This study reveals that jasmonate has a positive role in regulating freezing stress responses in Arabidopsis. JAZ proteins, the repressors of jasmonate signaling, physically interact with ICE1 and ICE2 transcription factors, thereby repressing the ICE-CBF/DREB1 cold signaling pathway.
Abstract
The INDUCER OF CBF EXPRESSION (ICE)–C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 (CBF/DREB1) transcriptional pathway plays a critical role in modulating cold stress responses in Arabidopsis thaliana. Dissecting crucial upstream regulatory signals or components of the ICE-CBF/DREB1 cascade will enhance our understanding of plant cold-tolerance mechanisms. Here, we show that jasmonate positively regulates plant responses to freezing stress in Arabidopsis. Exogenous application of jasmonate significantly enhanced plant freezing tolerance with or without cold acclimation. By contrast, blocking endogenous jasmonate biosynthesis and signaling rendered plants hypersensitive to freezing stress. Consistent with the positive role of jasmonate in freezing stress, production of endogenous jasmonate was triggered by cold treatment. In addition, cold induction of genes acting in the CBF/DREB1 signaling pathway was upregulated by jasmonate. Further investigation revealed that several JASMONATE ZIM-DOMAIN (JAZ) proteins, the repressors of jasmonate signaling, physically interact with ICE1 and ICE2 transcription factors. JAZ1 and JAZ4 repress the transcriptional function of ICE1, thereby attenuating the expression of its regulon. Consistent with this, overexpression of JAZ1 or JAZ4 represses freezing stress responses of Arabidopsis. Taken together, our study provides evidence that jasmonate functions as a critical upstream signal of the ICE-CBF/DREB1 pathway to positively regulate Arabidopsis freezing tolerance.
INTRODUCTION
Temperatures outside an organism’s optimal tolerance range are regarded as a major environmental stress. Extreme low temperature disrupts cellular homeostasis and severely impairs plant growth and development. To tolerate cold stress, plants have evolved sophisticated mechanisms involving altered physiological and biochemical processes. Previous studies have revealed that the INDUCER OF CBF EXPRESSION (ICE)–C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 (CBF/DREB1) transcriptional cascade plays a critical role in the cold-response pathways in Arabidopsis thaliana (Thomashow, 1999; Chinnusamy et al., 2007). ICE1, a basic helix-loop-helix (bHLH) transcription factor, directly binds to cis-elements (CANNTG) in the CBF3/DREB1a promoter (Chinnusamy et al., 2003). Expression of CBF3/DREB1a and its downstream target genes is downregulated in ice1 mutants; ice1 plants consequently display significantly reduced cold acclimation-induced freezing tolerance (Chinnusamy et al., 2003). Similarly, ICE2, the homolog of ICE1, influences the expression of CBF1/DREB1b and participates in regulating plant cold stress responses (Fursova et al., 2009). Moreover, the SMALL UBIQUITIN-RELATED MODIFIER E3 ligase SIZ1 (SAP and Miz) and the ubiquitin E3 ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 have been shown to modify ICE1 posttranslationally and function in the ICE-CBF/DREB1 signaling pathway (Dong et al., 2006; Miura et al., 2007). However, whether there are other crucial regulators acting upstream of the ICE transcription factors needs to be investigated.
In addition to the ICE-CBF/DREB1 regulatory pathway, plant hormones have been demonstrated to modulate cold stress responses. For example, low temperature induces a transient increase in levels of the endogenous stress hormone abscisic acid (ABA), and exogenous application of ABA enhances plant cold tolerance (Lång et al., 1994; Mäntylä et al., 1995). Jeon et al. (2010) showed that cytokinin receptors Arabidopsis HISTIDINE KINASE2 (AHK2) and AHK3 and type-A Arabidopsis RESPONSE REGULATOR (ARR) proteins play regulatory roles in cold stress signaling via inhibition of ABA responses. Recently, ethylene signaling was shown to negatively regulate plant freezing stress responses by repressing the expression of CBF/DREB1 and type-A ARR genes in Arabidopsis (Shi et al., 2012).
The phytohormone jasmonate acts as an important regulatory signal to influence multiple plant processes. In Arabidopsis, jasmonate is directly involved in root growth (Staswick et al., 1992; Feys et al., 1994; Pauwels et al., 2010), male fertility (McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000; Cheng et al., 2009), anthocyanin accumulation (Franceschi and Grimes, 1991; Shan et al., 2009), senescence (Ueda and Kato, 1980; Schommer et al., 2008; Shan et al., 2011), and defense responses (Howe et al., 1996; McConn et al., 1997; Reymond and Farmer, 1998; Vijayan et al., 1998; Farmer, 2001; Farmer et al., 2003; Browse, 2009). Jasmonate is perceived by the F-box protein CORONATINE INSENSITIVE1 (COI1), which subsequently facilitates the degradation of JAZ proteins via the SCFCOI1-26S proteasome pathway (Xu et al., 2002; Chini et al., 2007; Thines et al., 2007; Yan et al., 2009; Sheard et al., 2010). JAZ proteins, harboring the Jas domain, act as repressors of jasmonate signaling via their physical interactions with a wide array of transcription factors; their degradation releases these factors and subsequently activates downstream signal cascades. The bHLH transcription factors JASMONATE INSENSITIVE1 (JIN1/MYC2), MYC3, and MYC4 function as direct targets of JAZ proteins to modulate a subset of jasmonate-regulated responses, such as the inhibition of root elongation and defense responses (Boter et al., 2004; Lorenzo et al., 2004; Chini et al., 2007; Dombrecht et al., 2007; Fernández-Calvo et al., 2011). The essential components of WD-repeat/bHLH/MYB transcriptional complexes, including bHLH proteins GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3), and TRANSPARENT TESTA8 and MYB domain proteins GLABRA1 (GL1) and MYB75, also act as JAZ targets to mediate jasmonate-regulated biological processes (Qi et al., 2011). In addition, two other MYB transcription factors (MYB21 and MYB24) have recently been identified as JAZ targets to regulate jasmonate-mediated anther development and filament elongation (Song et al., 2011).
Although jasmonate has been implicated in cold storage of several tropical and subtropical fruits (González-Aguilar et al., 2000, 2004; Cao et al., 2009; Mohammad et al., 2011; Zhao et al., 2013), the exact role played by jasmonate in mediating freezing responses and the molecular mechanisms of these regulatory responses remain to be elucidated. In this study, we undertook a molecular and genetic approach to investigate the role of jasmonate in plant freezing stress responses. We found that exogenous application of jasmonate significantly improved Arabidopsis freezing tolerance, while blocking jasmonate biosynthesis and signaling decreased plant freezing tolerance. Mechanism investigation revealed that several JAZ proteins physically interact with ICE1 and ICE2 and repress their transcriptional functions and that overexpression of JAZ1 or JAZ4 represses responses to freezing stress. Our results thus provide evidence that jasmonate acts as an upstream signal of the ICE-CBF/DREB1 transcriptional pathway to positively regulate freezing stress responses of Arabidopsis.
RESULTS
Exogenous Application of Jasmonate Enhances Plant Freezing Tolerance
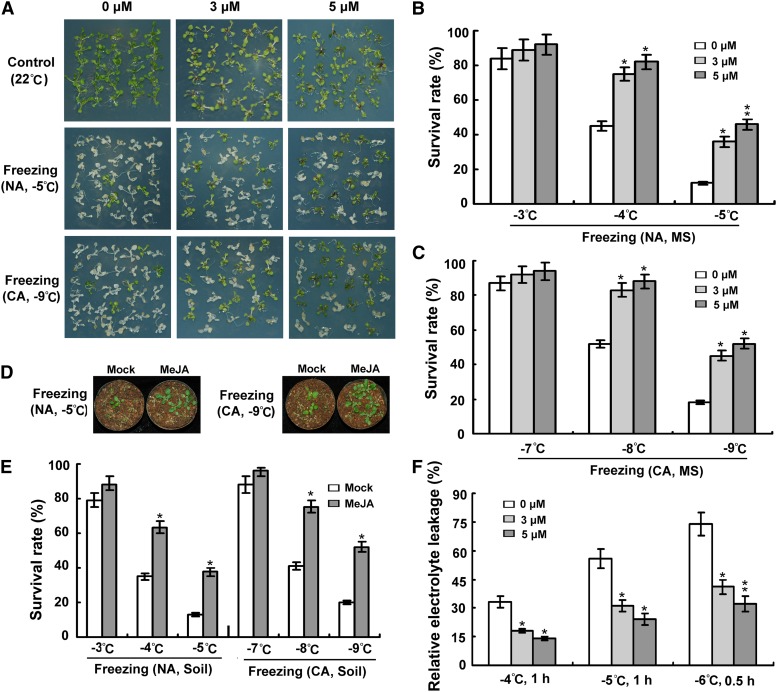
To explore the regulatory role of jasmonate in cold stress responses, we first examined whether the freezing tolerance of wild-type plants was affected by exogenous application of jasmonate under both nonacclimated and cold-acclimated conditions. In the presence of methyl jasmonate (MeJA; 3 and 5 μM), the wild-type plants were slightly smaller in size and accumulated more anthocyanin compared with nontreated plants (Figure 1A). Without cold acclimation, the survival rates of MeJA-treated plants were significantly higher than those of control plants after exposure to freezing temperatures of −4 and −5°C (Figures 1A and 1B). Similarly, under cold-acclimated conditions (2 weeks at 4°C), MeJA-treated plants also exhibited enhanced tolerance of freezing stress; their survival rates after freezing treatments (−8 and −9°C) were dramatically higher than those of control plants (Figures 1A and 1C). To further confirm the role of exogenous jasmonate on freezing tolerance, we analyzed the performances of soil-grown wild-type plants with or without MeJA treatments in response to freezing temperatures. As shown in Figures 1D and 1E, MeJA-treated soil-grown plants also displayed substantially increased tolerance to freezing stress under both nonacclimated and cold-acclimated conditions. Consistent with this, lower levels of relative electrolyte leakage, a parameter for evaluation of cold-induced membrane injury (Lyons, 1973), were observed in MeJA-treated plants than in control plants subjected to freezing temperatures (Figure 1F). These results suggested that jasmonate may have a positive role in modulating Arabidopsis freezing tolerance with or without cold acclimation.
Figure 1.
Effect of Exogenous Jasmonate on Plant Freezing Tolerance.
(A) Freezing phenotypes of 12-d-old wild-type seedlings grown on MS medium or MS medium containing 3 or 5 μM MeJA. Nonacclimated (NA) seedlings were treated at −5°C for 1 h, and cold-acclimated seedlings (CA; 2 weeks at 4°C) were exposed to −9°C for 1 h, followed by recovery at 22°C for 4 d. Experiments were performed three times with similar results.
(B) Survival rates of nonacclimated plants after exposure to indicated freezing temperatures. Error bars show sd from three replicates. MS, seedlings grown on MS medium.
(C) Survival rates of cold-acclimated (2 weeks at 4°C) plants after exposure to indicated freezing temperatures. Error bars show sd from three replicates.
(D) Freezing phenotypes of soil-grown plants with or without jasmonate treatment. Eighteen-day-old soil-grown plants were treated with water (Mock) or 30 μM MeJA. Nonacclimated plants were treated at −5°C for 1.5 h, and cold-acclimated plants (2 weeks at 4°C) were exposed to −9°C for 1.5 h, followed by recovery at 22°C for 7 d. Experiments were performed three times with similar results using over 100 plants per treatment.
(E) Survival rates of nonacclimated and cold-acclimated (2 weeks at 4°C) soil-grown plants with or without jasmonate treatment after exposure to the indicated freezing temperatures. Error bars show sd from three replicates. Soil, plants grown in soil.
(F) Ion leakage assays of plants treated with indicated freezing temperatures. Error bars show sd from three replicates. *Differences between MeJA-treated plants and control plants are significant (P < 0.05). **Differences between MeJA-treated plants and control plants are highly significant (P < 0.01).
Blocking Jasmonate Biosynthesis and Signaling Renders Plants Hypersensitive to Freezing Stress
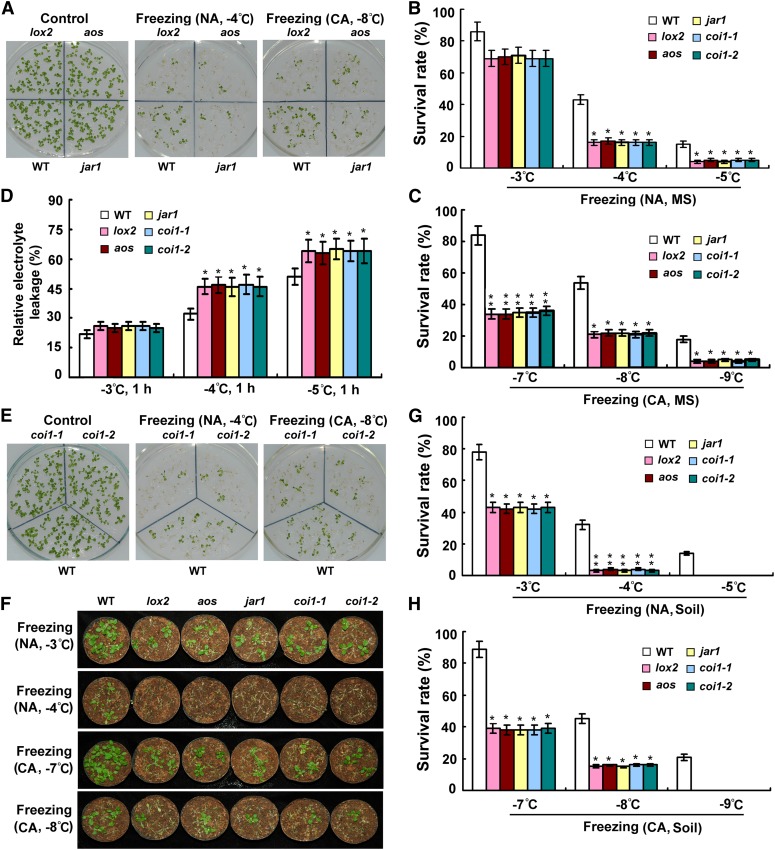
To further determine jasmonate’s role in mediating freezing tolerance, we tested the involvement of jasmonate biosynthesis and signaling in freezing stress responses. ALLENE OXIDE SYNTHASE (AOS) and LIPOXYGENASE2 (LOX2) are key enzymes in jasmonate synthesis, whereas JASMONATE RESISTANT1 (JAR1) carries out a jasmonate conjugation/activation reaction (Song et al., 1993; Bell et al., 1995; Staswick et al., 2002). With or without acclimation, loss-of-function mutants lox2, aos, and jar1 displayed substantially decreased tolerance of freezing temperatures compared with wild-type plants (Figure 2A); their survival rates in response to freezing temperatures (nonacclimated plants to −4 and −5°C and cold-acclimated plants to −7, −8, and −9°C) were dramatically lower than those of wild-type plants (Figures 2B and 2C). Consistent with these phenotypes, the relative electrolyte leakage levels following freezing (−4 and −5°C) were significantly higher in lox2, aos, and jar1 mutants than in control plants (Figure 2D). The F-box protein COI1 is the jasmonate receptor and a key positive regulator in the jasmonate signaling pathway (Xie et al., 1998; Yan et al., 2009). Freezing tolerance assays showed that disruption of COI1 rendered plants hypersensitive to freezing stress under both nonacclimated and cold-acclimated conditions compared with wild-type plants (Figures 2B to 2E). To further corroborate the biological role of jasmonate in modulating freezing tolerance, we analyzed the performances of soil-grown mutants under freezing stress. As shown in Figures 2F to 2H, with or without cold acclimation, the soil-grown mutants were also more sensitive to freezing temperatures than the wild-type plants. Taken together, these observations indicate that jasmonate biosynthesis and signaling positively regulate Arabidopsis freezing tolerance under both nonacclimated and cold-acclimated conditions.
Figure 2.
Freezing Tolerance of Mutants Involved in Jasmonate Biosynthesis and Signaling.
(A) Freezing phenotypes of MS medium–grown lox2, aos, and jar1 mutant plants with or without cold acclimation. Nonacclimated (NA) 12-d-old seedlings were treated at −4°C for 1 h, and cold-acclimated seedlings (2 weeks at 4°C) were exposed to −8°C for 1 h, followed by recovery at 22°C for 4 d. Experiments were performed three times with similar results. WT, the wild type.
(B) and (C) Survival rates of various nonacclimated (B) and cold-acclimated (C) mutant plants after exposure to indicated freezing temperatures. Error bars show sd from three replicates. CA, cold-acclimated (2 weeks at 4°C); MS, seedlings grown on MS medium; NA, nonacclimated.
(D) Ion leakage assays of mutants treated with indicated freezing temperatures. Error bars show sd from three replicates.
(E) Freezing phenotypes of coi1-1 and coi1-2 mutants with or without cold acclimation. Nonacclimated 12-d-old seedlings were treated at −4°C for 1 h, and cold-acclimated (2 weeks at 4°C) seedlings were exposed to −8°C for 1 h, followed by recovery at 22°C for 4 d. Experiments were performed three times with similar results.
(F) Freezing phenotypes of various soil-grown mutants with or without cold acclimation. Nonacclimated 18-d-old soil-grown seedlings were treated at −3 or −4°C for 1.5 h, and cold-acclimated seedlings (2 weeks at 4°C) were exposed to −7 or −8°C for 1.5 h, followed by recovery at 22°C for 7 d. Experiments were performed three times with similar results using over 100 plants per treatment.
(G) and (H) Survival rates of various nonacclimated (G) and cold-acclimated (H) soil-grown mutant plants after exposure to indicated freezing temperatures. Error bars show sd from three replicates. CA, cold-acclimated (2 weeks at 4°C); NA, nonacclimated; Soil, seedlings grown in soil. *Differences between the mutant and the wild type are significant (P < 0.05). **Differences between the mutant and the wild type are highly significant (P < 0.01).
Cold Stress Induces an Increase in Endogenous Jasmonate Levels
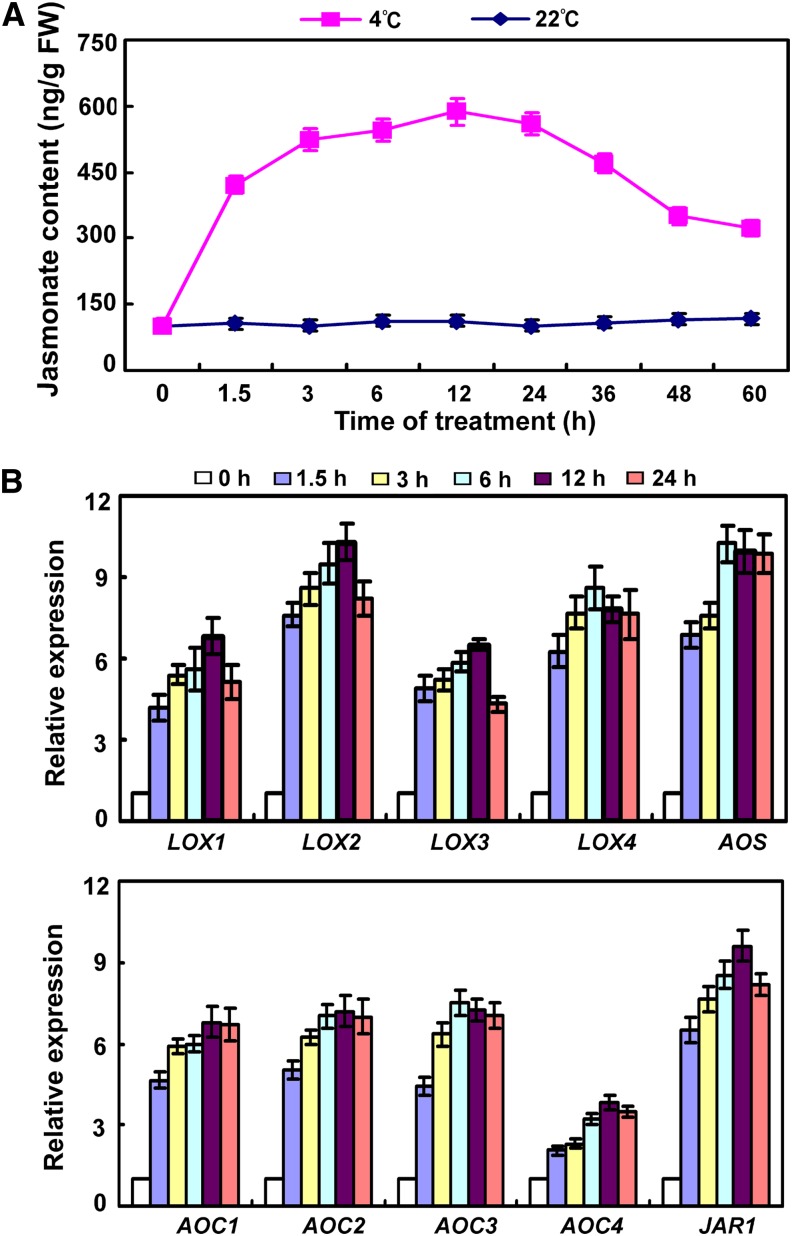
Having ascertained that jasmonate is involved in plant freezing tolerance, we then asked whether endogenous jasmonate production is affected by cold stress. To test this, we measured jasmonate production in wild-type plants following cold treatment. As shown in Figure 3A, endogenous jasmonate production was induced by cold treatment. To further understand the effect of cold stress on jasmonate production, we examined the expression of several genes encoding critical enzymes for jasmonate biosynthesis. As shown in Figure 3B, expression of analyzed genes was upregulated by cold stress. These results indicate that cold stress triggers the accumulation of endogenous jasmonate in Arabidopsis.
Figure 3.
Effect of Cold Stress on Production of Endogenous Jasmonate.
(A) Cold-stress induction of endogenous jasmonate production. Eighteen-day-old soil-grown wild-type plants were treated at 4°C or kept at 22°C. Endogenous jasmonate levels were measured at various time points. FW, fresh weight.
(B) Expression of genes involved in jasmonate biosynthesis under cold stress. Eighteen-day-old wild-type plants were treated at 4°C for the indicated time periods. Transcripts were measured by qRT-PCR. Error bars show sd from three independent RNA extractions.
Cold Induction of CBF/DREB1 and Their Targets Is Upregulated by Jasmonate
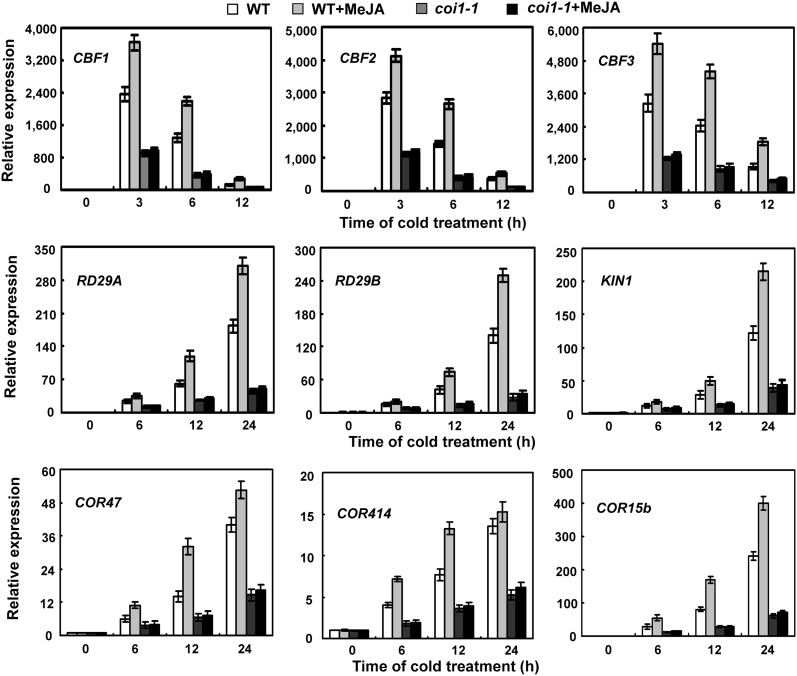
Currently, the best understood freezing tolerance pathway is the CBF/DREB1 transcriptional regulatory cascade. As jasmonate is involved in plant freezing stress responses, we queried whether jasmonate regulates the CBF/DREB1 cold signaling pathway. Initially, we analyzed expression levels of CBF/DREB1 genes and several of their targets, including RESPONSIVE TO DESSICATION29A (RD29A), RD29B, COLD REGULATED15B (COR15b), COR414, COR47, and KIN1, in response to jasmonate treatment under cold stress. The results shown in Figure 4 demonstrate that cold-induced expression levels of CBF/DREB1 and their target genes were significantly upregulated by jasmonate. To confirm the regulatory effect of jasmonate on the CBF/DREB1 pathway, we also detected expression of CBF/DREB1 and their target genes in coi1-1 mutants. The base expression levels of these cold-responsive genes, compared with wild-type plants, were generally low and unaltered in coi1-1 mutants (Figure 4). Under cold stress, however, transcripts of CBF/DREB1 and their target genes were reduced in coi1-1 mutants compared with those in wild-type plants (Figure 4; see Supplemental Figure 1 online). To further investigate whether jasmonate affects all cold-regulated gene expression, we analyzed the expression of several cold-responsive genes acting in ABA or cytokinin signaling pathway and those associated with the synthesis of soluble sugars (functioning as cryoprotectant molecules) in coi1-1 mutants, such as ABSCISIC ACID RESPONSIVE ELEMENT BINDING FACTOR1 (ABF1), ARR5, and SUCROSE SYNTHASE. As shown in Supplemental Figure 2 online, their transcripts were not affected in coi1-1 mutants under cold stress, compared with those in wild-type plants. Taken together, these results indicate that jasmonate positively regulates the CBF/DREB1 transcriptional regulatory pathway in Arabidopsis.
Figure 4.
Expression of CBF/DREB1 and Their Regulons in Response to Jasmonate under Cold Stress.
Eighteen -day-old wild-type (WT) and coi1-1 mutant plants were treated at 4°C with water or 100 μM MeJA for the indicated time periods. Error bars show sd from three independent RNA extractions.
JAZ Proteins Physically Interact with ICE1 and ICE2
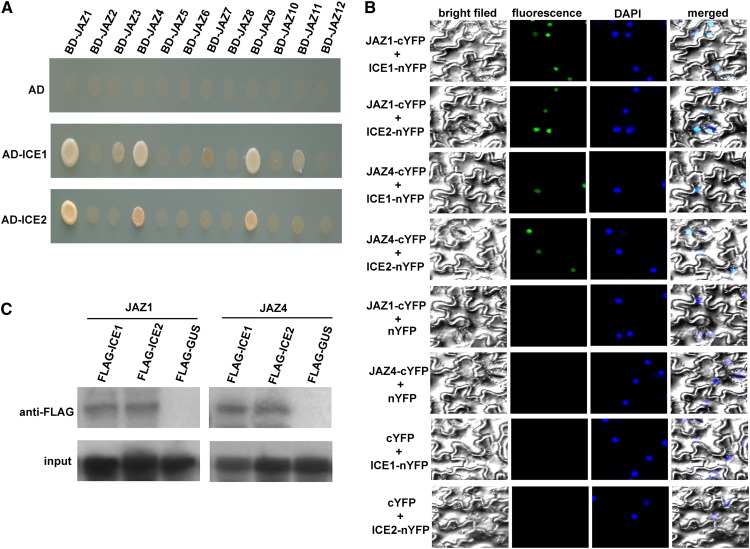
To understand how jasmonate modulates the CBF/DREB1 cascade and freezing tolerance, we used the yeast two-hybrid system to identify potential downstream transcription factors of JAZ repressors. The full-length of JAZ1 was fused to the Gal4 DNA binding domain of the bait vector (BD-JAZ1). After screening, three independent clones encoding ICE1 were identified by prototrophy for His and Ade. To confirm the interaction, the full-length coding sequence (CDS) of ICE1 was cloned and introduced into the prey vector (AD-ICE1). The bait and prey vectors were cotransformed into yeast and the protein–protein interaction was reconstructed (Figure 5A). To confirm whether JAZ1 specifically interacts with ICE1, we also analyzed its interaction with the homolog of ICE1, ICE2. As shown in Figure 5A, JAZ1 also interacts with ICE2 in the yeast two-hybrid system. We further investigated interactions of ICE1 and ICE2 with all 12 Arabidopsis JAZ proteins in the yeast two-hybrid system. Besides JAZ1, ICE1 also strongly interacted with JAZ4 and JAZ9, and slightly interacted with JAZ3 and JAZ11 (Figure 5A). Similarly, ICE2 also interacted with JAZ4 and JAZ9 in yeast (Figure 5A). As a control, we examined the expression levels of the 12 BD-JAZ fusion proteins by immunoblot analysis, finding that all BD-JAZ fusion proteins were expressed in yeast (see Supplemental Figure 3 online).
Figure 5.
Interactions between JAZ Repressors and ICE Transcription Factors.
(A) Yeast two-hybrid assay analysis. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The GAL4 activation domain expressed by pGADT7 (shown as AD) was used as negative controls.
(B) BiFC analysis. Fluorescence was observed in nuclear compartments of N. benthamiana leaf epidermal cells; the fluorescence resulted from complementation of the N-terminal portion of YFP fused to ICE factors (ICE-nYFP) with the C-terminal portion of YFP fused to JAZ proteins (JAZ-cYFP). No signal was observed from negative controls. DAPI, 4′,6-diamidino-2-phenylindole.
(C) CoIP analysis. MYC-fused JAZ proteins were immunoprecipitated using an anti-MYC antibody, and coimmunoprecipitated ICE factors were then detected using an anti-FLAG rabbit antibody. Protein input for MYC-JAZ proteins in immunoprecipitated complexes was also detected and shown.
Interactions of JAZ proteins with ICE1 and ICE2 in planta were further corroborated by bimolecular fluorescence complementation (BiFC) and coimmunoprecipitation (CoIP) assays. JAZ1 and JAZ4 were used as representatives in the BiFC and CoIP assays. For the BiFC assays, JAZ proteins were fused to the C-terminal yellow fluorescent protein (YFP) fragment (JAZ-cYFP) and ICE factors to the N-terminal YFP fragment (ICE-nYFP). When fused JAZ1-cYFP was coexpressed with ICE1-nYFP or ICE2-nYFP in leaves of tobacco (Nicotiana benthamiana), the YFP signal was detected in the nuclear compartment of transformed cells, as revealed by staining with 4′,6-diamidino-2-phenylindole (Figure 5B). Similar results were also observed for interactions of JAZ4 with ICE1 and ICE2 (Figure 5B). No fluorescence was detected in negative control experiments in which either JAZ-cYFP was coexpressed with unfused nYFP or unfused cYFP was coexpressed with ICE-nYFP (Figure 5B). In addition to the BiFC assays, JAZ–ICE interaction was verified by CoIP assays using plant total protein (Figure 5C). Taken together, these results demonstrate that ICE1 and ICE2 interact with JAZ proteins in plant cell nuclei, implying that ICE1 and ICE2 function as direct targets of JAZ proteins.
The C-Terminal Fragment of ICE1 and the Jas Domain of JAZ1 Are Responsible for the Interaction
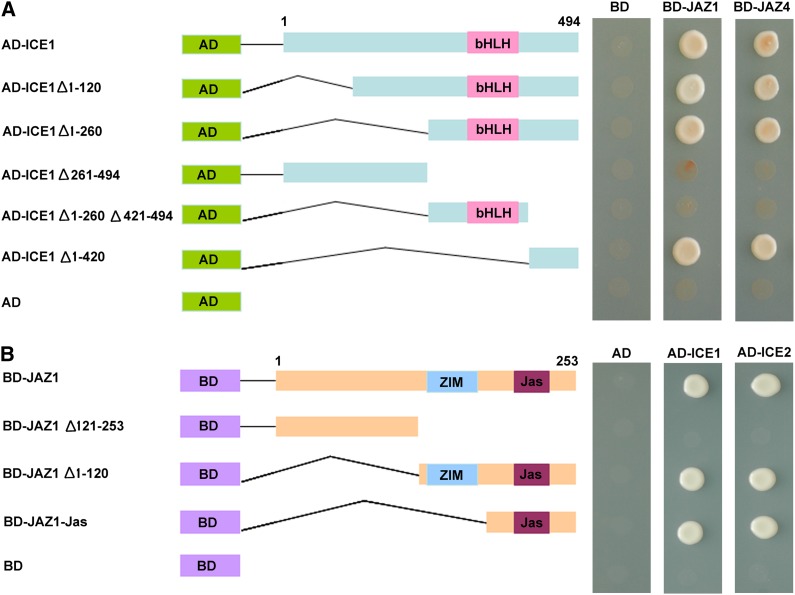
To investigate which region of ICE1 is required for interaction with JAZ proteins, we fused five truncated ICE1 variants to the Gal4 activation domain of the prey vector. The interaction between these derivatives and JAZ proteins was then assayed using the yeast two-hybrid system. As shown in Figure 6A, deletion of the N-terminal 260 residues of ICE1 (AD-ICE1, Δ1–260) did not affect its interactions with JAZ1 and JAZ4. By contrast, deletion of the C-terminal of ICE1 (AD-ICE1, Δ261–494) completely eliminated the interactions between ICE1 and JAZ proteins (Figure 6A; see Supplemental Figure 4A online). This finding demonstrates that the C-terminal domain of ICE1 is essential for the interaction with JAZ proteins. Further mapping revealed that the 74 amino acids at the C-terminal end are specifically responsible for the interaction because a derivative with amino acids 261 to 420 deleted from the C terminus of ICE1 could still interact with JAZ proteins (Figure 6A; see Supplemental Figure 4A online).
Figure 6.
Yeast Two-Hybrid Assay for Identifying ICE1 and JAZ1 Regions Required for Their Interaction.
(A) The C-terminal fragment of ICE1 is involved in the interaction of ICE1 with JAZ1 and JAZ4. Left: Diagram of full-length and truncated ICE1 constructs with specific deletions. Right: Interactions were indicated by the ability of yeast cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The empty pGADT7 prey vector was used as a negative control.
(B) The JAZ1 Jas domain is responsible for interaction of JAZ1 with ICE1 and ICE2. Left: Diagram of full-length and truncated JAZ1 constructs with specific deletions. Right: Interactions were indicated by the ability of yeast cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The empty pGBKT7 bait vector was used as a negative control.
To identify the JAZ1 region responsible for the interaction with ICE1 and ICE2, we performed additional directed yeast two-hybrid analysis. JAZ1 was divided into N-terminal (BD-JAZ1, Δ121–253) and C-terminal (BD-JAZ1, Δ1–120) portions, and the C-terminal part was further truncated into the Jas domain (BD-JAZ1-Jas). The result showed that ICE1 and ICE2 interacted with both the C-terminal portion and the Jas domain of JAZ1 in yeast (Figure 6B; see Supplemental Figure 4B online), indicating that the Jas domain of JAZ1 is required for the interaction with ICE1 and ICE2.
JAZ Proteins Repress the Transcriptional Function of ICE1
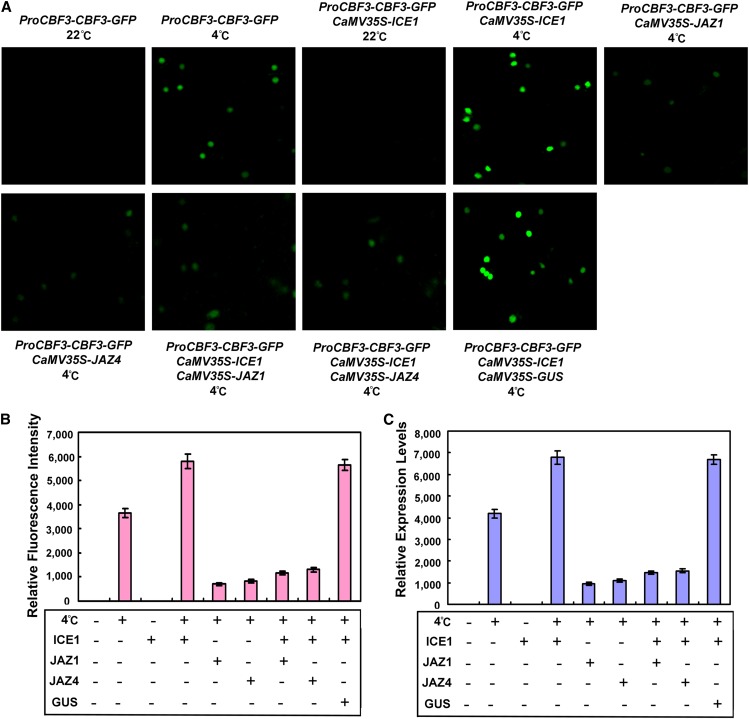
Because JAZ proteins directly interact with ICE1 and ICE2, we hypothesized that the physical interactions might interfere with transcriptional function of these factors. To test this possibility, we generated and analyzed a CBF3 promoter-driven CBF3-green fluorescent protein (GFP) fused protein (ProCBF3-CBF3-GFP) in a transient expression assay. When the reporter construct was transformed into the leaves of N. benthamiana and kept at 22°C, no fluorescence signal was observed (Figures 7A and 7B). However, if the reporter construct was induced at 4°C, fluorescence signals were detected in the nucleus (Figures 7A and 7B). When ProCBF3-CBF3-GFP was coinfiltrated into N. benthamiana leaves along with ICE1 driven by the 35S cauliflower mosaic virus promoter (CaMV35S-ICE1), much stronger fluorescence signals were observed after 4°C treatment (Figures 7A and 7B). However, coinfiltration of ProCBF3-CBF3-GFP with CaMV35S-JAZ1 or CaMV35S-JAZ4 generated dramatically lower fluorescence levels at 4°C (Figures 7A and 7B). In addition, coinfiltration of ProCBF3-CBF3-GFP with CaMV35S-JAZ1 or CaMV35S-JAZ4 and CaMV35S-ICE1 also generated much weaker fluorescence signals in comparison with infiltration of ProCBF3-CBF3-GFP alone or coinfiltration of ProCBF3-CBF3-GFP with CaMV35S-ICE1 after cold treatment (Figures 7A and 7B). As a control, coinfiltration of ProCBF3-CBF3-GFP with CaMV35S-GUS and CaMV35S-ICE1 was performed, but no obvious differences in fluorescence signals were observed compared with coinfiltration of ProCBF3-CBF3-GFP with CaMV35S-ICE (Figures 7A and 7B). Taken together, these results demonstrate that JAZ proteins repress the transcriptional function of ICE1.
Figure 7.
JAZ1 and JAZ4 Repress ICE1 Transcriptional Function.
(A) Transient expression assays showed that JAZ1 and JAZ4 repress transcriptional activation of ICE1. GFP fluorescence was detected 48 h after coinfiltration with the indicated constructs after cold treatment (4°C for 4 h). The experiment was repeated three times with similar results.
(B) Quantitative analysis of GFP fluorescence intensity in the transformed and cold-treated plants. Fifty independent fluorescent spots were assessed for fluorescence intensity. Error bars represent se.
(C) qRT-PCR analysis of the accumulation of CBF3-GFP transcripts. Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with combinations of various constructs in (A). The N. benthamiana ACTIN gene was used as an internal control. Error bars show sd from three independent RNA extractions.
To further verify the effect of JAZ proteins on the transcriptional function of ICE1, we analyzed relative expression of CBF3-GFP in N. benthamiana leaves in response to low temperature. As shown in Figure 7C, we detected high levels of CBF3-GFP transcripts in ProCBF3-CBF3-GFP and CaMV35S-ICE1 coinfiltrated N. benthamiana leaves after cold treatment. By contrast, coexpression of JAZ proteins with ICE1 significantly suppressed the accumulation of CBF3-GFP transcripts (Figure 7C). These results further support the notion that JAZ proteins repress the transcriptional function of ICE1.
Overexpression of JAZ1 or JAZ4 Represses Freezing Tolerance
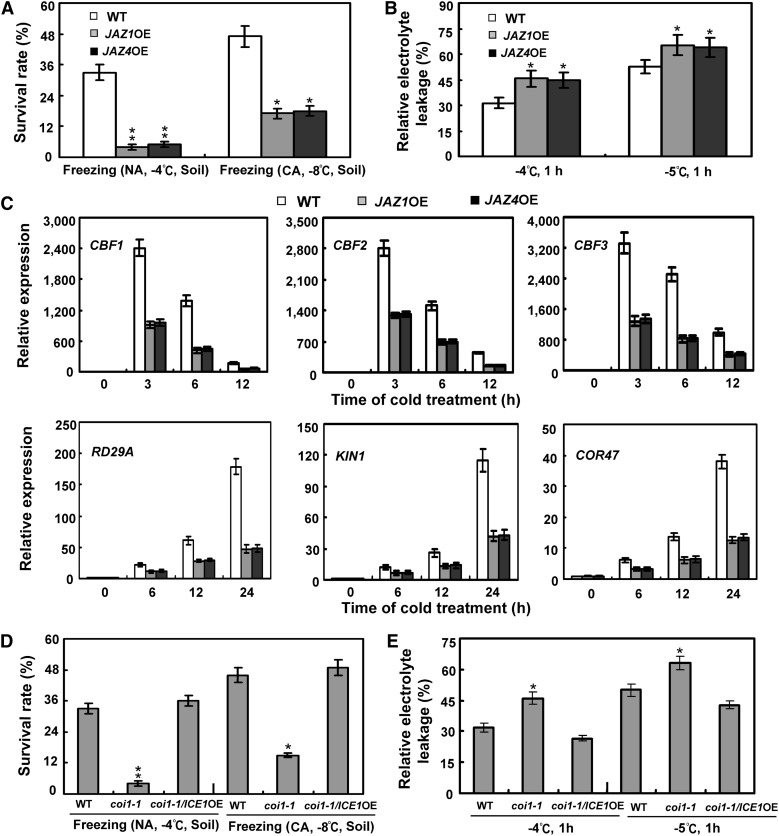
As several JAZ repressors interact with ICE factors and modulate the transcriptional function of ICE1, we queried whether disruption or overexpression of these JAZ proteins affected Arabidopsis freezing stress responses. To test this possibility, we first analyzed the performances of mutant plants in response to freezing stress. The tested jaz4 and jaz9 single and the jaz4 jaz9 double mutants behaved similarly to the wild-type plants after treatments, showing similar survival rates and induced expression levels of cold-responsive genes (see Supplemental Figure 5 online). However, overexpression of JAZ1 or JAZ4 rendered transgenic plants (JAZ1OE or JAZ4OE) sensitive to freezing stress (Figures 8A and 8B). Consistent with this, transcripts of CBF/DREB1 and their downstream targets were reduced in JAZ1OE and JAZ4OE under cold stress (Figure 8C). These results indicated that overexpression of JAZ1 or JAZ4 represses the ICE-ICBF/DREB1 signaling pathway and freezing stress responses in Arabidopsis.
Figure 8.
Phenotypic Characterization of the JAZ1 and JAZ4 Overexpression Plants.
(A) Survival rates of the JAZ1 and JAZ4 overexpression plants (JAZ1OE and JAZ4OE) with or without cold acclimation. Nonacclimated (NA) 18-d-old soil-grown plants were treated at −4°C for 1.5 h, and cold-acclimated seedlings (CA; 2 weeks at 4°C) were exposed to −8°C for 1.5 h, followed by recovery at 22°C for 7 d. Error bars show sd from three replicates. Soil, plants grown in soil. *Differences between overexpression plants and the wild type (WT) are significant (P < 0.05). **Differences between overexpression plants and the wild type are highly significant (P < 0.01).
(B) Ion leakage assays of the JAZ1 and JAZ4 overexpression plants (JAZ1OE and JAZ4OE) with indicated freezing temperatures. Error bars show sd from three replicates. *Differences between overexpression plants and wild type are significant (P < 0.05).
(C) Expression of CBF/DREB1 and their regulons in the JAZ1 and JAZ4 overexpression plants (JAZ1OE and JAZ4OE) under cold stress. Eighteen-day-old soil-grown plants were treated at 4°C for the indicated time periods. Error bars show sd from three independent RNA extractions.
(D) Survival rates of coi1-1/ICE1OE plants with or without cold acclimation. Nonacclimated 18-d-old soil-grown plants were treated at −4°C for 1.5 h, and cold-acclimated seedlings (2 weeks at 4°C) were exposed to −8°C for 1.5 h, followed by recovery at 22°C for 7 d. Error bars show sd from three replicates.
(E) Ion leakage assays of coi1-1/ICE1OE treated with freezing temperatures. Error bars show sd from three replicates. coi1-1/ICE1OE, overexpression of ICE1 in coi1-1 background. Soil, plants grown in soil. *Differences between mutant plants and the wild type are significant (P < 0.05). **Differences between mutant plants and the wild type are highly significant (P < 0.01).
Previous studies have shown that disruption of the jasmonate receptor COI1 protein permits JAZ repressors to accumulate in the nucleus (Chini et al., 2007; Thines et al., 2007), thereby repressing downstream transcription factors. To further corroborate the regulatory effect of JAZ proteins on the ICE–CBF/DREB1 cascade in Arabidopsis, we investigated whether overexpression of ICE1 could restore the freezing-sensitive phenotype of coi1-1 mutants. As shown in Figures 8D and 8E, transgenic expression of ICE1 was able to rescue the phenotype of coi1-1 mutants in response to freezing stress. This observation further supports the idea that JAZ proteins repress the ICE-CBF/DREB1 transcriptional cascade in Arabidopsis.
Expression of Several CBF/DREB1 Pathway-Independent Genes Is Reduced in coi1-1
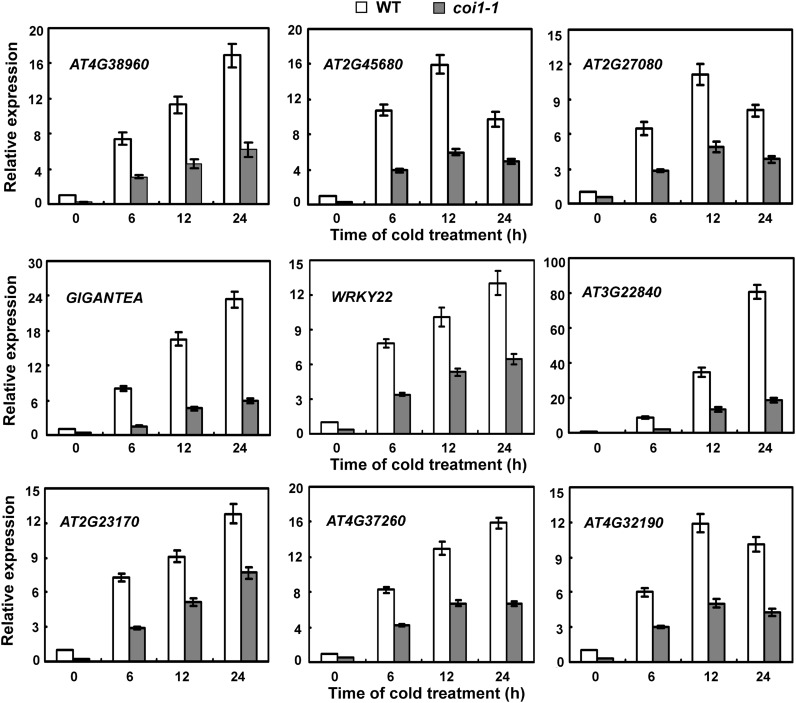
Our expression analysis showed that cold induction of well-known genes acting in the CBF/DREB1 signaling pathway was upregulated by jasmonate (Figure 4). To further investigate whether jasmonate modulates Arabidopsis cold acclimation–induced freezing tolerance exclusively through the CBF/DREB1 pathway, we profiled the transcriptomes of coi1-1 mutant plants after 4°C treatments for 6 or 24 h by microarray analysis. As shown in Supplemental Data Set 1 online, more than 50 cold-responsive genes were downregulated in coi1-1 mutants under cold stress, compared with wild-type plants. As expected, among the jasmonate-regulated genes were members of the CBF/DREB1 signaling pathway. In addition, several cold-responsive genes (such as AT4G38960, AT2G27080, and GIGANTEA) that are not parts of the CBF/DREB1 pathway were also affected by jasmonate signaling under cold stress (see Supplemental Data Set 1 online). To confirm the reliability of the microarray data, we examined the cold-induced expression of several genes by quantitative RT-PCR (qRT-PCR) analysis. Consistent with the microarray results, the induced expression levels of these selected genes were lower in coi1-1 mutants than in the wild type (Figure 9; see Supplemental Figure 6 online). These results indicated that jasmonate may also modulate plant cold acclimation-induced freezing tolerance through the CBF/DREB1-independent pathways.
Figure 9.
Induced Expression of Genes Responsive to Cold but Independent of the CBF/DREB1 Signaling Pathway in Soil-Grown coi1-1 under Cold Stress.
Eighteen-day-old soil-grown wild-type (WT) and coi1-1 mutant plants were treated at 4°C for the indicated time periods. Error bars show sd from three independent RNA extractions used for qRT-PCR.
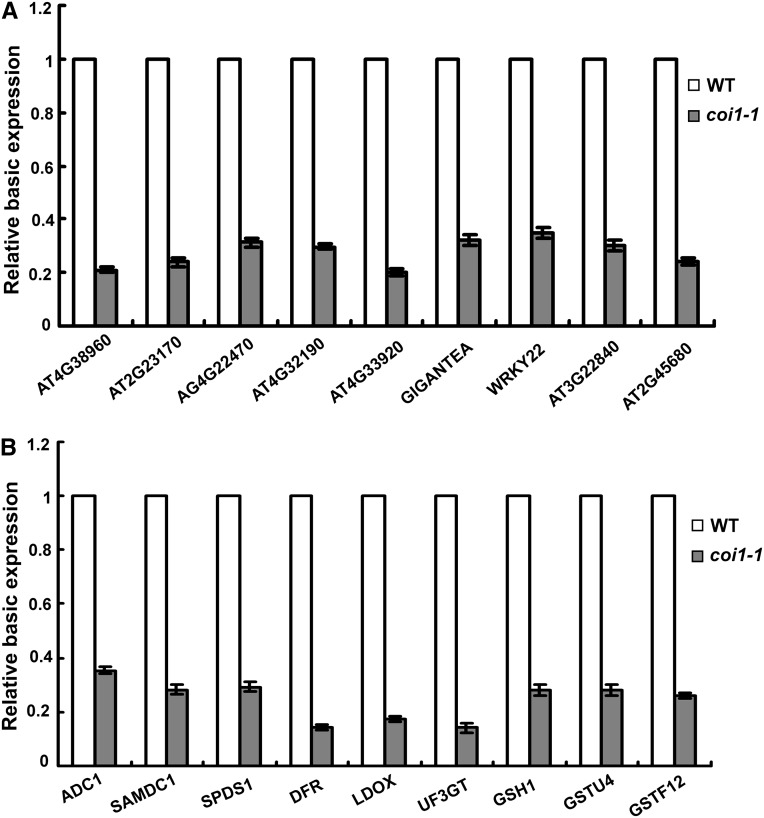
In addition to the cold acclimation–induced freezing tolerance, our results revealed that jasmonate also positively regulated Arabidopsis constitutive freezing tolerance. Under nonacclimated conditions, the jasmonate biosynthesis- and signaling-related mutant plants exhibited reduced freezing tolerance compared with wild-type plants (Figures 2), indicating that the constitutive levels of jasmonate are required for normal levels of Arabidopsis constitutive freezing tolerance. However, our expression analysis showed that the basic expression levels of CBF/DREB1 and their target genes were not affected by jasmonate signaling (Figure 4). These results indicated that jasmonate may modulate Arabidopsis constitutive freezing tolerance through CBF/DREB1-independent pathways. To understand how jasmonate modulates Arabidopsis constitutive freezing tolerance, we compared the transcriptomes of wild-type and the coi1-1 mutant plants grown under normal growth conditions. By comparing the microarray results, we identified several cold-responsive but CBF/DREB1 pathway-independent genes (such as GIGANTEA, AT4G38960, and AT3G22840) that showed lower expression levels in coi1-1 mutant plants (Figure 10A; see Supplemental Data Set 1 and Supplemental Figure 7A online). Moreover, a number of genes encoding enzymes for synthesis of secondary metabolites (such as polyamine, glutathione, and anthocyanins) were also affected by jasmonate signaling (Figure 10B; see Supplemental Data Set 1 and Supplemental Figure 7B online). These results suggested that jasmonate may regulate Arabidopsis constitutive freezing tolerance through modulating the base expression of some CBF/DREB1 pathway-independent cold-responsive genes and the synthesis of cold stress–related secondary metabolites.
Figure 10.
Base Expression of Genes Responsive to Cold but Independent of the CBF/DREB1 Signaling Pathway and Genes for the Synthesis of Secondary Metabolites in Soil-Grown coi1-1.
(A) Base expression of genes responsive to cold but independent of the CBF/DREB1 signaling pathway in 18-d-old soil-grown coi1-1. WT, the wild type.
(B) Base expression of genes encoding enzymes for the synthesis of polyamine, anthocyanins, and glutathione in 18-d-old soil-grown coi1-1. Error bars show sd from three independent RNA extractions used for qRT-PCR.
DISCUSSION
The plant hormone jasmonate is ubiquitous in the plant kingdom and is required for regulation of multiple physiological processes. In this study, we investigated jasmonate’s role in freezing stress in Arabidopsis by examining the effect of exogenous application of jasmonate on plant freezing tolerance, the effect of impaired jasmonate biosynthesis and signaling on freezing stress, and the changes in endogenous jasmonate levels in response to cold. Our results demonstrate that jasmonate has a positive role in both constitutive and cold acclimation–induced freezing tolerance of Arabidopsis. Treating wild-type plants with exogenous MeJA significantly enhanced tolerance to freezing stress (Figure 1). By contrast, blocking endogenous jasmonate biosynthesis and signaling conferred decreased freezing tolerance under both nonacclimated and cold-acclimated conditions (Figure 2). Furthermore, endogenous jasmonate production was triggered by cold treatment (Figure 3). We therefore conclude that jasmonate positively regulates Arabidopsis constitutive and cold acclimation–induced freezing tolerance.
Previous studies indicated that jasmonate-induced chilling tolerance of several fruits may be associated with the increase of cryo-protective compounds (González-Aguilar et al., 2000, 2004; Cao et al., 2009; Mohammad et al., 2011; Zhao et al., 2013). However, the exact molecular mechanisms of jasmonate-mediated cold stress responses remained unclear. Activation of jasmonate-regulated responses requires a profound transcriptional reprogramming of cellular genetic programs involved in the complex interplay between negative and positive regulators (e.g., JAZ repressors and downstream transcription factors). JAZ proteins repress jasmonate-mediated responses through interaction and attenuation of their downstream transcription factors. The bHLH transcription factors MYC2, MYC3, and MYC4, essential components of the WD-repeat/bHLH/MYB transcriptional complexes such as GL3, EGL3, and GL1, MYB21 and MYB24, and ETHYLENE INSENSITIVE3 (EIN3), were recently identified as direct targets of JAZ proteins mediating various jasmonate-regulated processes (Chini et al., 2007; Fernández-Calvo et al., 2011; Qi et al., 2011; Song et al., 2011; Zhu et al., 2011). In this study, we found that the ICE1 and ICE2 transcription factors also function as targets of JAZ proteins (Figures 5 and 6) and demonstrated that JAZ proteins repress the transcriptional function of ICE1 (Figure 7). Consistent with these findings, overexpression of JAZ1 or JAZ4 repressed the cold-induced expression of CBF/DREB1 and their regulons, thereby rendering transgenic plants sensitive to freezing (Figure 8). Moreover, overexpression of ICE1 was able to rescue freezing sensitive phenotype of coi1-1 mutant plants (Figure 8).
The transcription factors ICE1 and ICE2 can activate expression of downstream CBF3/DREB1a and CBF1/DREB1b genes, respectively, in response to cold stress (Chinnusamy et al., 2003; Fursova et al., 2009). Our expression analysis showed that CBF3/DREB1a, CBF1/DREB1b, and their targets were induced by MeJA treatment under cold stress (Figure 4), consistent with the enhanced freezing tolerance of MeJA-treated wild-type plants and the repressive effect of JAZ proteins on ICE1 and ICE2 transcription factors (Figures 1 and 7). CBF2/DREB1c expression in response to cold stress was also upregulated by MeJA treatment (Figure 4). This was surprising because previous studies have suggested that CBF2/DREB1c expression is not influenced by ICE1 and ICE2 (Chinnusamy et al., 2003; Fursova et al., 2009); furthermore, cold-induced expression of CBF2/DREB1c was not found to be associated with CBF3/DREB1a and CBF1/DREB1b in a previous study (Novillo et al., 2007). Further research is thus required to investigate how jasmonate signaling affects CBF2/DREB1c expression under cold stress. Recently, Zhao et al. (2013) showed that two MYC2 proteins of banana (Musa acuminata) interact with ICE1. This suggests that banana MYC2 proteins may participate in regulating the expression of cold-responsive genes. However, in our study, the expression of three CBF/DREB1 genes was not affected in the myc2-2 mutant compared with the wild type under cold stress (see Supplemental Figure 8 online). This observation suggests that MYC2 may be not involved in cold stress responses in Arabidopsis.
Recently, Shi et al. (2012) demonstrated that ethylene signaling negatively regulates freezing tolerance in Arabidopsis. Further genetic and biochemical analysis in their study revealed that EIN3 and EIL1 directly bind CBF/DREB1 promoters and negatively regulate expression of these cold-induced genes. EIN3 and EIL1 were also identified in another study as direct targets of JAZ repressors mediating jasmonate-regulated responses (Zhu et al., 2011). Consequently, jasmonate signaling appears to negatively modulate CBF/DREB1 gene expression via EIN3 and EIL1. We speculate that the dual regulations of the CBF/DREB1 signaling pathway by jasmonate may be a balancing mechanism between establishment of the appropriate stress tolerance and minimization of detrimental effects on plant growth and development. Nevertheless, the final outcome of jasmonate regulation is enhanced plant adaptation to cold stress (Figures 1 and 2). Interestingly, a similar dual regulation was observed previously: EIN3 and MYC2 act antagonistically to mediate jasmonate-regulated resistance against the necrotrophic fungal pathogen Botrytis cinerea (Berrocal-Lobo et al., 2002; Lorenzo et al., 2004). Further characterization of these dual regulations may shed new light on the molecular basis of tight regulation and fine-tuning of jasmonate signaling pathways.
In addition to the CBF/DREB1 signaling pathway, our results revealed that jasmonate also regulates expression of several genes independent of the CBF/DREB1 signaling pathway under cold stress (see Supplemental Data Set 1 online; Figure 9). These findings indicate that jasmonate also modulates plant cold acclimation through the CBF/DREB1-independent pathways. Fowler and Thomashow (2002) showed that only 12% of the cold-responsive genes were regulated by CBF/DREB1 transcription factors, suggesting that the CBF/DREB1-independent pathways may also play important roles in mediating plant cold stress responses. Interestingly, endogenous jasmonate has a peak of induction by low temperature at 12 h (Figure 3), which appears to be inconsistent with the results that the cold-induced expression of CBF/DREB1 genes peaks at 3 h and decreases at 12 h of cold treatment (Figure 4). It is possible that the high levels of endogenous jasmonate at 12 to 24 h may be involved in the induction of CBF/DREB1-independent genes. Consistent with this possibility, most of the selected genes independent of the CBF/DREB1 signaling pathway were strongly induced at 12 to 24 h by cold treatment (Figure 9). Further investigation of these jasmonate-affected CBF/DREB1-independent genes may enhance our understanding of the regulatory mechanisms of plant cold acclimation processes. Besides the cold acclimation-induced freezing tolerance, jasmonate is also required for Arabidopsis constitutive freezing tolerance (Figure 2). To understand how jasmonate modulates Arabidopsis constitutive freezing tolerance, we compared the transcriptomes of wild-type and coi1-1 mutant plants grown under control conditions and identified several cold-responsive but CBF/DREB1 pathway-independent genes (see Supplemental Data Set 1 online; Figure 10A). In addition, a number of genes required for synthesis of secondary metabolites (such as polyamine, glutathione, and anthocyanins) were also regulated by jasmonate signaling (see Supplemental Data Set 1 online; Figure 10B). Consistent with this, accumulating evidence has demonstrated that these secondary metabolites are involved in plant cold stress responses (Christie et al., 1994; Hummel et al., 2004; Lei et al., 2004; Cuevas et al., 2008; Kovács et al., 2010; Alcázar et al., 2011; Bai et al., 2012; Xie et al., 2012).
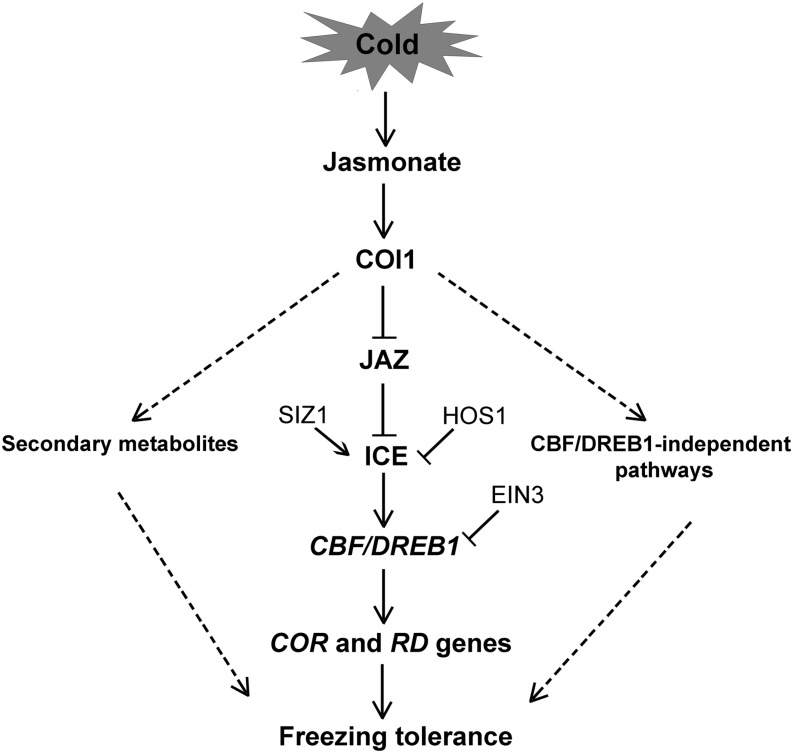
To elucidate the molecular mechanism of jasmonate-regulated cold stress responses in Arabidopsis, we propose the following simplified model (illustrated in Figure 11). Under normal growth conditions, JAZ repressors physically interact with ICE1 and ICE2 transcription factors (Figures 5 and 6), attenuate their transcriptional functions (Figure 7), and thereby repress expression of downstream cold-responsive genes. However, under cold conditions, production of endogenous jasmonate, perceived by the receptor COI1 (Yan et al., 2009), is triggered instead (Figure 3). COI1 then recruits JAZ proteins for degradation (Xie et al., 1998; Thines et al., 2007), and ICE1 and ICE2 are released to activate the expression of CBF/DREB1 genes and their targets. Based on the findings that the production of endogenous jasmonate is rapidly triggered under cold stress and the induction of CBF/DREB1 genes peaks after 3 h of cold exposure (Figures 3 and 4), we speculate that jasmonate may regulate CBF/DREB1 expression by activating ICE transcription factors at the early stage of cold stress. Altogether, jasmonate acts as a crucial upstream signal of the ICE-CBF/DREB1 signaling pathway to positively modulate Arabidopsis freezing tolerance under cold-acclimated conditions. In addition, jasmonate also regulates Arabidopsis constitutive and cold acclimation–induced freezing tolerance through mediating the synthesis of secondary metabolites and some CBF/DREB1-independent pathways (Figures 9 and 10; see Supplemental Data Set 1 online).
Figure 11.
Model for Jasmonate-Regulated Cold Signaling Pathway in Arabidopsis.
Under normal growth conditions, JAZ repressors interact with ICE transcription factors. The physical interactions attenuate transcriptional functions of these downstream transcription factors. Under cold conditions, production of endogenous jasmonate is induced. The COI1 receptor perceives jasmonate and targets JAZ proteins for degradation. Upon degradation of JAZ proteins, the ICE factors are subsequently released to activate CBF/DREB1 gene expression and their downstream cold-responsive genes. Jasmonate also regulates Arabidopsis constitutive and cold acclimation–induced freezing tolerance by influencing the synthesis of secondary metabolites and via some CBF/DREB1-independent pathways. The final outcome of jasmonate regulation is the enhancement of plant tolerance to freezing stress.
METHODS
Materials and Plant Growth Conditions
The plant hormone MeJA was purchased from Sigma-Aldrich. Taq DNA polymerases were purchased from Takara Biotechnology, and other common chemicals were obtained from Shanghai Sangon. Arabidopsis thaliana plants were grown in an artificial growth chamber at 22°C under a 10-h-light/14-h-dark photoperiod. Wild-type and mutant plants used in this study were in the Columbia-0 genetic background. The mutants coi1-1 (Xie et al., 1998) and coi1-2 (Xu et al., 2002) were described as previously. Other mutants used in this study are listed as follows: lox2 (CS3748), aos (CS6149), jar1 (CS8072), and myc2-2 (SALK_083483). To generate the overexpression transgenic plants, the full-length cDNA of JAZ1, JAZ4, or ICE1 were cloned into the pOCA30 vector in the sense orientation behind the CaMV 35S promoter (Hu et al., 2013). All primers used for clones are listed in Supplemental Table 1 online.
Freezing Tolerance Assays
The freezing tolerance assays were performed as described (Chinnusamy et al., 2003), with some modifications. Briefly, 12-d-old plants grown at 22°C on Murashige and Skoog (MS) agar medium with or without cold acclimation (2 weeks at 4°C) were placed in a freezing chamber set to −1°C and programmed to cool at −1°C per hour. Petri dishes of plants were removed after exposure to the desired temperatures. After the freezing treatment, plants were incubated at 4°C in the dark for 10 h and then transferred to light at 22°C. The survival rates of the seedlings were scored visually after 4 d. For soil-grown plants, 18-d-old plants with or without cold acclimation (2 weeks at 4°C) were placed in a freezing chamber set to 0°C and programmed to cool at −1°C per 1.5 h. Plants were removed after exposure to the desired temperatures. After the freezing treatment, plants were incubated at 4°C in the dark for 10 h and then transferred to light at 22°C. The survival rates of the seedlings were scored visually after 7 d. For jasmonate treatment, 3 or 5 μM MeJA was added to the MS agar medium. Seeds of the wild type were germinated on this MeJA-containing medium, and seedlings were grown at 22°C for 12 d. Soil-grown plants were sprayed with water or a 30 μM MeJA solution diluted from the stock. The water- and MeJA-treated plants were maintained in a small chamber with a transparent cover for 3 d before freezing treatment (under nonacclimated conditions). Under cold-acclimated conditions, plants were treated with MeJA at 4°C for 3 d and then were removed from the chamber and kept at 4°C for another 11 d.
Relative Electrolyte Leakage
To measure relative electrolyte leakage, 12-d-old Arabidopsis seedlings were treated at freezing temperatures. After treatment, entire seedlings were harvested and relative electrolyte leakage was measured according to Jiang et al. (2007).
Determination of Jasmonate Contents
To measure jasmonate contents, 18-d-old soil-grown wild-type plants were treated at 4°C or kept at 22°C. After treatment, entire seedlings were harvested and jasmonate was analyzed by ELISA using monoclonal antibodies of jasmonate according to Deng et al. (2008).
RNA Extraction and qRT-PCR
Total RNA was extracted from Arabidopsis seedlings using the Trizol reagent (Invitrogen). qRT-PCR was performed as described by Hu et al. (2012). Briefly, first-strand cDNA was synthesized from 1.5 μg DNase-treated RNA in a 20-μL reaction volumes using M-MuLV reverse transcriptase (Fermentas) with oligo(dT)18 primer. qRT-PCR was performed using 2× SYBR Green I master mix on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer’s instructions. At least three biological replicates for each sample were used for qRT-PCR analysis, and at least two technical replicates were analyzed for each biological replicate. The ACTIN2 gene was used as an internal control of gene expression. Gene-specific primers used to detect transcripts are listed in Supplemental Table 2 online.
Yeast Two-Hybrid Screening and Confirmation
The full-length JAZ1 CDS was cloned into the bait vector pGBKT7 and then transformed into the yeast strain Y2HGold (Clontech). The cDNA library was obtained from Clontech (catalog number 630487). Two-hybrid screening was performed via the mating protocol described in Clontech’s Matchmaker Gold Yeast Two-Hybrid user manual. To confirm protein–protein interactions, the full-length ICE1, ICE2, or truncated ICE1 CDS were cloned into the prey vector pGADT7. Primers used for amplifying these truncated or mutated fragments are listed in Supplemental Table 1 online.
BiFC Assays
cDNA sequences of the N-terminal, 173–amino acid, enhanced YFP (nYFP) and C-terminal, 64–amino acid (cYFP) fragments were PCR amplified and cloned into pFGC5941 to generate pFGC-nYFP and pFGC-cYFP, respectively (Kim et al., 2008). The full-length JAZ1 and JAZ4 CDS was inserted into pFGC-cYFP to generate C-terminal in-frame fusions with cYFP, while ICE1 and ICE2 CDS were introduced into pFGC-nYFP to form N-terminal in-frame fusions with nYFP. The resulting plasmids were introduced into Agrobacterium tumefaciens (strain GV3101), and infiltration of Nicotiana benthamiana was performed as described previously (Hu et al., 2013). Infected tissues were analyzed 48 h after infiltration. YFP and 4′,6-diamidino-2-phenylindole fluorescence were observed under a confocal laser scanning microscope (Olympus).
CoIP Assays
The full-length CDS of JAZ1, JAZ4, ICE1, and ICE2 were individually cloned into tagging plasmids behind the MYC or FLAG tag sequence in the sense orientation behind the CaMV 35S promoter. The constructs were transformed into Agrobacterium GV3101. MYC-fused JAZ1 or JAZ4 and FLAG-fused ICE1 or ICE2 were then transiently coexpressed in N. benthamiana. Infected leaves were sectioned 48 h after infiltration. CoIP assays were performed using leaf protein extracts as described by Shang et al. (2010). Briefly, MYC-fused JAZ1 and JAZ4 were immunoprecipitated using an anti-MYC antibody and the coimmunoprecipitated protein was then detected using an anti-FLAG rabbit antibody (Sigma-Aldrich).
Transient Expression Assays
The transient expression assays were performed in N. benthamiana leaves. The full-length CDS of CBF3 was fused with GFP reporter gene behind the native promoter of CBF3. The full-length CDSs of JAZ1, JAZ4, and ICE1 were driven by the CaMV 35S promoter. These constructs were then introduced into the Agrobacterium strain GV3101. The infiltration of N. benthamiana was performed as described previously (Kim et al., 2008). Plants were kept at 22°C for 48 h before induced by cold stress (4°C for 4 h). Infected leaves were observed under a confocal laser scanning microscope (Olympus). All experiments were repeated with five independent biological replicates with similar results.
Microarray Analysis
Eighteen-day-old soil-grown wild type and coi1-1 mutant plants were used for microarray analysis. Wild-type and coi1-1 mutant plants grown under normal growth conditions were treated at 4°C for 6 or 24 h. Total RNA was isolated from three replicates of control or treated plants using the Trizol reagent (Invitrogen). RNA quantity was assessed using the NanoDrop ND-1000 and the integrity was assessed using standard denaturing agarose gel electrophoresis. The microarray analysis was performed by the Shanghai Kangchen Biological Technology Company. For microarray analysis, the Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols. Briefly, total RNA from each sample was amplified and transcribed into fluorescent complementary RNA using Agilent’s Quick Amp Labeling protocol (version 5.7; Agilent Technologies). The labeled cRNAs were hybridized onto the Whole Genome Oligo Microarray (4x44K; Agilent Technologies). After the slides were washed, the arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). Differentially expressed genes were identified through fold change filtering.
Accession Numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: LOX1, AT1G55020; LOX2, AT3G45140; LOX3, AT1G17420; LOX4, AT1G72520; AOS, AT5G42650; AOC1, AT3G25760; AOC2, AT3G25770; AOC3, AT3G25780; AOC4, AT1G13280; JAR1, AT2G46370; COI1, AT2G39940; JAZ1, AT1G19180; JAZ2, AT1G74950; JAZ3, AT3G17860; JAZ4, AT1G48500; JAZ5, AT1G17380; JAZ6, AT1G72450; JAZ7, AT2G34600; JAZ8, AT1G30135; JAZ9, AT1G70700; JAZ10, AT5G13220; JAZ11, AT3G43440; JAZ12, AT5G20900; ICE1, AT3G26744; ICE2, AT1G12860; CBF1, AT4G25490; CBF2, AT4G25470; CBF3, AT4G25480; RD29A, AT5G52310; RD29B, AT5G52300; KIN1, At5g15960; COR47, At1g20440; COR414, At1g29395; COR15b, At2g42530; ADC1, AT2G16500; SAMDC, AT3G02470; SPDS1, AT1G23820; DFR, AT5G42800; LDOX, AT4G22880; UF3GT, AT5G54060; GSH1, AT4G23100; GSTu4, AT2G29460; GSTF12, AT5G17220; GIGANTEA, AT1G22770; WRKY22, AT4G01250; and ACTIN2, AT3G18780.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cold-Induced Expression of CBF/DREB1 and Their Regulons in Response to Jasmonate in MS Medium–Grown Plants.
Supplemental Figure 2. Expression of Cold-Responsive Genes Acting in the ABA or Cytokinin Pathway and Those Associated with the Synthesis of Soluble Sugars.
Supplemental Figure 3. Immunoblot Analysis of JAZ Proteins Expressed in Yeast.
Supplemental Figure 4. Immunoblot Analysis of the Truncated ICE1 and JAZ1 Proteins Expressed in Yeast.
Supplemental Figure 5. Freezing Tolerance of jaz4 and jaz9 Mutants.
Supplemental Figure 6. Induced Expression of Genes Responsive to Cold but Independent of the CBF/DREB1 Signaling Pathway in MS Medium–Grown coi1-1 under Cold Stress.
Supplemental Figure 7. Base Expression of Genes Responsive to Cold but Independent of the CBF/DREB1 Signaling Pathway and Genes for the Synthesis of Secondary Metabolites in MS Medium–Grown coi1-1.
Supplemental Figure 8. Expression of CBF/DREB1 Genes in myc2-2 Mutant Plants under Cold Stress.
Supplemental Table 1. Primers Used for Generating Clones.
Supplemental Table 2. Primers Used for qRT-PCR.
Supplemental Data Set 1. MS Excel File with Probe Names, Fold-Change Information, AGI Numbers, and Annotation of Cold-Regulated Genes Affected by Jasmonate Signaling.
Acknowledgments
We thank Zhixiang Chen (Purdue University), Daoxin Xie (Tsinghua University, China), Chengwei Yang (South China Normal University, China), Xiaoya Chen (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences), and Xiangyang Hu (Kunming Institute of Botany, Chinese Academy of Sciences) for sharing research materials. We also thank Weiqi Li and Faqing Tao (Kunming Institute of Botany, Chinese Academy of Sciences) for their assistance in measuring jasmonate contents and the anonymous reviewers for their arduous work and instructive advice. This work was supported by the Natural Science Foundation of China (U1202264 and 31171183) and the Science Foundation of the Chinese Academy of Sciences (KSCX3-EW-N-07 and the CAS 135 program XTBG-F04).
AUTHOR CONTRIBUTIONS
Y.H. designed and performed experiments, analyzed data, and wrote the article. L.J. and F.W. performed experiments and helped to analyze data. D.Y. designed experiments and helped to interpret data and edit the article. All authors read and approved the final article.
Glossary
- bHLH
basic helix-loop-helix
- ABA
abscisic acid
- MeJA
methyl jasmonate
- CDS
coding sequence
- BiFC
bimolecular fluorescence complementation
- CoIP
coimmunoprecipitation
- qRT-PCR
quantitative RT-PCR
- MS
Murashige and Skoog
References
- Alcázar R., Cuevas J.C., Planas J., Zarza X., Bortolotti C., Carrasco P., Salinas J., Tiburcio A.F., Altabella T. (2011). Integration of polyamines in the cold acclimation response. Plant Sci. 180: 31–38 [DOI] [PubMed] [Google Scholar]
- Bai X.G., Chen J.H., Kong X.X., Todd C.D., Yang Y.P., Hu X.Y., Li D.Z. (2012). Carbon monoxide enhances the chilling tolerance of recalcitrant Baccaurea ramiflora seeds via nitric oxide-mediated glutathione homeostasis. Free Radic. Biol. Med. 53: 710–720 [DOI] [PubMed] [Google Scholar]
- Bell E., Creelman R.A., Mullet J.E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Cao S.F., Zheng Y.H., Wang K.T., Jin P., Rui H.J. (2009). Methyl jasmonate reduces chilling injury and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chem. 115: 1458–1463 [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J.K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12: 444–451 [DOI] [PubMed] [Google Scholar]
- Christie P.J., Alfenito M.R., Walbot V. (1994). Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194: 541–549 [Google Scholar]
- Cuevas J.C., López-Cobollo R., Alcázar R., Zarza X., Koncz C., Altabella T., Salinas J., Tiburcio A.F., Ferrando A. (2008). Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 148: 1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng A., Tan W., He S., Liu W., Nan T., Li Z., Wang B., Li Q.X. (2008). Monoclonal antibody-based enzyme linked immunosorbent assay for the analysis of jasmonates in plants. J. Integr. Plant Biol. 50: 1046–1052 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E. (2001). Surface-to-air signals. Nature 411: 854–856 [DOI] [PubMed] [Google Scholar]
- Farmer E.E., Alméras E., Krishnamurthy V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Thomashow M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi V.R., Grimes H.D. (1991). Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc. Natl. Acad. Sci. USA 88: 6745–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova O.V., Pogorelko G.V., Tarasov V.A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429: 98–103 [DOI] [PubMed] [Google Scholar]
- González-Aguilar G.A., Fortiz J., Cruz R., Baez R., Wang C.Y. (2000). Methyl jasmonate reduces chilling injury and maintains postharvest quality of mango fruit. J. Agric. Food Chem. 48: 515–519 [DOI] [PubMed] [Google Scholar]
- González-Aguilar G.A., Tiznado-Hernández M.E., Zavaleta-Gatica R., Martínez-Téllez M.A. (2004). Methyl jasmonate treatments reduce chilling injury and activate the defense response of guava fruits. Biochem. Biophys. Res. Commun. 313: 694–701 [DOI] [PubMed] [Google Scholar]
- Howe G.A., Lightner J., Browse J., Ryan C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen L., Wang H., Zhang L., Wang F., Yu D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Hu Y., Dong Q., Yu D. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185-186: 288–297 [DOI] [PubMed] [Google Scholar]
- Hummel I., El-Amrani A., Gouesbet G., Hennion F., Couée I. (2004). Involvement of polyamines in the interacting effects of low temperature and mineral supply on Pringlea antiscorbutica (Kerguelen cabbage) seedlings. J. Exp. Bot. 55: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Jeon J., Kim N.Y., Kim S., Kang N.Y., Novák O., Ku S.J., Cho C., Lee D.J., Lee E.J., Strnad M., Kim J. (2010). A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 285: 23371–23386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Yang B., Harris N.S., Deyholos M.K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58: 3591–3607 [DOI] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács Z., Simon-Sarkadi L., Szucs A., Kocsy G. (2010). Differential effects of cold, osmotic stress and abscisic acid on polyamine accumulation in wheat. Amino Acids 38: 623–631 [DOI] [PubMed] [Google Scholar]
- Lång V., Mäntylä E., Welin B., Sundberg B., Palva E.T. (1994). Alterations in water status, endogenous abscisc acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 104: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X.Y., Zhu R.Y., Zhang G.Y., Dai Y.R. (2004). Attenuation of cold-induced apoptosis by exogenous melatonin in carrot suspension cells: the possible involvement of polyamines. J. Pineal Res. 36: 126–131 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J.M. (1973). Chilling injury in plants. Annu. Rev. Plant Physiol. 24: 445–466 [Google Scholar]
- Mäntylä E., Lång V., Palva E.T. (1995). Role of abscisic acid in drought-induced freezing tolerance, cold acclimation and accumulation of LTI78 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 107: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Creelman R.A., Bell E., Mullet J.E., Browse J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S., Mesbah B., Siamak K., Domingo M.R., Fabián G., María S., Daniel V. (2011). Vapour treatments with methyl salicylate or methyl jasmonate alleviated chilling injury and enhanced antioxidant potential during postharvest storage of pomegranates. Food Chem. 124: 964–970 [Google Scholar]
- Novillo F., Medina J., Salinas J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 104: 21002–21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P., Farmer E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Wang J., Chua L., Jiang D., Peng W., Xie D. (2011). The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60: 3849–3860 [DOI] [PubMed] [Google Scholar]
- Shang Y., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., Yang S. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.C., Funk C.D., Brash A.R. (1993). Molecular cloning of an allene oxide synthase: A cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc. Natl. Acad. Sci. USA 90: 8519–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W.P., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Ueda J., Kato J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66: 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P., Shockey J., Lévesque C.A., Cook R.J., Browse J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7209–7214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xie X.B., Li S., Zhang R.F., Zhao J., Chen Y.C., Zhao Q., Yao Y.X., You C.X., Zhang X.S., Hao Y.J. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.L., Wang J.N., Shan W., Fan J.G., Kuang J.F., Wu K.Q., Li X.P., Chen W.X., He F.Y., Chen J.Y., Lu W.J. (2013). Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 36: 30–51 [DOI] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]