Elevations in extracellular Ca2+ stimulate stomatal closure, though the underlying mechanisms remain elusive. This study reports the isolation and functional characterization of cau1, a novel allele of PRMT5/SKB1, using forward genetics, and reveals that CAU1/PRMT5/SKB1 synchronizes extracellular Ca2+ levels and stomatal closure via dynamic histone methylation of the CAS chromatin.
Abstract
Elevations in extracellular calcium ([Ca2+]o) are known to stimulate cytosolic calcium ([Ca2+]cyt) oscillations to close stomata. However, the underlying mechanisms regulating this process remain largely to be determined. Here, through the functional characterization of the calcium underaccumulation mutant cau1, we report that the epigenetic regulation of CAS, a putative Ca2+ binding protein proposed to be an external Ca2+ sensor, is involved in this process. cau1 mutant plants display increased drought tolerance and stomatal closure. A mutation in CAU1 significantly increased the expression level of the calcium signaling gene CAS, and functional disruption of CAS abolished the enhanced drought tolerance and stomatal [Ca2+]o signaling in cau1. Map-based cloning revealed that CAU1 encodes the H4R3sme2 (for histone H4 Arg 3 with symmetric dimethylation)-type histone methylase protein arginine methytransferase5/Shk1 binding protein1. Chromatin immunoprecipitation assays showed that CAU1 binds to the CAS promoter and modulates the H4R3sme2-type histone methylation of the CAS chromatin. When exposed to elevated [Ca2+]o, the protein levels of CAU1 decreased and less CAU1 bound to the CAS promoter. In addition, the methylation level of H4R3sme2 decreased in the CAS chromatin. Together, these data suggest that in response to increases in [Ca2+]o, fewer CAU1 protein molecules bind to the CAS promoter, leading to decreased H4R3sme2 methylation and consequent derepression of the expression of CAS to mediate stomatal closure and drought tolerance.
INTRODUCTION
Cytosolic calcium ([Ca2+]cyt) plays a pivotal role as a second messenger in plant development and interactions with the environment (Helpler and Wayne, 1985; Poovaiah and Reddy, 1993; Bush, 1995). Various signals, including biotic stress, abiotic stress, hormones, and mechanical disturbance, are sensed by plant cells and elicit repetitive oscillation or spiking of [Ca2+]cyt, which are termed Ca2+ signatures and regulated by Ca2+ channels, pumps, or carriers localized at the plasma membrane or in the membranes of organelles (Bush, 1995; Thuleau et al., 1998; Rudd and Franklin-Tong, 1999; Sanders et al., 1999; Allen et al., 2000, 2001; Harper, 2001). These Ca2+ signatures vary in frequency and amplitude in response to different stimuli and thus are believed to encode stimulus-specific information that can be transduced and sensed by downstream Ca2+ sensors, including calmodulins, calcium-dependent proteins, and calcineurin B-like proteins (Zielinski, 1998; Luan et al., 2002; Sanders et al., 2002). These calcium sensors decode Ca2+ signatures by binding Ca2+ ions via conserved EF-hand domains and transduce them into specific cellular responses, such as altered phosphorylation and expression of target genes, to bring about physiological responses corresponding to the original stimuli (Luan et al., 2002; Sanders et al., 2002).
In contrast with intracellular Ca2+, the role of extracellular Ca2+ ([Ca2+]o) remains to be determined. In mammals and other animals, parathyroid cells can alter the secretion of parathyroid hormone in response to serum Ca2+ levels, which in turn modulates Ca2+ uptake from the intestine, net release from the skeleton and its conservation efficiency in the kidneys, thus returning serum Ca2+ to a baseline level (Shoback et al., 1983, 1984; Brown, 1991). It was later proposed that Ca2+ can actually serve as an extracellular first messenger (Brown, 1991), as parathyroid gland cells are able to sense [Ca2+]o levels in the serum and produce intracellular Ca2+ signals through the activation of CaR, a G protein–coupled Ca2+-sensing receptor, and the phospholipase C/inositol-1,4,5-trisphosphate (IP3)/Ca2+ signaling pathway (Brown et al., 1987, 1993; Shoback et al., 1988).
In plants, evidence indicating a similar function for [Ca2+]o has been mainly provided by studies of stomatal guard cells (Schwartz, 1985; MacRobbie, 1992; Allen et al., 2000, 2001; Han et al., 2003). When exposed to [Ca2+]o, [Ca2+]cyt oscillation was observed in guard cells, and the oscillation patterns varied with different extracellular Ca2+ levels (McAinsh et al., 1995; Allen et al., 2000). Abortion or alteration in [Ca2+]cyt oscillations failed to induce stomatal closure in response to [Ca2+]o (Allen et al., 2000, 2001). These observations suggested that [Ca2+]o signals can be perceived by cells and are encoded in the intracellular Ca2+ signatures. The isolation of CAS and identification of its downstream signaling components provided further evidence for this hypothesis (Han et al., 2003; Hetherington and Brownlee, 2004; Tang et al., 2007, Wang et al., 2012a). However, given that CAS localizes exclusively to chloroplasts instead of the cell surface, where [Ca2+]o sensing is supposed to occur (Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008), we postulate that other regulatory components must exist upstream of CAS if the hypothesis should stand.
In this study, a Ca2+ underaccumulation mutant, cau1, was identified in a large-scale genetic screen. The mutant showed enhanced stomatal closure and drought tolerance. Further analysis showed that CAU1 encodes an H4R3sme2-type histone methylase and acts as an immediate upstream suppressor of the CAS gene. Elevated [Ca2+]o decreases CAU1 protein levels and consequently the H4R3sme2 methylation level in the CAS chromatin, thus derepressing CAS expression to close stomata.
RESULTS
Isolation and Characterization of the Calcium Underaccumulation Mutant cau1
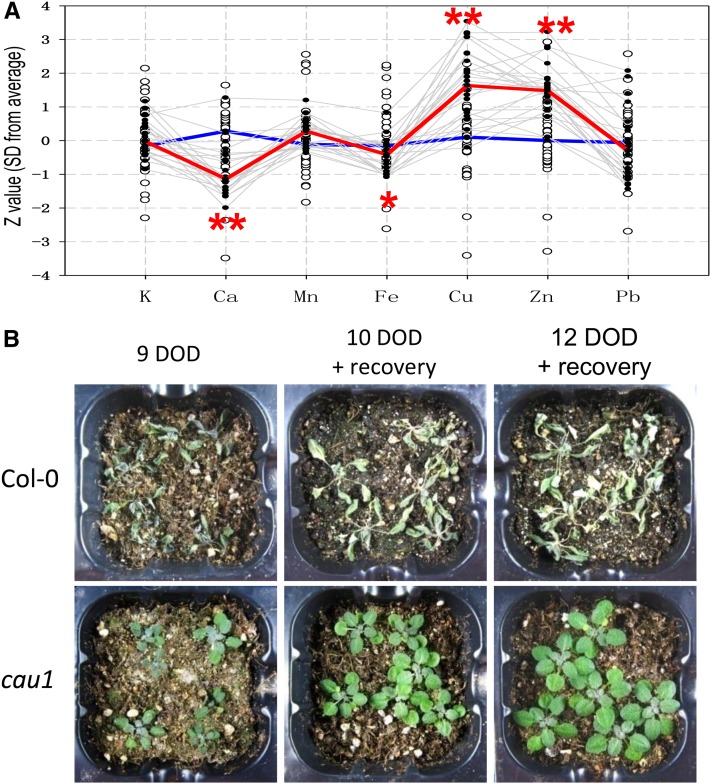
In a large-scale screen of Ca2+ accumulation mutants, a Ca2+ underaccumulation mutant, cau1, was identified (Figure 1A). Compared with the wild type Columbia-0 (Col-0), cau1 plants accumulated less Ca2+ and Fe in rosette leaves, but their copper and zinc2+ accumulation was significantly increased (Figure 1A). Further analysis showed that Ca2+ accumulation was decreased in the shoots of cau1 when treated with 0.5 mM Ca2+ (see Supplemental Figure 1A online). These results demonstrated that CAU1 preferentially regulates Ca2+ accumulation in the aerial parts.
Figure 1.
The Calcium Underaccumulation Mutant cau1 with Enhanced Drought Tolerance.
(A) Ion accumulation profile of cau1. Seven cationic elements in cau1 M3 mutant plants were measured by ICP-MS. Open black circles represent Z values (sd from the average value of the wild type control, y axis) of the individual wild-type controls, and gray lines highlight the ion profile data of cau1 mutants. Median Z values of the wild type (n > 30) and cau1 mutants (n > 30) were calculated, and the resultant ion profiles were outlined by the blue line and red line, respectively.
(B) Wild-type and cau1 mutant plants subjected to drought stress for 9 to 12 d. Plants were exposed to 9 d of drought (DOD) treatment or treated for 10 or 12 d and subsequently reirrigated and allowed to recover for 1 week. *P < 0.05 and **P < 0.01.
Interestingly, enhanced drought tolerance was also observed to cosegregate with the ionomic phenotype of cau1. When exposed to drought stress, Col-0 was severely dehydrated and did not recover upon reirrigation. By contrast, cau1 did not show any significant dehydration after 9 d of drought treatment and was able to fully recover from 10 or 12 d of drought (Figure 1B). Other visual phenotypes, including partial and degenerate chlorosis in young leaves, increased lateral shoots with as many as 10 cauline leaves, and late flowering, were also observed in cau1 mutant plants (see Supplemental Figure 2 online).
As Ca2+ accumulation fluctuates with plant ages, and cau1 mutant plants were late flowering, we further analyzed the Ca2+ levels from plants of different ages. As expected, Ca2+ accumulation did vary from 21 to 50 d of age, though overall significantly lower Ca2+ levels were detected in cau1 compared with Col-0 at all analyzed ages (see Supplemental Figure 1B online). The Ca2+ levels in cau1 were also consistently lower than those of RRS-7 and Lov-1, which were randomly selected as representative late-flowering ecotypes (see Supplemental Figure 1C online) and later found to show no changes in CAS expression (see Supplemental Figure 1D online). These results suggested that CAU1 functions to regulate ion homeostasis and plant development independently.
Genetic Analysis of the cau1 Mutant and Map-Based Cloning of the CAU1 Gene
After backcrossing the homozygous cau1 to Col-0, a phenotypic assay was performed in the derived F1 and F2 generations. All F1 progenies grew and developed similarly to the wild type. In the F2 generation, the segregation of 221 wild-type plants and 75 cau1 mutant corresponded to the expected ratio of 3:1 (χ2 = 0.018, P = 0.893), indicating that cau1 carries a single recessive mutation.
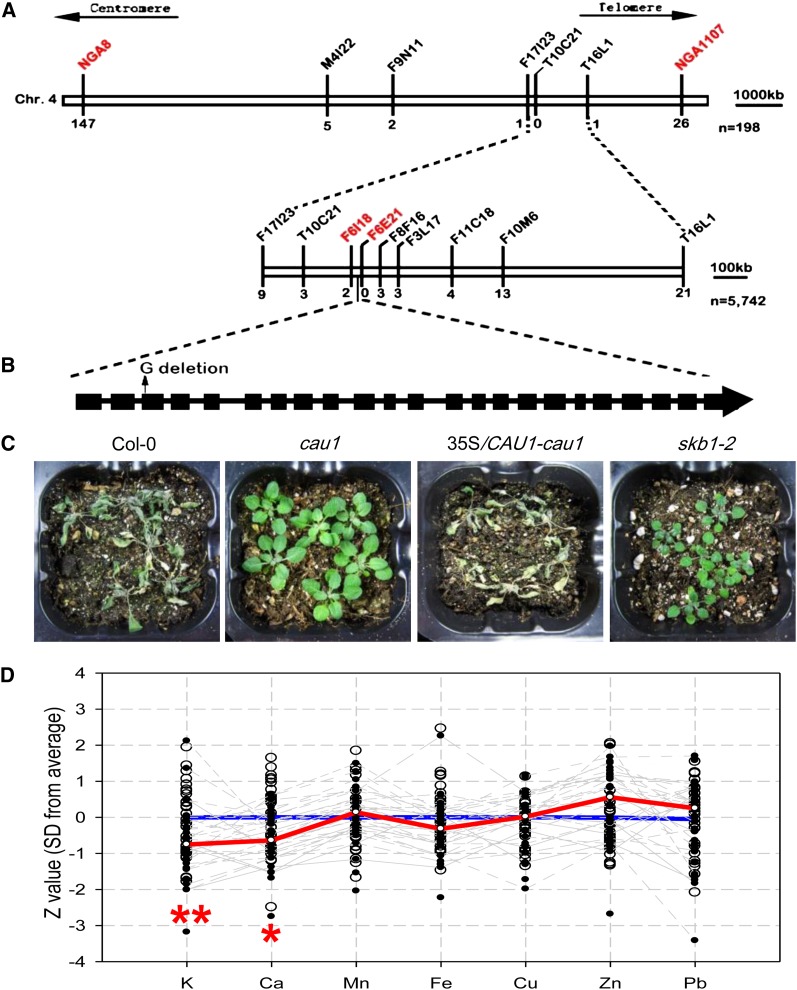
To isolate the mutant gene, cau1 was outcrossed to the ecotype Landsberg erecta and the F1 progenies were self-pollinated to generate an F2 mapping population. Bulked segregant analysis using 198 F2 mutants roughly mapped the cau1 locus to the distal arm of chromosome 4, between the simple sequence length polymorphism (SSLP) markers NGA8 and NGA1107 (Figure 2A). Fine mapping using 5742 F2 mutant plants further localized the cau1 locus to a 43-kb genomic region between markers F6I18 and F6E21 (Figure 2A). Fifteen genes were predicted in this region, and sequencing of these candidate genes revealed a G nucleotide deletion in the third exon of the At4g31120 locus (Figure 2B), which resulted in a shift of the open reading frame and, hence, a premature stop codon 40 bp downstream of the mutation site. A complementation assay using the coding sequence of the At4g31120 locus successfully restored drought tolerance (Figure 2C), ion accumulation (see Supplemental Figure 3A online), and morphological alteration (see Supplemental Figures 3C to 3F online) to wild-type levels. RT-PCR further showed that the mRNA level of At4g31120 was decreased in cau1 (see Supplemental Figure 3B online).
Figure 2.
Molecular Identification of the cau1 Gene.
(A) Map-based cloning of the cau1 locus on chromosome 4.
(B) Structure of the CAU1 gene and the mutation in the cau1 allele. Black boxes represent exons, and black lines represent introns. The Arabidopsis Genome Initiative number for CAU1 is At4g31120.
(C) Complementation assay of drought tolerance in cau1. Wild-type, cau1 mutant, 35S/CAU1-cau1 (cau1 harboring construct 35S/CAU1-pBI121), and skb1-2 plants were subjected to dehydration for 12 d, reirrigation, and recovery for 1 week.
(D) Ion accumulation profile of skb1-2. Seven cationic elements in skb1-2 mutant plants were measured by ICP-MS. Open black circles represent Z values (y axis) of individual wild-type controls, and gray lines highlight ion profiles of skb1-2 individuals. Median Z values of the wild type (n > 30) and skb1-2 mutant (n > 30) were calculated, and the consequent ion profiles were outlined by the blue line and the red line, respectively. *P < 0.05 and **P < 0.01.
At4g31120 was previously identified as PRMT5/SKB1 (Pei et al., 2007; Wang et al., 2007), and we analyzed drought tolerance and ion accumulation in the T-DNA insertion mutant line skb1-2 (Salk_095085). As expected, comparable phenotypes for both drought tolerance (Figure 2C) and Ca2+ underaccumulation (Figure 2D) were observed in the cau1 and skb1-2 mutant lines. These data demonstrated that the mutation in At4g31120 is responsible for the observed cau1 phenotypes.
CAS Expression Is Steadily Elevated in the cau1 Mutant
CAU1/PRMT5/SKB1 has been identified as a type II methyltransferase, regulating pleiotropic physiological processes by either H4R3sme2 regulation and/or pre-mRNA splicing (Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008; Deng et al., 2010; Hong et al., 2010; Sanchez et al., 2010; Zhang et al., 2011). To investigate how CAU1 regulates Ca2+ level and drought tolerance, microarray analyses were performed. Among the genes with significantly altered expression, FLC was upregulated in cau1 (see Supplemental Table 1 online), an observation that is consistent with the late flowering phenotype observed in this (see Supplemental Figures 2D to 2F online) and previous studies (Wang et al., 2007).
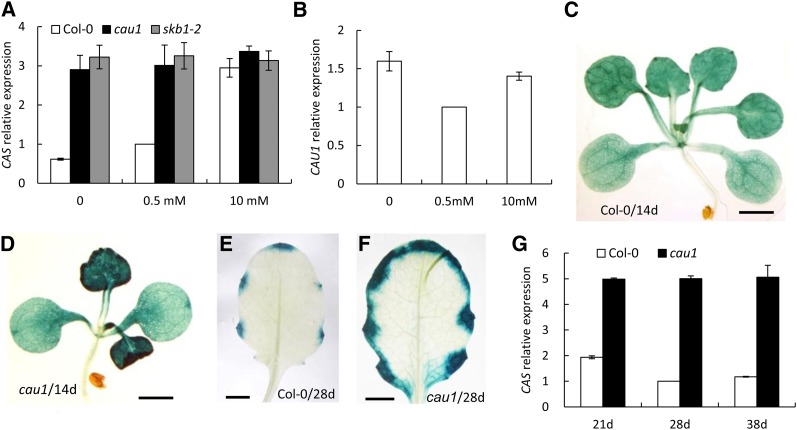
More significantly, the expression of CAS was enhanced in cau1 compared with the wild-type Col-0 (see Supplemental Table 1 online). Given that CAS mediates [Ca2+]o signaling (Han et al., 2003; Tang et al., 2007; Nomura et al., 2008; Weinl et al., 2008) and Ca2+ levels were altered in cau1, we performed quantitative RT-PCR and found that upon exposure to increased external Ca2+ concentrations, the expression levels of CAS were upregulated in the wild-type Col-0, in contrast with the constitutively high levels in the mutants cau1 and skb1-2 (Figure 3A). Note that Ca2+ treatments did not significantly affect CAU1 expression (Figure 3B).
Figure 3.
CAS Expression Is Enhanced in cau1 Plants.
(A) and (B) Three-week-old plants grown hydroponically were treated with 0, 0.5, and 10 mM Ca2+ for 4 d. The expression levels of CAS (A) or CAU1 (B) in shoots were determined by RT-PCR. Actin2 was used as an internal control. Values are mean ± se, n = 3.
(C) to (F) Histochemical localization of GUS driven by the CAS promoter in 14-d-old wild-type (C) and cau1 mutant seedlings (D) or 28-d-old wild-type (E) and cau1 leaves (F). Bars = 0.25 cm.
(G) CAS expression in wild-type and cau1 plants grown in soil to the indicated ages.
In addition, histochemical analyses showed that β-glucuronidase (GUS) activity driven by the CAS promoter was higher in cau1 compared with Col-0 at both seedling (Figures 3D versus 3C) and mature stages (Figure 3F versus 3E). RT-PCR analysis further revealed that although CAS levels fluctuated in Col-0 at 21, 28, and 38 d of age, a steadily enhanced expression was observed in cau1 (Figure 3G). CAS expression was restored to wild-type levels in the complemented lines (see Supplemental Figure 4G online). Taken together, these results indicate that regardless of the external Ca2+ levels or plant age, CAS expression was consistently higher in cau1.
In addition to CAS and FLC, several genes involved in iron uptake and metabolism were also upregulated in cau1 (see Supplemental Table 1 online), which might explain the chlorosis observed in young leaves (see Supplemental Figures 2A to 2C, 2G, and 2H online) and the iron underaccumulation phenotype (Figure 1A).
CAU1 Mediates Drought Tolerance and Stomatal Closure through CAS
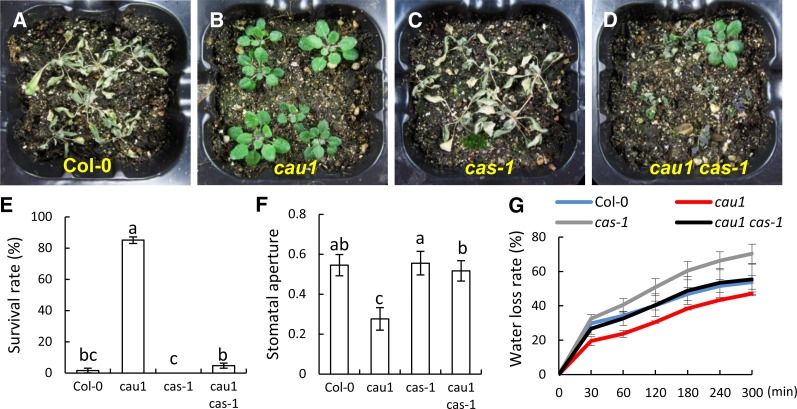
CAS has been shown to mediate [Ca2+]o signaling in guard cells. Given that the Ca2+ underaccumulation mutant cau1 is drought tolerant and shows constitutively enhanced CAS expression, we sought to determine whether these phenotypes are genetically correlated with CAS. The results showed that Col-0 (Figures 4A and 4E) and cas-1 (Figures 4C and 4E) are sensitive to drought stress. By contrast, significant tolerance was observed in cau1 plants (Figures 4B and 4E). However, in the double mutant cau1 cas-1 plants (Figures 4D and 4E), which show growth rates similar to cau1, drought tolerance conferred by the cau1 mutation was abolished. A similar phenotype was observed in the complementation line 35S/CAU1-cau1 (see Supplemental Figures 4A to 4D online). These results indicated that CAS is genetically downstream of CAU1 in mediating drought tolerance.
Figure 4.
Increased Drought Tolerance and Stomatal Closure in cau1 Is Attributed to CAS.
(A) to (D) Drought tolerance in wild-type (A), cau1 (B), cas-1 (C), and cau1 cas-1 (D) plants. Watering of 14-d-old plants was withheld for 12 d, and then plants were rewatered and allowed to recover for 1 week.
(E) Survival rates of plants exposed to drought stress in (A) to (D). Values are mean ± sd from three independent experiments.
(F) Stomatal assay of epidermis peeled directly from 22-d-old rosette leaves. Stomatal apertures were evaluated by stomata width/length. Values are mean ± sd (n ≥ 30). Different lowercase letters above the bars in (E) and (F) indicate statistically different averages at P < 0.05 in t tests.
(G) Water loss rate of 22-d-old wild-type, cau1, cas-1, and cau1 cas-1 plants. Values are mean ± sd calculated from three independent experiments, each containing data from six leaves.
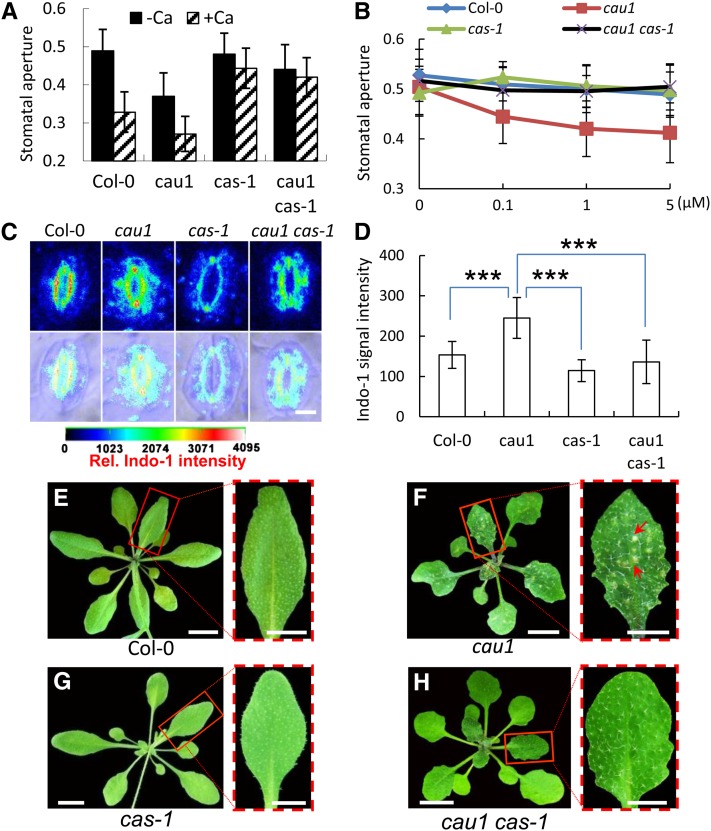
Stomatal apertures were also determined using fresh epidermis peeled directly from leaves. As shown in Figure 4F, increased stomatal closure was observed in cau1 compared with Col-0, while cas-1 and Col-0 showed similar stomatal apertures. In the double mutant cau1 cas-1, stomatal apertures were comparable to those of the wild type and cas-1 mutants (Figure 4F). A previous study showed that overexpression of CAS led to increased stomatal closure (Nomura et al., 2008), equivalent to the phenotype observed in cau1. The water loss rate was consistently increased in the cas-1 mutant and decreased in cau1 (Figure 4G). Crossing cas-1 to cau1 successfully restored the water loss rate to levels comparable to those of the wild type (Figure 4G). In the complementation line 35S/CAU1-cau1, both the stomatal aperture and water loss rate were restored to wild-type levels (see Supplemental Figures 4E and 4F online). These results suggest that CAS provides an essential contribution to the observed increased stomatal closure and the consequent drought tolerance and acts genetically downstream of CAU1. Water loss rates in the cau1 cas-1 double mutant were not as high as in the cas-1 mutant. CAU1 may function pleiotropically (see Supplemental Figure 2 online); thus, components other than CAS might also be involved in the control of water loss.
CAU1 Is an Upstream Regulator of the CAS-Mediated [Ca2+]o Signaling in Guard Cells
To further determine whether CAU1 regulates the [Ca2+]o signaling in guard cells, a stomatal closure assay was performed. As expected, apparent stomatal closure was triggered by [Ca2+]o in the wild-type Col-0 and cau1 mutant and disrupted in cas-1 and the double mutant cau1 cas-1 where CAS is functionally disrupted (Figure 5A). Interestingly, the starting stomatal apertures prior to [Ca2+]o application were much smaller in cau1 than in Col-0, implying that cau1 guard cells might be oversensitive to extracellular Ca2+. To test this hypothesis, EGTA was added to the opening buffer to chelate the trace amount of free Ca2+. Stomatal apertures in Col-0 and cau1 were comparable in the absence of extracellular Ca2+, and upon exposure to 0.1 to 5 µM Ca2+, significant stomatal closure was observed in cau1 but not in Col-0, cas-1, or cau1 cas-1 (Figure 5B). The stomatal apertures of cau1 were restored in the complementation assay (see Supplemental Figures 5A and 5B online). Taking into account that CAS expression levels were consistently higher in cau1 compared with the wild type (Figure 3), we concluded that enhanced CAS expression led to the increased [Ca2+]o sensitivity observed in cau1 and that CAU1 is an upstream regulator of the CAS-mediated [Ca2+]o signaling in guard cells.
Figure 5.
CAU1 Regulates Guard Cell [Ca2+]o Signaling and Ca2+ Starvation Tolerance via CAS.
(A) and (B) [Ca2+]o-induced stomatal closure in plants. As described in Methods, 2 mM Ca2+ (A) or a final concentration of 0, 0.1, 1, or 5 μM Ca2+ (B) was added, and stomatal aperture was calculated as the ratio of pore width/length. n > 120 stomata per Ca2+ treatment, and values are mean ± sd.
(C) Representative imaging of [Ca2+]cyt in stomata of the wild type, cau1, cas-1, and double mutant cau1 cas-1 by Indo-1 staining and UV confocal assay. The first row shows fluorescence images, and the second row shows merged fluorescence and bright-field images. [Ca2+]cyt was calibrated and pseudo-color-coded according to the color scale shown.
(D) Quantification of the relative Indo-1 fluorescence intensities indicating [Ca2+]cyt levels in stomata of the wild type, cau1, cas-1, and cau1 cas-1. Values are mean ± sd, n = 46. ***P < 0.001 in t tests.
(E) to (H) Chlorotic lesions in leaves of the wild type (E), cau1 (F), cas-1 (G), and cau1 cas-1 (H) exposed to calcium starvation. Each red dash-framed panel represents a scale-up of one corresponding leaf from (E) to (H).
Bars = 5 µM in (C), 1 cm in (E) to (H), and 0.5 cm in the dash-framed panels.
Consistent with this, [Ca2+]cyt was significantly increased in cau1 guard cells compared with the wild-type Col-0, while in cas-1, the level was ∼50% lower than in cau1 and significantly lower than in Col-0 (Figures 5C and 5D). In the double mutant cau1 cas-1, [Ca2+]cyt levels were significantly lower than in cau1 and comparable to cas-1 (Figures 5C and 5D), consistent with the model proposed for CAS (Han et al., 2003; Tang et al., 2007; Nomura et al., 2008; Weinl et al., 2008). [Ca2+]cyt was restored to wild-type levels in the cau1 complemented lines (see Supplemental Figures 5C and 5D online). Further analysis showed that 4 d of Ca2+ starvation resulted in significant necrotic lesions, a typical symptom indicating plant Ca2+ deficiency, in cau1 leaves (Figure 5F, indicated by red arrows), but not in Col-0 (Figure 5E), cas-1 (Figure 5G), or the double mutant cau1 cas-1 (Figure 5H), indicating that the increased sensitivity to Ca2+ starvation in cau1 was essentially attributable to elevated CAS expression.
Taken together, these results (Figures 1A and 5A to 5H) demonstrated that the mutation in CAU1 derepressed CAS expression, thus enhancing CAS-mediated [Ca2+]o signaling and stomatal closure, which subsequently decreased the transpiration rate (Figure 4G) and led to a variety of physiological changes, including drought tolerance (Figure 4B), Ca2+ underaccumulation (Figure 1A), and, hence, sensitivity to Ca2+ starvation (Figures 5E to 5H).
CAU1 Mediates CAS Expression by H4R3sme2 Methylation in the Promoter Region of CAS
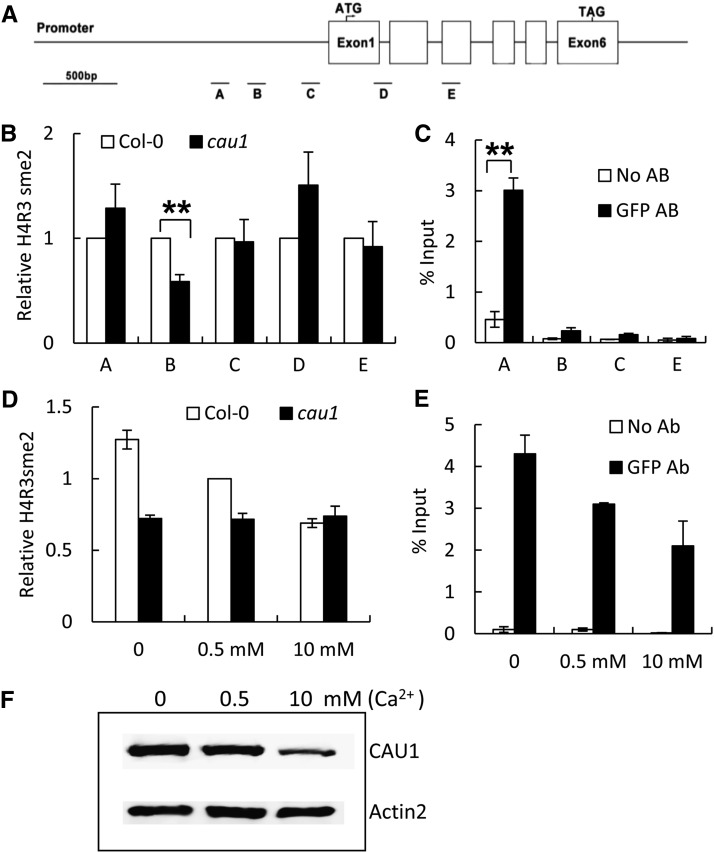
CAU1 is the Arabidopsis thaliana homolog of PRMT5, a mammalian methyltransferase marking histone H4 Arg 3 with symmetric dimethylation (H4R3sme2) (Pollack et al., 1999; Bedford and Richard, 2005; Ancelin et al., 2006). To determine whether CAU1 regulates CAS expression via a similar mechanism, we performed a chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) assay to analyze the H4R3sme2 levels in the CAS promoter region, using the H4R3sme2 antibody (Figures 6A and 6B). Our results showed that the H4R3me2 level in region B of the CAS promoter was significantly reduced in cau1 (Figures 6A and 6B), suggesting that CAU1 regulates CAS transcription through histone methylation.
Figure 6.
CAU1 Regulates the Histone Methylation of the CAS Chromatin and the Consequent CAS Expression in Response to [Ca2+]o.
(A) A diagram of the genomic structure of CAS. Genomic regions A to E subjected to ChIP assay are indicated.
(B) and (C) ChIP assay with antibodies against H4R3sme2 (B) to determine histone methylation status or against GFP (C) to determine if CAU1 binds to the CAS promoter using 35S/EYFP:CAU1-cau1 plants. TUB8 was used as an internal control in (B). Three independent experiments were performed, and values are mean ± se.
(D) and (E) ChIP assay with antibodies against H4R3sme2 within the B region (D) or against GFP within the A region (E) of CAS. Wild-type and cau1 plants were treated with calcium as indicated. Three independent experiments were performed, and TUB8 was used as an internal control in (D). Values are mean ± se.
(F) Immunoblotting analysis of CAU1 levels in response to [Ca2+]o. Twenty-four-day-old plants were exposed to 0, 0.5, or 10 mM Ca2+ treatment for 4 d. Immunoblotting was performed as described in Methods. **P < 0.01.
A ChIP-qPCR assay using a green fluorescent protein (GFP) antibody was also performed to determine whether CAU1 binds to the CAS chromatin (Figures 6A and 6C). CAU1 strongly associated with region A of the CAS promoter, whereas a similar CAU1-CAS interaction was not detected in regions B, C, and E, indicating that CAU1 binds to the CAS chromatin in region A and functions to mediate histone methylation levels in region B (Figure 6B).
Given that CAS expression was upregulated in response to elevated [Ca2+]o (Figure 3A), we then determined the correlation between [Ca2+]o and H4R3sme2 methylation in the CAS promoter. As shown in Figure 6D, elevated [Ca2+]o significantly decreased the H4R3sme2 levels in region B of the CAS promoter from the wild type, while no change was observed in the cau1 mutant. Further analysis showed that the binding of CAU1 to the CAS promoter (Figure 6E) as well as CAU1 protein levels decreased significantly in response to elevated [Ca2+]o (Figure 6F). These data suggest that increased [Ca2+]o decreases CAU1 protein levels and its binding to the CAS promoter, thus decreasing H4R3sme2 methylation of the CAS chromatin and enhancing the expression of CAS.
CAU1 Mediates the Expression of Genes Involved in Ca2+ Signaling and Homeostasis
PRMT5/SKB1/CAU1 has been proposed to mediate flowering time, the circadian clock or salt stress by histone methylation, and/or nonhistone methylation mechanisms; the nonhistone modification mechanism causes further modifications of mRNA splicing (Deng et al., 2010; Hong et al., 2010; Sanchez et al., 2010; Zhang et al., 2011). To test whether a pre-mRNA splicing mechanism was involved in the CAU1-regulated [Ca2+]o signaling, we examined the transcripts of key genes in the CAS-IP3 pathway (Han et al., 2003; Tang et al., 2007). We failed to detect any alternatively spliced transcripts by both RT-PCR amplification and sequencing, even though some of these genes were previously predicted to have alternative splicing, and we performed RT-PCR using specifically designed primers for them (Deng et al., 2010; see Supplemental Figure 6 online).
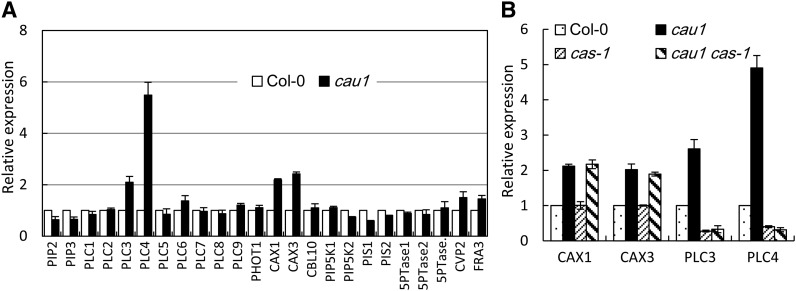
Instead, the expression levels of several genes were significantly altered in cau1. Expression of PLC3 and PLC4, two essential genes mediating IP3 production, markedly increased in cau1 compared with the wild-type Col-0 (Figure 7A), while it decreased in cas-1 and cau1 cas-1 mutant plants (Figure 7B), supporting a model wherein CAU1 functions upstream of the CAS-IP3 cascade (Figure 8). CAX1 and CAX3, which encode tonoplast-localized Ca2+ transporters, also showed higher expression levels in cau1 (Figure 7A), though the upregulation was not affected by CAS (Figure 7B). Given that the overall Ca2+ content decreased in cau1 shoots (Figure 1A; see Supplemental Figure 1A online), while the cytosolic Ca2+ concentration was increased (Figures 5C and 5D), we assume that the enhanced expression of CAX1 and CAX3 might merely be a local response to the increased requirement to move excess [Ca2+]cyt to vacuoles. This assumption helps to explain the observations that less Ca2+ accumulated in cau1 and cax1/cax3 shoots (Figure 1A; see Supplemental Figure 1A online; Cheng et al., 2005; Conn et al., 2011). Note that we do not exclude the possibility that CAX1 and CAX3 may be actively involved in the [Ca2+]o signaling pathway via complex mechanisms such as reported recently (Cho et al., 2012).
Figure 7.
Analysis of Expression Levels of Ca2+ Homeostasis or Signaling Pathway Genes.
(A) Quantitative determination of expression levels of genes involved in the IP3 pathway or Ca2+ homeostasis.
(B) Expression of CAX1, CAX3, PLC3, and PLC4 in cau1, cas-1, and cau1 cas-1 plants. Plants were grown to 28 d old in soil.
The y axes show RNA levels normalized to those of ACTIN2. Values are mean ± se, n = 3.
Figure 8.
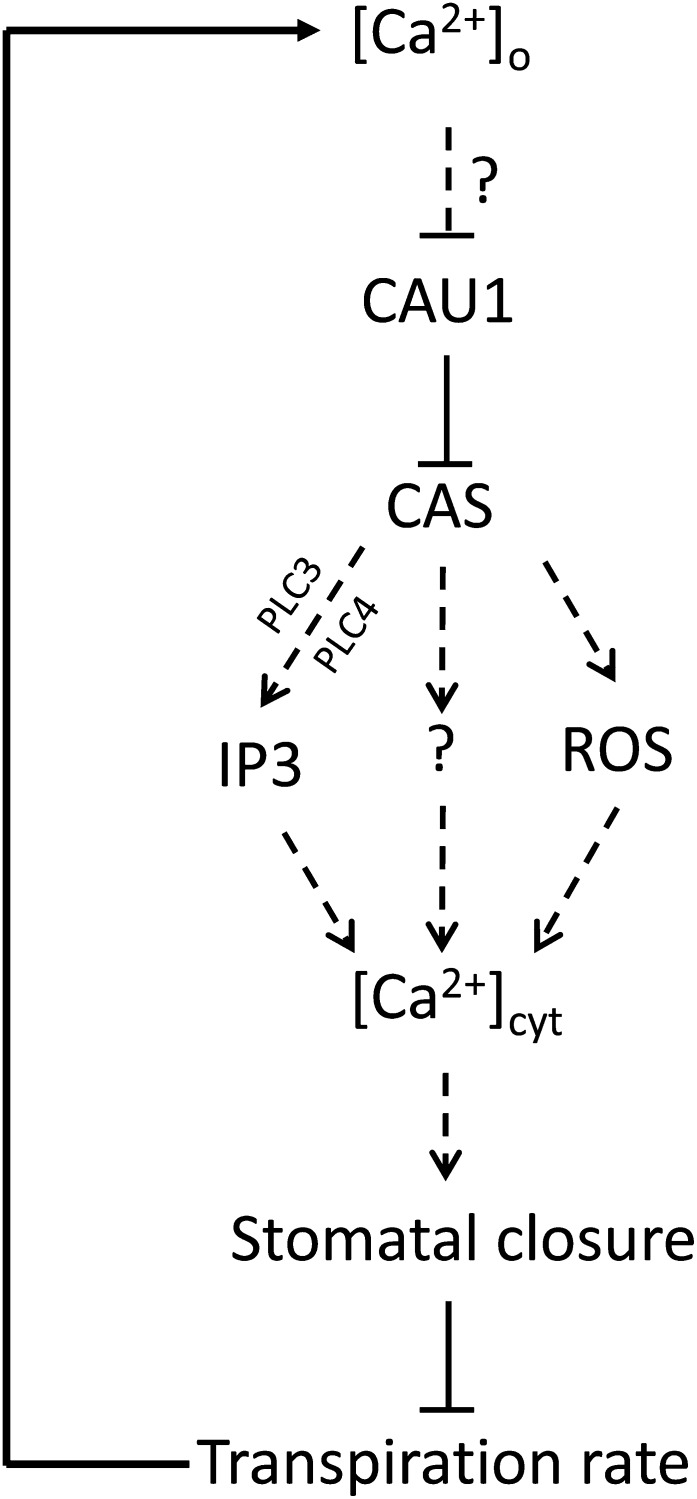
A Model for CAU1/CAS-Mediated [Ca2+]o Signaling in Stomatal Closure.
Elevated [Ca2+]o reduces the CAU1 association with the CAS promoter, which consequently decreases histone methylation of the CAS chromatin and lifts the repression of CAS. The resulting upregulation of CAS further evokes cytosolic Ca2+ ([Ca2+]cyt) and the subsequent stomatal closure (Tang et al., 2007), possibly through IP3 (Tang et al., 2007), ROS (Wang et al., 2012a), or mechanisms that are yet to be identified. Stomatal closure decreases the transpiration rate, leading to feedback inhibition of Ca2+ translocation to the plant aerial parts. Thus, a dynamic synchronization between [Ca2+]o and stomatal closure is established and functions to mediate plant adaptation to the environment. The question marks and dashed lines represent possible unidentified steps or steps that have been identified but are not shown here.
cau1 Is Oversensitive to Abscisic Acid
Abscisic acid (ABA) is an essential plant hormone regulating [Ca2+]cyt oscillations and the derived stomatal closure. Interestingly, we found that cau1 was oversensitive to ABA in both seed germination and stomatal closure, and crossing cas-1 to cau1 fully or partially restored these phenotypes observed in cau1 (see Supplemental Figure 7 online), which appear to indicate that in addition to [Ca2+]o, the CAU1–CAS interaction also responds to ABA. However, further analysis did not reveal significant differences in stomatal closure or seed germination between cas-1 and the wild-type Col-0 (see Supplemental Figure 7 online). Taken together, these results suggest that ABA interacts with CAU1 to mediate stomatal closure, whereas the role of CAS in this process remains to be determined.
DISCUSSION
Elevations in [Ca2+]o stimulate stomatal closure; however, the mechanism involved in [Ca2+]o signal sensing and transduction remains elusive. In this study, we demonstrated that CAU1/PRMT5/SKB1, an H4R3sme2-type histone methylase, mediates [Ca2+]o signaling by histone methylation of the CAS chromatin. Functional disruption of CAU1 led to steady upregulation of CAS and, consequently, enhanced stomatal closure and drought tolerance, consistent with the model that CAU1 acts as an epigenetic repressor of CAS to mediate [Ca2+]o signaling in stomatal closure.
CAU1 Is an Essential Component in [Ca2+]o Signaling
It is known that [Ca2+]o elicits intracellular [Ca2+]cyt oscillations to close stomata (McAinsh et al., 1995; Allen et al., 2000, 2001), and CAS was proposed to be a [Ca2+]o sensing receptor in this process to synchronize [Ca2+]o and [Ca2+]cyt oscillations (Han et al., 2003; Tang et al., 2007). Further research suggested that the IP3 cascade functions downstream of CAS in the [Ca2+]o signaling pathway (Tang et al., 2007). Together, these discoveries imply that the [Ca2+]o signaling mechanism might be highly conserved in plants and animals. However, the accuracy of this hypothesis remains an open question, mainly because CAS is exclusively localized to chloroplasts instead of the cell surface (Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008). This leaves a wide gap to be filled before it can be determined whether [Ca2+]o serves as an extracellular first messenger in plant cells as previously proposed (Han et al., 2003; Tang et al., 2007) or acts in conjunction with other molecular machineries.
In a large-scale screening of Ca2+ accumulation mutants, we unexpectedly found that the histone methylase CAU1 functions as an upstream suppressor of CAS to mediate [Ca2+]o sensing in guard cells (Figures 3 to 6 and 8). In response to [Ca2+]o signals, both the protein levels of CAU1 and its association with CAS chromatin are reduced, consequently reducing histone methylation in the promoter region of CAS and, hence, derepressing CAS expression. The promoted CAS expression can then initiate intracellular Ca2+ signaling, leading to stomatal closure, which in return decreases the transpiration rate and Ca2+ translocation to the aerial parts. This model (Figure 8) fits well with the observations that the loss-of-function mutant cau1 shows constantly high expression levels of CAS, a decreased transpiration rate and Ca2+ underaccumulation in leaves, as well as enhanced drought tolerance compared with the wild-type controls.
CAU1 was identified as SKB1/PRMT5 in previous work, where CAU1/SKB1/PRMT5 was found to act as an upstream suppressor of either FLC to mediate flowering time (Wang et al., 2007; Pei et al., 2007; Schmitz et al., 2008; Deng et al., 2010), or regulators other than FLC to mediate salt tolerance (Zhang et al., 2011), or the circadian clock (Hong et al., 2010; Sanchez et al., 2010). These observations suggest that CAU1 functions pleiotropically but mediates different biological processes via specific downstream target gene(s). Consistent with this postulation, our data demonstrated that CAU1 serves as an essential player in the [Ca2+]o-mediated stomatal closure and drought tolerance via its interaction with CAS. Furthermore, our data suggest that this process might at least be independent of plant development or flowering transition, as cau1 cas-1 behaved similarly to cau1 except for the stomatal closure and drought tolerance.
Although CAU1 mediates [Ca2+]o signaling by the epigenetic modulation of CAS expression, it is probably not a direct extracellular Ca2+ sensor because of the following considerations: (1) CAU1/PRMT5 localizes to either the nucleus or the cytoplasm (Hong et al., 2010) rather than the cell surface, where [Ca2+]o sensing occurs; and (2) the EF-hand domain for Ca2+ binding is not observed in CAU1. We therefore hypothesize that additional components might act upstream of CAU1 and CAS in the [Ca2+]o signaling pathway. Furthermore, CAU1 also interacts with ABA to mediate stomatal closure (see Supplemental Figure 7 online), implying a potentially more complex role of CAU1 in Arabidopsis Ca2+ signaling.
CAU1 Mediates [Ca2+]o Signaling Preferentially via Histone Modification of Its Downstream Target CAS
CAU1/PRMT5/SKB1 was shown to encode a histone methylase, though its effects on target genes are mediated by histone methylation, pre-mRNA splicing, or a combination of both (Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008; Deng et al., 2010; Hong et al., 2010; Sanchez et al., 2010; Zhang et al., 2011). In this study, however, we did not detect any alternative transcripts of CAS (see Supplemental Figure 6 online). By contrast, a significant increase in CAS mRNA was observed in cau1 (Figure 3), consistent with the fact that under elevated [Ca2+]o treatments, histone methylation was significantly decreased in CAS chromatin (Figure 6).
Given that CAU1/SKB1/PRMT5 methylates the small nuclear ribonucleoprotein LSM4, thus regulating pre-mRNA splicing of a group of stress-related genes (Zhang et al., 2011), we then analyzed transcripts of additional genes believed to be involved in [Ca2+]o signaling and intracellular Ca2+ homeostasis. We could not detect any alternative transcripts of these genes in the cau1 mutant but observed an increase in PLC3 and PLC4 (Figure 7A), suggesting that CAU1 might mediate [Ca2+]o sensing primarily through histone modification of the CAS chromatin. However, we do not exclude the possibility that alternative splicing mechanisms might also be involved because CBL1, which has been implicated in guard cell function (Kim et al., 2010), appeared to show alternative splicing affected by CAU1/PRMT5/SKB1 (Zhang et al., 2011).
Possible Downstream Regulatory Mechanisms in the CAU1-CAS–Mediated [Ca2+]o Signaling in Stomatal Closure
It is generally believed that [Ca2+]o mediates stomatal closure by evoking [Ca2+]cyt (McAinsh et al., 1995; Allen et al., 2000, 2001), and CAS is required to synchronize [Ca2+]o and [Ca2+]cyt oscillations despite its subcellular localization to chloroplasts (Han et al., 2003; Tang et al., 2007; Nomura et al., 2008; Weinl et al., 2008). In agreement with these observations, our study showed that in cau1, CAS expression positively correlated with [Ca2+]cyt levels and that the functional disruption of CAS significantly decreased the [Ca2+]cyt levels (Figures 5C and 5D), supporting the model that the [Ca2+]o signal is transduced and encoded in [Ca2+]cyt oscillations through the interaction of CAU1 with CAS (Figure 8), though it remains to be determined how exactly CAU1-CAS translates the signal to [Ca2+]cyt oscillation.
IP3 may act downstream of CAU1-CAS (Figure 8). In animal cells, it is well established that a [Ca2+]o signal is perceived by the extracellular calcium receptor CaR and then transduced to the downstream IP3/Ca2+ cascade to modulate [Ca2+]cyt (Brown et al., 1987, 1993; Shoback et al., 1988; Hofer 2005). The [Ca2+]o-induced IP3 generation was observed in plant guard cells in a CAS-dependent manner, and inhibition of IP3 synthesis resulted in failure to evoke [Ca2+]cyt oscillations (Tang et al., 2007). Consistent with these findings, our research also found that the expression of PLC3 and PLC4, two essential genes in regulating IP3 biosynthesis, showed a significant increase in cau1 compared with the wild type (Figure 7A), and this increase was dependent on CAS (Figure 7B).
An alternative model may involve reactive oxygen species (ROS) signaling (Figure 8). H2O2 has been shown to elicit [Ca2+]cyt (Allen et al., 2000; Pei et al., 2000). In a more recent study, a correlation between H2O2/NO and CAS was established; H2O2 and NO accumulation were observed in guard cells exposed to [Ca2+]o, and the production of H2O2 and NO were dependent on CAS. More importantly, stomatal closure in the functional disruption mutants CASas and noa1 was insensitive to [Ca2+]o treatments (Wang et al., 2012a). All these observations appear to support the hypothesis that the ROS pathway might act downstream of CAU1-CAS to mediate the [Ca2+]o signaling in guard cells.
Additionally, other unknown mechanisms might also exist (Figure 8). The rationale for this hypothesis is that pre-mRNA splicing mediated by CAU1/SKB1/PRMT5 appears to be a common mechanism in regulating diverse biological processes (Deng et al., 2010; Hong et al., 2010; Sanchez et al., 2010; Zhang et al., 2011). However, we were not able to detect any alternative splicing in those genes already correlated to IP3/Ca2+ signaling or Ca2+ homeostasis (see Supplemental Figure 6 online). This may be simply due to the fact that unknown genes that are yet to be associated with this pathway exist, an idea that is supported by the observation that CBL1 showed altered transcript splicing in skb1 (Zhang et al., 2011).
In summary, the functional characterization of CAU1 revealed that CAU1/PRMT5/SKB1 serves as an epigenetic suppressor of CAS and hence plays an essential role in [Ca2+]o signaling in guard cells to mediate stomatal closure and the consequent drought tolerance.
METHODS
Plant Materials, Growth Conditions, and Physiological Analyses
Arabidopsis thaliana plants were grown in soil to the indicated ages at 22°C with 16-h-light/8-h-dark cycles. At 14 d of age, drought stress was induced by withholding watering for 9 to 14 d, and the survival rate was scored 7 d after reirrigation. Alternatively, as indicated, rosette leaves of 22-d-old plants were collected, and their fresh weights were used as a measure at indicated time points to determine the water loss rate (Vartanian et al., 1994; Pei et al., 1998). Stomatal assays were performed as previously described for the 2 mM Ca2+ treatment (Pei et al., 2000) or with minor modifications; the abaxial epidermis was peeled from rosette leaves of 22-d-old plants to determine stomatal aperture by direct imaging. When indicated, the epidermal peels were put in an EGTA-containing buffer (10 mM KCl, 6 mM EGTA, and 10 mM MES-Tris, pH 6.15) and incubated under light conditions for 2 h to allow the stomata to open. Subsequently, the epidermal peels were transferred to the same buffer supplemented with 32.68 μM, 340 μM, or 1.3 mM CaCl2, corresponding to 0.1, 1, or 5 μM free Ca2+ according to Maxchelator (http://www.stanford.edu/~cpatton/webmaxc/webmaxcS.htm), and incubated under light conditions for 2 h. Stomatal apertures were determined by measuring the pore widths and lengths with a digital ruler in Image-pro plus 6.0 (Media Cybernetics).
Alternatively, plants were grown in quarter-strength hydroponics as previously described (Arteca and Arteca, 2000; Gong et al., 2003). At ∼3 weeks of age, plants were treated with or without 0.5 or 10 mM Ca2+ for 4 d. Rosette leaves and roots were sampled and subjected to further analyses as indicated.
The skb1-2 mutant was ordered from the SALK T-DNA collection (Salk_095085) from the ABRC. Isolation of ion accumulation mutants was performed according to the previously described screen at the University of California at San Diego (Lahner et al., 2003), with minor modification: fast neutron-mutagenized M2 seeds (population M2F-02-01; Lehle Seeds) were planted in peat moss soil (Pindstrup) premixed with 5 PPM of aluminum, calcium, iron, potassium, manganese, lead, and zinc. Mature dried cauline leaves were sampled and subjected to inductively coupled plasma-mass spectrometry (ICP-MS) analysis of ion accumulation as indicated.
Map-Based Cloning, Sequencing, and Genetic Complementation Assay
The Ca2+ underaccumulation mutant cau1 in Col-0 background was crossed to the ecotype Landsberg erecta to generate a mapping population. SSLP markers were designed according to The Arabidopsis Information Resource (http://www.Arabidopsis.org). Bulked segregant analysis was performed to determine the genetic linkage between cau1 and SSLP markers (Michelmore et al., 1991). Fine mapping of cau1 was performed using DNA from 5742 F2 mutants as previously described (Lukowitz et al., 2000). Mutations in CAU1 were determined by PCR amplification and sequencing using CAU1-1 primers (see Supplemental Table 2 online).
For the complementation of cau1, direct transformation of CAU1-containing constructs into cau1 was not used because cau1 is hypersensitive to bacteria. Instead, transgenic plants harboring 35S/CAU1-pBI121 or 35S/EYFP:CAU1-pMON530 were crossed to the cau1 mutant. The derived F2 progenies were then screened for kanamycin resistance, and cau1 homozygous plants were determined by PCR amplification and sequencing using CAU1-2 primers (see Supplemental Table 2 online).
DNA Constructs and Plant Transformation
The CAU1 cDNA was amplified by RT-PCR. The two restriction sites for BamHI and SacI were introduced using CAU1-3 primers; XhoI and EcoRI sites were introduced using CAU1-4 primers (primers are listed in Supplemental Table 2 online). The resulting fragments were confirmed by sequencing and then subcloned into the binary vector pBI121 (predigested with BamHI and SacI) or 35S/EYFP-pMON530 (predigested with XhoI and EcoRI). The generated constructs 35S/CAU1-pBI121 and 35S/EYFP:CAU1-pMON530, together with the construct 35S/EYFP-pMON530, were transformed into Col-0 by floral dip (Clough and Bent, 1998). Transgenic lines with a segregation rate of 3:1 grown on kanamycin plates were used for further homozygote and strong allele screenings.
Microarray Analysis
Total RNA was isolated from rosette leaves of 4-week-old Col-0 and cau1 mutant plants using TRIzol reagent (Invitrogen). Microarray hybridization to the Arabidopsis whole-genome ATH1 arrays was performed by Shanghai OE Biotech. Data were extracted and normalized according to the manufacturer’s standard protocol.
RT-PCR/Quantitative RT-PCR
Total RNA from plants grown under the indicated conditions was isolated using TRIzol reagent. First-strand cDNA synthesis, RT-PCR, and quantitative RT-PCR were performed as previously described (Li et al., 2010). CAU1-5, CAS-RT, and Actin2 primers were used in regular RT-PCR. CAS-QP and Actin2-QP primers were used in quantitative RT-PCR (see Supplemental Table 2 online).
Histochemical Analysis
Transgenic plants expressing the construct pCAS/GUS-pBI101 (Han et al., 2003) were crossed to cau1 mutant plants. Plants showing the cau1 phenotype in the F2 population were subjected to histochemical analysis as previously described (Li et al., 2010).
Isolation of cas-1 and cau1 cas-1 Mutant Plants and Introduction of 35S/CAU1-pBI121 into cau1
The T-DNA insertion line SALK_070416 obtained from the ABRC was screened for the homozygous knockout mutant cas-1 as previously described (Weinl et al., 2008). To generate the cau1 cas-1 double mutant, cau1 was crossed to cas-1 to make an F2 population; cau1-like plants were further analyzed to isolate the genotype cas-1/cas-1 using the PCR primers CAS-salk and LBa1 as previously described (Krysan et al., 1999). The same strategy was used to introduce the construct 35S/CAU1-pBI121 into cau1: Col-0 plants containing 35S/CAU1-pBI121 were crossed to cau1, and further PCR analysis was performed using primers mentioned above for homozygosity screening.
[Ca2+]cyt Imaging in Guard Cells
Fluorescence imaging of [Ca2+]cyt was performed according to Legué et al. (1997) and Wymer et al. (1997) with minor modifications: Abaxial epidermis peels taken from 22-d-old soil-grown plants were perfused with 20 µM Indo-1 solution (in 10 mM dimethylglutaric acid, pH 4.5) and incubated in the dark for 1 h. Then, the peels were washed with liquid Murashige and Skoog medium followed by imaging using confocal laser scanning microscopy (Carl Zeiss; LSM 510 Meta) (364-nm excitation and 400- to 435-nm and 480 ± 20-nm emission). Calcium levels were calculated and pseudocolored according to the color scale as described (Wang et al., 2012b). Briefly, pseudocolor ratio images of calcium level were calculated with the Olympus Fluoview ver. 1.7a viewer tool (Olympus) to measure fluorescence intensities. White color indicates ∼2 times the calcium level of that indicated by blue color and 1.5 times of that indicated by yellow color.
Chromatin Immunoprecipitation Assay
Twenty-day-old Col-0, cau1, and 35S/EYFP:CAU1-cau1 plants grown under long-day conditions were harvested for the ChIP assay (Aggarwal et al., 2010). Total proteins extracted from plants of Col-0 and cau1 were immunoprecipitated with antisymmetric dimethyl-H4R3 antibody (Abcam), and proteins from cau1 and 35S/EYFP:CAU1-cau1 were immunoprecipitated with an anti-GFP antibody (Invitrogen). Primers used for ChIP-qPCR are as follows: region A (CAS-9), region B (CAS-11), region C (CAS-14), region D (CAS-17), and region E (CAS-19). TUB8 was used as a control (Mathieu et al., 2005). The primer sequences are given in Supplemental Table 2 online.
Protein Gel Blotting Analysis
Transgenic 35S/EYFP:CAU1-cau1 plants were grown in hydroponics to 24 d of age and then exposed to Ca2+ treatments at the indicated concentrations for 4 d. Total protein were extracted from leaf samples using buffer E (125 mM Tris-HCl, pH 8.0, 1% [w/v] SDS, 10% [v/v] glycerol, and 50 mM NaS2O5). Thirty micrograms of total proteins of each sample were separated on 12% SDS-PAGE gel and analyzed by protein gel blot according to the manufacturer’s instructions. Mouse anti-Actin2 and -GFP (Abmart; at 1:5000 dilution) were used as primary antibodies. The membranes were visualized using a Super-Signal West Pico Chemiluminescent Substrate Kit (Thermo Scientific) according to the manufacturer’s instructions.
Statistical Analysis
Two-tailed Student’s t tests were performed. Differences were deemed significant at P < 0.05 and extremely significant at P < 0.01.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CAU1 (At4g31120), CAS (At5g23060), CAX1 (At2g38170), CAX3 (At3g51860), PLC3 (At5g58670), PLC4 (At5g58670), and Actin2 (At3g18780). Additional sequence data are available in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. CAU1 Affects Calcium Accumulation.
Supplemental Figure 2. cau1 Mutant Shows Pleiotropic Phenotypes.
Supplemental Figure 3. Complementation Assay of the cau1 Mutant.
Supplemental Figure 4. Stomatal Closure, Drought Tolerance, and CAS Expression in 35S/CAU1-cau1.
Supplemental Figure 5. CAU1 Complements Stomatal Closure and [Ca2+]o Signaling Phenotype in the cau1 Mutant.
Supplemental Figure 6. Analysis of Alternative Splicing of Ca2+ Homeostasis or Signaling Pathway Genes.
Supplemental Figure 7. cau1 Is Hypersensitive to ABA.
Supplemental Table 1. Transcriptomic Analysis of Ca2+ Homeostasis Genes, Ca2+ Signaling Pathway Genes, and Genes Known to Be Regulated by CAU1.
Supplemental Table 2. List of Primer Sequences.
Acknowledgments
We thank Zhenming Pei (Duke University) for kindly providing the seeds of pCAS/GUS; Julian Schroeder (University of California at San Diego), Xiao-Feng Cao (Chinese Academy of Sciences), and Hai Huang (Chinese Academy of Sciences) for helpful discussion; Zhenbiao Yang and Irene Lavagi (University of California at Riverside) for careful reading of the article; and Xiao-Shu Gao (Shanghai Institutes for Biological Sciences) for helping with confocal microscopy. This research was supported by the National Science Foundation of China (30900785 and 31121063), the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams, the Chinese Academy of Sciences program for Creative Basic Research (KSCX2-EW-J-12), and in part by the Sanofi-Aventis-Shanghai Institutes for Biological Sciences Scholarship Program.
AUTHOR CONTRIBUTIONS
J.-M.G. and Y.-L.F. designed the research. Y.-L.F., G.-B.Z., X.-F.L., Y.G., and H-Y.Y. performed the research. J.-M.G. and Y.-L.F. analyzed the data and wrote the article.
Glossary
- IP3
inositol-1,4,5-trisphosphate
- Col-0
Columbia-0
- SSLP
simple sequence length polymorphism
- ChIP-qPCR
chromatin immunoprecipitation quantitative PCR
- ABA
abscisic acid
- ROS
reactive oxygen species
- ICP-MS
inductively coupled plasma–mass spectrometry
References
- Aggarwal P., Vaites L.P., Kim J.K., Mellert H., Gurung B., Nakagawa H., Herlyn M., Hua X., Rustgi A.K., McMahon S.B., Diehl J.A. (2010). Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.J., Chu S.P., Harrington C.L., Schumacher K., Hoffmann T., Tang Y.Y., Grill E., Schroeder J.I. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Chu S.P., Schumacher K., Shimazaki C.T., Vafeados D., Kemper A., Hawke S.D., Tallman G., Tsien R.Y., Harper J.F., Chory J., Schroeder J.I. (2000). Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science 289: 2338–2342 [DOI] [PubMed] [Google Scholar]
- Ancelin K., Lange U.C., Hajkova P., Schneider R., Bannister A.J., Kouzarides T., Surani M.A. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Arteca R.N., Arteca J.M. (2000). A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol. Plant. 108: 188–193 [Google Scholar]
- Bedford M.T., Richard S. (2005). Arginine methylation an emerging regulator of protein function. Mol. Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Brown E., Enyedi P., LeBoff M., Rotberg J., Preston J., Chen C. (1987). High extracellular Ca2+ and Mg2+ stimulate accumulation of inositol phosphates in bovine parathyroid cells. FEBS Lett. 218: 113–118 [DOI] [PubMed] [Google Scholar]
- Brown E.M. (1991). Extracellular Ca2+ sensing, regulation of parathyroid cell function, and role of Ca2+ and other ions as extracellular (first) messengers. Physiol. Rev. 71: 371–411 [DOI] [PubMed] [Google Scholar]
- Brown E.M., Gamba G., Riccardi D., Lombardi M., Butters R., Kifor O., Sun A., Hediger M.A., Lytton J., Hebert S.C. (1993). Cloning and characterization of an extracellular Ca(2+)-sensing receptor from bovine parathyroid. Nature 366: 575–580 [DOI] [PubMed] [Google Scholar]
- Bush D.S. (1995). Calcium regulation in plant cells and its role in signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 95–122 [Google Scholar]
- Cheng N.H., Pittman J.K., Shigaki T., Lachmansingh J., LeClere S., Lahner B., Salt D.E., Hirschi K.D. (2005). Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 138: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D., Villiers F., Kroniewicz L., Lee S., Seo Y.J., Hirschi K.D., Leonhardt N., Kwak J.M. (2012). Vacuolar CAX1 and CAX3 influence auxin transport in guard cells via regulation of apoplastic pH. Plant Physiol. 160: 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conn S.J., et al. (2011). Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J.M., Lee D.A., Schroeder J.I. (2003). Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10118–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Tang R., Anderson L.K., Woerner T.E., Pei Z.M. (2003). A cell surface receptor mediates extracellular Ca(2+) sensing in guard cells. Nature 425: 196–200 [DOI] [PubMed] [Google Scholar]
- Harper J.F. (2001). Dissecting calcium oscillators in plant cells. Trends Plant Sci. 6: 395–397 [DOI] [PubMed] [Google Scholar]
- Helpler P.K., Wayne R.O. (1985). Calcium and plant development. Annu. Rev. Plant Physiol. 36: 397–439 [Google Scholar]
- Hetherington A.M., Brownlee C. (2004). The generation of Ca(2+) signals in plants. Annu. Rev. Plant Biol. 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hofer A.M. (2005). Another dimension to calcium signaling: A look at extracellular calcium. J. Cell Sci. 118: 855–862 [DOI] [PubMed] [Google Scholar]
- Hong S., Song H.R., Lutz K., Kerstetter R.A., Michael T.P., McClung C.R. (2010). Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21211–21216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P.J., Young J.C., Sussman M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B., Gong J., Mahmoudian M., Smith E.L., Abid K.B., Rogers E.E., Guerinot M.L., Harper J.F., Ward J.M., McIntyre L., Schroeder J.I., Salt D.E. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Legué V., Blancaflor E., Wymer C., Perbal G., Fantin D., Gilroy S. (1997). Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol. 114: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S., Kudla J., Rodriguez-Concepcion M., Yalovsky S., Gruissem W. (2002). Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14 (suppl.): S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie E.A.C. (1992). Calcium and ABA-induced stomatal closure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 338: 5–18 [Google Scholar]
- Mathieu O., Probst A.V., Paszkowski J. (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh M.R., Webb A., Taylor J.E., Hetherington A.M. (1995). Stimulus-induced oscillations in guard cell cytosolic free calcium. Plant Cell 7: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R.W., Paran I., Kesseli R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H., Komori T., Kobori M., Nakahira Y., Shiina T. (2008). Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 53: 988–998 [DOI] [PubMed] [Google Scholar]
- Pei Y., Niu L., Lu F., Liu C., Zhai J., Kong X., Cao X. (2007). Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Ghassemian M., Kwak C.M., McCourt P., Schroeder J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287–290 [DOI] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Pollack B.P., Kotenko S.V., He W., Izotova L.S., Barnoski B.L., Pestka S. (1999). The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 274: 31531–31542 [DOI] [PubMed] [Google Scholar]
- Poovaiah B.W., Reddy A.S. (1993). Calcium and signal transduction in plants. CRC Crit. Rev. Plant Sci. 12: 185–211 [DOI] [PubMed] [Google Scholar]
- Rudd J.J., Franklin-Tong V.E. (1999). Calcium signaling in plants. Cell. Mol. Life Sci. 55: 214–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Sanders D., Brownlee C., Harper J.F. (1999). Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 (Suppl): S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Sung S., Amasino R.M. (2008). Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A. (1985). Role of Ca2+ and EGTA on stomatal movements in Commelina communis L. Plant Physiol. 79: 1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoback D., Thatcher J., Leombruno R., Brown E. (1983). Effects of extracellular Ca++ and Mg++ on cytosolic Ca++ and PTH release in dispersed bovine parathyroid cells. Endocrinology 113: 424–426 [DOI] [PubMed] [Google Scholar]
- Shoback D.M., Membreno L.A., McGhee J.G. (1988). High calcium and other divalent cations increase inositol trisphosphate in bovine parathyroid cells. Endocrinology 123: 382–389 [DOI] [PubMed] [Google Scholar]
- Shoback D.M., Thatcher J.G., Brown E.M. (1984). Interaction of extracellular calcium and magnesium in the regulation of cytosolic calcium and PTH release in dispersed bovine parathyroid cells. Mol. Cell. Endocrinol. 38: 179–186 [DOI] [PubMed] [Google Scholar]
- Tang R.H., Han S., Zheng H., Cook C.W., Choi C.S., Woerner T.E., Jackson R.B., Pei Z.M. (2007). Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315: 1423–1426 [DOI] [PubMed] [Google Scholar]
- Thuleau P., Schroeder J.I., Ranjeva R. (1998). Recent advances in the regulation of plant calcium channels: Evidence for regulation by G-proteins, the cytoskeleton and second messengers. Curr. Opin. Plant Biol. 1: 424–427 [DOI] [PubMed] [Google Scholar]
- Vainonen J.P., Sakuragi Y., Stael S., Tikkanen M., Allahverdiyeva Y., Paakkarinen V., Aro E., Suorsa M., Scheller H.V., Vener A.V., Aro E.M. (2008). Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J. 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Vartanian N., Marcotte L., Giraudat J. (1994). Drought rhizogenesis in Arabidopsis thaliana (differential responses of hormonal mutants). Plant Physiol. 104: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.H., Yi X.Q., Han A.D., Liu T.W., Chen J., Wu F.H., Dong X.J., He J.X., Pei Z.M., Zheng H.L. (2012a). Calcium-sensing receptor regulates stomatal closure through hydrogen peroxide and nitric oxide in response to extracellular calcium in Arabidopsis. J. Exp. Bot. 63: 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., Chong K. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26: 1934–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chu Y.J., Xue H.W. (2012b). Inositol polyphosphate 5-phosphatase-controlled Ins(1,4,5)P3/Ca2+ is crucial for maintaining pollen dormancy and regulating early germination of pollen. Development 139: 2221–2233 [DOI] [PubMed] [Google Scholar]
- Weinl S., Held K., Schlücking K., Steinhorst L., Kuhlgert S., Hippler M., Kudla J. (2008). A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol. 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Wymer C.L., Bibikova T.N., Gilroy S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12: 427–439 [DOI] [PubMed] [Google Scholar]
- Zhang Z., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23: 396–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski R.E. (1998). Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 697–725 [DOI] [PubMed] [Google Scholar]