Functional overlap between homologous genes in plants is considered a substantial limitation for identifying gene functions. A large online searchable collection of gene families targeting artificial microRNAs (amiRNAs) and 10 functionally pooled amiRNA libraries for large and targeted small scale screens are resources developed here as tools to explore novel overlapping gene functions.
Abstract
Traditional forward genetic screens are limited in the identification of homologous genes with overlapping functions. Here, we report the analyses and assembly of genome-wide protein family definitions that comprise the largest estimate for the potentially redundant gene space in Arabidopsis thaliana. On this basis, a computational design of genome-wide family-specific artificial microRNAs (amiRNAs) was performed using high-performance computing resources. The amiRNA designs are searchable online (http://phantomdb.ucsd.edu). A computationally derived library of 22,000 amiRNAs was synthesized in 10 sublibraries of 1505 to 4082 amiRNAs, each targeting defined functional protein classes. For example, 2964 amiRNAs target annotated DNA and RNA binding protein families and 1777 target transporter proteins, and another sublibrary targets proteins of unknown function. To evaluate the potential of an amiRNA-based screen, we tested 122 amiRNAs targeting transcription factor, protein kinase, and protein phosphatase families. Several amiRNA lines showed morphological phenotypes, either comparable to known phenotypes of single and double/triple mutants or caused by overexpression of microRNAs. Moreover, novel morphological and abscisic acid–insensitive seed germination mutants were identified for amiRNAs targeting zinc finger homeodomain transcription factors and mitogen-activated protein kinase kinase kinases, respectively. These resources provide an approach for genome-wide genetic screens of the functionally redundant gene space in Arabidopsis.
INTRODUCTION
Large gene families containing genes with highly similar or even identical sequences are common in various organisms and statistically more abundant in plants, including Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000). Genetic redundancy can include partially overlapping functions of gene family members (Tautz, 2000; Briggs et al., 2006; Kafri et al., 2009) and can be tightly connected to mechanistic robustness of cellular networks (Wagner, 2005). Functional overlap and partial or complete redundancy between different family members are considered to be reasons for the lack of observable phenotypes in single-gene deletion mutants, while at the same time higher order mutants show increasing severity of phenotypes (Arabidopsis Genome Initiative, 2000; Bouché and Bouchez, 2001; Cutler and McCourt, 2005; Kwak et al., 2003; Park et al., 2009).
Traditional forward genetic screens using diverse mutagens are limited in the targeted identification of functional overlapping (partially) redundant genes. A recent comprehensive study of phenotypes in Arabidopsis (Lloyd and Meinke, 2012) shows that only ∼591 (partially) redundant genes have been described to date. These studies underline the potential for novel tools that enable screens of the functionally redundant gene space. Different overexpression strategies (Tautz, 2000; Weigel et al., 2000; Briggs et al., 2006; Ichikawa et al., 2006; Kafri et al., 2009; Mitsuda et al., 2011) and RNA interference–based strategies (Abbott et al., 2002; Ott et al., 2005; Wagner, 2005) have been applied to address functional overlap. However, overexpression strategies may cause neomorphic and pleiotropic phenotypes that are difficult to interpret. RNA interference approaches (Abbott et al., 2002; Ott et al., 2005) may suffer from off-target effects as the processing of one double-stranded RNA generates multiple small RNAs with potentially undesired targets (Jackson et al., 2003; Wickham, 2009). An alternative in plants are artificial microRNAs (amiRNAs) (Schwab et al., 2005; Ossowski et al., 2008). AmiRNA precursors can be computationally designed to target a specific group of potentially redundant genes, and the amiRNA precursor is specifically processed to give one mature amiRNA (Schwab et al., 2006). However, so far, amiRNAs have not been designed on a genome-wide scale to target potentially redundant genes. Therefore family-targeting amiRNA libraries, which would enable assessment of redundancy in large-scale forward genetic screens, are presently not available.
Here, we report the genome-wide assembly of gene family definitions that include 22,020 proteins of the Arabidopsis proteome that encode gene family members. Using the derived integrated gene family definitions, we next computationally designed of over 2,000,000 multi-gene targeting amiRNAs against protein families in Arabidopsis using a high-performance computing resource (Triton Compute Cluster, University of California San Diego [UCSD]). The analyses are searchable online in a Web resource that integrates data from other large-scale data sets, including subcellular localization, transcriptome, interactome, and phylogenetic data. In addition, a subset of 22,000 amiRNAs in 10 sublibraries targeting over 18,000 genes has been computationally assembled for amiRNA library synthesis and was subsequently cloned as pools into a plant expression vector. A proof of concept forward genetic screen using over 120 amiRNAs shows the validity of the approach and identifies known and novel gene family-linked phenotypes. Considering the many large-scale seed germination screens for abscisic acid (ABA) insensitivity, it is remarkable that we found in our concept screen a novel ABA-insensitive mutant. The resources represent a new generation of potent tools for screening or targeted analysis of a yet poorly understood functionally redundant gene space in Arabidopsis.
RESULTS
Analysis of Genome-Wide Protein Families
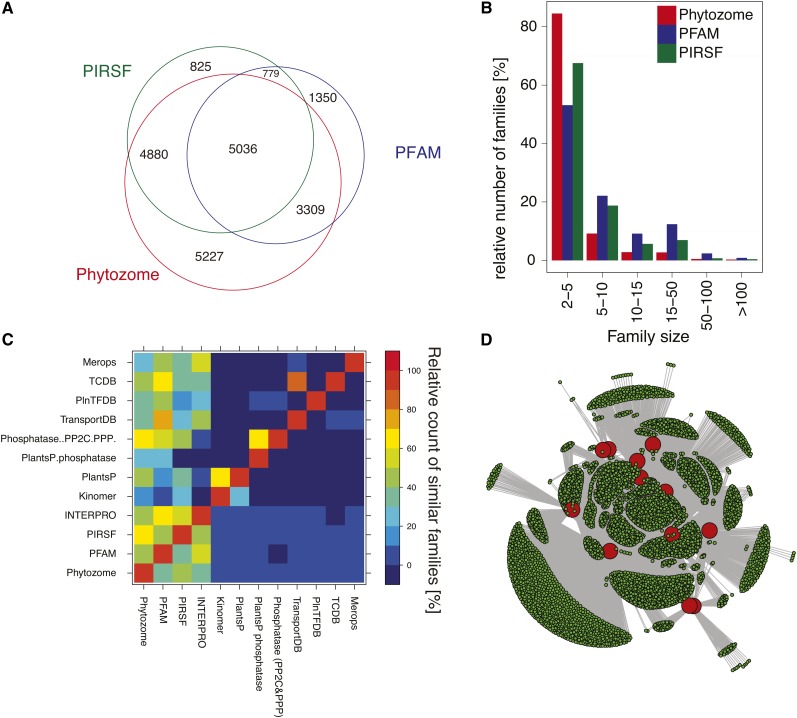
There are two main genetic mechanisms considered responsible for robustness against mutations (i.e., the lack of measurable phenotypes in loss of function mutants under diverse conditions). These are either buffering through alternative pathways or (partial) redundancy originating from gene duplication and coexpression of homologous genes (Hetherington, 2001; Leonhardt et al., 2004; Briggs et al., 2006; Delattre and Félix, 2009). The latter is relevant for our chosen amiRNA-based approach. Protein families and superfamilies are by definition groups of homologous sequences, which may be considered to contain all potential functionally redundant homologous proteins. There are several available data sets of Arabidopsis gene families, hereafter referred as family definitions, and it was not evident a priori whether one of these family definitions would be best for our novel amiRNA-based strategy. Therefore, we analyzed 16 existing gene family definitions of Arabidopsis (Figure 1; see Supplemental Table 1 online). Overall, 22,020 genes (80% of all protein coding genes) belong to families with at least two members (see Supplemental Figure 1 online). The majority of the families contain between two and five proteins (Figure 1B). However, the overlap of gene families among individual family definitions is relatively small (Figures 1C and 1D; see Supplemental Table 2 online). The largest groups of genes, which are found in the same families independent of the family definition, are F-box (98 genes), lipase (94 genes), and UDP-glucosyltransferase (93 genes) genes. Our analyses show that no single all-encompassing family definition exists. Phytozome is the most encompassing family definition, but inclusion of 11 additional family definitions is necessary to substantially increase combinatorial genome-wide coverage (Figure 1C; see Supplemental Table 2 online).
Figure 1.
Analysis of Publicly Available Family Definitions for the Proteome of Arabidopsis.
(A) Venn diagram of protein-coding loci in the three largest family definitions (Phytozome, PIRSF, and PFAM).
(B) The bar graph shows the relative distribution of family size in the Phytozome, PIRSF, and PFAM family definitions (100% is the total number of families in the individual family definition). The families are categorized in six groups according to their size.
(C) Heat map of a comparative analysis of family definitions. Note that all of the 12 depicted gene family definitions contribute additional gene families that were included for the genome-wide amiRNA design. The color scale represents the relative similarity between the family definitions (see Supplemental Figure 1 and Supplemental Tables 1 and 2 online for further details).
(D) Integrated view of 12 genome-wide Arabidopsis gene family definitions (red circles) with their associated gene families (green circles). Gray lines (edges) connect the 12 family definitions with their associated families. Gray lines also connect families that share members.
Genome-Wide Design of an AmiRNA Library
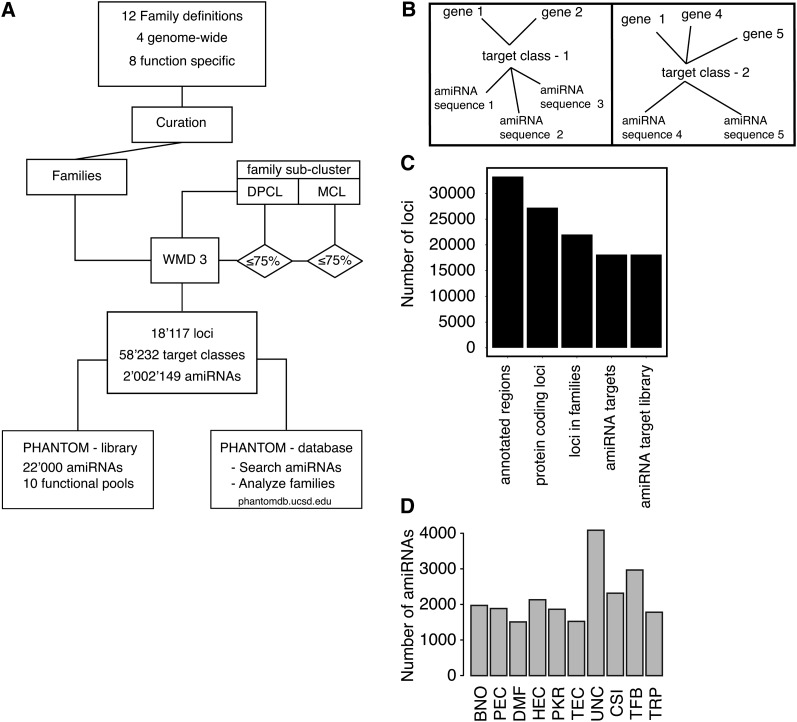
The design of amiRNAs against families defined in the selected family definitions was performed using the publicly available Web microRNA designer (WMD; Ossowski et al., 2008) in an iterative approach shown in Figure 2A and described in detail in Methods. The computational design of genome-wide amiRNAs targeting gene families required over 90,000 h of CPU time on a high-performance cluster (2048 CPUs and 256 nodes; Triton Compute Cluster, UCSD). Each family of the 12 individual definitions (Figures 1C and 1D; see Supplemental Table 1 online) was used as input for the computational design of amiRNAs. This resulted in a first set of amiRNAs with variable coverage depending on the family size and on sequence diversity within the family as well as the presence of potential off-target sites. In a second phase, an iterative approach was used to increase the coverage for all those families not covered to at least 75% of the family members by any combination of amiRNAs (Figure 2A). The families were first subclustered using a Dirichlet process clustering algorithm (DPCL; Brown, 2008), and these subclusters then were used as input. In cases where the coverage did not reach at least 75% by taking the amiRNAs generated in the first and second round into account, we used clusters obtained with the Markov chain clustering algorithm (MCL; Enright et al., 2002), with a configuration that led to smaller subclusters. Then, amiRNAs were designed against these smaller subclusters. In total, 2,002,149 unique amiRNAs were designed targeting 18,117 genes in 58,232 target classes (Figures 2B and 2C). A target class is a term for a group of genes targeted by one or several amiRNAs (Figure 2B). All amiRNAs target 18,117 of the 22,020 genes that belong to gene families consisting of at least two genes. Thus, the computationally designed amiRNAs correspond to 80% of all protein coding loci in the analyzed family definitions. Note that the lack of sufficient conservation of nucleotide sequence within members of a targeted gene family or the presence of similar nucleotide sequences in genes, which are not part of the targeted family (i.e., potential off-targets) precluded a complete coverage of all gene family members.
Figure 2.
Design of a Genome-Wide AmiRNA Collection Targeting Gene Families.
(A) Simplified schematic illustration of the workflow used for the computational iterative design of family-targeting amiRNAs. Starting with the analysis of family definitions, the amiRNA design was performed using whole families and using subclusters for increasing the number of targets per family. The resulting amiRNAs were assembled in a Web-accessible database. A selection of 22,000 amiRNAs was chosen for synthesis (see Results for details).
(B) Schematic representation of the relation between target classes, amiRNAs that target specific genes, and the targeted genes. Shown are two target classes that target two (left) or three (right) genes (targets). The target class is a term for a group of genes targeted by one or several amiRNAs.
(C) Overview of the total number of genes in the relevant groups (x axis from left to right): all annotated loci in the genome of Arabidopsis, all protein coding loci, all loci in the family definitions analyzed (see Supplemental Table 1 online), all loci targeted by any designed amiRNA, and all loci targeted by the 10 PHANTOM sublibraries.
(D) Size distribution of the functional classified amiRNA sublibrary pools represented in the designed PHANTOM library. TFB, transcription factors and other RNA and DNA binding proteins; PKR, protein kinases, protein phosphatases, receptors, and their ligands; HEC, hydrolytic enzymes (enzyme classification [EC] class 3), excluding protein phosphatases; CSI, proteins that form or interact with protein complexes including stabilization of those; TEC, metabolic and other enzymes catalyzing transfer reactions (EC class 2); PEC, catalytic active proteins, mainly enzymes; BNO, proteins binding small molecules; TRP, proteins that transport organic and inorganic molecules across membranes; DMF, protein with diverse functional annotation not found in the other categories; UNC, genes for which the function is not known or cannot be inferred.
This large-scale collection of family specific amiRNAs is available as a searchable resource targeting families that are part of the redundant gene space in Arabidopsis. Development of similar resources will prove useful for other organisms with a large redundant gene space. As a user-friendly resource, we created a website named PHANTOM DB (http://phantomdb.ucsd.edu) providing an interface to search the entire collection of amiRNAs. In addition, all 16 family definitions and family subclusters analyzed in this work are integrated, allowing the user to quickly identify closely related groups of genes. Data on subcellular localization (SUBA II; Heazlewood et al., 2007), gene expression levels from microarray experiments, interaction data (STRING; Szklarczyk et al., 2011), and approximate phylogenetic trees along with the alignment are provided as help for deciding which amiRNA is optimal for the intended experiment.
Assembly of a Library for Forward Genetic Screening
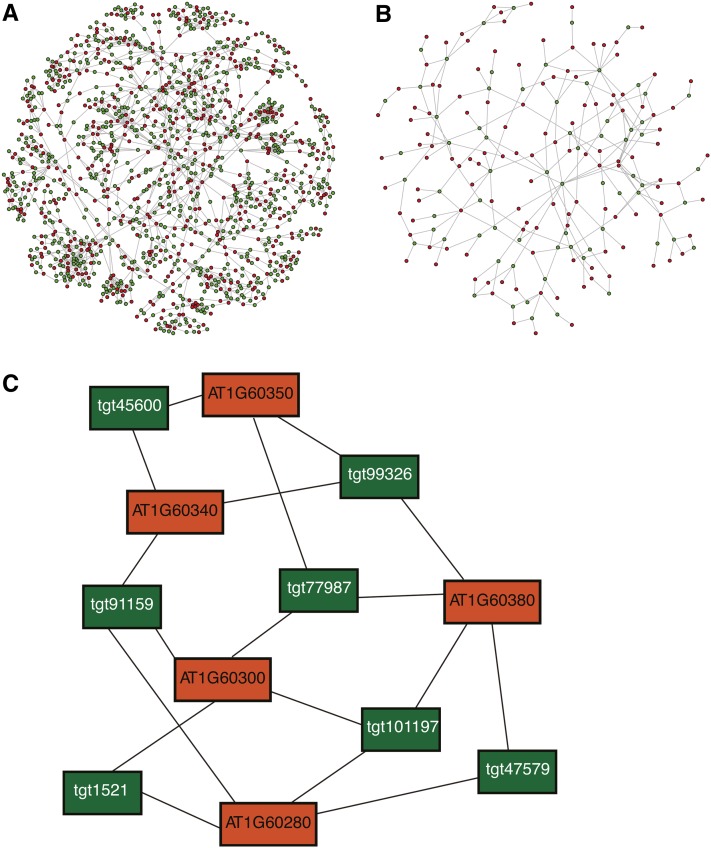
It is evident that a data set consisting of over 2,000,000 amiRNAs is very useful for targeted analyses of functionally redundant gene families but is not of interest for screening purposes in most research areas or when considering, for example, a saturating screen. Therefore, we selected a subset of 22,000 amiRNA sequences targeting the same 18,117 loci with 22,000 target classes based on criteria as follows. In brief, after filtering out amiRNAs targeting more than 16 genes, amiRNAs were selected based on different amiRNA properties, including the score assigned by WMD (for details, see Methods). In our algorithm, while maintaining the total number of targeted genes, no target class is chosen twice, but overlapping target classes were chosen to optimize silencing of the same genes with more than one amiRNA. This leads to a high number of different target gene combinations (Figure 3A). Furthermore, the portion of overlapping target gene sets (cf. Figures 3A and 3B), the number of gene combinations, and the robustness of the screens performed needed to be considered. Figure 3C illustrates the robustness of our combinatorial design, whereby five homologous genes (Figure 3C, orange boxes) are cotargeted by seven distinct amiRNAs (Figure 3C, green boxes), such that even if any single amiRNA is less effective, other amiRNAs overlap in their target genes. Overall, this approach leads to a robust and diverse library, hereafter named the PHANTOM library. The designed library contains mainly (96%) amiRNAs that target two to five genes (see Supplemental Figures 2D and 2E online). Targeting fewer genes is advantageous when considering dose dependency of amiRNA efficiency (Arvey et al., 2010) but even more so when considering follow-up experiments to identify the relevant genes.
Figure 3.
Visualization of Relationships between AmiRNAs (Green) and Targeted Genes (Red).
(A) The network shown visualizes the largest connected unit in the PHANTOM library. The library assembly procedure maximizes the number of unique groups of targeted genes. Thus, the connectivity and complexity of the network is high. The target genes are part of the Kinomer TKL (Tyr kinase-like kinases) family, by far the largest group of kinases in land plants (Martin et al., 2009). AmiRNAs (green circles, nodes) are connected by edges (gray lines) to their target genes (red circles, nodes). Green circles represent one amiRNA sequence.
(B) The network shown visualizes the connected unit in an alternative assembly procedure, which was tested for an initial PHANTOM library design but not pursued further (see Results). This alternative assembly procedure maximizes the number of amiRNAs with a minimal set of groups of targeted genes. This reduces the connectivity and complexity of the network. The target genes are part of the AP2 transcription factor family (Kim et al., 2006). AmiRNAs (green circles, nodes) are connected by edges (gray lines) to their target genes (red circles, nodes). Green circles represent here more than one amiRNA sequence.
(C) Example of a typical target class target gene network in the PHANTOM amiRNA library representing members of the NAC transcription factor family. Orange rectangles represent target genes, while green rectangles represent amiRNAs labeled by an identifier. Note that most gene pairs are targeted by two or more amiRNAs, and each amiRNA targets a separate group of genes, thus enhancing robustness and gene combinations for screening.
While 22,000 amiRNAs in one pool might be of interest for a large-scale screen, dividing the amiRNAs into smaller pools grouped according to protein functions increases the flexibility and allows smaller, more targeted screens. We reduced the GO-SLIM classification available from The Arabidopsis Information Resource (TAIR; Garcia-Hernandez et al., 2002) from 12 down to 10 classes (see Supplemental Table 3 online) and classified first the targeted loci and in a second step the target classes into these groups. In summary, the functionally subpooled PHANTOM library represents a robust and flexible set of amiRNAs targeting over 18,000 genes in different gene families (e.g., transporters [4632 genes and 1777 amiRNAs], transcription factors, and other DNA/RNA binding proteins [8611 genes and 2964 amiRNAs]) and even protein families of unknown function (11,554 genes and 4082 amiRNAs) (Figure 2D; see Supplemental Table 3 online).
Bar-coded oligonucleotides containing fragments of the amiRNA precursor were synthesized in situ (Cleary et al., 2004), and the recovered amiRNA precursors were cloned as described in Methods. A high-copy Gateway-compatible vector, which allows flexible and simple transfer into compatible plasmids of choice, was used as a backbone for the 10 amiRNA-precursor sublibraries. In a second set of 10 plasmid pools, the amiRNA precursors were transferred into a standard plant expression vector. To assess the quality of our library, we performed high-throughput sequencing analyses for both sets of the 10 sublibraries (20 and 21 bp amiRNA libraries [see Methods for details]). All 20 library pools contained ≥ 95% of all sequences designed for each sublibrary (see Supplemental Figure 3 online). All libraries are publicly available via the ABRC.
Design and Screen of a Small AmiRNA Library as Proof of Concept
We constructed 126 amiRNAs targeting transcription factors, protein phosphatases, and kinases (TPK library; Table 1; see Supplemental Figure 4 online) using the WMD algorithm for the design of amiRNAs described above. In total, 122 amiRNAs were transformed in Arabidopsis Columbia-0 (Col-0) harboring a pRAB18:GFP (for green fluorescent protein) reporter. The selected T1 generation was screened for visible morphological phenotypes, and the T2 generation was screened for ABA insensitivity during seed germination. In summary, for 122 transformed amiRNAs, 121 transformed T1 lines could be recovered. For one construct, no transformants could be isolated in several attempts (amiRNA-GRF: growth-regulating factor 1 transcription factors; see Supplemental Table 4 online). In three cases, the phenotype in the T1 generation was too variable between different T1 plants and due to the lack of sufficient seeds from independent T1 transformants could not be confirmed in independent T2 transformants (amiRNA-G2-like: G2-like transcription factor, eight targets; amiRNA-MYB1: c-MYB homolog transcription factor; amiRNA-MYB2: c-MYB homolog transcription factor; see Supplemental Table 4 online). The T2 generation of 116 amiRNA lines (at least three independent lines each) was screened for ABA insensitivity in seed germination. In total, 10 lines exhibited either a visible morphological phenotype distinct from the wild type (Figures 4 and 5; see Supplemental Tables 5 and 6 online) or showed a reduced ABA sensitivity in seed germination (amiRNA-M3K; Figures 5C, 5F, and 5G; see Supplemental Tables 5 and 6 online).
Table 1. Summary of AmiRNAs and Targets in the TPK Library.
| Function of Target Genes | AmiRNA | Families Targeted | Loci Targeted |
|---|---|---|---|
| Transcription factor | 86 | 44 | 568 |
| Protein kinase | 33 | 24 | 216 |
| Protein phosphatase | 3 | 3 | 13 |
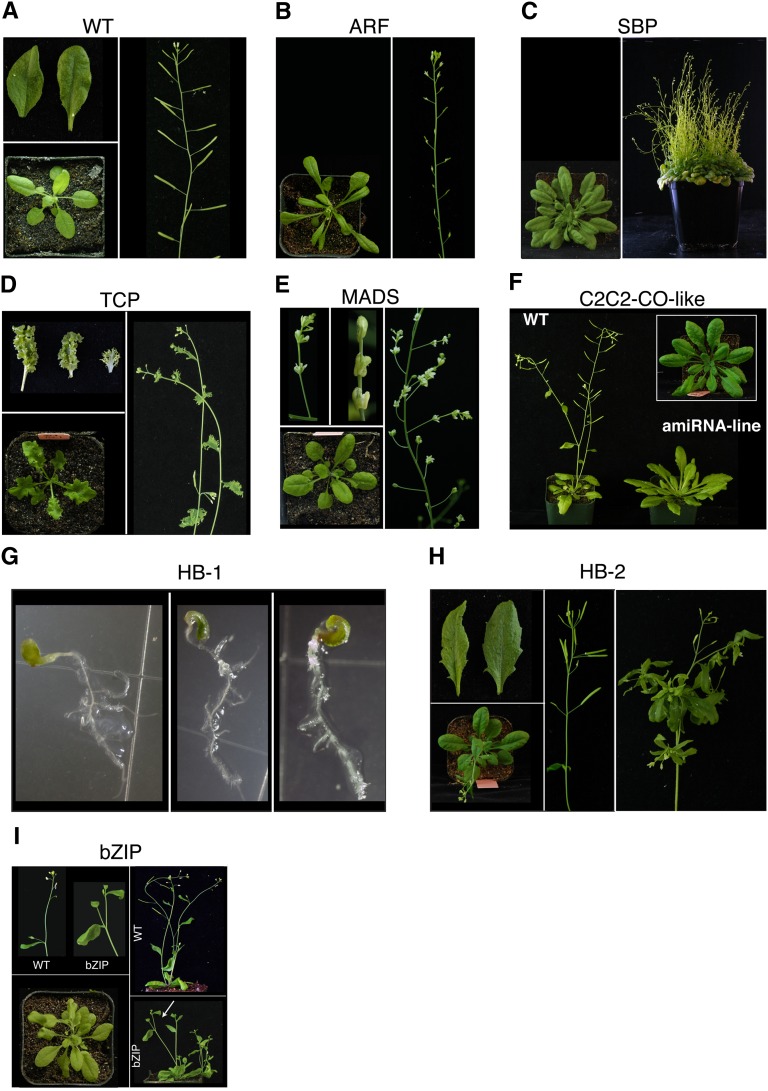
Figure 4.
Phenotypes of AmiRNA Lines Obtained with the AmiRNAs of the TPK Library, Which Are Related to Known Morphological Phenotypes.
(A) Rosette, leaves, and inflorescence of a representative untransformed wild-type control (Col-0 pRAB18:GFP) plant.
(B) Rosette, leaves, and inflorescence of a representative line transformed with amiRNA-ARF (predicted targets: ARF1, ARF3, ARF4, ARF5, ARF6, and ARF8; see Supplemental Table 5 online) with similar phenotype to miRNA167 overexpression (Wu et al., 2006) or arf6 arf8 double mutants (Nagpal et al., 2005).
(C) Rosette and whole plant of a representative line transformed with amiRNA-SBP (predicted targets: SPL2, SPL3, SPL4, SPL6, SPL9, SPL10, SPL11, SPL13B, SPL13A, and SPL15; see Supplemental Table 5 online) with similar phenotype to miRNA 156b overexpression plants (Schwab et al., 2005).
(D) Rosette, leaves, and inflorescence of a representative line transformed with amiRNA-TCP (TB1, CYC, and PCF family; predicted targets: TCP2, TCP4, TCP10, TCP13, and TCP24; see Supplemental Table 5 online) with similar phenotype to miRNA319a overexpression plants (Palatnik et al., 2003).
(E) Rosette and inflorescence of a representative line transformed with amiRNA-MADS (MCM1, AGAMOUS, DEFICIENS, SRF family; predicted targets: AG, AGL3, AGL6, AGL7, AGL10, AGL13, AGL15, AGL25, and AGL72; see Supplemental Table 5 online) with similar phenotype to agamous alleles (Bowman et al., 1991).
(F) Rosette and inflorescence of a representative line transformed with amiRNA-C2C2-CO-like [C(2)-C(2) zinc finger constans like; predicted targets: CO, COL1, COL2, COL4, and COL5; see Supplemental Table 5 online] with similar phenotypes to CONSTANS alleles (Koornneef et al., 1991).
(G) Whole seedling of three independent transformants obtained with amiRNA-HB-1 (predicted targets: AT1G05230, ATML1, HDG11, HDG3, HDG9, and PDF2; see Supplemental Table 5 online). Comparable to pdf2 atml1 double mutants (Abe et al., 2003), these plants also do not develop further than shown in the image (see Supplemental Tables 5 and 6 online for details).
(H) Rosette, leaves, inflorescence, and aerial rosette of a representative line transformed with amiRNA-HB-2 (predicted targets: BEL1, BLH2, BLH3, BLH4, BLH5, PNF, ATH1, BLH11, and PETB; see Supplemental Table 5 online). While the leaf borders share similarity with a sawtooth-1 sawtooth-2 double mutant (Kumar et al., 2007), the internode patterning of the siliques and the aerial rosette resemble ath1-1 pnf pny triple mutants (Rutjens et al., 2009) thereby possibly showing additive effects of the targets.
(I) Rosette and whole plant of a representative line transformed with amiRNA-bZIP (basic region/Leu zipper motif transcription factor; predicted targets: PAN, TGA1, TGA1, TGA2, TGA4, TGA5, and TGA6; see Supplemental Table 5 online). The plant leaves seem to be paler, and the distance between the last cauline leaf and the flower is shortened compared with wild-type plants (white arrow marks inflorescence shown in the top left of the panel).
Figure 5.
Phenotypes of AmiRNA Lines Obtained with the AmiRNAs of the TPK Library, Which Show Gene Family–Linked Phenotypes Not Previously Described (See Supplemental Tables 5 and 6 Online for Details).
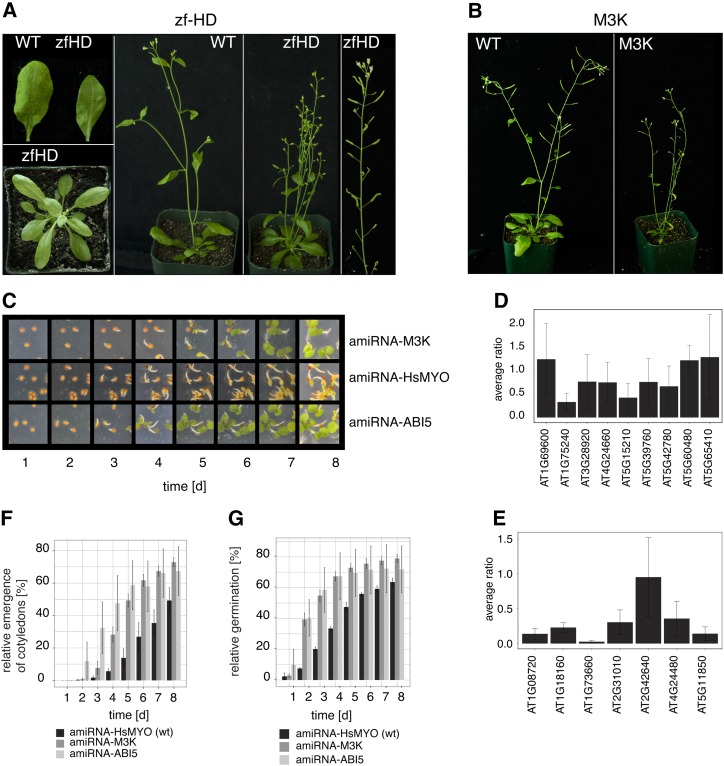
(A) Rosette, leaves, whole plant, and inflorescence of a representative line transformed with amiRNA-zfHD (predicted targets: ATHB22, ATHB23, ATHB25, ATHB26, ATHB27, ATHB29, ATHB30, ATHB33, and ATHB34; see Supplemental Table 5 online). The larger number of stems, larger number of flowers, and short siliques are typical for this amiRNA.
(B) Whole plant of a representative line transformed with amiRNA-M3K (predicted targets: AtMAP3Kδ1, AtMAP3Kδ3, AtMAP3Kδ5, AtMAP3Kθ1, AtMAP3Kθ2, AT1G73660, and AT1G18160; see Supplemental Table 5 online). In general, those plants were smaller than wild-type plants of the same age.
(C) Images of seeds germinating on plates containing 2 μM ABA at the indicated time points. Shown are representative lines of an amiRNA line targeting a set of seven M3Ks (amiRNA-M3K; top row). Wild-type control amiRNA targeting human myosin 2 (amiRNA-HsMYO, no target in Arabidopsis; middle row) and one amiRNA line targeting ABI5 (amiRNA-ABI5) as reference for insensitivity is shown in the other two rows.
(D) Average ratio of amiRNA-zfHD target gene expression levels in the wild type versus amiRNA-zfHD expressing plants as determined by quantitative real-time PCR (n = 4; the bars represent average ± sd).
(E) Average ratio of amiRNA-M3K target gene expression levels of amiRNA-M3K–expressing plants compared with the wild type as determined by quantitative real-time PCR (n = 4; the bars represent average ± sd).
(F) Time course of hypocotyl emergence in the presence of ABA for wild-type control (two lines; amiRNA targeting human myosin 2, no target in Arabidopsis; amiRNA-HsMYO), one amiRNA targeting ABI5 (four lines amiRNA-ABI5) as a reference for insensitivity, and amiRNA lines targeting seven mitogen-activated kinases (five lines). The bars represent average ± se for each amiRNA in percentage relative to the total number of seeds.
(G) Time course of germination in the presence of ABA for amiRNA lines targeting seven mitogen-activated kinases (amiRNA-M3K; five lines), one amiRNA targeting ABI5 (amiRNA-ABI5; four lines) as a reference for insensitivity and wild-type control (amiRNA targeting human myosin 2 [amiRNA-HsMYO], no target in Arabidopsis, two lines). The bars represent average ± se for each amiRNA in percentage relative to the total number of seeds.
A comparative analysis of predicted target genes, phenotypes, and published data showed a connection for several of the observed phenotypes. Transformants containing p35S:amiRNA-ARF (for Auxin Response Factor) show leaf shape and inflorescence morphologies (Figure 4B) similar to published miRNA 167 overexpression lines (Wu et al., 2006). The morphology of plants transformed with p35S:amiRNA-SBP (for SQUAMOSA-PROMOTER BINDING PROTEIN; Figure 4C) resemble miRNA 156b overexpression plants (Schwab et al., 2005), while p35S:amiRNA-TCP transformants (Figure 4D) resemble miRNA 319a overexpression plants (Palatnik et al., 2003). The amiRNAs target the same gene families, but different combinations of family members compared with the natural miRNA. Lines obtained with p35S:amiRNA-MADS (Figure 4E) or p35S:amiRNA-C2C2-CO-like (CONSTANS like; Figure 4F) reflected known single-gene mutant alleles included in the target class of these amiRNAs. In the case of p35S:amiRNA-MADS, agamous alleles (e.g., agamous2; Yanofsky et al., 1990; Bowman et al., 1991) show similar flowers (Figure 4E). The late-flowering phenotype and the increased plant size observed with p35S:amiRNA-C2C2-CO-like (Figure 4F) are typical for CONSTANS alleles (Koornneef et al., 1991).
AmiRNAs Cause Higher Order Mutant Phenotypes
The amiRNA screen identified phenotypes of higher order mutants. The phenotype of p35S:amiRNA-ARF mentioned above resembles arf6 arf8 double mutant plants (Nagpal et al., 2005). A growth arrest response at the early seedling stage (Figure 4G) was observed with plants transformed with p35S:amiRNA-HB-1 (homeobox domain). A similar phenotype is also found in pdf2 atml1 double mutants, both targets of this amiRNA (Abe et al., 2003). A more complex case was obtained with p35S:amiRNA-HB-2 (Figure 4H). On the one hand, the plants showed to a certain extent jagged leaf borders reminiscent of sawtooth-1 sawtooth-2 double mutant plants (Kumar et al., 2007). On the other hand, internode patterning of siliques and aerial rosettes (Figure 4H) resembled ath1-1 pnf pny triple mutants (Rutjens et al., 2009). In total, nine genes, including four of the ones mentioned above, are targeted by p35S:amiRNA-HB-2 (see Supplemental Table 5 online). p35S:amiRNA-bZIP (Figure 4I) caused partially curled leaves, initially shorter plants, and a smaller distance between the last cauline leaf and flowers (Jakoby et al., 2002). This is likely a multifactor effect due to the central role of the targeted seven TGA transcription factors that are known to be involved in regulation of salicylic acid response and floral development (see Supplemental Table 5 online; Hepworth et al., 2005; Murmu et al., 2010; Xu et al., 2010; Rivas-San Vicente and Plasencia, 2011).
New Leaf/Inflorescence Development and ABA Response Mutants
Two lines revealed phenotypes not described previously (Figures 5A and 5C). Plants expressing p35S:amiRNA-zfHD (zinc finger homeodomain transcription factor) showed narrower and elongated leaf planes. The glossy surface of the leaves was not due to the lack of trichomes based on microscopy analyses (see Supplemental Figure 5B online). More impressively, reduced apical dominance and evidently shorter siliques were characteristic for this amiRNA line (Figure 5A; see Supplemental Figure 5A online). Quantitative real-time PCR data indicate that transcriptional repression of the zfHD transcription factors for two transcripts (AT1G75240 and AT5G15210) is stronger than twofold (Figure 5D).
The p35S:amiRNA-M3K targeting mitogen-activated protein kinase kinase kinases (M3K) was the only amiRNA in the TPK library causing a reduced ABA sensitivity in seed germination compared with the wild type (2 µM ABA; Figures 5C and 5G). The amiRNA line also exhibited enhanced seedling growth and cotyledon emergence in the presence of ABA (Figures 5C and 5F). The level of reduced ABA insensitivity is approximately comparable to the control amiRNA line used in this screen to silence the single gene ABI5 (Figures 5C, 5F, and 5G; Finkelstein, 1994). The analysis of target gene expression changes with quantitative real-time PCR indicates transcriptional repression of several M3K genes (Figure 5E), including a gene (AT1G73660) that has been reported to be a negative regulator of salt tolerance (Gao and Xiang, 2008). Overall, the p35S:amiRNA-M3K plants showed a slightly reduced growth phenotype compared with the wild type grown in parallel (Figure 5B). In summary, our results show the potential of our approach, by reproducing known phenotypes, in particular of known double mutants and the potential to identify candidate genes causing previously undescribed gene family-linked phenotypes.
DISCUSSION
Measurable phenotypes of loss-of-function mutants provide an approach for analyzing and assigning gene functions. The large number of gene families in Arabidopsis with functionally overlapping members is considered to be a substantial source of genetic redundancy (Cutler and McCourt, 2005; O’Malley and Ecker, 2010). Only a small fraction of (partially) redundant genes has been studied to date (Lloyd and Meinke, 2012). These studies underline the potential for novel tools that enable screens of the functionally redundant gene space. Our approach that combines amiRNA technology with a comprehensive computational analysis of gene families represents such a tool. The newly created PHANTOM DB Web resource provides a database of over two million amiRNAs that target potential functionally overlapping genes. The PHANTOM DB further enables the parallel viewing of up to 16 differing family definitions along with a selection of gene expression data (Leonhardt et al., 2004; Schmid et al., 2005), subcellular localization (Heazlewood et al., 2007), protein domain architecture (Mulder et al., 2003), and interaction data (Szklarczyk et al., 2011). The PHANTOM DB facilitates the decision that genes are preferential targets.
Combinatorial Design of the Library for Enhancing Coverage and Robustness
Based on our large family specific pool of computationally designed amiRNAs, we pursued a combinatorial selection algorithm of the most promising amiRNAs for synthesis of our PHANTOM library. Instead of simply selecting the smallest set of ∼10,000 target classes and then adding amiRNA sequences of the same target classes (Figure 3B), we added new target classes giving rise to a larger combinatorial coverage of the redundant gene space (e.g., Figure 3A versus 3B). As a result the network, consisting of all target classes and targeted genes as nodes (e.g., Figure 3C), contains for the smallest set 28,432 nodes (10,317 target classes and 18,117 targets). However, for our current combinatorial approach, the network contains 40,117 nodes (22,000 target classes and 18,117 targets). This difference in complexity is also reflected in the number of clusters in the networks (1502 versus 3123 clusters) and the size of the largest clusters (Figure 3A versus 3B). Considering that only silencing of certain combinations leads to a measurable phenotype, our overlapping combinatorial approach should increase the probability of including a large portion of such pairs within the group of targeted genes (e.g., Figure 3C). Furthermore each of the 10 sublibraries consists of two separately synthesized libraries (see Supplemental Figure 3 online) containing the 21- or 20-mer amiRNA (Ossowski et al., 2008), which is likely to further enhance robustness of forward genetic screening.
Proof of Concept Screen with Transcription Factor, Protein Kinase, and Phosphatase Families as AmiRNA Targets
In order to test our strategy, we constructed the TPK library, targeting transcription factors, protein phosphatases, and protein kinases. Remarkably, with only 122 constructs, we found 10 causing phenotypes different from the wild type in at least three independent T1 transformants and the following T2 generation (see Supplemental Table 5 online). From these, three resembled known miRNA overexpression phenotypes (Schwab et al., 2005; Wu et al., 2006; Schwarz et al., 2008), two were similar to single mutant alleles (C2C2-CO-like and MADS; Koornneef et al., 1991; Ledger et al., 2001), and three shared aspects of higher order mutants (HB1, HB2, bZIP, and ARF; Abe et al., 2003; Nagpal et al., 2005; Kumar et al., 2007; Rivas-San Vicente and Plasencia, 2011), while the remaining two were novel. Overall the phenotypes were stable and consistent among independent transformants in subsequent generations. However, in three cases (see Supplemental Table 4 online), the strength of phenotypical properties was weaker or lost in parts of the subsequent generation. This described variability of the phenotypic response in independent lines has been previously reported (Alvarez et al., 2006). In an activation tagging mutagenesis approach, certain phenotypes, such as sterility, reduced apical dominance, or paleness, were observed in ∼1% of lines (Weigel et al., 2000). In the proof-of-concept screen with the TPK library, in total from the 10 lines with phenotypes, two from this category exhibit sterility (amiRNA-MADS) or reduced apical dominance (amiRNA-zfHD), suggesting a frequency that may be in this range or higher than 1%. A comparison between the knockout data published by Lloyd and Meinke (2012) and the entire set of genes targeted by amiRNAs showed that the amiRNA approach missed potential phenotypes (e.g., the ABA hyposensitivity of perk4 [Bai et al., 2009] and mpk6 [Xing et al., 2009] mutants), but also described new phenotypes. The large synthesized set of amiRNAs in our PHANTOM library can compensate for individual amiRNAs that may be less effective.
Developmental and ABA Sensitivity Phenotypes
To our knowledge, the phenotypes of the amiRNAs targeting nine zinc finger homeodomain transcription factors (amiRNA-zfHD; Figure 5A) and that of seven M3Ks (amiRNA-M3K; Figure 5C) have not previously been linked to any of the targeted genes. Single-gene deletions in six of the nine targets of the amiRNA-zfHD showed no apparent effects (Tan and Irish, 2006). The negative regulation of ATHB33 by ARF2 (Wang et al., 2011) indicates that auxin misregulation might contribute to the alteration of the inflorescence morphology, which partially resembles the amiRNA-ARF phenotype (Figure 4B).
The amiRNA-M3K targeting seven M3Ks was identified as the only amiRNA causing reduced ABA sensitivity in seed germination (Figures 5C, 5F, and 5G). Control amiRNA-ABI5 lines that silence the ABI5 transcription factor (Finkelstein and Lynch, 2000) and showed a phenotype were used as reference for ABA insensitivity (Figures 5C, 5F, and 5G). Identification of a new ABA-insensitive mutant in this small screen is remarkable since even large-scale activation tagging screens of ∼66,000 independent lines (∼106 seeds screened) did not identify a single robust ABA-insensitive mutant (Kuhn et al., 2006). Several reports suggest that during seed germination, mitogen-activated protein kinase cascades are part of the ABA signaling network (Lu et al., 2002; Jammes et al., 2009; Liu, 2012). MPK6, MPK9, MPK10 (Jammes et al., 2009; Xing et al., 2009), and At-MKK1 (Xing et al., 2009) proteins were reported to play a role in ABA signaling. Although, to date, no M3K has been shown to reduce ABA signaling, single mutants in two amiRNA targets (Wawrzynska et al., 2008; Gao and Xiang, 2008) were found to be ABA hypersensitive or indistinguishable from the wild type, respectively. Further studies are necessary to elucidate the underlying mechanisms of M3K and zinc finger homeodomain transcription factor amiRNA phenotypes.
The strength of repression of the target transcript has been reported to depend on sequence composition of (a)miRNA and targets, the target gene expression level, mRNA turnover rate, and additional unknown factors (Brodersen et al., 2008; Arvey et al., 2010; Larsson et al., 2010; Schwab et al., 2010). Note that the amiRNA score includes some of these parameters; however, by including specificity-related information, such as number of possible off-targets, the predictive power of the score for the efficiency is limited and therefore was not used here (Ossowski et al., 2008). The use of amiRNAs designed here could yield data contributing to statistical improvement in the prediction of parameters that would enhance amiRNA efficiency. We analyzed our small-scale TPK data to determine whether we could find indications for effects originating from physicochemical amiRNA properties. Comparing amiRNAs from plants with phenotypes to the remainder of the TPK library (see Supplemental Figures 6A to 6C online) suggests a relevance of higher binding energy (see Supplemental Figure 6A online), less mismatches (see Supplemental Figure 6B online), and the location of the amiRNA target site (see Supplemental Figure 6C online). Similarly, a set of quantitative real-time PCR data (see Supplemental Figures 6D to 6F and Supplemental Table 5 online) suggest a linear correlation between strength of transcriptional repression and Gibbs-free binding energy and number of mismatches (see Supplemental Figures 6A and 6B online) but no linear correlation with the location of the amiRNA binding site within the target transcript (see Supplemental Figure 6C online).
In conclusion, based on a genome-wide assembly of protein family definitions, we computationally created a Web-accessible resource of family targeting amiRNAs and have designed and synthesized 10 expression libraries as novel tools for addressing functional overlap in the Arabidopsis genome. It might well be that the amiRNAs could also be used in other related plant species as described previously (Alvarez et al., 2006). In a proof-of-principle screen, we provide evidence for the potential of our approach by identifying known and new amiRNA-linked phenotypes. The created resources represent a novel approach, which enables addressing of functional genetic redundancy in large and small-scale screens and in targeted analyses.
METHODS
Analyses of Protein Family Definitions
Family definitions were derived from the databases listed in Supplemental Table 1 either by downloading from the websites listed in Supplemental Table 1 with the exception of three data sets obtained either directly from the authors (PlantTribes, Kerr Wall; Transorgalin, Pascal Mäser) or curators (Robert Finn, PFAM). Sequences were downloaded from TAIR (Garcia-Hernandez et al., 2002). The annotation versions were always kept separate, and nonexisting loci were excluded from further analyses. Where necessary the identifiers were mapped from Uniprot/Trembl identifiers to Arabidopsis thaliana gene identifiers using three data sources in hierarchical order: (1) mapping available at TAIR (Garcia-Hernandez et al., 2002), (2) mapping available through Integr8 (Kersey et al., 2005), and (3) mapping through protein identifier cross-reference service (Côté et al., 2007). This minimized the “loss” of Arabidopsis gene identifiers due to redundant sequence versions. In rare cases where a protein was assigned to two different families, the identifiers were collected in a new family named by the combined string of the original family identifiers. The Kinomer definition was reproduced using the HMM model (Martin et al., 2009) for searching the two annotation versions (Garcia-Hernandez et al., 2002) to avoid potential erroneous mapping. For PlantsP, the downloaded file and the Web content were merged. In the next step, all singletons (i.e., families consisting of one locus) were removed from the datasets. In order to break down large families into subfamilies, DPCL (Brown, 2008), MCL (Enright et al., 2002), and BLAST cluster (Altschul et al., 1990) algorithms were applied to protein sequences and in two cases (MCL and BLAST cluster) also to nucleotide sequences. Procedures were automated using scripts written in Python (van Rossum and de Boer, 1991), and SQLite (http://www.sqlite.org) was used for pre- and postprocessing and storage of the data, respectively. The similarity heat map (Figure 1C) was generated as follows. Each family in each family definition was compared with each family in all of the other family definitions. If the proportion of common genes was ≥75%, the family was counted as similar. In this way, a score is assigned to every family definition, which is the sum of all counted similar families (see Supplemental Table 2 online). The counts were the converted into relative values by setting the highest count in each column of Figure 1C to 100%.
Design of Family Member–Targeting AmiRNAs, the PHANTOM Database, and Assembly of the Pooled Library
All genome-wide amiRNA designs were run on the UCSD Triton Compute Cluster after the public release of AmiRNA/WMD3 Version 3.1 (Ossowski et al., 2008) software. The preset AmiRNA/WMD3 settings were used for all procedures as follows. A maximum of five mismatches in a gap-less alignment are allowed for initial target identification, and one hit per gene and one isoform only are considered in the evaluation. The binding energies were calculated using RNAcofold (Lorenz et al., 2011) at a temperature of 23°C. No off-targets and minimal two targets were required for all amiRNAs. Further details of the AmiRNA/WMD3 algorithm are described by Ossowski et al. (2008) and the documentation of the software. Note that the current AmiRNA/WMD3 target search may contain few false negative/positive predicted amiRNA targets (Schwab et al., 2005), which can be minimized when new target search algorithms become available. Scripts written in Python were used for preprocessing, parallel processing (Dalcin et al., 2008), and postprocessing of the data. Large-scale designs were run on a high-performance cluster (2048 CPUs and 256 nodes; Triton Resource; San Diego Supercomputer Center; University of California San Diego). For each family in the family definitions used (Interpro, Kinomer, Phytozome, PFAM, PIRSF, Merops, PlantsP, PP, TCDB, TransportDB, and TransfacDB; see Supplemental Table 1 online for explanations of the abbreviations and relevant references), the design procedure was first run against all members in a family. When the designed amiRNAs covered 75% or more of the family members, the design was considered sufficient. For ≤75% family member coverage, the design was repeated, now using first subfamilies as defined by the DPCL algorithm (Brown, 2008). If again 75% coverage was not reached, the design was repeated against subfamilies as defined by MCL algorithm of cDNA sequences. This resulted in coverage of each family where most families are covered more than 75% (see Supplemental Figure 2A online). The amiRNA sequences were loaded into a SQLite database (http://sqlite.org/) that was integrated as a searchable Web resource (PHANTOM database; http://phantomdb.ucsd.edu) together with the analyzed family definitions and other publicly available data, including microarray-derived gene expression, subcellular localization, protein domain, protein interaction, and phylogenetic data using tools documented in the “about” section of the PHANTOM database. All quantitative and network graphs and analyses were generated using R (R Development Core Team, 2008) and additional packages (Csardi and Nepusz, 2006; Wickham, 2009; Kalinka and Tomancak, 2011). For the PHANTOM library synthesis, 22,000 amiRNA sequences were selected as follows. All amiRNAs targeting more than 16 loci were filtered out. Then a new score, Ns, was calculated for all amiRNAs according to the equation Ns = Ms − As + Si(tN), where As is the original score as provided by AmiRNA/WMD3, Ms is the maximum score observed in all amiRNAs designed, and Si(tN) is a weighting factor. The value of Si(tN) is a function of the number of targets, tN, and has the following discrete values (listed in the format tN:Si): 2:5.0, 3:40.0, 4:50.0, 5:60.0, 6:65.0, 7:60.0, 8:55.0, 9:50.0, 10:45.0, 11:40.0, 12:35.0, 13:30.0, 14:25.0, 15:20.0, and 16:15. This formula prefers amiRNAs with six targets, and large score values correlate with better predicted amiRNA properties. AmiRNAs targeting two genes were weighted less, as even with this low weight there is a large proportion of amiRNAs that target two genes (see Supplemental Figure 2E online). The highest scoring amiRNAs, which include unique loci not targeted by other amiRNAs, were chosen, and then additional amiRNAs were added provided that they added new targets to the set. This procedure was repeated until at least 22,000 amiRNAs were collected, and the worst scoring amiRNAs were removed to reach exactly 22,000 amiRNAs. This procedure leads to unique target classes; therefore, the same genes are targeted by amiRNAs of different target classes, thus enhancing the combinatorial coverage of the gene space (for example, see Figure 3C). Note that the amiRNA score, which is based on findings from a finite number of validated genes (Schwab et al., 2005), was used here solely for ranking purposes. The score value is limited in its predictive power for amiRNA efficiency and specificity, and further data, including findings from the use of the PHANTOM library, could allow improvement in the scoring scheme.
Classification of the PHANTOM AmiRNAs According to Target Function
The 12 GO-SLIM definitions (Garcia-Hernandez et al., 2002) were compressed into 10 groups to divide the amiRNAs into pools (see Supplemental Table 3 online) and the targeted loci classified according to the Gene Ontology term “molecular function.” The following procedure was applied to equalize the pool size, in particular to reduce the larger nonclassified group: (1) members of family definitions with functionally distinct groups (TransfacDB, PlantsP, Kinomer, TCDB, and TransportDB), and enzymes classified by AraCyc (Zhang et al., 2005) were moved into the corresponding groups (see Supplemental Table 3 online); (2) nonclassified loci were rearranged if TAIR10 GO-SLIM had a function; (3) nonclassified loci were rearranged if a MapMan (Thimm et al., 2004) function was found (see Supplemental Table 3 for details). Based on this locus classification, the amiRNAs were functionally pooled using a majority-based rule (i.e., most frequent class with a function in a group of targets determines the pool of the amiRNA). When this approach did not resolve the proper subpool of an amiRNA, the amiRNA was added to the pool named DMF (for diverse molecular functions).
PHANTOM AmiRNA Library Construction
The 172-bp oligonucleotides containing the amiRNA (amir*/amir) and part of the mir319 backbone were synthesized in situ as described previously (Agilent; Cleary et al., 2004). As originally amiRNAs including those in the TPK library (see below) were constructed by adding one base to the precursor stem (Ossowski et al., 2008) and on the other hand this extension in some cases might influence the amiRNA properties, all 22,000 amiRNAs were constructed in two backbones differing in length by two bases (i.e., the original mir319 has been replaced either by 20- or by 21-bp amir sequences) (Ossowski et al., 2008). All DNA fragments were purified at UCSD by agarose gel electrophoresis before subsequent reactions. First, the amiRNA oligonucleotides were PCR amplified using pool-specific primers (0.3 µM each) and KOD polymerase (Novagen) according to the manufacturer’s recommendations and the following amplification protocol: initial denaturation 95°C, 2 min; 25 cycles with 95°C, 30 s; 55°C, 30 s; and 70°C, 30 s. To minimize potential uneven template sequence dependent amplification, separate reactions were performed in the presence and in the absence of 5% DMSO and pooled before purification by gel electrophoresis and subsequent extraction (QiaexII; Qiagen). The purified PCR products were cut in two reactions either with MfeI-HindIII or with MfeI-BsaI, purified, and ligated overnight into the EcoRI-HindIII digested pFH0315. The 40 ligations (10 pools, two different backbones, and two different enzyme combinations) were individually transformed by electroporation into library-competent DH10B cells (Invitrogen) and plated on total 800 plates (150 mm diameter). The plasmid library of each pool was isolated from the harvested cells using a Maxiprep kit (Tip 500; Qiagen) and digested with BbsI. The BbsI loop fragment obtained from pFH0313 was then ligated into the digested plasmid library pools. As before, ligations were transformed and plated, transformed cells harvested, and plasmids isolated. Stock cultures were separated and planned for donation to the ABRC for distribution. Subsequently, the plasmid library pools were recombined into the plant expression vector pFH0032 (see Supplemental Table 7 online) using LR Clonase (Invitrogen). Quality control was done on the one hand by isolating plasmids from a total 190 colonies and subsequent sequencing for 13 pools. In these sublibraries, 60 to 93% of all sequenced clones were correct (i.e., 467 bp spanning the entire amiRNA precursor correspond to one of the expected sequences in the pools). On the other hand, the content and complexity of each pool was analyzed by deep sequencing. This was achieved by Illumina sequencing of adapted (SBS3 and Truseq) amplicons containing the guide (mir) and flanking sequences (see Supplemental Table 7 online). Samples were multiplexed and sequenced with an Illumina MiSeq sequencer using a custom recipe (30 dark cycles, 22 read cycles, and seven index reads cycles). On average, the enrichment factor (number of reads for an amiRNA in the pool of interest divided by total reads of this amiRNA in all pools) of amiRNAs in each pools was >0.94 (94%) for any of the pools.
Transcription Factor, Protein Phosphatase, and Kinase Library Construction
To test the amiRNA design in the context of ABA signal transduction, we designed a set of amiRNAs targeting transcription factor, protein phosphatase, and protein kinase families and subfamilies (i.e., groups of proteins most likely involved in signaling). AmiRNAs were designed against filtered (see computational analyses for details) transcription factor, protein phosphatase, and protein kinase families and subfamilies using the publicly available AmiRNA/WMD (versions 2 and 3.1 through the website) (Ossowski et al., 2008; Schwab et al., 2010) with the default parameters, minimal two targets and in general no off-targets. In case of subfamilies where no amiRNA could be found, the remaining family members were allowed as off-targets. In general, amiRNAs were initially chosen for cloning according to the following criteria in decreasing order of importance: they target five or more genes, add more coverage with minimal overlap to already generated amiRNAs in the TPK library, contain largest group of targets whose expression is affected based on microarray data from ABA-treated plants (Leonhardt et al., 2004; Goda et al., 2008; Yang et al., 2008), or target three or more genes of families not covered by any amiRNA so far. Primer design (RS300 setting on AMIRNA/WMD3) and overlapping PCR were done using publicly available tools and protocols from WMD (Ossowski et al., 2008) except that the A and B primers were replaced with 5′-caccGAATTCCTGCAGCCCcaaacacacg-3′ and 5′-GGATCCCCCcatggcgatgcctta-3′, respectively. All amiRNA PCR products were cloned into pENTR D TOPO (Invitrogen). Using LR Clonase, the amiRNA was transferred into the vector pFH0032 (see Supplemental Table 7 online). Clones in pENTR D TOPO and pFH0032 were verified by sequencing. The verified plasmids were individually electroporated into a pSOUP containing Rhizobium radiobacter GV3101(pMP90), and the presence of the plasmid was confirmed by PCR as described elsewhere (Weigel and Glazebrook, 2006).
Plant Material, Growth Conditions, and Transformation
Surface-sterilized seeds (10 to 20 min; 70% ethanol and 0.1% SDS; three to four washes with ∼100% ethanol) of Arabidopsis were cold-treated for 2 to 5 d at 4°C and germinated on half-strength Murashige and Skoog basal medium for 5 to 7 d. The medium was supplemented with Gamborg's vitamins (Sigma-Aldrich; Murashige and Skoog, 1962; Gamborg et al., 1968), 1% Suc, 0.8% Phytoagar (Difco), and pH adjusted (pH 5.8; 4-morpholinoethane sulfonic acid [2.6 mM; Sigma-Aldrich] titrated with potassium hydroxide). Plants were transferred to plastic pots containing sterilized premixed soil (Sunshine Professional Blend; McConkey) supplemented with an appropriate amount of fungicide (Cleary) and insecticide (Marathon) and propagated under the following conditions: long day (16 h light/8 h dark); 23 to 27°C, 20 to 70% humidity, 60 to 100 mmol m−2 s−1 light. Plant transformation by floral dip was essentially performed as described elsewhere (Clough and Bent, 1998) with the following modifications: Rhizobium radiobacter GV3101(pMP90) (Koncz and Schell, 1986) was grown under selection of all markers (i.e., genomic, Ti-plasmid, pSOUP, and T-DNA plasmid). The infiltration medium for resuspension of the bacteria and floral dip contained 5% Suc (w/v) and 0.02% (v/v) Silwet L-77 (Clough and Bent, 1998). When appropriate, media for growth of bacteria or plant selection contained the following concentrations of antibiotics (mg mL−1): carbenicillin 100, kanamycin 30, rifampicin 50, spectinomycin 100, tetracyclin 10, and phosphinotricin 15.
Screen with the TPK Library
Homozygous transgenic Arabidopsis Col-0 lines harboring a promoter RAB18:GFP fusion (Kim et al., 2011) were transformed as described above. T1 plants (on average 10 lines) were selected and seeds from individual lines collected. Seeds from individual T2 lines were screened on plates (see above) for insensitivity to ABA (2 μM; Sigma-Aldrich) during seed germination. For each amiRNA, at least three lines were tested along with the controls on one ABA containing plate. As control for ABA insensitivity, lines containing one of two different amiRNAs targeting ABI5 were used, and the Col-0 pRAB18:GFP line (Kim et al., 2011) was used as wild-type control. In parallel, for all amiRNA lines, the resistance against phosphinotricine was tested again. The putative ABA insensitivity was scored in a binary manner for similarity to the amiRNA-ABI5 lines after 5 to 10 d using green cotyledons as indicator (Kuhn et al., 2006). For lines that showed a putative ABA insensitivity, the seed germination assay was repeated, this time imaging both plates (±ABA) daily for at least 8 d and manually counting emergence of radicles and cotyledons using Fiji (Schindelin et al., 2012).
Quantitative RNA Analyses
Plant tissue (seedlings, leaves, and inflorescences) for RNA preparation was frozen in liquid nitrogen and stored at −80°C. For RNA isolation, 100 to 300 mg of the frozen tissue was homogenized using a cryo bead mill (2 × 1 min at 30 Hz; Retsch), and ∼100 mg of the ground tissue was processed using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions. The spectrophotometrically quantified RNA was DNaseI treated (TURBO DNA-free kit; Ambion/Applied Biosystems) according to the manufacturer’s instructions and after quantification the integrity analyzed by standard gel electrophoresis (Maniatis et al., 1982). RNA (1 to 5 μg) was reverse transcribed using either the first-strand cDNA synthesis kit (GE) or Superscript III (Invitrogen) with oligo(dT) primers at the conditions indicated in the manual. Quantitative real-time PCR was performed using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) with a CFX Real-Time PCR cycler (Bio-Rad) with the following conditions: 95°C, 3-min initial denaturation, followed by 40 cycles of 95°C for 10 s, 55°C for 10 s, 72°C for 30 s; 10 to 15 μL reaction volume in duplicates. A melting curve analysis was performed between 65 and 95°C with increments of 0.5°C for 5 s. Primers were designed using QuantPrime (Arvidsson et al., 2008) with the default settings or taken from Czechowski et al. (2004). For normalization, three reference genes (AT4G05320, AT1G13320, and AT1G58050) were used with primers reported elsewhere (Czechowski et al., 2005). Analysis of the quantitative real-time PCR data was performed using the methods available in the qPCR package (version 1.3.6; relevant parameters: data were normalized and the background subtracted; starting fit model: l4; efficiency estimation: cpD2; outlier check: uni2; uncertainties: propagate; refmean: True; Ritz and Spiess, 2008) and the dixon test available in the outliers package (Komsta, 2011) and python tools (http://pythonhosted.org/uncertainties/; http://pyqpcr.sourceforge.net) to summarize the data. Correlation analysis between physicochemical amiRNA properties and observed regulation was performed graphically using a glm fit available in ggplot2 (Wickham, 2009) and statistically using global validation of linear model assumptions implemented in the gvlma (Peña and Slate, 2006) package in R with a level of significance of 0.05.
Accession Numbers
The PHANTOM library is publicly available through the ABRC under the following accession numbers: CD4-62, CD4-63, CD4-64, CD4-65, CD4-66, CD4-67, CD4-68, CD4-69, CD4-70, CD4-71, CD4-72, CD4-73, CD4-74, CD4-75, CD4-76, CD4-77, CD4-78, CD4-79, CD4-80, CD4-81, and CD4-82. Accession numbers for targeted genes, amiRNA sequences, and pool names can be found in Supplemental Table 5 and Supplemental Data Set 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Overlap and Family Size in the Family Definitions Analyzed for the AmiRNA Design.
Supplemental Figure 2. Detailed Overview of the Complete Set of AmiRNAs and Target Loci.
Supplemental Figure 3. Summary of Illumina High-Throughput Sequencing of the AmiRNA Library Pools.
Supplemental Figure 4. Percentage of Genes in Each Family Targeted by One or Several AmiRNAs in the TPK (Transcription Factors, Protein Phosphatases, and Kinases) Library.
Supplemental Figure 5. Supplemental Data for AmiRNA-zfHD.
Supplemental Figure 6. Distributions of AmiRNA and Target Properties.
Supplemental Table 1. Databases and Other Original Sources of the Family Definitions Used.
Supplemental Table 2. Comparative Analysis of Family Definitions.
Supplemental Table 3. Detailed Description of the Functional Classes That Form the Pools in the AmiRNA Library.
Supplemental Table 4. Target Families, Loci, and Description of Genes Targeted by an AmiRNA for Which Either No Transformants (GRF) or Variable Phenotypes Were Obtained.
Supplemental Table 5. Target Families, Loci, and Description of Genes Targeted by an AmiRNA Listed in Supplemental Table 6 and Have a Morphological Phenotype or Show a Partial Insensitivity to Abscisic Acid in Seed Germination When Compared with Wild Type.
Supplemental Table 6. Targeted Family and Corresponding AmiRNA Sequence of Constructs Listed in Supplemental Table 5.
Supplemental Table 7. Comprehensive List of Relevant Plasmids and Illumina Primers Used in This Study.
Supplemental Data Set 1. List of Target Genes, AmiRNA Sequences Designed for the TPK Library and of Target Genes, AmiRNA Sequences, and Pool Names Designed for the PHANTOM Library.
Acknowledgments
We thank Rebecca Schwab for providing the plasmids pRS1102 and pRS300, Detlef Weigel for discussions and comments on the article, Kerr Wall, Pascal Mäser, and the curator team of PFAM for providing access to raw data, and numerous others for supporting this work with publicly available software and data. We thank the UCSD Super Computer Center for access to the new Triton Compute Cluster. F.H. was a recipient of a one-year Swiss National Science Foundation fellowship (PBEZA-121224; in 2008/2009). This research was supported by the National Science Foundation (IOS-1025837; MCB0918220) and by the National Institutes of Health (R01GM060396-ES010337 to J.I.S.). Analyses of TPK library phenotypes were in part supported by a grant from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (DE-FG02-03ER15449) (J.I.S.). Development of WMD software by S.O. and J.F. was supported by Max Planck Society funds to D. Weigel. U.D. was supported by a German Academic Exchange Service (DAAD) fellowship.
AUTHOR CONTRIBUTIONS
J.I.S. proposed the research project. Most of the research was conducted by F.H. (analysis and assembly of gene family definitions, development of scripts, computational design and construction of amiRNAs, plant transformation and phenotyping of TPK amiRNA plants, amiRNA library synthesis, and interactive website design). F.H. cloned the libraries with technical support from U.D. G.J.H. and K.C. conducted automated oligonucleotide synthesis for the PHANTOM library generation and conducted Hi-seq quality control. J.F. and S.O. programmed and publicly released AmiRNA/WMD3. F.H. developed procedures and the PHANTOM DB website with the help of W.C. F.H. and J.I.S. analyzed data and wrote the article.
Glossary
- amiRNA
artificial microRNA
- ABA
abscisic acid
- WMD
Web microRNA designer
- DPCL
Dirichlet process clustering algorithm
- MCL
Markov chain clustering algorithm
- TAIR
The Arabidopsis Information Resource
- Col-0
Columbia-0
Footnotes
Online version contains Web-only data.
References
- Abbott J.C., Barakate A., Pinçon G., Legrand M., Lapierre C., Mila I., Schuch W., Halpin C. (2002). Simultaneous suppression of multiple genes by single transgenes. Down-regulation of three unrelated lignin biosynthetic genes in tobacco. Plant Physiol. 128: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M., Katsumata H., Komeda Y., Takahashi T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arvey A., Larsson E., Sander C., Leslie C.S., Marks D.S. (2010). Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 6: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riaño-Pachón D.M., Mueller-Roeber B. (2008). QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Zhang G., Zhou Y., Zhang Z., Wang W., Du Y., Wu Z., Song C.-P. (2009). Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 60: 314–327 [DOI] [PubMed] [Google Scholar]
- Bouché N., Bouchez D. (2001). Arabidopsis gene knockout: Phenotypes wanted. Curr. Opin. Plant Biol. 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Briggs G.C., Osmont K.S., Shindo C., Sibout R., Hardtke C.S. (2006). Unequal genetic redundancies in Arabidopsis—A neglected phenomenon? Trends Plant Sci. 11: 492–498 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brown D.P. (2008). Efficient functional clustering of protein sequences using the Dirichlet process. Bioinformatics 24: 1765–1771 [DOI] [PubMed] [Google Scholar]
- Cleary M.A., et al. (2004). Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat. Methods 1: 241–248 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Côté R.G., Jones P., Martens L., Kerrien S., Reisinger F., Lin Q., Leinonen R., Apweiler R., Hermjakob H. (2007). The Protein Identifier Cross-Referencing (PICR) service: Reconciling protein identifiers across multiple source databases. BMC Bioinformatics 8: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi, G. and Nepusz, T. (2006). The igraph software package for complex network research. http://igraph.sf.net Accessed August 15, 2013.
- Cutler S., McCourt P. (2005). Dude, where’s my phenotype? Dealing with redundancy in signaling networks. Plant Physiol. 138: 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Bari R.P., Stitt M., Scheible W.-R., Udvardi M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalcin L., Paz R., Storti M., D'Elia J. (2008). MPI for Python: Performance improvements and MPI-2 extensions. J. Parallel Distrib. Comput. 68: 655–662 [Google Scholar]
- Delattre M., Félix M.-A. (2009). The evolutionary context of robust and redundant cell biological mechanisms. Bioessays 31: 537–545 [DOI] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5: 765–771 [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Gao L., Xiang C.-B. (2008). The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 67: 125–134 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M., et al. (2002). TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genomics 2: 239–253 [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 35 (Database issue): D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Zhang Y., McKim S., Li X., Haughn G.W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington A.M. (2001). Guard cell signaling. Cell 107: 711–714 [DOI] [PubMed] [Google Scholar]
- Ichikawa T., et al. (2006). The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 48: 974–985 [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., Parcy, F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis Trends Plant Sci. 7: 106–111. [DOI] [PubMed]
- Jammes F., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R., Springer M., Pilpel Y. (2009). Genetic redundancy: New tricks for old genes. Cell 136: 389–392 [DOI] [PubMed] [Google Scholar]
- Kalinka A.T., Tomancak P. (2011). linkcomm: An R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics 27: 2011–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P., et al. (2005). Integr8 and Genome Reviews: Integrated views of complete genomes and proteomes. Nucleic Acids Res. 33 (Database issue): D297–D302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Soltis P.S., Wall K., Soltis D.E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23: 107–120 [DOI] [PubMed] [Google Scholar]
- Kim T.H., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komsta, L. (2011). Outliers: Tests for Outliers (Contributed R Package). http://CRAN.R-project.org/package=outliers Accessed August 15, 2013.
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Genet. Genomics 204: 383–396 [Google Scholar]
- Koornneef M., Hanhart C.J., van der Veen J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kuhn J.M., Boisson-Dernier A., Dizon M.B., Maktabi M.H., Schroeder J.I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Kushalappa K., Godt D., Pidkowich M.S., Pastorelli S., Hepworth S.R., Haughn G.W. (2007). The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.-M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D.G., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E., Sander C., Marks D. (2010). mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol. 6: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger S., Strayer C., Ashton F., Kay S.A., Putterill J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26: 15–22 [DOI] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. (2012). Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Lloyd J., Meinke D. (2012). A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 158: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. (2011). ViennaRNA package 2.0. Algorithms Mol. Biol. 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Han M.-H., Guevara-Garcia A., Fedoroff N.V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 99: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E., and Sambrook, J. (1982). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY; Cold Spring Harbor Laboratory Press). [Google Scholar]
- Martin D.M.A., Miranda-Saavedra D., Barton G.J. (2009). Kinomer v. 1.0: A database of systematically classified eukaryotic protein kinases. Nucleic Acids Res. 37 (Database issue): D244–D250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Matsui K., Ikeda M., Nakata M., Oshima Y., Nagatoshi Y., Ohme-Takagi M. (2011). CRES-T, an effective gene silencing system utilizing chimeric repressors. Methods Mol. Biol. 754: 87–105 [DOI] [PubMed] [Google Scholar]
- Mulder N.J., et al. (2003). The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Murmu J., Bush M.J., DeLong C., Li S., Xu M., Khan M., Malcolmson C., Fobert P.R., Zachgo S., Hepworth S.R. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Ecker J.R. (2010). Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Ott T., van Dongen J.T., Günther C., Krusell L., Desbrosses G., Vigeolas H., Bock V., Czechowski T., Geigenberger P., Udvardi M.K. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Park S.-Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña E.A., Slate E.H. (2006). Global validation of linear model assumptions. J. Am. Stat. Assoc. 101: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2008). R: A language and environment for statistical computing. http://www.R-project.org/ Accessed August 15, 2013.
- Ritz C., Spiess A.-N. (2008). qpcR: An R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 24: 1549–1551 [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Rutjens B., Bao D., van Eck-Stouten E., Brand M., Smeekens S., Proveniers M. (2009). Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 58: 641–654 [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Warthmann N., Weigel D. (2010). Directed gene silencing with artificial microRNAs. Methods Mol. Biol. 592: 71–88 [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L.J., von Mering C. (2011). The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39 (Database issue): D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q.K.-G., Irish V.F. (2006). The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 140: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. (2000). A genetic uncertainty problem. Trends Genet. 16: 475–477 [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- van Rossum, G., and de Boer, J. (1991). Interactively testing remote servers using the python programming language. CWI Quarterly 4: 283–303. [Google Scholar]
- Wagner A. (2005). Distributed robustness versus redundancy as causes of mutational robustness. Bioessays 27: 176–188 [DOI] [PubMed] [Google Scholar]
- Wang L., Hua D., He J., Duan Y., Chen Z., Hong X., Gong Z. (2011). Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynska A., Christiansen K.M., Lan Y., Rodibaugh N.L., Innes R.W. (2008). Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D. and Glazebrook, J. (2006). Transformation of Agrobacterium using electroporation. CSH Protoc. 2006: pdb.prot4665. [DOI] [PubMed]
- Wickham, H. ggplot2: Elegant graphics for data analysis. New York: Springer, 2009. [Google Scholar]
- Wu M.-F., Tian Q., Reed J.W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2009). AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol. Biol. 70: 725–736 [DOI] [PubMed] [Google Scholar]
- Xu M., Hu T., McKim S.M., Murmu J., Haughn G.W., Hepworth S.R. (2010). Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 63: 974–989 [DOI] [PubMed] [Google Scholar]
- Yang Y., Costa A., Leonhardt N., Siegel R.S., Schroeder J.I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Zhang P., Foerster H., Tissier C.P., Mueller L., Paley S., Karp P.D., Rhee S.Y. (2005). MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 138: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]