any of the predicted motifs are solvent accessible or reside in a secondary structure element not readily recognized by the protease.
Experimental Validation of MC9 Protein Substrates
For the experimental validation of the MC9 substrates, we prioritized all in vivo singleton peptides together with those proteins bearing cleavage sites identified in more than one proteomic analysis. In this manner, 99 sites in 74 different proteins were selected for cleavage validation (Table 1), from which 18 sites were identical in the three different setups, 35 sites in both in vivo analyses, 19 sites between the mc9/Col-0 in vivo and the in vitro analysis, and finally 15 sites between the mc9/35S:MC9 in vivo and the in vitro analysis. The remaining 12 sites were reported as in vivo singleton peptides (Table 1; see Supplemental Figure 2A online).
Table 1. Selection of Neo-N-Terminal Peptides Identified in at Least Two Independent Analyses.
| No. of Cleavage Sites | Accession | P4-P3′ | Sequence | Description |
|---|---|---|---|---|
| Identified in all three analyses | ||||
| 1 | AT1G12000 | AVTR(10)DLT | DLTAVGSPENAPAKGR | Phosphofructokinase family protein |
| 2 | AT1G49240 | MNQK(53)DAY | DAYVGDEAQSKR | Actin 8 |
| 3 | AT2G21060 | HMAR(150)ECS | ECSQGGGGYSGGGGGGR | Gly-rich protein 2B |
| 4 | AT2G42560 | VGSK(454)AVD | AVDLTKEKAAVAADTVVGYTAR | LEA domain–containing protein |
| 5 | AT3G02480 | MKEK(51)AQG | AQGAADVVKDKTGMNKSH | LEA family protein |
| 6 | AT3G09840 | KSKK(15)DFS | DFSTAILER | Cell division cycle 48 |
| 7 | AT3G17520 | EKAK(184)EAK | EAKEAAKR | LEA family protein |
| 8 | AKDK(262)ASQ | ASQSYDSAAR | ||
| 9 | AT3G53040 | QKTK(111)ETA | ETADYTADKAR | Putative/LEA protein |
| 10 | DKTK(155)ETA | ETAEYTAEKAR | ||
| 11 | AT4G02930 | GKAK(99)AIA | AIAFDEIDKAPEEKKR | GTP binding Elongation factor Tu family protein |
| 12 | AT4G21020 | EKAK(156)DYA | DYAEDTMDNAKEKAR | LEA family protein |
| 13 | AT4G37870 | SFPK(20)GPV | GPVMPKITTGAAKR | PEPCK1 |
| 14 | AT5G16050 | EAPK(254)EVQ | EVQKVDEQAQPPPSQ | General regulatory factor 5 |
| 15 | AT5G44310 | ERTK(70)DYA | DYAEQTKNKVNEGASR | LEA family protein |
| 16 | EKTK(110)DYA | DYAEEAKDKVNEGASR | ||
| 17 | EKTK(179)NYA | NYAEQTKDKVNEGASR | ||
| 18 | AT5G47210 | RYAK(341)EAA | EAAAPAIGDTAQFPSLG | Hyaluronan/mRNA binding family |
| Identified in both in vivo analyses | ||||
| 1 | AT1G04750 | VLDR(156)GEK | GEKIELLVDKTENLR | Vesicle-associated membrane protein 721 |
| 2 | AT1G07780 | SVAK(109)DIS | DISKVAR | Phosphoribosylanthranilate isomerase 1 |
| 3 | AT1G09310 | TEAK(165)EAV | EAVAIKEAVAVKEAA | Protein of unknown function, DUF538 |
| 4 | AT1G11480 | ARPR(404)ELV | ELVLKER | Eukaryotic translation initiation factor-related |
| 5 | AT1G29350 | QNAR(525)ELD | ELDFQYSPFSAQQSMQSR | Kinase-related protein of unknown function (DUF1296) |
| 6 | AT1G47540 | AYDR(31)KCL | KCLKEYGGDVGFSYCAPR | Trypsin inhibitor 2 |
| 7 | AT1G49240 | SIEK(241)NYE | NYELPDGQVITIGAER | Actin 8 |
| 8 | AT1G49970 | FEKR(363)DYD | DYDGTLAQR | CLP protease proteolytic subunit 1 |
| 9 | AT1G51710 | VATR(157)ELF | ELFGELDR | Ubiquitin-specific protease 6 |
| 10 | AT1G55490 | TVAR(114)EVE | EVELEDPVENIGAKLVR | Chaperonin 60β |
| 11 | AT1G56070 | ICLK(544)DLQ | DLQDDFMGGAEIIKSDPVVSFR | Ribosomal protein S5/Elongation factor G/III/V family protein |
| 12 | AT2G01250 | VESK(6)VVV | VVVPESVLKKR | Ribosomal protein L30/L7 family protein |
| 13 | AT2G18960 | GSYR(902)ELS | ELSEIAEQAKR | H(+)-ATPase 1 |
| 14 | AT2G19730 | SVNK(88)SIL | SILKKEFPR | Ribosomal L28e protein family |
| 15 | AT2G26250 | IVNR(18)GIE | GIEPSGPNAGSPTFSVR | 3-Ketoacyl-CoA synthase 10 |
| 16 | AT2G28000 | PRGR(82)NVV | NVVLDEFGSPKVVNDGVTIAR | Chaperonin-60α |
| 17 | TIAR(103)AIE | AIELPNAMENAGAALIR | ||
| 18 | AT2G32240 | VKSR(709)DID | DIDLSFSSPTKR | Unknown protein |
| 19 | AT2G36620 | NIPK(149)SAA | SAAPKAAKMGGGGGR | Ribosomal protein L24 |
| 20 | AT2G42560 | ARAK(324)DYT | DYTLQKAVEAKDVAAEKAQR | LEA domain–containing protein |
| 21 | AT3G13300 | VGER(708)NLD | NLDVSSVEEISR | Transducin/WD40 repeat-like superfamily protein |
| 22 | AT3G53040 | TVLK(449)EAD | EADQMTGQTFNDVGEIDDEEKVR | Putative/LEA protein |
| 23 | AT3G61260 | SLDR(92)DVK | DVKLADLSKEKR | Remorin family protein |
| 24 | AT4G15410 | LRSR(109)GGA | GGAGENKETENPSGIR | Ser/Thr protein phosphatase 2A |
| 25 | AT4G20360 | ERAR(127)GIT | GITINTATVEYETENR | RAB GTPase homolog E1B |
| 26 | AT4G27440 | YITK(337)GYV | GYVSETESGKR | Protochlorophyllide oxidoreductase B |
| 27 | AT4G31700 | IVKK(119)GEN | GENDLPGLTDTEKPR | Ribosomal protein S6 |
| 28 | AT5G04200 | LFGR(216)DAG | DAGLKFR | Metacaspase 9 |
| 29 | AT5G06140 | EQPR(9)NIS | NISGSMQSPR | Sorting nexin 1 |
| 30 | AT5G12140 | GGVR(15)DID | DIDANANDLQVESLAR | Cystatin-1 |
| 31 | AT5G44310 | EKTK(150)DFA | DFAEETKEKVNEGASR | LEA family protein |
| 32 | AT5G52440 | TLER(145)EIG | EIGLDDISTPNVYNQNR | Bacterial sec-independent translocation protein mtta/Hcf106 |
| 33 | AT5G54190 | YITK(341)GYV | GYVSESEAGKR | Protochlorophyllide oxidoreductase A |
| 34 | AT5G56000 | SSKK(604)TME | TMEINPENSIMDELR | Heat shock protein 81.4 |
| 35 | AT5G62575 | TREK(42)ALL | ALLAEDSALKR | Unknown protein |
| Identified in the mc9/35S:MC9 and the in vitro analysis | ||||
| 1 | AT1G13270 | RKMR(78)ELE | ELETKSKVR | Met aminopeptidase 1B |
| 2 | AT1G54870 | EKIK(288)NFG | NFGSEVPMKR | Aldehyde reductase |
| 3 | AT1G72370 | LGTK(33)NCN | NCNYQMER | 40s ribosomal protein SA |
| 4 | AT2G21660 | KAMK(64)DAI | DAIEGMNGQDLDGR | Gly-rich, RNA binding protein atgrp7 |
| 5 | AT2G42560 | MRER(214)EGK | EGKESAGGVGGR | LEA domain–containing protein |
| 6 | ATEK(268)GKE | GKEAGNMTAEQAAR | ||
| 7 | EKGK(270)EAG | EAGNMTAEQAAR | ||
| 8 | VEAK(335)DVA | DVAAEKAQR | ||
| 9 | AT3G17520 | ESAK(260)DKA | DKASQSYDSAAR | LEA family protein |
| 10 | AT3G22640 | TRSK(231)EIG | EIGQGIIR | Cupin family protein |
| 11 | AT3G51160 | TAPK(20)ADS | ADSTVVEPR | Gdp-d-mannose-4,6-dehydratase 2, gmd2 |
| 12 | AT3G53040 | DKTK(133)ETA | ETADYAAEKAR | Putative/LEA protein |
| 13 | AT4G37870 | SLTR(102)ESG | ESGPKVVR | PEPCK1 |
| 14 | AT4G39260 | DLQR(25)TFS | TFSQFGDVIDSKIINDR | Cold, circadian rhythm, and RNA binding 1 |
| 15 | AT5G10360 | IVKK(119)GVS | GVSDLPGLTDTEKPR | Ribosomal protein s6e |
| Identified in the mc9/Col-0 and the in vitro analysis | ||||
| 1 | AT1G47540 | CAPR(49)IFP | IFPTFCDQNCR | Trypsin inhibitor 2 |
| 2 | AT1G55490 | VLTK(382)ETS | ETSTIVGDGSTQDAVKKR | Chaperonin 60β |
| 3 | AT1G65090 | TSER(162)TLQ | TLQDDKKSGNAKSEEVQEQPEKR | Unknown protein |
| 4 | AT2G21660 | TDDR(23)ALE | ALETAFAQYGDVIDSKIINDR | Gly-rich, RNA binding protein atgrp7 |
| 5 | AT2G36640 | QKTR(338)EST | ESTESGAQKAEETKDSAAVR | Embryonic cell protein 63 |
| 6 | AT2G40490 | VTER(47)KVS | KVSATSEPLLLR | Uroporphyrinogen decarboxylase |
| 7 | AT2G42560 | QTTK(607)NIV | NIVIGDAPVR | LEA domain–containing protein |
| 8 | AT3G02480 | QQMK(49)EKA | EKAQGAADVVKDKTGMNKSH | LEA family protein |
| 9 | AT3G11710 | QTTK(11)ALS | ALSELAMDSSTTLNAAESSAGDGAGPR | Lysyl-tRNA synthetase 1 |
| 10 | AT3G12960 | EKGK(58)EQS | EQSAASGDQTQIQR | Unknown protein |
| 11 | AT3G13470 | VLTK(378)EMT | EMTTIVGDGTTQEAVNKR | TCP-1/cpn60 chaperonin family protein |
| 12 | AT3G15280 | TEYR(135)GVE | GVEDLHQQTGGEVKSP | Unknown protein |
| 13 | AT3G15670 | IKNK(134)AQD | AQDAAQYTKETAQGAAQYTKETAEAGR | LEA family protein |
| 14 | QYTK(143)ETA | ETAQGAAQYTKETAEAGR | ||
| 15 | AT3G48870 | ISDR(501)FLP | FLPDKAIDLIDEAGSR | Clp ATPase |
| 16 | AT4G02510 | VSSR(607)EFS | EFSFGGKEVDQEPSGEGVTR | Translocon at the outer envelope membrane of chloroplasts 159 |
| 17 | AT4G21020 | EKAK(116)DTA | DTAYNAKEKAKDYAER | LEA family protein |
| 18 | ESTK(218)NAA | NAAQTVTEAVVGPEEDAEKAR | ||
| 19 | AT4G38630 | QKDK(258)DGD | DGDTASASQETVAR | Regulatory particle non-ATPase 10 |
| Fraction of singleton neo-N-terminal peptides that were solely found in one of the in vivo analyses | ||||
| 1 | AT1G02150 | VTSK(474)EAV | EAVLELLR | Pentatricopeptide repeat–containing protein |
| 2 | AT1G55090 | NSNK(710)EIG | EIGVVAANSGDPSAGL | NAD synthetase |
| 3 | AT3G23990 | YAAK(35)EIK | EIKFGVEAR | Heat shock protein 60 |
| 4 | AT3G45190 | VHDR(558)DYD | DYDLAGLANNLNQFR | SIT4 phosphatase-associated family protein |
| 5 | AT2G42790 | KEDK(86)GLK | GLKLYDPGYLNTAPVR | Citrate synthase 3, CSY3 |
| 6 | AT2G47470 | YIEK(320)GSD | GSDYASKETER | Thioredoxin family protein |
| 7 | AT3G23990 | LMLK(49)GVE | GVEDLADAVKVTMGPKGR | Heat shock protein 60 |
| 8 | AT4G05180 | DQAR(110)DFS | DFSLALKDR | Photosystem II subunit Q-2 |
| 9 | AT4G34450 | GIEK(27)GAV | GAVLQEAR | Coatomer γ-2 subunit, putative |
| 10 | AT5G16130 | LTGK(181)DVV | DVVFEYPVEA | Ribosomal protein S7e family protein |
| 11 | AT5G35910 | ESTR(578)DLI | DLIMGAANTNEGR | Polynucleotidyl transferase |
| 12 | AT5G54310 | QSGK(469)DFD | DFDFSSLMDGMFTKH | ARF-GAP domain 5 |
The equivalent experiments, peptide sequence, accession number, and protein description are given. Amino acid positions of the P1' sites are indicated in parentheses in the P4-P3′ column.
Two independent validation approaches were chosen. First, 18–amino acid peptides harboring the identified cleavage sites were synthesized for six randomly selected candidate protein substrates and used for in vitro rMC9 cleavage assays. Second, in vitro–transcribed and translated, radiolabeled (35S-Met) proteins, generated from available cDNA clones (ABRC), were incubated with rMC9 and their digestion products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
After incubation with increasing concentrations of rMC9 (0, 31, 125, and 500 nM), synthetic peptides and their digestion products were separated by reverse-phase HPLC and analyzed by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (for a detailed method description, see Supplemental Methods 4 online). Peptides spanning the MC9 cleavage sites identified in trypsin inhibitor 2 (AT1G47540), general regulatory factor 5 (GRF5; AT5G16050), Gly-rich RNA binding protein (GRP7; AT2G21660), and protochlorophyllide oxidoreductase B (PORB; AT4G27440) were cleaved by rMC9 at the sites identified by COFRADIC. The degree of peptide processing increased with increasing rMC9 concentrations (see Supplemental Figure 5 online).
For proteins containing more than one identified cleavage site, such as the late-embryogenesis abundant (LEA) domain–containing protein (AT2G42560), PEPCK1 (AT4G37870), and GRP7, two synthetic peptides were tested (see Supplemental Figure 6 online). All these peptides were cleaved by rMC9 at the sites identified by COFRADIC. However, additional alternative cleavage sites were observed as well, probably due to sequence similarities with the COFRADIC-deduced sites. For instance, the additional cleavage site R212↓E in the LEA domain–containing protein precursor was identical to the COFRADIC-deduced site R214↓E. As for GRP7, the peptide precursor was originally expected to be cleaved at the MK64↓D site but was additionally processed at EK61↓A, a similar site, because in both cases, K was adjacent to an acidic residue (Asp or Glu). Furthermore, the consensus sequence for Lys-cleaved peptides supported the preference for acidic residues either at the N-terminal or C-terminal side of the cleaved Lys.
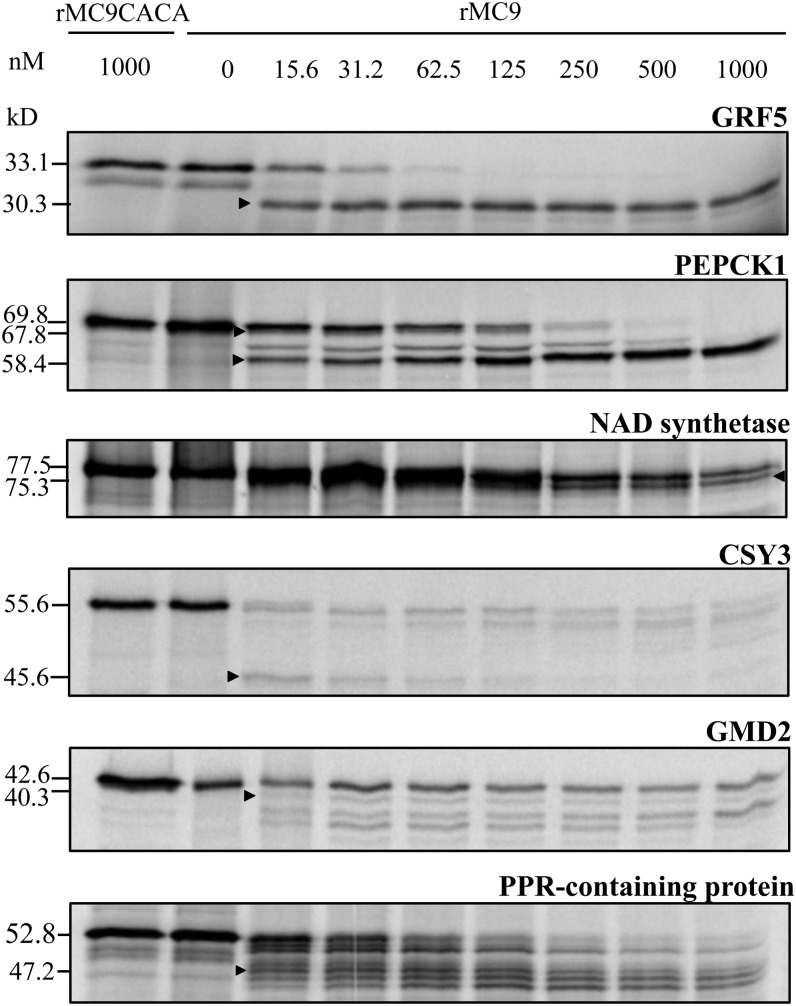
To validate experimentally the substrate cleavages at the protein level, we employed in vitro transcription and translation coupled assays. In total, we successfully translated 16 proteins, all of which were cleaved by rMC9. Although the precursor protein levels clearly decreased or even were completely negligible after the rMC9 treatment, for some substrates, cleavage fragments were not always observed. It is possible that the substrate fragments were not detectable by autoradiography because they did not contain a radiolabeled Met or that proteolytic fragments <20 kD might have been too small for proper separation and visualization by the SDS-PAGE system used. The substrates that were processed into fragments of correctly predicted molecular weight based on the cleavage site(s) previously identified by COFRADIC and MS, were GRF5, PEPCK1, nicotinamide adenine dinucleotide (NAD) synthetase (AT1G55090), GDP-mannose 4,6-dehydratase 2 (GMD2; AT3G51160), pentatricopeptide repeat (PPR)–containing protein (AT1G02150), and citrate synthase3 (CSY3; AT2G42790) (Figure 4). Furthermore, these proteins were not cleaved by the inactive rMC9 mutant, rMC9C147AC29A (Belenghi et al., 2007), indicating that the observed fragments were indeed due to rMC9 and not to other (contaminating) proteases present in the assay mixture.
Figure 4.
In Vitro–Transcribed and Translated Proteins Incubated with Wild-Type Recombinant MC9 Protease and the Catalytically Inactive Mutant rMC9CACA.
Arrowheads indicate the proteolytic fragments with molecular masses matching those identified by COFRADIC.
MC9 Cleaves PEPCK1 in Vivo, Leading to Increased PEPCK1 Activity
PEPCK, which catalyzes the reversible ATP-dependent decarboxylation of oxaloacetate to phosphoenolpyruvate, is involved in gluconeogenesis, photosynthetic CO2-concentrating mechanisms during C4 photosynthesis (Edwards et al., 1971), Crassulacean acid metabolism (Dittrich et al., 1973), and nitrogenous compound metabolism (Leegood and Walker, 2003). As PEPCK is posttranslationaly regulated by reversible phosphorylation (Walker and Leegood, 1995) and prone to rapid proteolytic cleavage in planta (Walker et al., 1995, 1997; Malone et al., 2007), which gives rise to fragments with sizes that correlate to the cleavage sites observed by N-terminal COFRADIC, we focused on the MC9-dependent PEPCK1 cleavage. To this end, we assessed whether MC9 was responsible for in planta cleavage of PEPCK1, which is highly active during germination at the 2-d-old seedling stage of the studied proteome (Rylott et al., 2003).
PEPCK1 polypeptides (74 and 73 kD) were previously immunodetected in Arabidopsis cotyledons with a polyclonal antiserum raised against PEPCK from Guinea grass (Panicum maximum) leaves (Malone et al., 2007). Similarly, in pea (Pisum sativum) cotyledons, besides the mature 72-kD form, a smaller 68-kD PEPCK polypeptide was detected that was attributed either to a genuine isoform or to a regulatory protein processing event (Delgado-Alvarado et al., 2007). Recently, the pineapple (Ananas comosus) PEPCK had been found to have two active polypeptide variants of 74 and 65 kD in vivo (Martín et al., 2011). In cucumber (Cucumis sativus), an in vivo phosphorylation site is located in the proteolytically removed N-terminal fragment (Walker and Leegood, 1995). In Guinea grass, the activity of the full-length phosphorylated PEPCK was lower than that of the full-length or truncated dephosphorylated PEPCK, indicating that removal of the phosphorylation site(s)–containing N-terminal region might restore the PEPCK activity (Walker et al., 2002).
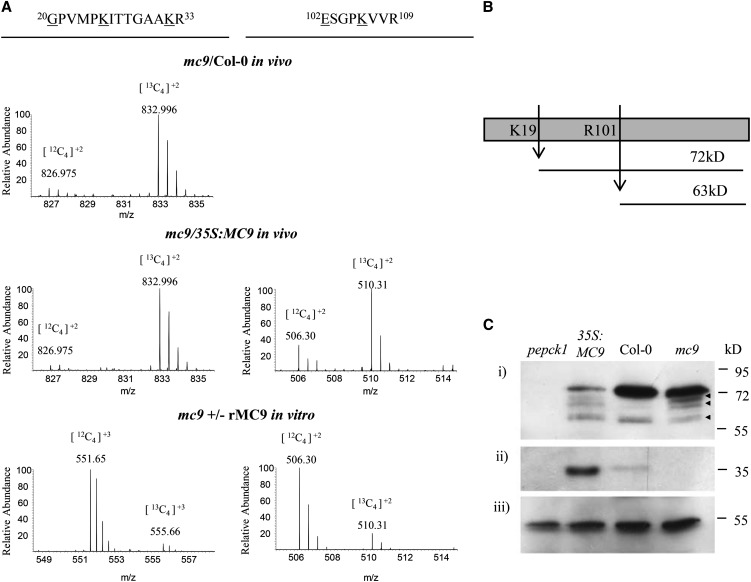
Cleavage of PEPCK1 by MC9 in vivo and in vitro at Lys-19 and Arg-101 (Figures 4 and 5A; see Supplemental Figure 6 online) led to C-terminal fragments of 72 and 63 kD, respectively (Figure 5B). With rabbit polyclonal antibodies raised against the recombinant Arabidopsis PEPCK1, we monitored the PEPCK1 processing in MC9 gain-of-function and loss-of-function mutants. Protein extracts of mc9, Col-0, and 35S:MC9 from 2-d-old seedlings were prepared under postlytic proteolysis-inhibiting conditions, directly mixed with the SDS-PAGE loading buffer, and heated before protein gel blot analysis with anti-PEPCK1. The 74-kD precursor protein was processed in the 35S:MC9 extracts, whereas in Col-0 or mc9 extracts, it remained predominantly intact (Figure 5C). Smaller cross-reactive fragments of 57 to 68 kD were equally abundant in mc9, Col-0, and 35S:MC9 extracts, implying that they could result from another protease activity or degradation. Taken together, our data indicate that overexpression of MC9 increased the PEPCK1 processing in vivo.
Figure 5.
Cleavage of the PEPCK1 Protein as Deduced from the COFRADIC Data and in Vivo Validated by the PEPCK1 Immunodetection in the Wild Type and the MC9 Gain- and Loss-of-Function Mutant Extracts.
(A) Mass spectra of the PEPCK1 proteolysis reporter peptides as identified in mc9/Col-0, mc9/35S:MC9, and the in vitro studies. Mass/charge values for each isotope are given on top of each peptide ion. Butyrylated residues are underlined.
(B) Schematic representation of PEPCK1 proteolysis by MC9 at the identified cleavage sites and generation of fragments with variable size.
(C) Immunodetection of (i) PEPCK1 protein in extracts of 2-d-old seedlings from pepck1, 35S:MC9, Col-0, and mc9, (ii) full-length MC9 zymogen, and (iii) CAT2 (AT4G35090) loading gel control. The PEPCK1 proteolytic fragments of 57 to 68 kD are marked with arrowheads.
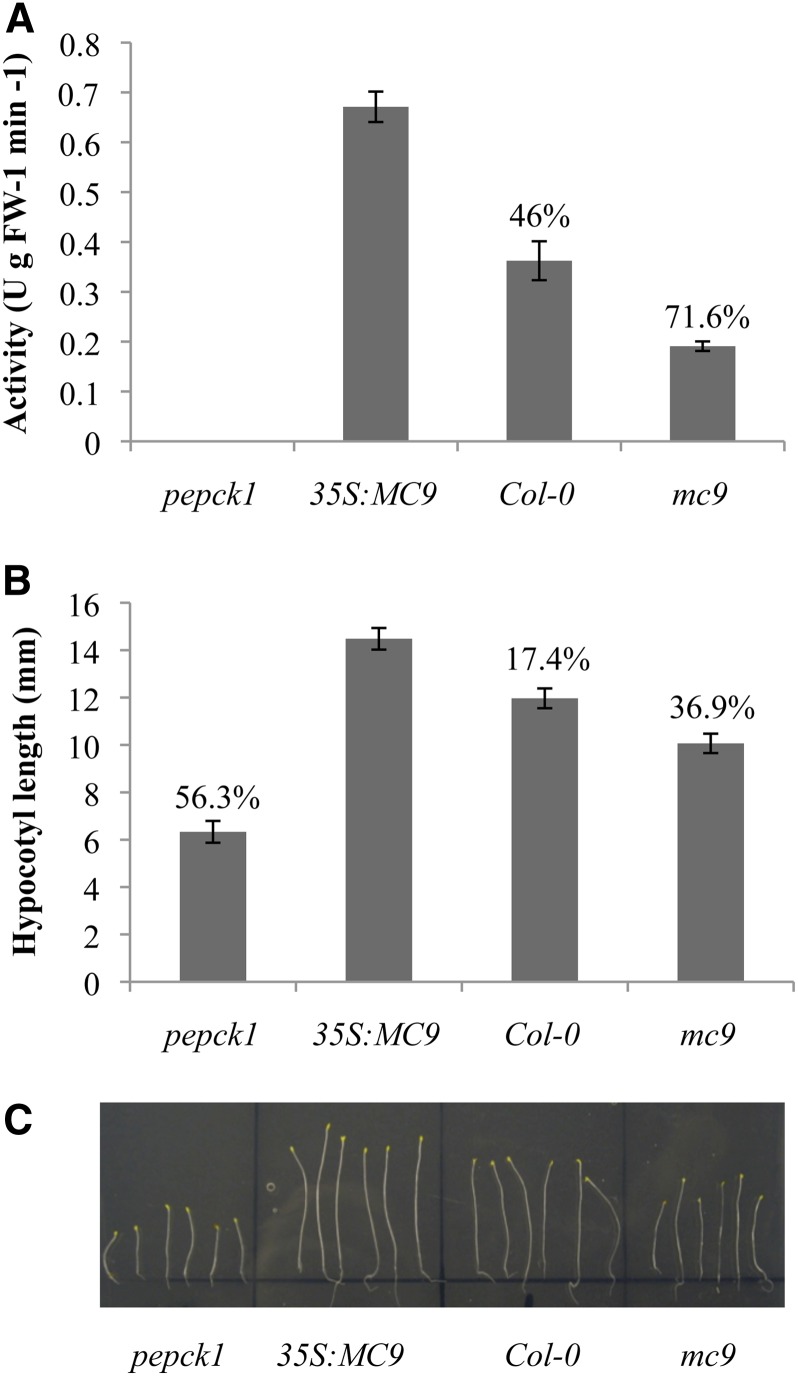
To assess the effect of the MC9-dependent PEPCK1 cleavage, we measured the carboxylation activity of PEPCK1 in young seedling extracts of Col-0, pepck1 (Penfield et al., 2004), and MC9 gain-of-function and loss-of-function mutants (Figure 6A). PEPCK1 activity was the highest in 35S:MC9 plant extracts (46 and 71% higher than that of Col-0 and mc9, respectively), revealing that MC9 cleavage enhanced PEPCK1 activity and that proteolysis might remove the putative PEPCK1 inhibitory phosphorylation site (Ser-62). As PEPCK1 is involved in Suc synthesis from lipids during oilseed plant germination, the pepck1 mutants are impaired in hypocotyl elongation when grown in the dark in Suc-deprived growth media (Rylott et al., 2003; Penfield et al., 2004). Hence, under these conditions, we monitored the hypocotyl elongation of pepck1, mc9, Col-0, and 35S:MC9 for a period of 10 d after germination. As expected, pepck1 hypocotyls were the smallest and the mc9 hypocotyl growth was significantly compromised (P value < 0.05) in comparison to that of Col-0. Reciprocally, 35S:MC9 hypocotyls were the longest (17.4, 36.9, and 56.3% longer than Col-0, mc9, and pepck1 hypocotyls, respectively) (Figures 6B and 6C). These results are indicative of a positive regulatory effect of MC9 on the PEPCK1 activity during gluconeogenesis.
Figure 6.
Involvement of MC9 in Gluconeogenesis.
(A) PEPCK activity as measured in Arabidopsis seedling extracts from pepck1, 35S:MC9, Col-0, and mc9. One unit of PEPCK activity corresponded to the production of 1 μmol product per min at 25°C. Values represent the mean of five replicate measurements ±95% confidence interval. Percentages indicate the observed decrease in PEPCK1 activity relative to 35S:MC9 extracts. FW, fresh weight.
(B) Hypocotyl length of the same plant lines grown in the dark without Suc. Values represent the mean length of more than 100 plants per line ±95% confidence interval. Percentages indicate the observed decrease in hypocotyl length relative to the 35S:MC9 plants.
(C) Dark-grown pepck1, 35S:MC9, Col-0, and mc9 plants without Suc for 10 d.
[See online article for color version of this figure.]
MC9 Subcellular Localization
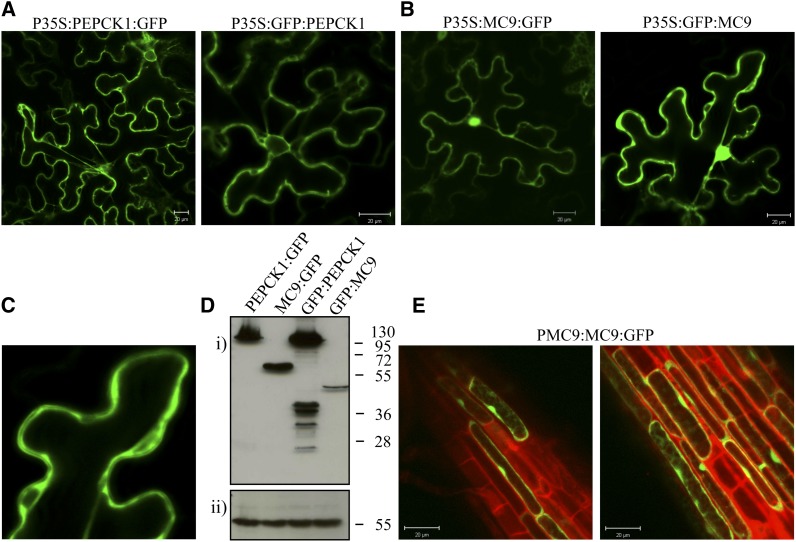
Because of the apparent discrepancy between the cytosolic localization of PEPCK1 and the detection of MC9 in the apoplast of 35S:MC9 lines (Vercammen et al., 2006), we reassessed the subcellular localizations of MC9 and PEPCK1 by transiently expressing N- and C-terminal green fluorescent protein (GFP)–tagged MC9 and PEPCK1 in tobacco (Nicotiana benthamiana) leaves (for a detailed description, see Supplemental Methods 5 online). PEPCK1 fusion proteins were exclusively localized in the cytosol (Figure 7A) and N- and C-terminal MC9 fusion proteins in the nucleocytoplasm (Figures 7B and 7C), but not in the apoplast, possibly due to the GFP instability in the acidic apoplast environment (Ward, 1981; Patterson et al., 1997). To exclude that the nuclear GFP signal was due to diffusion of free GFP after proteolytic processing of the fusion protein, we evaluated the integrity of the MC9 fusion proteins by protein gel blot analysis (Figure 7D). An ∼45-kD GFP:MC9 protein was found that corresponded with the 25-kD GFP protein fused with the large p20 subunit of MC9 (20 kD). In this case, the small p10 subunit (15 kD) was removed after autocatalytic activation of the protease (Vercammen et al., 2004), indicating that MC9 was active in the observed subcellular compartments. For the C-terminal MC9:GFP fusion, an ∼60-kD protein correlated with the full length of the fusion protein, revealing that the GFP fusion at the C terminus of MC9 might have interfered with the proteolytic separation of the two subunits. However, MC9 localized in the same cellular compartments irrespective of the GFP fusion conformations.
Figure 7.
Subcellular Localization of MC9 and PEPCK1.
(A) Localization of PEPCK1 with C-terminal and N-terminal GFP fusions under the control of the cauliflower mosaic virus 35S promoter. Bars = 20 μm.
(B) Localization of MC9 in cytosol and nucleus of tobacco leaf cells under the control of the 35S promoter. Bars = 20 μm.
(C) Detail of the MC9 cytosolic localization.
(D) Immunodetection of MC9 and PEPCK1 via their GFP tag (i) and of catalase 2 (CAT2) as gel loading control (ii).
(E) Localization of MC9 in cytosol and nucleus of Arabidopsis root epidermal cells under the control of its native promoter. Bars = 20 μm.
To confirm the nucleocytoplasmic localization of MC9, we additionally analyzed Arabidopsis roots of the ProMC9:MC9:GFP reporter lines. Interestingly, the localization of MC9 in the nucleus and cytoplasm of epidermal cells was confirmed (Figure 7E). Altogether, our localization study demonstrated that MC9, besides its apoplastic location, is also present in nucleus and cytoplasm.
DISCUSSION
To date, our knowledge on metacaspase substrates is limited to the At Serpin 1 inhibitor of At MC9 (Vercammen et al., 2004), the udor staphylococcal nuclease (TSN) target of Norway spruce mcII-pa (Sundström et al., 2009), the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) target of YCA1 (Silva et al., 2011), the metacaspase Tb MCA4 target of Tb MCA3 (Proto et al., 2011), and the Escherichia coli elongation factor (EF-Tu) target of Tb MCA2 (Moss et al., 2007). As EF-Tu was cleaved upon the bacterial production of the recombinant Tb MCA2, the EF-Tu cleavage had not been demonstrated to be a physiologically relevant proteolytic event (Moss et al., 2007). Interestingly, we also identified an elongation factor Tu family protein (AT4G02930) to be cleaved in vitro and in vivo by MC9 (Table 1). Furthermore, although the cleavage sites were not identical as reported previously, the Arabidopsis TSN-1 (AT5G07350), TSN-2 (AT5G61780), and GAPDH B subunit (AT1G42920), a protein sharing 37% amino acid sequence similarity to the YCA1 substrate GAPDH, were all cleaved by rMC9 in vitro at conserved Arg or Lys residues (see Supplemental Data Set 1C online). Therefore, it is tempting to hypothesize that metacaspase substrates are conserved across kingdoms, a criterion proposed as a possible parameter to distinguish physiologically relevant substrates (Crawford and Wells, 2011). An alternative strategy to prioritize protease substrates in downstream detailed functional analyses would be the combination of selected reaction monitoring with positional proteomics, an approach recently used to determine the kinetics of caspase-mediated proteolysis (Agard et al., 2012). In that study, the catalytic efficiencies for hundreds of substrates provided a quantitative understanding of proteolysis in biological processes.
Here, we report on the proteome-wide identification of substrates for a plant protease by means of positional proteomics. The number of cleaved proteins identified in the in vitro analysis was clearly much higher than that in vivo, possibly because within an in vitro setting the substrate/protease colocalization prerequisite for cleavage is abolished and because the recombinant MC9 level is higher than the cellular one. By imposing the established Arg/Lys specificity, the combined in vitro and in vivo approaches have revealed a set of 74 potential physiological MC9 substrates (Table 1) and have led to two revised cleavage signatures of MC9 for either cleavage after Arg or Lys, containing basic residues (Arg or Lys) at substrate position P3 and acidic residues (Asp or Glu) at position P1′. To exclude the possibility that the MC9 substrate selection was biased by stress responses induced by the MC9 mutation in the transgenic plant lines and led to differential protein upregulation or downregulation, we examined the expression levels of the endoplasmic reticulum stress response marker genes (Liu et al., 2007) (for the method description, see Supplemental Methods 2 online). This stress response, also known as the unfolded protein response, triggers the protein folding activities in the endoplasmic reticulum by the induction of marker genes. As shown in Supplemental Figure 7 online, none of these marker genes was induced in the MC9 gain-of-function and loss-of-function lines, indicating that the mc9 mutation did not provoke a stress response.
By two different in vitro approaches, we validated the MC9 cleavage of GRF5, PEPCK1, CSY3, GMD2, GRP7, PORB, trypsin inhibitor-2, NAD synthetase, a pentatricopeptide repeat–containing protein, and a LEA domain–containing protein. The functional importance of PEPCK1 cleavage by MC9 was demonstrated in planta because the PEPCK1 activity was enhanced by MC9 cleavage and it translated into improved seedling establishment. Until now, the functionality of MC9 remained elusive, mostly due to the lack of phenotypic aberrations in MC9 gain-of-function and loss-of-function mutants. Stirred by the discovery of the MC9-driven cleavage of PEPCK1 and its role in seed germination and early seedling development, we report a hitherto unknown phenotypic aberration caused by a perturbation in MC9 expression. We believe that further confirmation of the metacaspase functionalities implied by this MC9 degradome study will greatly improve our knowledge of plant proteases.
A Role for MC9-Dependent Proteolysis in Seed Development and Metabolic Processes
Within the MC9 degradome, proteins of various functional categories are present, supporting the proposed multifunctionality of plant metacaspases (Tsiatsiani et al., 2011). In analogy with caspases, multiple transcription and translation elongation factors, ribosomal proteins, and structural components are cleaved by MC9 (Crawford et al., 2012). Moreover, proteins involved in seed development, such as several LEA proteins, are frequently found among the cleaved proteins. LEA or LEA domain–containing proteins are small proteins encoded by multigene families in plants. They accumulate during late stages of seed development, are very hydrophilic, thermostable, and unfolded in solution, and might gain a considerable degree of ordered structures upon water loss. Moreover, they dissociate aggregated proteins and exhibit an overall protein stabilization function that could protect cell membranes during seed dehydration and osmotic stress (Galau et al., 1987; Chakrabortee et al., 2007; Boucher et al., 2010). Recently, certain LEA and other chaperone proteins from Medicago truncatula, such as a Gly-rich RNA binding protein similar to the GRP7 MC9 substrate, have been associated with seed longevity and protection against desiccation-induced protein aggregation (Chatelain et al., 2012). Possibly, MC9 cleavage of LEA proteins during water stress could induce the conformational changes necessary for LEA protein folding and their protective functions. This hypothesis is supported by the cleavage patterns of LEA proteins from which the MC9-dependent processing could not be deduced as a destructive event.
The MC9-dependent PEPCK1 proteolysis indicated its involvement in gluconeogenesis by regulating PEPCK1 activity. Although Arabidopsis PEPCK1 phosphorylation has not been demonstrated yet, the putative Ser-62 phosphosite that is phosphorylated in tomato (Solanum lycopersicum), cucumber, and Guinea grass is conserved in the Arabidopsis protein sequence (Leegood and Walker, 2003; Bailey et al., 2007). Hence, cleavage of PEPCK1 at the identified Arg-101 cleavage site removes the N-terminal fragment containing this putative inhibitory phosphorylated Ser residue and thus recovers the PEPCK1 activity. Despite the N-terminomics data indicating that the intensity of the PEPCK1 fragments was higher in Col-0 or 35S:MC9 than in mc9 (Figure 5A), the PEPCK1 precursor was reduced strikingly only in the 35S:MC9 extract, without the appearance of discrete proteolytic fragments (Figure 5C), possibly because cleavage fragments at the COFRADIC-identified sites are unstable or processed further downstream, thereby losing the necessary epitopes for antibody recognition. Although high-quality MS/MS data from single peptides accurately identify protein N termini and sites of proteolytic processing, they cannot always be detected by protein gel blot analysis or other gel-based validation methods (Timmer et al., 2009).
Another MC9 substrate involved in metabolic processes is CSY3, an enzyme catalyzing the conversion of acetyl-CoA and oxaloacetate to citrate at the onset of the glyoxylate cycle in germinating seeds. Notably, the glyoxylate cycle and gluconeogenesis are consecutive steps in the catabolism of fatty acids into sugars during seed germination and seedling establishment suggesting that this pathway regulation by MC9 may happen at different levels.
Metacaspase Maturation
As MC9 is autocatalytically activated by proteolytic separation of its p20 and p10 subunits (Vercammen et al., 2004), it was not surprisingly detected in the in vivo degradome (Table 1). However, the autoprocessing site Arg-183 found upon bacterial overproduction was not identified. The neo-N-terminal MC9 peptide identified in both 35S:MC9 and Col-0 N-terminomes was generated after cleavage at Arg-214, which is the first Arg residue following the previously reported autoprocessing site of rMC9 (Arg-183). It is possible that only the neo-N-terminal peptide 215DAGLKFR221 and not the corresponding peptide after cleavage at Arg-183 and Arg-214 has been detected by MS/MS because the latter is too long. A double processing step during zymogen maturation has also been reported for most procaspases that are processed at two Asp residues (Fuentes-Prior and Salvesen, 2004). Similarly, the Norway spruce metacaspase mcII-Pa is processed at Arg-188 and Lys-269 (Bozhkov et al., 2005). Therefore, we suggest that MC9 maturation involves processing at two sites, at Arg-183 and Arg-214, but whether cleavage at Arg-214 is necessary for proteolytic activity remains to be established. The recombinant MC4 is also processed at another site besides the primary site at Lys-225 (Watanabe and Lam, 2011b), whereas Arg-190, a site that is conserved in MC5, corresponds to one of the two processing sites in mcII-Pa (Bozhkov et al., 2005).
MC9 Specificity Properties
Thanks to the analysis of the identified cleavage sites in the in vivo and in vitro MC9 degradomes, we could refine the MC9 cleavage signature. We demonstrated that Lys is relatively preferred over Arg at the P1 site, a finding not completely in agreement with the obtained PS-SCL results (Vercammen et al., 2006). Additionally, we observed differences in the specificity derived from the in vitro COFRADIC and the PS-SCL data concerning the P2 site. One obvious explanation might be that native proteins were analyzed with COFRADIC and that the protease interaction with its substrates took place under physiological conditions, whereas with PS-SCL, small synthetic tetrapeptides were used (Turk et al., 2012). Moreover, in the calculation of the PS-SCL scores, the natural amino acid occurrence was not taken into account. Due to the limitation of PS-SCL in screening only sequences N-terminal to the cleavage bond, the role played by the C-terminal sequences in steering the MC9 specificity could not have been detected.
We demonstrated that the P1′ site plays an important role in determining the enzyme specificity and is often occupied by acidic amino acids. This property also might differentiate metacaspases from caspases, with the latter generally preferring small amino acid residues, such as Ser and Ala at P1′ (Timmer and Salvesen, 2007), besides the strong metacaspase preference for basic residues at P1. Another important observation in accordance with the known tetrapeptide MC9 specificity (Vercammen et al., 2006) is the MC9 preference for basic P3 and P1 residues. In this way, KxK, KxR, RxR, and RxK combinations as in VRPR-AMC were very frequently cleaved in native seedling proteins by MC9. Although this result could hint at subsite cooperativity (Schilling and Overall, 2008) between P3 and P1 for protease specificity, such a conclusion remains premature without supporting structural and kinetics data.
The crystal structure of the T. brucei type-I metacaspase MCA2 has recently been resolved shown to act as monomer activated in the presence of its substrate and calcium upon removal of a N-terminal domain normally blocking substrate access to the active site (McLuskey et al., 2012). As type-II metacaspases are found in plants only and, in contrast with type-I metacaspases, their processing is required for their activity (Vercammen et al., 2007), we assume there is a different activation mechanism for type-II metacaspases. Therefore, resolution of a type-II metacaspase structure is highly anticipated because it will help explain how substrates are recognized and bound.
Perspectives
The comprehensive catalog of MC9 substrates presented here will help follow-up studies on the functionality and relevance of metacaspase-dependent proteolysis. Production and characterization of gain-of-function and loss-of-function mutants, in which either the selected substrate levels are perturbed or the metacaspase cleavage sites are mutated, will provide more detailed insights into the specific roles of plant metacaspases in various biological processes. Indeed, activities and expression of metacaspases have been reported, for example, during xylem differentiation (Bollhöner et al., 2013). In addition, similar degradome studies of other Arabidopsis metacaspases, such as MC4, will unravel further the cleavage specificity of plant metacaspases and the potential redundancy or exclusivity in their substrates.
METHODS
MC9 Expression Analysis
ProMC9:GUS reporter lines were generated as described in Supplemental Methods 1 online, and plants at different stages were used for the expression analysis of MC9. In summary, whole plants or separate organs were incubated in 90% (v/v) acetone for 10 min at room temperature, washed in PBS, pH 7.4, and then incubated in 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 2 mM ferricyanide, and 0.5 mM ferrocyanide in 100 mM phosphate buffer, pH 7.4, at 37°C in the dark for 4 h to overnight. The material was cleared with 85% lactate solution and observed under stereomicroscopy and conventional light microscopy. Total RNA was extracted from 2-d-old seedlings as described (Meng and Feldman, 2010) with modifications. cDNA was prepared with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Real-time quantitative PCR was performed in a final volume of 20 µL, including 5 μL of eightfold diluted cDNA, 250 nM of each primer, and 50% (v/v) SYBR® green mix (Invitrogen), with an iCycler instrument (Bio-Rad), according to the manufacturer’s instructions. The housekeeping gene UBIQUITIN C (AT5G25760) was used as internal reference and for normalization of transcript levels. The gene-specific primer sequences used for the quantitative RT-PCR are listed in Supplemental Table 1 online. For a detailed description, see Supplemental Methods 2 online.
N-Terminal COFRADIC: Sample Preparation, Peptide Identification, and Quantification
Seeds of Arabidopsis thaliana, accession Col-0, the MC9 T-DNA insertion line (GK-506H04), and the MC9 overexpression line (Vercammen et al., 2006) were placed in round plastic plates containing wet Whatman paper for stratification at 4°C in the dark. After 3 d, seeds were transferred to 21°C with a 16-h-light/8-h-dark photoperiod with 80 to 100 µmol m−2 s−1 light intensity and left to germinate for 48 h. Young seedlings, including seed coats, were collected, frozen in liquid nitrogen, and ground into a fine powder with mortar and pestle. To achieve a total protein content of ∼4 mg, 0.5 g frozen ground tissue was resuspended in 1 mL buffer containing 1% (w/v) 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.5% (w/v) deoxycholate, 0.1% (w/v) SDS, 5 mM ethylenediaminetetraacetic acid, and 10% glycerol in PBS, pH 7.5, further containing the suggested amount of protease inhibitor mixture (one tablet/10 mL buffer) according to the manufacturer’s instructions (Roche Applied Science). The sample was centrifuged at 16,000g for 10 min at 4°C, and guanidinium hydrochloride was added to the cleared supernatant to reach a final concentration of 4 M.
For the in vitro COFRADIC analysis, protein extracts were prepared in the MC9 optimal activity buffer consisting of 50 mM MES, pH 5.5, 150 mM NaCl, 10% (w/v) Suc, 0.1% (w/v) CHAPS, and 10 mM dithiothreitol. Four milligrams of total seedling protein extract (48 h after germination) was incubated at 30°C with recombinant MC9 (Vercammen et al., 2004) for a total duration of 1 h; every 20 min, 10 μg of fresh protease (855 units) was preactivated in the same optimal activity buffer at room temperature for 15 min and then added to the protein extract to a total amount of 30 μg protease. Enzyme activity was 57 units μL−1, and one unit corresponded to the enzyme activity that catalyzes the hydrolysis of 1 μmol VRPR-AMC per min at 30°C. The same procedure was used for the control sample, but without addition of rMC9. By means of the gel analysis option in the ImageJ software (Abramoff et al., 2004), the level of endogenous MC9 protein in Col-0 extracts was estimated to be 14-fold lower than that in 35S:MC9 extracts and ∼104-fold lower than the total amount of recombinant MC9 (30 µg) used in the in vitro COFRADIC analysis. Protein concentrations were measured with the DC protein assay (Bio-Rad), and protein extracts were further modified for N-terminal COFRADIC analysis as described (Staes et al., 2011).
All α-free amines, either preexisting or generated upon proteolysis, and all ε-amines (Lys residues) were blocked with N-hydroxysuccinimide esters of butyric acid–stable isotopic variants. For the analysis of the in vivo–generated samples, primary amines in the mc9 proteome were 12C4-butyrylated, whereas Col-0 or 35S:MC9 primary amines were 13C4-butyrylated. For the in vitro analysis, primary amines in the rMC9-treated sample were 13C4-butyrylated and the untreated proteome was 12C4-butyrylated. After equal amounts of the labeled proteomes had been mixed, tryptic digestion generated internal, non-N-terminal peptides that were removed by strong cation exchange at low pH (Staes et al., 2011). After reversed phase-high performance liquid chromatography peptide separations with 2,4,6-trinitrobenzene sulfonic acid modification, internal peptides were further depleted and N-terminal peptides were isolated. Finally, liquid chromatography–MS/MS analysis was used for peptide identification and quantification (for a more detailed description, see Supplemental Methods 3 online).
Given that Lys residues were all butyrylated and, therefore, uncleavable by trypsin (Gevaert et al., 2003; Van Damme et al., 2005, 2010; Staes et al., 2011), we expected to identify only tryptic peptides generated after Arg (Arg-C specificity; C-terminal to Arg). Therefore, in the search settings with one missed cleavage allowance, endoproteinase semi-Arg C/P was used that allows the identification of neo-N-terminal peptides generated after Arg-C cleavage at one terminus (using the “semi” setting) and cleavage events between Arg and Pro as well. Variable modifications were set to pyroglutamate formation of N-terminal Gln and fixed modifications included Met oxidation to its sulfoxide derivative, S-carbamidomethylation of Cys, and butyrylation (12C4 or 13C4) of the Lys side chain and peptide N termini. Identified peptides were quantified with the Mascot Distiller Toolbox (version 2.3.2.0; Matrix Science) in the precursor mode.
Assessment of Protein Cleavage
cDNA clones received from the ABRC were used as templates for in vitro transcription and translation (T7/T3 Coupled Reticulocyte Lysate System; Promega) of 35S-radiolabeled proteins, according to the manufacturer’s instructions. Translation products were prepared in 50-μL reactions, and one-tenth was used for each rMC9 digestion. The production of the recombinant MC9 protease has been described previously (Vercammen et al., 2004), and a range of concentrations (0 to 1000 nM) prepared in the MC9 activity buffer were included in the digestions. The reactions were incubated at 30°C for 30 min. The inactive MC9 mutant (rMC9C147AC29A) was used as a negative control (Belenghi et al., 2007). One-tenth of the digestion assay products was separated with SDS-PAGE (Laemmli, 1970). These gels were dried and exposed to phospho-imaging plates and scanned by a phosphor imager (445SI; Molecular Dynamics). PageRuler prestained protein ladder was used as protein marker (Thermo Scientific), and apparent molecular weight was calculated with ImageJ software (Abramoff et al., 2004)
Recombinant PEPCK1 Production
The Arabidopsis PEPCK1 (AT4G37870) open reading frame was amplified by PCR from cDNA of Col-0 seeds, extended to complete attB1 and attB2 Gateway cloning sites, and cloned into the bacterial protein expression vector pDEST17 (primer sequences are listed in Supplemental Table 1 online). Bacterial (BL21-DE3-pLysE) protein production was induced in Luria Broth–growing cultures of successfully transformed bacterial clones with 0.2 mM isopropyl β-d-1-thiogalactopyranoside at 20°C overnight. Denatured PEPCK1 protein was isolated from protein inclusion bodies and subsequently purified on HIS-Select Nickel Affinity Gel resin (Sigma-Aldrich) in a buffer consisting of 50 mM sodium phosphate, 0.3 mM NaCl, and 4 M urea. For the production of the Arabidopsis PEPCK1 antibodies, 0.2 mg of purified recombinant protein was injected four times as antigen into each rabbit (Eurogentec). The rabbit antiserum was used at a 1:5000 dilution for PEPCK1 immunodetection in plant extracts.
Protein Immunodetection
Proteins were immunodetected in 2-d-old Arabidopsis seedling extracts (or infiltrated tobacco [Nicotiana benthamiana] leaves) prepared in 1% (w/v) CHAPS, 0.5% (w/v) deoxycholate, 0.1% (w/v) SDS, 5 mM ethylenediaminetetraacetic acid, and 10% glycerol in PBS at pH 7.5 and containing the suggested amount of protease inhibitor mixture (one tablet/10 mL buffer) according to the manufacturer’s instructions (Roche Applied Science). Extracts were centrifuged at 16,000g for 10 min at 4°C, and cleared supernatants were immediately mixed with SDS-PAGE loading buffer (Laemmli, 1970) and heated at 95°C for 5 min. Of the protein extract, 40 µg was used for the immunodetection of PEPCK1, MC9, CAT2 (AT4G35090), or GFP. For reprobing purposes, Immobilon P (Millipore) membranes were stripped with 0.1 M 2-mercapoethanol, 2% SDS, and 62.5 mM Tris-HCl, pH 6.7. Immunoreactive proteins were visualized with a chemiluminescence kit (Western Lightning Plus-ECL; Perkin-Elmer).
PEPCK1 Activity Assay
Seeds were set to germinate on wet Whatman paper for 6 d at 21°C after a 3-d stratification period at 4°C in the dark. Sampling included the remaining seed coats. The carboxylation activity of PEPCK1 was measured spectrophotometrically at 340 nm as previously described (Malone et al., 2007) on UV-Star 96-well plates (Greiner Bio-One). One unit of PEPCK1 activity corresponded to the production of 1 μmol product per min at 25°C. The values represent the mean of five measurements ±95% confidence interval (Cumming et al., 2007).
Hypocotyl Elongation Assay
During the hypocotyl elongation experiments, seeds were sown on half-strength Murashige and Skoog salts without Suc and then stratified for 3 d at 4°C in the dark. Prior to transfer to a Lovibond incubator (24 h in the dark at 21°C), seeds were exposed for 30 min to light. Hypocotyl lengths were measured to the nearest millimeter with the ImageJ software.
Subcellular Protein Localization
N- and C-terminal GFP fusions of MC9 and PEPCK1 were generated as described in Supplemental Methods 5 online. For imaging, a confocal microscope 100M with software package LSM 510 version 3.2 (Zeiss) was used. Excitation was done with the 488-nm line of an argon laser. Emission fluorescence for GFP and for propidium iodide was captured via a 500- to 550-nm band-pass filter and a 585-nm long-pass filter, respectively.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT5G04200, AT4G37870, AT1G79340, AT1G47540, AT5G16050, AT2G21660, AT4G27440, AT2G42560, AT1G55090, AT3G51160, AT1G02150, AT2G42790, AT4G02930, AT5G07350, AT5G61780, AT1G42920, AT1G02170, AT4G25110, AT5G64240, AT1G79330, AT1G79320, AT1G79310, and AT1G16420. Furthermore, the complete set of accession numbers listed in this study is included in the Supplemental Data Set 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. General Workflow of the Positional Proteomics Methodology and Experimental Setups.
Supplemental Figure 2. Venn Diagram Generated by VENNY (Oliveros, 2007).
Supplemental Figure 3. MC9 Specificity Monitored via Its in Vitro Proteome-Wide Substrate Profiling.
Supplemental Figure 4. iceLogo Illustrating the Amino Acid Frequencies in Substrate Positions P4-P3′ (Colaert et al., 2009).
Supplemental Figure 5. Reverse-Phase HPLC UV Absorbance Chromatogram Profiles of the Trypsin Inhibitor 2, GRF5, GRP7, and PORB Oligopeptides with the MC9 Cleavage Sites.
Supplemental Figure 6. Reverse-Phase HPLC UV Absorbance Chromatogram Profiles of the PEPCK1, GRP7, and a LEA Domain–Containing Protein Oligopeptides with the MC9 Cleavage Sites.
Supplemental Figure 7. Expression of the UPR Marker Genes in the Wild-Type (Col-0), the Loss-of-Function (mc9), and the Gain-of-Function (35S:MC9) Transgenic Plant Lines.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplemental Methods 1. Generation of Reporter Lines and MC9 Tissue Expression Analysis.
Supplemental Methods 2. RNA Extraction and qRT-PCR Gene Expression Assays.
Supplemental Methods 3. Peptide Identification and Quantification.
Supplemental Methods 4. Assessment of Synthetic Peptide Cleavage.
Supplemental Methods 5. Subcellular Protein Localization.
Supplemental Data Set 1A. List of Potential Endogenous MC9 Substrates Identified in the Col-0 Proteome.
Supplemental Data Set 1B. List of Potential Endogenous MC9 Substrates Identified in the 35S:MC9 Proteome.
Supplemental Data Set 1C. List of Potential Endogenous MC9 Substrates Generated by the rMC9-Driven Proteolysis Added to the mc9 Proteome.
Supplemental Data Set 1D. Arabidopsis Proteins That Contain the Suggested MC9 Specificity Consensus Sequences.
Acknowledgments
We thank Richard Leegood for useful discussion and the kind gift of the pepck1 T-DNA insertional mutant seeds, Davy Maddelein for the generation of heat maps from the PS-SCL data of Figure 3C, Pierre Hilson for fruitful comments, and Martine De Cock for help in preparing the article. This work was supported by the Research Foundation-Flanders (Grant G.0038.09N) and Ghent University (Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” project; grant 01MRP510W). L.T. was the recipient of a VIB International PhD Program predoctoral fellowship. D.V. was and P.V.D. is a Postdoctoral Fellow of the Research Foundation-Flanders. S.S. is indebted to the Special Research Fund of Ghent University for a postdoctoral fellowship.
AUTHOR CONTRIBUTIONS
L.T., K.G., and F.V.B. conceived the project. L.T., D.V., and P.V.D. designed the experiments. L.T., E.T., P.-J.D.B., T.B., and S.S. performed the research. L.T. and A.S. analyzed the data. B.v.d.C. and M.G. contributed to the development of analytic tools. L.T., with the help of K.G. and F.V.B., wrote the article.
Glossary
- COFRADIC
combined fractional diagonal chromatography
- Col-0
Columbia-0
- GUS
β-glucuronidase
- MS/MS
tandem mass spectrometry
- PS-SCL
positional scanning of a synthetic combinatorial library
- AMC
7-amino-4-methylcoumarin
- LEA
late-embryogenesis abundant
- NAD
nicotinamide adenine dinucleotide
- GFP
green fluorescent protein
References
- Abramoff M., Magalhães P., Ram S. (2004). Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Agard N.J., Mahrus S., Trinidad J.C., Lynn A., Burlingame A.L., Wells J.A. (2012). Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc. Natl. Acad. Sci. USA 109: 1913–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambit A., Fasel N., Coombs G.H., Mottram J.C. (2008). An essential role for the Leishmania major metacaspase in cell cycle progression. Cell Death Differ. 15: 113–122 [DOI] [PubMed] [Google Scholar]
- Aravind L., Dixit V.M., Koonin E.V. (1999). The domains of death: Evolution of the apoptosis machinery. Trends Biochem. Sci. 24: 47–53 [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (2002). Classification of the caspase-hemoglobinase fold: Detection of new families and implications for the origin of the eukaryotic separins. Proteins 46: 355–367 [DOI] [PubMed] [Google Scholar]
- Avci U., Petzold H.E., Ismail I.O., Beers E.P., Haigler C.H. (2008). Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 56: 303–315 [DOI] [PubMed] [Google Scholar]
- Bailey K.J., Gray J.E., Walker R.P., Leegood R.C. (2007). Coordinate regulation of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase by light and CO2 during C4 photosynthesis. Plant Physiol. 144: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenghi B., Romero-Puertas M.C., Vercammen D., Brackenier A., Inzé D., Delledonne M., Van Breusegem F. (2007). Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 282: 1352–1358 [DOI] [PubMed] [Google Scholar]
- Bollhöner B., Zhang B., Stael S., Denancé N., Overmyer K., Goffner D., Van Breusegem F., Tuominen H. (July 8, 2013). Post mortem function of AtMC9 in xylem vessel elements. New Phytol.http//dx..org/10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- Bonneau L., Ge Y., Drury G.E., Gallois P. (2008). What happened to plant caspases? J. Exp. Bot. 59: 491–499 [DOI] [PubMed] [Google Scholar]
- Boucher V., Buitink J., Lin X., Boudet J., Hoekstra F.A., Hundertmark M., Renard D., Leprince O. (2010). MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 33: 418–430 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov P.V., Suarez M.F., Filonova L.H., Daniel G., Zamyatnin A.A., Jr, Rodriguez-Nieto S., Zhivotovsky B., Smertenko A. (2005). Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc. Natl. Acad. Sci. USA 102: 14463–14468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahomchuen T., Akiyama K., Sekito T., Sugimoto N., Okabe M., Nishimoto S., Sugahara T., Kakinuma Y. (2009). Tributyltin induces Yca1p-dependent cell death of yeast Saccharomyces cerevisiae. J. Toxicol. Sci. 34: 541–545 [DOI] [PubMed] [Google Scholar]
- Chakrabortee S., Boschetti C., Walton L.J., Sarkar S., Rubinsztein D.C., Tunnacliffe A. (2007). Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl. Acad. Sci. USA 104: 18073–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain E., Hundertmark M., Leprince O., Le Gall S., Satour P., Deligny-Penninck S., Rogniaux H., Buitink J. (2012). Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 35: 1440–1455 [DOI] [PubMed] [Google Scholar]
- Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009). Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6: 786–787 [DOI] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Crawford E.D., Seaman J.E., Barber A.E., II, David D.C., Babbitt P.C., Burlingame A.L., Wells J.A. (2012). Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death Differ. 19: 2040–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford E.D., Wells J.A. (2011). Caspase substrates and cellular remodeling. Annu. Rev. Biochem. 80: 1055–1087 [DOI] [PubMed] [Google Scholar]
- Cumming G., Fidler F., Vaux D.L. (2007). Error bars in experimental biology. J. Cell Biol. 177: 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Alvarado A., Walker R.P., Leegood R.C. (2007). Phosphoenolpyruvate carboxykinase in developing pea seeds is associated with tissues involved in solute transport and is nitrogen-responsive. Plant Cell Environ. 30: 225–235 [DOI] [PubMed] [Google Scholar]
- Dittrich P., Campbell W.H., Black C.C., Jr (1973). Phosphoenolpyruvate carboxykinase in plants exhibiting Crassulacean acid metabolism. Plant Physiol. 52: 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G.E., Kanai R., Black C.C. (1971). Phosphoenolpyruvate carboxykinase in leaves of certain plants which fix CO2 by the C4-dicarboxylic acid cycle of photosynthesis. Biochem. Biophys. Res. Commun. 45: 278–285 [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P., Salvesen G.S. (2004). The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384: 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G.A., Bijaisoradat N., Hughes D.W. (1987). Accumulation kinetics of cotton late embryogenesis-abundant mRNAs and storage protein mRNAs: Coordinate regulation during embryogenesis and the role of abscisic acid. Dev. Biol. 123: 198–212 [DOI] [PubMed] [Google Scholar]
- Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G.R., Vandekerckhove J. (2003). Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21: 566–569 [DOI] [PubMed] [Google Scholar]
- González I.J., Desponds C., Schaff C., Mottram J.C., Fasel N. (2007). Leishmania major metacaspase can replace yeast metacaspase in programmed cell death and has arginine-specific cysteine peptidase activity. Int. J. Parasitol. 37: 161–172 [DOI] [PubMed] [Google Scholar]
- Hamann A., Brust D., Osiewacz H.D. (2007). Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina. Mol. Microbiol. 65: 948–958 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I., Hatsugai N. (2011). The role of vacuole in plant cell death. Cell Death Differ. 18: 1298–1304 Erratum. Cell Death Differ. 18: 1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms M.J., Ambit A., Appleton P., Tetley L., Coombs G.H., Mottram J.C. (2006). Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J. Cell Sci. 119: 1105–1117 [DOI] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K.A., Maldener C., Fröhlich K.-U., Wissing S., Büttner S., Fehr M., Sigrist S., Madeo F. (2004). Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164: 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen P.F., Overall C.M. (2012). N- and C-terminal degradomics: New approaches to reveal biological roles for plant proteases from substrate identification. Physiol. Plant. 145: 5–17 [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Hardwick J.M. (2005). Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 170: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Laverrière M., Cazzulo J.J., Alvarez V.E. (2012). Antagonic activities of Trypanosoma cruzi metacaspases affect the balance between cell proliferation, death and differentiation. Cell Death Differ. 19: 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.E.C., Brunette S., Puente L.G., Megeney L.A. (2010). Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 107: 13348–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.E.C., Puente L.G., Kærn M., Megeney L.A. (2008). A non-death role of the yeast metacaspase: Yca1p alters cell cycle dynamics. PLoS ONE 3: e2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood R.C., Walker R.P. (2003). Regulation and roles of phosphoenolpyruvate carboxykinase in plants. Arch. Biochem. Biophys. 414: 204–210 [DOI] [PubMed] [Google Scholar]
- Liu J.-X., Srivastava R., Che P., Howell S.H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Herker E., Maldener C., Wissing S., Lächelt S., Herlan M., Fehr M., Lauber K., Sigrist S.J., Wesselborg S., Fröhlich K.-U. (2002). A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9: 911–917 [DOI] [PubMed] [Google Scholar]
- Malone S., Chen Z.-H., Bahrami A.R., Walker R.P., Gray J.E., Leegood R.C. (2007). Phosphoenolpyruvate carboxykinase in Arabidopsis: Changes in gene expression, protein and activity during vegetative and reproductive development. Plant Cell Physiol. 48: 441–450 [DOI] [PubMed] [Google Scholar]
- Martens L., Vandekerckhove J., Gevaert K. (2005). DBToolkit: Processing protein databases for peptide-centric proteomics. Bioinformatics 21: 3584–3585 [DOI] [PubMed] [Google Scholar]
- Martín M., Rius S.P., Podestá F.E. (2011). Two phosphoenolpyruvate carboxykinases coexist in the Crassulacean Acid Metabolism plant Ananas comosus. Isolation and characterization of the smaller 65 kDa form. Plant Physiol. Biochem. 49: 646–653 [DOI] [PubMed] [Google Scholar]
- McLuskey K., Rudolf J., Proto W.R., Isaacs N.W., Coombs G.H., Moss C.X., Mottram J.C. (2012). Crystal structure of a Trypanosoma brucei metacaspase. Proc. Natl. Acad. Sci. USA 109: 7469–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Feldman L. (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol. J. 5: 183–186 [DOI] [PubMed] [Google Scholar]
- Meslin B., Beavogui A.H., Fasel N., Picot S. (2011). Plasmodium falciparum metacaspase PfMCA-1 triggers a z-VAD-fmk inhibitable protease to promote cell death. PLoS ONE 6: e23867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss C.X., Westrop G.D., Juliano L., Coombs G.H., Mottram J.C. (2007). Metacaspase 2 of Trypanosoma brucei is a calcium-dependent cysteine peptidase active without processing. FEBS Lett. 581: 5635–5639 [DOI] [PubMed] [Google Scholar]
- Ojha M., Cattaneo A., Hugh S., Pawlowski J., Cox J.A. (2010). Structure, expression and function of Allomyces arbuscula CDP II (metacaspase) gene. Gene 457: 25–34 [DOI] [PubMed] [Google Scholar]
- Oliveros, J.C. (2007). VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- Overall C.M., Tam E.M., Kappelhoff R., Connor A., Ewart T., Morrison C.J., Puente X., López-Otín C., Seth A. (2004). Protease degradomics: Mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol. Chem. 385: 493–504 [DOI] [PubMed] [Google Scholar]
- Patterson G.H., Knobel S.M., Sharif W.D., Kain S.R., Piston D.W. (1997). Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Rylott E.L., Gilday A.D., Graham S., Larson T.R., Graham I.A. (2004). Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E. (2012). Plant proteases - From detection to function. Physiol. Plant. 145: 1–4 [DOI] [PubMed] [Google Scholar]
- Proto W.R., Castanys-Munoz E., Black A., Tetley L., Moss C.X., Juliano L., Coombs G.H., Mottram J.C. (2011). Trypanosoma brucei metacaspase 4 is a pseudopeptidase and a virulence factor. J. Biol. Chem. 286: 39914–39925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J. (1993). Evolutionary families of peptidases. Biochem. J. 290: 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie D.L., Miley M.D., Bhabhra R., Robson G.D., Rhodes J.C., Askew D.S. (2007). The Aspergillus fumigatus metacaspases CasA and CasB facilitate growth under conditions of endoplasmic reticulum stress. Mol. Microbiol. 63: 591–604 [DOI] [PubMed] [Google Scholar]
- Rylott E.L., Gilday A.D., Graham I.A. (2003). The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol. 131: 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I., Berger A. (1967). On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27: 157–162 [DOI] [PubMed] [Google Scholar]
- Schilling O., Overall C.M. (2008). Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26: 685–694 [DOI] [PubMed] [Google Scholar]
- Silva A., Almeida B., Sampaio-Marques B., Reis M.I.R., Ohlmeier S., Rodrigues F., do Vale A., Ludovico P. (2011). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase. Biochim. Biophys. Acta 1813: 2044–2049 [DOI] [PubMed] [Google Scholar]
- Staes A., Impens F., Van Damme P., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., Gevaert K. (2011). Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 6: 1130–1141 [DOI] [PubMed] [Google Scholar]
- Sundström J.F., et al. (2009). Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Timmer J.C., Salvesen G.S. (2007). Caspase substrates. Cell Death Differ. 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Timmer J.C., Zhu W., Pop C., Regan T., Snipas S.J., Eroshkin A.M., Riedl S.J., Salvesen G.S. (2009). Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 16: 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L., Gevaert K., Van Breusegem F. (2012). Natural substrates of plant proteases: How can protease degradomics extend our knowledge? Physiol. Plant. 145: 28–40 [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P.V. (2011). Metacaspases. Cell Death Differ. 18: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B., Turk D., Turk V. (2012). Protease signalling: The cutting edge. EMBO J. 31: 1630–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren A.G., O’Rourke K., Aravind L., Pisabarro M.T., Seshagiri S., Koonin E.V., Dixit V.M. (2000). Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6: 961–967 [DOI] [PubMed] [Google Scholar]
- Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. (2005). Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2: 771–777 [DOI] [PubMed] [Google Scholar]
- Van Damme P., Maurer-Stroh S., Hao H., Colaert N., Timmerman E., Eisenhaber F., Vandekerckhove J., Gevaert K. (2010). The substrate specificity profile of human granzyme A. Biol. Chem. 391: 983–997 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Vartapetian A.B., Tuzhikov A.I., Chichkova N.V., Taliansky M., Wolpert T.J. (2011). A plant alternative to animal caspases: Subtilisin-like proteases. Cell Death Differ. 18: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D., Belenghi B., van de Cotte B., Beunens T., Gavigan J.-A., De Rycke R., Brackenier A., Inzé D., Harris J.L., Van Breusegem F. (2006). Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J. Mol. Biol. 364: 625–636 [DOI] [PubMed] [Google Scholar]
- Vercammen D., Declercq W., Vandenabeele P., Van Breusegem F. (2007). Are metacaspases caspases? J. Cell Biol. 179: 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D., van de Cotte B., De Jaeger G., Eeckhout D., Casteels P., Vandepoele K., Vandenberghe I., Van Beeumen J., Inzé D., Van Breusegem F. (2004). Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 279: 45329–45336 [DOI] [PubMed] [Google Scholar]
- Walker R.P., Acheson R.M., Técsi L.I., Leegood R.C. (1997). Phosphoenolpyruvate carboxykinase in C4 plants: Its role and regulation. Aust. J. Plant Physiol. 24: 459–468 [Google Scholar]
- Walker R.P., Chen Z.-H., Acheson R.M., Leegood R.C. (2002). Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant Guinea grass. Plant Physiol. 128: 165–172 [PMC free article] [PubMed] [Google Scholar]
- Walker R.P., Leegood R.C. (1995). Purification, and phosphorylation in vivo and in vitro, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett. 362: 70–74 [DOI] [PubMed] [Google Scholar]
- Walker R.P., Trevanion S.J., Leegood R.C. (1995). Phosphoenolpyruvate carboxykinase from higher plants: Purification from cucumber and evidence of rapid proteolytic cleavage in extracts from a range of plant tissues. Planta 196: 58–63 [Google Scholar]
- Ward, W.W. (1981). Properties of the coelenterate green-fluorescent proteins. In Bioluminescence and Chemiluminescence: Basic Chemistry and Analytical Applications, M.A. DeLuca and W.D. McElroy, eds (New York: Academic Press), pp. 235–242. [Google Scholar]
- Watanabe N., Lam E. (2005). Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 280: 14691–14699 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2011a). Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 66: 969–982 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2011b). Calcium-dependent activation and autolysis of Arabidopsis metacaspase 2d. J. Biol. Chem. 286: 10027–10040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalila H., González I.J., El-Fadili A.K., Delgado M.B., Desponds C., Schaff C., Fasel N. (2011). Processing of metacaspase into a cytoplasmic catalytic domain mediating cell death in Leishmania major. Mol. Microbiol. 79: 222–239 [DOI] [PMC free article] [PubMed] [Google Scholar]