The molecular mechanisms by which sHSPs are involved in cell protection remain unknown. This study reports that chloroplast HSP21 is essential for chloroplast development by maintaining plastid-encoded RNA polymerase function under heat stress.
Abstract
Compared with small heat shock proteins (sHSPs) in other organisms, those in plants are the most abundant and diverse. However, the molecular mechanisms by which sHSPs are involved in cell protection remain unknown. Here, we characterized the role of HSP21, a plastid nucleoid-localized sHSP, in chloroplast development under heat stress. We show that an Arabidopsis thaliana knockout mutant of HSP21 had an ivory phenotype under heat stress. Quantitative real-time RT-PCR, run-on transcription, RNA gel blot, and polysome association analyses demonstrated that HSP21 is involved in plastid-encoded RNA polymerase (PEP)–dependent transcription. We found that the plastid nucleoid protein pTAC5 was an HSP21 target. pTAC5 has a C4-type zinc finger similar to that of Escherichia coli DnaJ and zinc-dependent disulfide isomerase activity. Reduction of pTAC5 expression by RNA interference led to similar phenotypic effects as observed in hsp21. HSP21 and pTAC5 formed a complex that was associated mainly with the PEP complex. HSP21 and pTAC5 were associated with the PEP complex not only during transcription initiation, but also during elongation and termination. Our results suggest that HSP21 and pTAC5 are required for chloroplast development under heat stress by maintaining PEP function.
INTRODUCTION
The small heat shock proteins (sHSPs) and the related α-crystallins are virtually ubiquitous proteins that are strongly induced not only by heat stress but also by a variety of other stresses in prokaryotic and eukaryotic cells (Sun et al., 2002; Basha et al., 2012). The sHSPs are characterized by a core α-crystallin domain of ∼100 amino acids, which is flanked by an N-terminal arm of variable length and divergent sequence and a short C-terminal extension (Haslbeck et al., 2005). Although sHSP monomers are relatively small, ranging in size from ∼15 to 42 kD, the majority of these proteins exist as oligomers of between 12 and >48 subunits in their native state (Lambert et al., 2011; Basha et al., 2012). Moreover, sHSPs vary at the secondary, tertiary, and quaternary levels of protein organization, with dynamic exchange of subunits between sHSP oligomers (Stengel et al., 2010; Baldwin et al., 2011). In addition, sHSPs show extensive sequence variation and evolutionary divergence unlike other families of HSPs, such as the HSP90 and HSP70 chaperone families (Basha et al., 2012). Although the molecular mechanisms by which sHSPs and α-crystallins are involved in cell protection in many organisms remain largely unknown, many studies have demonstrated that both mammalian and plant sHSPs act as ATP-independent molecular chaperones by binding proteins that are unfolding or denaturing and thereby preventing their aggregation and facilitating subsequent substrate refolding by ATP-dependent chaperone systems (Lee et al., 1997; Haslbeck et al., 2005; Sun and MacRae, 2005; McHaourab et al., 2009). Due to their molecular chaperone characteristics, sHSPs are considered important components of the protein quality control network (Basha et al., 2012).
Plant sHSPs constitute an abundant and diverse group, in contrast with those from mammals and other organisms. For example, Arabidopsis thaliana has 19 and rice (Oryza sativa) has 23 sHSPs compared with 10 in humans, four in Drosophila melanogaster, and one or two in bacteria (Haslbeck et al., 2005). Plants have a total of 11 sHSP subfamilies, five of which include proteins targeted to the cytosol, whereas the others localize to the nucleus, chloroplasts, mitochondria, endoplasmic reticulum, and peroxisomes (Waters et al., 2008; Siddique et al., 2008). Organelle-targeted sHSPs are unique to plants, with the exception of a mitochondrion-targeted sHSP in D. melanogaster (Wadhwa et al., 2010). However, little is known about why plants have unusually abundant sHSPs and the distinct functions of different sHSPs localized to different organelles.
Because of the abundance and diversity of sHSPs in plants, extensive studies have focused on the biological functions of plant sHSPs (Sun and MacRae, 2005). Most sHSPs are highly expressed during heat stress, and such expression often confers increased thermal tolerance by protecting proteins from irreversible denaturation (Sun et al., 2002; Sun and MacRae, 2005). Plant sHSPs also protect cells against other environmental stresses, such as heavy metals, drought, cold, and oxidative stress (Sun et al., 2002). Furthermore, some sHSPs have been suggested to be involved in embryogenesis, seed germination, and fruit maturation (Volkov et al., 2005; Chauhan et al., 2012).
HSP21, a nuclear-encoded chloroplast-localized sHSP, has been described for many plant species. In addition to two conserved regions (consensus regions I and II) found in all sHSPs, it has a unique amphipathic, Met-rich domain at its N terminus that is highly conserved among all identified chloroplast sHSPs but not found in other sHSPs (Chen and Vierling, 1991). Many studies have suggested that HSP21 plays an important role in protecting the thermolabile photosystem II (PSII) against heat stress (Heckathorn et al., 1998; Wang and Luthe, 2003; Shakeel et al., 2011). Several studies have also demonstrated that HSP21 protects PSII against oxidative stress (Härndahl et al., 1999; Kim et al., 2012). In addition, HSP21 has been suggested to have a dual role: protecting PSII from oxidative stress and promoting color changes during fruit maturation in tomato (Solanum lycopersicum; Neta-Sharir et al., 2005). Although there are extensive studies on the function of HSP21, the molecular mechanism of its chaperone activity in vivo and its physiological targets remain unknown.
Plastid transcription is mediated by two types of RNA polymerase: plastid-encoded RNA polymerase (PEP) and nuclear-encoded RNA polymerase (NEP). PEP is composed of four core subunits (α, β, β', and β”) and a promoter recognition subunit (σ factor). Genes for PEP core subunits, α, β, β', and β”, were retained in plastid genomes as rpoA, rpoB, rpoC1, and rpoC2, respectively, during plant evolution, whereas genes for σ-factors involved in transcription initiation have been transferred to the nuclear genome in order to allow the nucleus to control PEP transcription initiation in response to developmental and environmental cues (Allison, 2000; Lerbs-Mache, 2011). PEP is responsible for transcription of photosynthesis genes in chloroplasts, while housekeeping genes encoding PEP core subunits and ribosomal proteins are transcribed by NEP (Hajdukiewicz et al., 1997; Maliga, 1998; Bruce Cahoon and Stern, 2001).
Transcription in plastids is also mediated by a number of nuclear-encoded factors in addition to PEP and NEP (Pfalz et al., 2006; Pfalz and Pfannschmidt, 2013). Many attempts have been made to identify nuclear-encoded proteins involved in plastid transcription using different biochemical purification procedures to enrich more distinct RNA polymerase complexes from chloroplasts (Pfalz and Pfannschmidt, 2013). Basically, three major types of plastid RNA polymerase preparations can be distinguished: nucleoids, the insoluble RNA polymerase preparation called plastid transcriptionally active chromosome (pTAC), and the soluble RNA polymerase preparation. Although the subunits of PEP core are present in these three types of preparations, the additional subunit compositions of these three preparations seem to be different. In maize (Zea mays), 127 proteins have been reported as strong candidates for being components of nucleoids (Majeran et al., 2012). The pTAC preparations are purer than those of nucleoids with respect to protein composition and the highly purified pTAC preparations in mustard (Sinapis alba) and Arabidopsis have led to the identification of at least 35 proteins, with 18 components named as pTACs (pTAC1 to pTAC18; Pfalz et al., 2006). Highly purified soluble RNA polymerase preparations from tobacco (Nicotiana tabacum) and mustard have been shown to contain 13 and 14 subunits, respectively (Suzuki et al., 2004; Steiner et al., 2011). It is proposed that nucleoids and pTAC may have different functional subdomains, including plastid transcription, RNA maturation, DNA replication/inheritance, and translation (Pfalz and Pfannschmidt, 2013). The pTAC is likely to present an intermediate structure between the transcriptional subdomain and the nucleoid with additional gene expression subdomains. The subunit compositions of the nucleoid and pTAC suggest that chloroplasts have lost most prokaryote nucleoid proteins and acquired many eukaryotic-type chloroplast nucleoid proteins during evolution (Sato, 2001). However, the roles of the eukaryotic-type nucleoid proteins in plastid transcription are still largely unknown (Pfalz and Pfannschmidt, 2013).
Here, we characterized the molecular mechanism of HSP21 in Arabidopsis. We found that HSP21 was localized in plastid nucleoids. Loss-of-function analyses in Arabidopsis indicate that HSP21 is essential for chloroplast development under heat stress through maintaining PEP function. Furthermore, we show that HSP21 interacts with plastid nucleoid protein pTAC5, which is also essential for maintaining PEP function under heat stress. Our results define pTAC5 as a physiological target of HSP21 and describe a molecular mechanism for chloroplast development under heat stress.
RESULTS
Loss of HSP21 Function Strongly Affects Seedling Development under Heat Stress
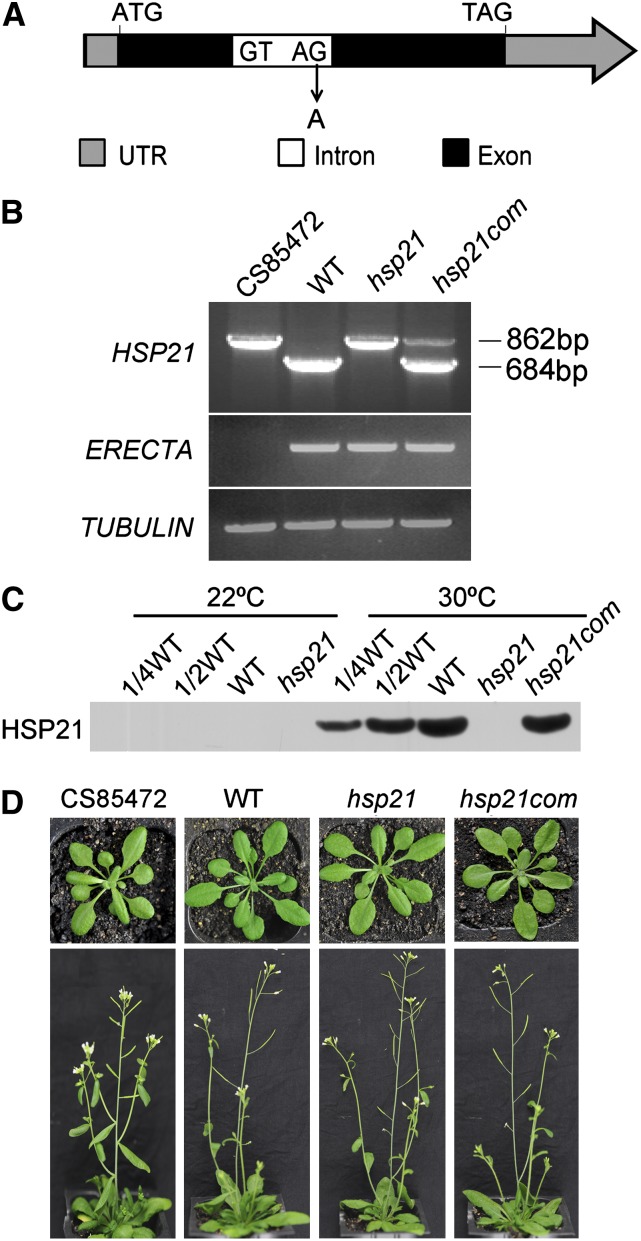
To investigate the function of HSP21, the CS85472 line mutated by ethyl methanesulfonate was obtained from the ABRC (http://abrc.osu.edu/). This line was found to contain three mutation sites in three different genes. One is a DNA rearrangement in the At2g26330 gene that encodes a putative receptor kinase ERECTA. Another is a synonymous mutation in the At2g46080 gene that encodes a protein related to BYPASS1. A third is a point mutation in the At4g27670 gene that encodes HSP21 (this study). To obtain an hsp21 single mutation line, CS85472 was backcrossed to the wild type (Columbia-0 ecotype) three times. The HSP21 single mutation line was verified by PCR, sequencing, immunoblot, and phenotype analyses (Figures 1A to 1C). The hsp21 single mutation was due to a G-to-A change at nucleotide position 414 bp in the right splice junction of the At4g27670 gene relative to the ATG codon. No HSP21 protein was detected in the hsp21 mutant, indicating that hsp21 is a knockout mutant.
Figure 1.
Characterization of the hsp21 Mutation.
(A) Schematic diagram of the HSP21 gene. Sequencing of the genomic sequence revealed a G-to-A mutation at nucleotide position 414 bp of the At4g27670 gene relative to the ATG codon. ATG and TAG indicate the start codon and the stop codon, respectively. The diagram is not drawn to scale. UTR, untranslated region.
(B) RT-PCR analysis of the CS85472 line, the wild type (WT), hsp21, and the complemented plant (hsp21com) grown for 5 d at 30°C using specific primers for At4g27670 (HSP21), At2g26330 (ERECTA), and TUBULIN. RT-PCR analysis shows the presence of one unspliced HSP21 transcript in the CS85472 line and the hsp21 mutant and the presence of correct HSP21 transcript in hsp21com, according to subsequent sequencing analysis. RT-PCR analysis also shows the presence of ERECTA transcript in the hsp21 mutant but not in the CS85472 line. In addition, sequencing of the genomic sequence reveals a synonymous mutation at nucleotide position 678 bp of the At2g46080 gene relative to the ATG codon in the CS85472 line, and this synonymous mutation was not found in the hsp21 mutant.
(C) Immunoblot analysis of HSP21 on the basis of equal total cotyledon proteins (15 μg) from wild-type, hsp21, and hsp21com seedlings grown for 5 d at 22 or 30°C.
(D) Phenotypes of the CS85472 line, wild-type, hsp21, and hsp21com plants grown for 4 or 6 weeks at 22°C. Arabidopsis was grown in soil with a 16-h photoperiod (100 μmol photons m−2 s−1) and 50% humidity.
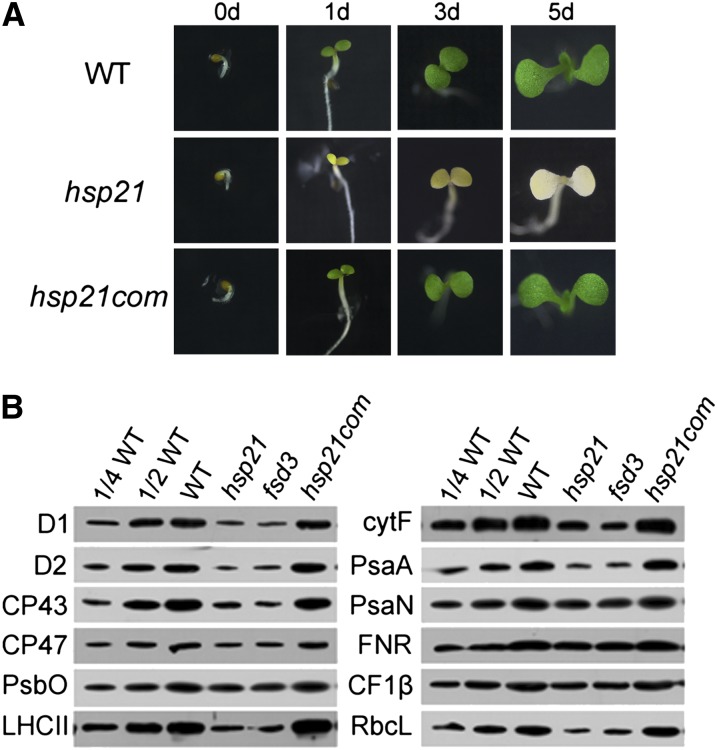
Under normal growth conditions (22°C), the hsp21 mutant grew well throughout the whole growth cycle and completed the life cycle similar to the wild type (Figure 1D). To investigate whether and how HSP21 affects seedling development under heat stress, dark-germinated seedlings at 22°C were subjected to 30°C for 1, 3, and 5 d (Figure 2A). Wild-type seedlings developed well with green cotyledons, whereas hsp21 seedlings exhibited an ivory phenotype. Moreover, the hsp21 seedlings showed an ivory phenotype after only 1 d at 30°C, suggesting that HSP21 plays an important role in the early development of seedlings under heat stress.
Figure 2.
Phenotype and Chloroplast Proteins in Wild-Type, Mutant (hsp21), and Complemented (hsp21com) Plants Grown at 30°C.
(A) Phenotypes of wild-type (WT), hsp21, and hsp21com seedlings. Dark germinated seedlings were grown at 30°C, and their phenotypes were observed at 0, 1, 3, and 5 d, respectively.
(B) Immunoblot analyses of chloroplast proteins on the basis of equal total proteins (15 μg) from the cotyledons of wild-type, hsp21, and hsp21com seedlings grown for 5 d at 30°C. The fsd3 seedlings were grown for 5 d at 22°C.
The ivory phenotype in hsp21 under heat stress suggests that HSP21 is required for chloroplast development. Thus, we investigated how the mutation of HSP21 affects the steady state levels of chloroplast proteins. We used antibodies for two members of the photosystem I complex (PsaA and PsaN), six members of the PSII complex (D1, D2, CP43, CP47, PsbO, and light-harvesting complex II [LHCII]), one member of ATP synthase (CF1β), one member of cytochrome b6f complex (cytF), ferredoxin NADP+ reductase (FNR), and the large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RbcL). Our results show that the levels of D1, D2, CP43, CP47, LHCII, cytF, PsaA, and RbcL in hsp21 were significantly reduced to ∼10 to 20% of those of the wild type. The levels of PsbO, PsaN, CF1β, and FNR showed around a 50% decrease in hsp21 compared with those in the wild type (Figure 2B). Since the fsd3 mutant (SALK_103228) has a similar ivory phenotype to that in hsp21 (Myouga et al., 2008; Pfalz and Pfannschmidt, 2013), we also made a comparison between the hsp21 mutant and the fsd3 mutant. The change in the level of LHCII in hsp21 was similar to that in fsd3 (Figure 2B).
To further prove that the lack of HSP21 was responsible for the ivory phenotype of the hsp21 mutant under heat stress, the full-length HSP21 open reading frame under the control of P35S was introduced into the hsp21 mutant. The complemented plant showed normal wild-type behavior under heat stress (Figures 1 and 2). Thus, it can be concluded that the lack of HSP21 is responsible for the observed hsp21 phenotypes under heat stress.
HSP21 Subcellular Location and HSP21 Gene Expression Patterns
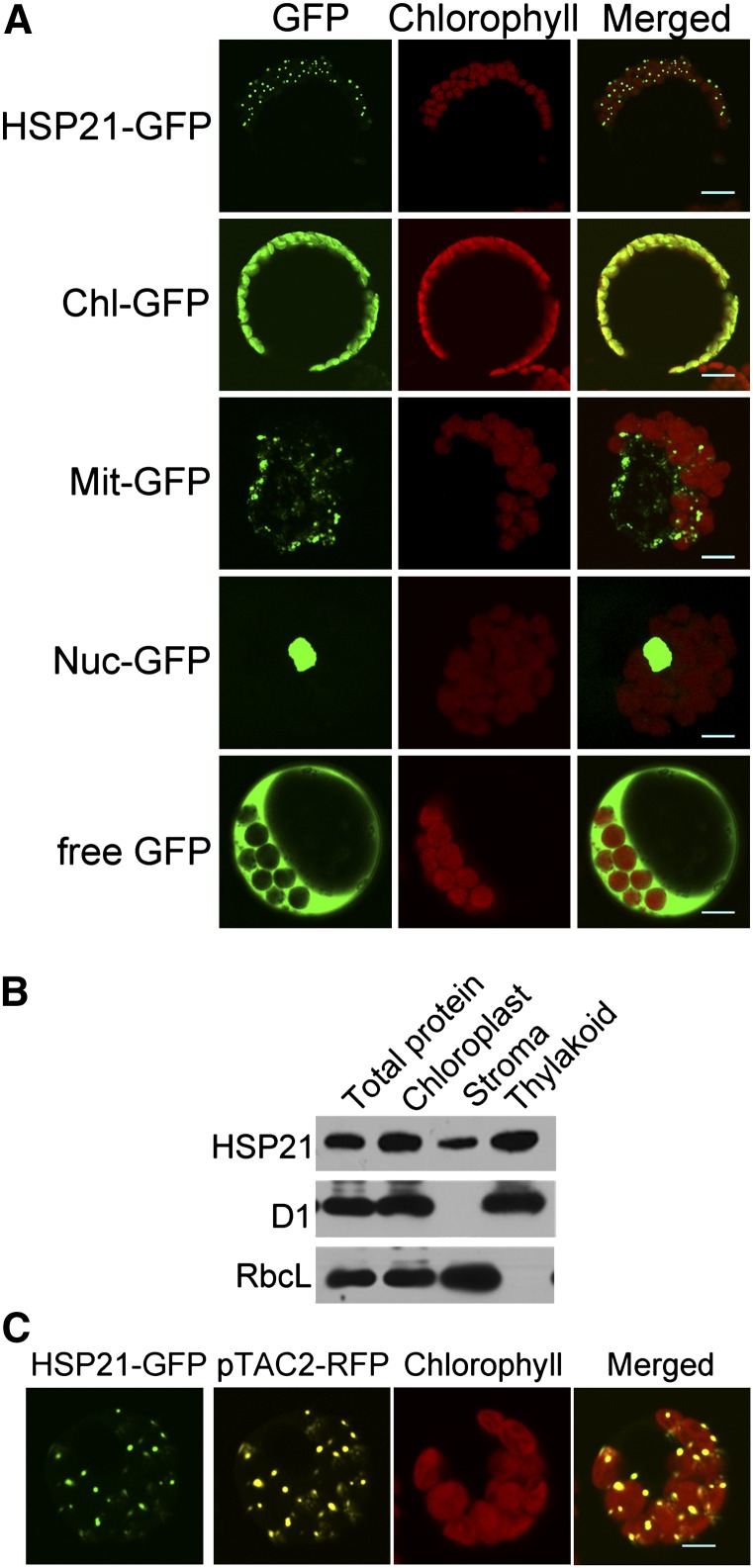
To determine the subcellular localization of HSP21, HSP21–green fluorescent protein (GFP) fusion was introduced into Arabidopsis protoplasts, and GFP fluorescence was found to be localized to chloroplasts (Figure 3A). The fluorescence pattern of HSP21 inside chloroplasts resembled the appearance of nucleoids, which are mostly associated with thylakoids. Thus, we further investigated whether HSP21 is associated with thylakoids (Figure 3B). The immunoblot analyses of stroma and membrane fractions from Percoll-purified chloroplasts demonstrated that HSP21 was mainly associated with thylakoid membranes. We further examined whether HSP21-GFP was colocalized with red fluorescent protein (RFP) fused with pTAC2, a well-characterized protein localized in nucleoids (Pfalz et al., 2006) (Figure 3C). The fluorescence signal overlay of HSP21-GFP and pTAC2-RFP in chloroplast nucleoids further indicates that HSP21 and pTAC2 colocalized in chloroplast nucleoids.
Figure 3.
Subcellular Localization of HSP21 Protein.
(A) Localization of HSP21 protein within the chloroplast by GFP assay. HSP21-GFP, HSP21-GFP fusion; Chl-GFP, chloroplast control; Mit-GFP, mitochondrial control; Nuc-GFP, nuclear control; free GFP, control with empty vector. Bars = 5 μm.
(B) HSP21 localizes mainly in the thylakoid membrane fractions. Intact chloroplasts were isolated from the cotyledons of wild-type seedlings grown for 5 d at 30°C and then separated into thylakoid membrane and stroma fractions. Polyclonal antisera were used against HSP21, the integral membrane protein D1, and the abundant stroma protein ribulose biphosphate carboxylase large subunit (RbcL). The amount of the total protein from cotyledons, the chloroplast protein, the stroma fraction, and the thylakoid fraction for each lane was 15, 7.5, 5, and 5 μg, respectively.
(C) Colocalization of HSP21-GFP (green) and pTAC2-RFP (yellow) fluorescence within chloroplast nucleoids. Bars = 5 μm.
We used β-glucuronidase (GUS) staining, quantitative RT-PCR, and immunoblot analyses to investigate the HSP21/HSP21 expression patterns (see Supplemental Figure 1 online). At 22°C, HSP21 was expressed only in pollen grains of budding flowers. After 2 h heat shock at 30°C, HSP21 was highly expressed in roots, stems, leaves, flowers, and immature and mature siliques but not in seeds. We further examined the HSP21 gene expression during early seedling development at 30°C. HSP21 was highly induced in young seedlings by heat stress. The increase in transcript and protein was visible within the first 15 min of heat stress treatment and both reached the highest level at around 1 h. These data indicate that the HSP21 gene is highly and quickly induced by heat stress during early seedling development.
Expression Analysis of Plastid-Encoded Genes
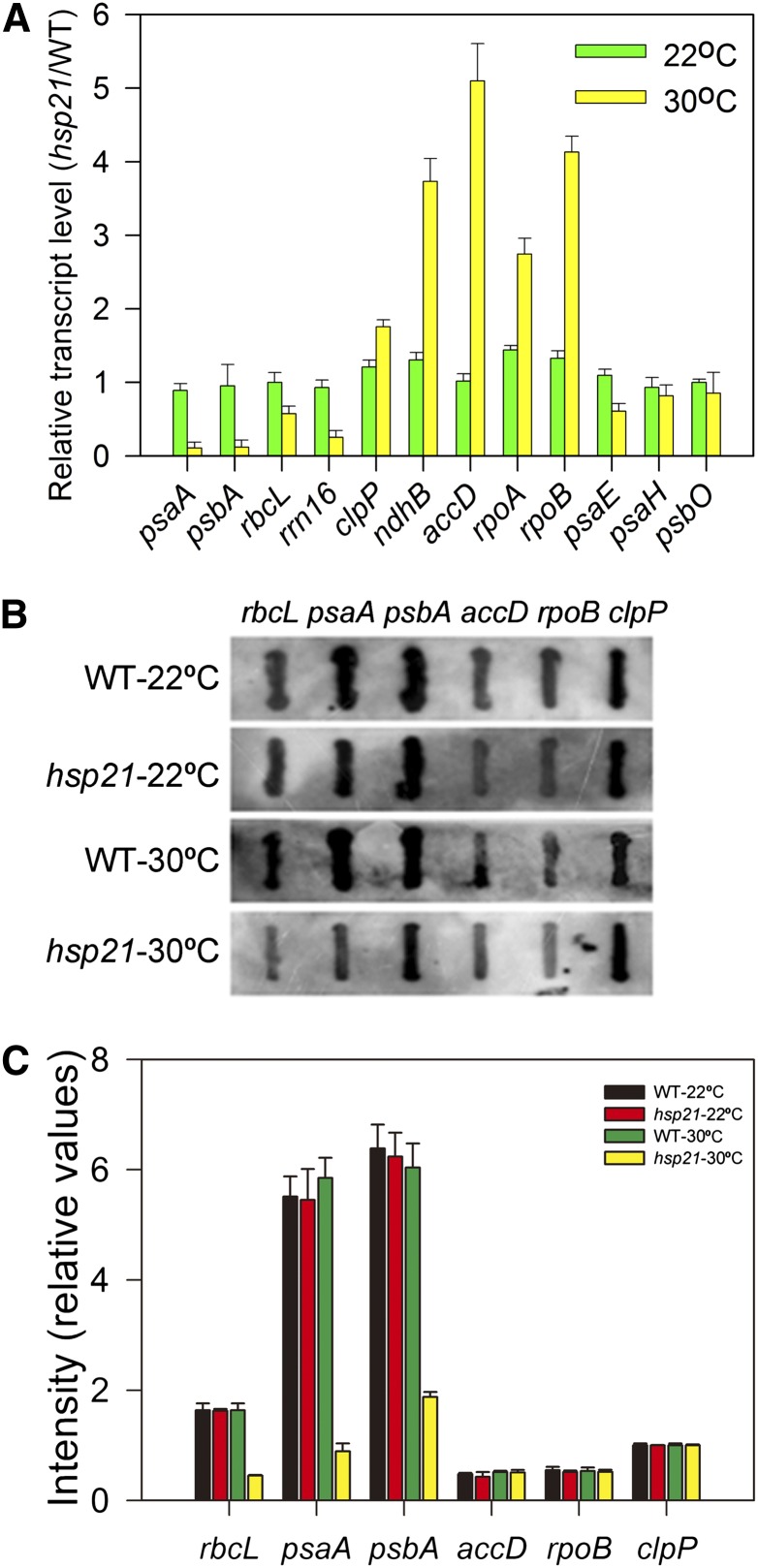
Since there was a significant decrease in the levels of plastid- and nuclear-encoded proteins in hsp21 compared with that in the wild type under heat stress (Figure 2B), we investigated whether such a decrease was associated with the expression of plastid- and nuclear-encoded genes. We used quantitative real-time PCR to analyze the levels of transcripts that encode several chloroplast proteins transcribed by PEP and/or NEP in the wild type and hsp21 (Figure 4A). psaA, psbA, and rbcL were selected as PEP-dependent genes (class I); accD, rpoA, and rpoB were selected as NEP-dependent genes (class III); and rrn16, clpP, and ndhB were chosen as both PEP and NEP-dependent genes (class II). At 22°C, no significant differences in transcript levels of class I, class II, and class III genes were observed between the wild type and hsp21. At 30°C, there was a consistent decrease in transcript levels of class I genes, while transcript levels for class III were enhanced in hsp21 compared with the wild type. In the cases of class II genes, transcript levels of clpP and ndhB were higher, but the level of rrn16 was lower in hsp21 than in the wild type. The expression of nuclear-encoded genes whose gene products are targeted to chloroplasts (psaE, psaH, and psbO) was largely unchanged in hsp21 compared with that in the wild type either at 22 or 30°C. The results from real-time PCR were further confirmed by RNA gel blots (see Supplemental Figure 2A online). It should be pointed out we did not observe any accumulation of larger transcripts for the detected genes, indicating that mRNA processing may not be defective in hsp21. These results suggest that PEP activity is decreased in hsp21 under heat stress.
Figure 4.
Chloroplast Gene Expression in the Wild Type and hsp21 Mutant Grown for 5 d at 22 or 30°C.
(A) Transcript abundance of plastid-encoded and nuclear-encoded genes measured by quantitative real-time RT-PCR. psaA, psbA, and rbcL were selected as PEP-dependent genes (class I); accD, rpoA, and rpoB were selected as NEP-dependent genes (class III); rrn16, clpP, and ndhB were chosen as class II genes; psaE, psaH, and psbO were chosen as nuclear-encoded genes. Error bars indicate sd (n = 3). WT, the wild type.
(B) Run-on transcription assay of chloroplast genes in the wild type and hsp21. Filters probed with run-on transcripts derived from chloroplasts isolated from wild-type and hsp21 cotyledons. Three independent experiments were performed, and one representative experiment is presented.
(C) Relative transcription rates of chloroplast genes in the wild type and hsp21. The signals were normalized to clpP signal intensity within the wild type and hsp21, respectively. Error bars indicate sd (n = 3).
To further demonstrate the decreased PEP activity in hsp21 under heat stress, we investigated the transcription rates of psaA, psbA, and rbcL in the wild type and hsp21 at 22 or 30°C. Run-on transcription assays show that the transcription rates of psaA, psbA, and rbcL were decreased significantly, while the transcription rates of accD, rpoB, and clpP were largely unaltered in hsp21 at 30°C (Figures 4B and 4C). However, the transcript levels of accD, rpoB, and clpP in hsp21 were upregulated at 30°C, as shown in Figure 4A. This inconsistency may suggest that the transcripts of these genes are posttranscriptionally stabilized in hsp21 under heat stress.
To further investigate the possibility that HSP21 may be involved in the translation of plastid-encoded mRNAs, the association of psaA and psbA with ribosomes in the wild type and hsp21 was compared (see Supplemental Figure 2B online). No significant differences in the association with polysomes between the wild type and hsp21 were observed for the psaA and psbA genes, suggesting that mRNA translation of the psaA and psbA genes proceeds at comparable efficiency and that HSP21 may be not involved in the translation of plastid-encoded mRNAs. Taken together, our results suggest that HSP21 is required for PEP-dependent transcription under heat stress.
Identification of HSP21 Target Proteins
To identify possible HSP21 target proteins, we generated transgenic Arabidopsis plants that express HSP21 protein carrying a His tag (HSP21-His). The possible HSP21 target proteins were affinity purified from HSP21-His plants. Total leaf protein extracts from wild-type and HSP21-His plants were incubated with anti-His MicroBeads to isolate the HSP21 complex. The purified proteins were separated by SDS-PAGE and further analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS). No HSP21 protein was purified from wild-type plants, which were used as negative controls (see Supplemental Figure 3 online), thus excluding the possibility that the HSP21 complex bound nonspecifically to the magnetic beads used in the affinity purification. The mass spectrometry data indicated that the protein encoded by At4g13670 was the top candidate HSP21-interacting protein because it had the largest number of matches of MS/MS spectra other than HSP21 (see Supplemental Table 1 online). The gene encodes pTAC5, which has been identified previously as a component of chloroplast nucleoids (Pfalz et al., 2006). The subcellular analysis confirmed that pTAC5 was indeed localized in chloroplast nucleoids (see Supplemental Figure 4 online).
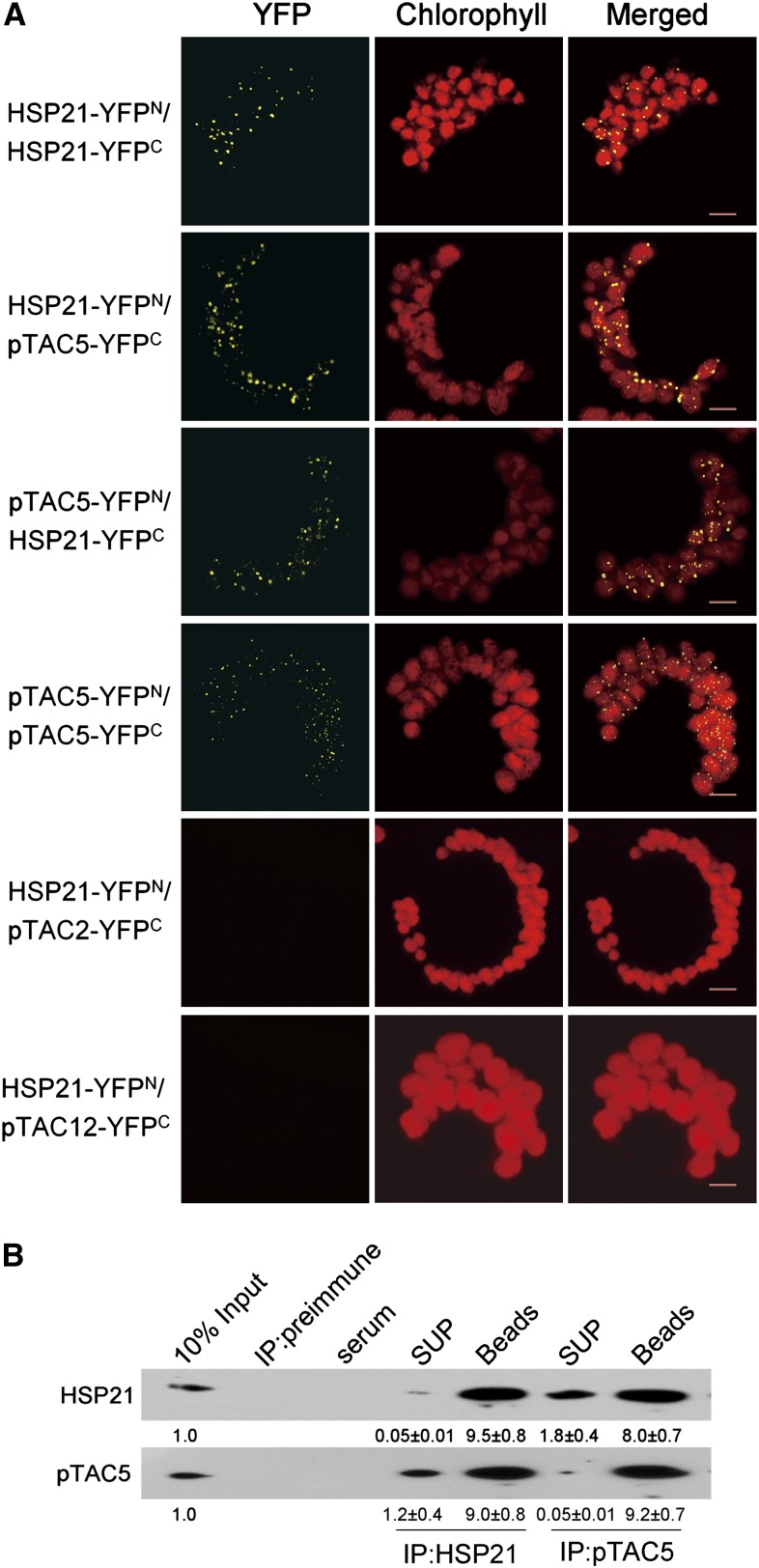
To confirm the interaction of HSP21 with pTAC5 in vivo, a bimolecular fluorescence complementation (BiFC) approach was performed in Arabidopsis protoplasts. A strong yellow fluorescent protein (YFP) fluorescence was observed when the combination of HSP21 and pTAC5 was expressed, demonstrating that HSP21 interacts with pTAC5. A strong YFP fluorescence was also observed in the combination of HSP21-YFPN and HSP21-YFPC or the combination of pTAC5-YFPN and pTAC5-YFPC, indicating that HSP21 and pTAC5 may potentially form homodimers or larger oligomers in chloroplast nucleoids. When the combination of HSP21-YFPN and pTAC12-YFPC or the combination of HSP21-YFPN and pTAC2-YFPC was cotransformed into protoplasts, no YFP fluorescence was observed, suggesting that HSP21 may not interact with pTAC2 and pTAC12 (Figure 5A). Coimmunoprecipitation experiments further confirmed that HSP21 interacts with pTAC5 in vivo (Figure 5B).
Figure 5.
In Vivo Interaction between HSP21 and pTAC5.
(A) In vivo interaction between HSP21 and pTAC5 examined by BiFC. YFP confocal microscopy images show Arabidopsis protoplasts transiently expressing constructs encoding the fusion proteins indicated. Each image is representative of at least three independent experiments. Bars = 5 μm.
(B) In vivo interaction between HSP21 and pTAC5 examined by coimmunoprecipitation analysis. Chloroplasts isolated from the cotyledons of wild-type seedlings grown for 5 d at 30°C were incubated with antibodies against HSP21 and pTAC5, respectively. The immunoprecipitates were then probed with antibodies against HSP21 and pTAC5. Three independent experiments were performed, and one representative experiment is presented. The relative protein levels shown below each lane were estimated by normalizing to the level of corresponding 10% input proteins. Values represent means ± sd of three independent experiments. X-ray films were scanned and analyzed using ImageMaster 2D Platinum software. SUP, supernatant.
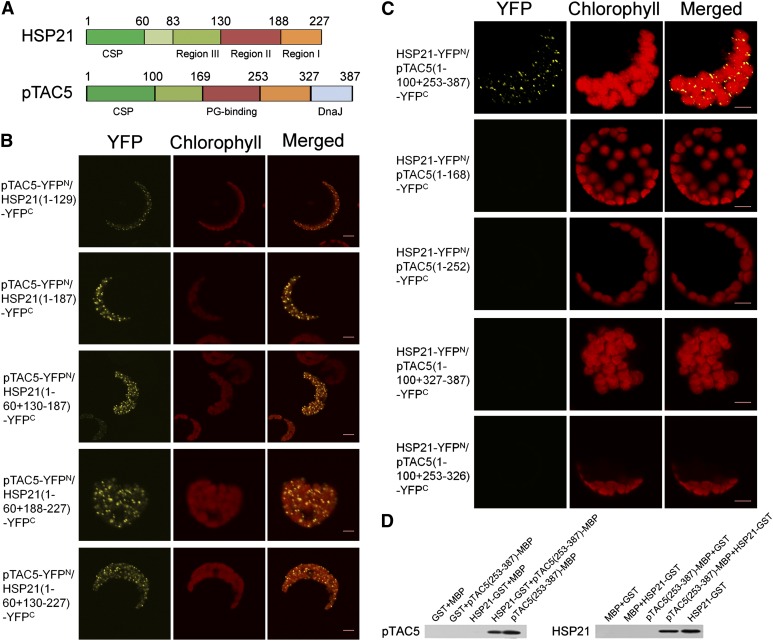
HSP21 has three conserved domains that were designated consensus regions I, II, and III (Chen and Vierling, 1991). In order to identify which domain in HSP21 is responsible for the interaction of HSP21 and pTAC5, we generated several HSP21 truncations (1-130-188-227 amino acids) (Figure 6A) and tested their ability to interact with pTAC5. BiFC analysis shows that each of three consensus regions was able to interact with pTAC5 (Figure 6B).
Figure 6.
Domain Deletion Analysis of HSP21 and pTAC5 Domains Involved in Protein Interactions.
(A) Structure sketches of HSP21 and pTAC5. In HSP21: CSP, chloroplast signal peptide, Region III, Met-rich domain; Region II and Region I, β-sheets of ACD domain. In pTAC5: CSP, PG binding, and DnaJ indicate chloroplast signal peptide, peptidoglycan binding domain, and DnaJ domain, respectively. Numbers indicate the positions of HSP21 and pTAC5 deletions for the protein interaction analysis used in this study.
(B) BiFC visualization of the interactions of full-length pTAC5 with different segments of HSP21 in Arabidopsis protoplasts, showing that regions I, II, and III all interacted with pTAC5. Three independent experiments were performed, and one representative experiment is presented. Bars = 5 μm.
(C) BiFC visualization of the interactions of full-length HSP21 with different segments of pTAC5 in Arabidopsis protoplasts, showing that only 253 to 387 amino acids of pTAC5 interacted with HSP21. Three independent experiments were performed, and one representative experiment is presented. Bars = 5 μm.
(D) Pull-down assay of the interaction of HSP21 with the C terminus (253 to 387 amino acids) of pTAC5. HSP21-GST and free GST proteins coupled to GST binding resin were incubated with pTAC5(253-387aa)-MBP and free MBP proteins, respectively (left). pTAC5(253-387aa)-MBP and free MBP proteins coupled to MBP binding resin were incubated with HSP21-GST and free GST proteins, respectively (right). Bound proteins were separated by SDS-PAGE and immunoblotted with pTAC5 or HSP21 antibodies. Similar results were obtained in two additional independent experiments.
pTAC5 contains a peptidoglycan binding-like domain (PG binding) and a DnaJ domain (DnaJ) (Pfalz et al., 2006). To examine whether PG binding and/or DnaJ interact with HSP21, four segments in pTAC5, corresponding to amino acids 1-169-253-327-387 (Figure 6A), were generated. BiFC analysis shows that none of these segments individually interacted with HSP21. However, the 253 to 387 amino acid region of pTAC5 containing DnaJ with its adjacent segment did interact with HSP21 in BiFC assays (Figure 6C), and this interaction was confirmed by pull-down assays (Figure 6D).
Reduction of pTAC5 Expression Leads to Similar Phenotypic Effects as Observed for Loss of HSP21 Function
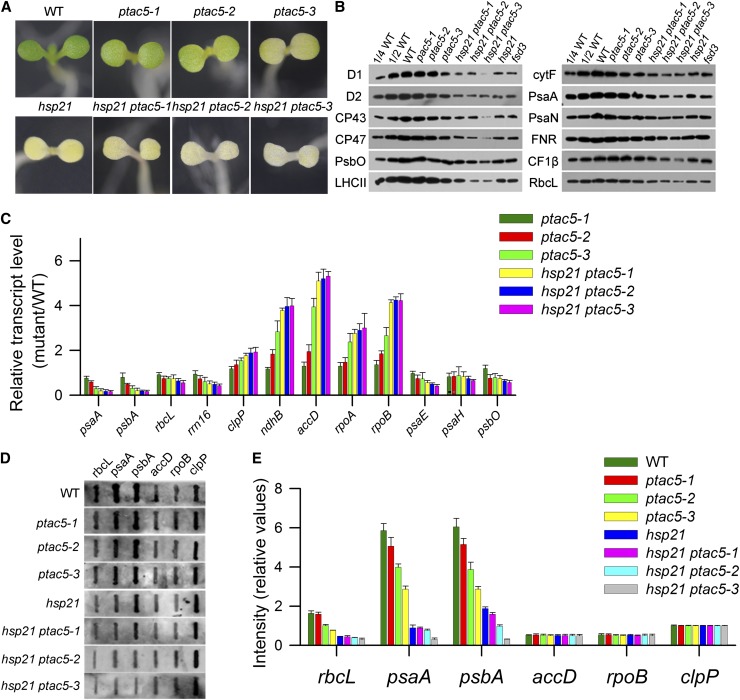
If pTAC5 is the main target protein for HSP21, we anticipated that reduction of pTAC5 expression would lead to similar phenotypic effects as observed for loss of HSP21 function. After inspection of the respective databases and analyses of the potential mutants of pTAC5 obtained from ABRC database and RIKEN Dissociation-tagged lines (Kuromori et al., 2004), we failed to identify knockout or knockdown mutants for pTAC5 at the time of this study. Therefore, we created transgenic Arabidopsis plants expressing a pTAC5 RNA interference (RNAi) construct. Three independent lines (ptac5-1, ptac5-2, and ptac5-3) were obtained with reduced contents of pTAC5. Both pTAC5 mRNA and pTAC5 protein in ptac5-1, ptac5-2, and ptac5-3 were decreased to ∼80, 50, and 10% of that in the wild type, respectively, either at 22 or 30°C (see Supplemental Figures 5A and 5B online). At 22°C, there was no visible difference in the appearance of the wild type, RNAi pTAC5 lines, and hsp21 ptac5 double mutants, and they had similar growth and developmental patterns until they reached maturity (see Supplemental Figures 5C and 5D online). However, after dark-germinated seedlings at 22°C were subjected to 30°C for 5 d, the seedlings of RNAi pTAC5 lines showed a yellowish phenotype with their cotyledons. The hsp21 ptac5 double mutants showed a more severe yellowish phenotype than their respective ptac5 line at 30°C (Figure 7A).
Figure 7.
Characterization of Transgenic Plants with Reduction of pTAC5 at 30°C.
(A) Phenotypes of wild-type (WT), ptac5, and hsp21 ptac5 seedlings grown for 5 d at 30°C.
(B) Immunoblot analysis of chloroplast proteins on the basis of equal total proteins (15 μg) from the cotyledons of wild-type, ptac5, and hsp21 ptac5 seedlings grown for 5 d at 30°C. The fsd3 seedlings were grown for 5 d at 22°C.
(C) Transcript abundance of plastid-encoded and nuclear-encoded genes in wild-type, ptac5, and hsp21 ptac5 seedlings grown for 5 d at 30°C. Data represent mean ± sd of three independent assays.
(D) Run-on transcription assay of chloroplast genes in wild-type, ptac5, and hsp21 ptac5 seedlings grown for 5 d at 30°C. Filters probed with run-on transcripts derived from chloroplast isolated from wild-type, ptac5, and hsp21 ptac5 cotyledons. Three independent experiments were performed, and one representative experiment is presented.
(E) Relative transcription rates of chloroplast genes in wild-type, ptac5, and hsp21 ptac5 seedlings grown for 5 d at 30°C. The signals were normalized to clpP signal intensity within wild-type, ptac5, and hsp21 ptac5, respectively. Error bars indicate sd (n = 3).
Given the yellowish phenotype of transgenic plants under heat stress, we performed a series of analyses in RNAi pTAC5 lines as described above for hsp21 (Figures 7B to 7E). There was a significant decrease in chloroplast proteins (e.g., D1, D2, LHCII, cytF, PsaA, CF1β, FNR, and RbcL) in RNAi pTAC5 lines. A consistent downregulation of PEP-dependent class I genes was observed in RNAi pTAC5 lines. The PEP transcription rate was also decreased significantly in RNAi pTAC5 lines. However, we observed that the transcript levels of clpP, accD, and rpoB were increased in RNAi pTAC5 lines compared with wild-type plants, while the transcription rates of these genes were less changed. A similar inconsistency has also been observed in another mutant that has decreased PEP activity (Chi et al., 2010). This may suggest that the transcripts of clpP,,, accD, and rpoB are posttranscriptionally stabilized in RNAi pTAC5 lines under heat stress conditions. RNA gel blot and polysome association analyses suggest that pTAC5 is also involved in PEP-dependent transcription (see Supplemental Figure 2 online).
Expression pattern analyses revealed that pTAC5 was expressed in stems, leaves, flowers, and young and mature siliques at 22°C and its expression abundance was induced by 2-h heat shock treatment at 30°C, with the highest level in leaves (see Supplemental Figure 6A online). pTAC5 mRNA and pTAC5 protein were induced within 15 min by heat stress at 30°C and gradually saturated at around 1 h (see Supplemental Figures 6B and 6C online).
We further analyzed the developmental expression of pTAC5 during greening of etiolated seedlings either at 22 or 30°C (see Supplemental Figures 6D and 6E online). Wild-type plants were grown in continuous darkness for 5 d and subsequently exposed to light for 1, 3, or 6 h. At either 22 or 30°C, both pTAC5 mRNA and pTAC5 protein were induced by light within 1 h. However, the induction levels of pTAC5 mRNA and pTAC5 protein were greater at 30°C than at 22°C, suggesting pTAC5 may play an important role in PEP-dependent transcription and be essential for light-induced chloroplast development under heat stress. The CP43 protein, which is a marker for light-dependent plastid proteins, was also induced by light. Taken together, these data indicate that pTAC5 is involved in PEP-dependent transcription in chloroplasts in a way similar to that observed for its interaction partner HSP21.
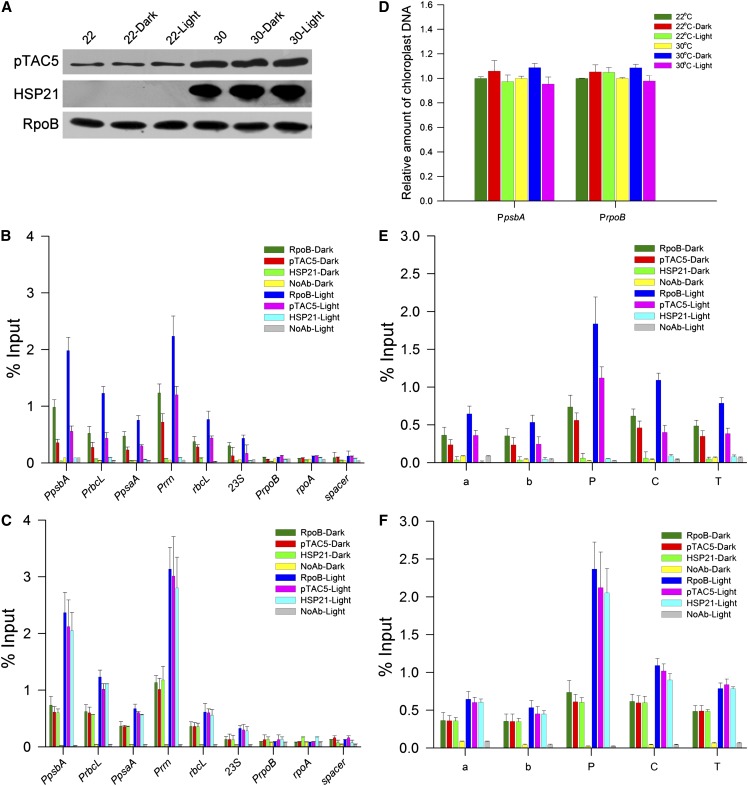
Association of HSP21 and pTAC5 with Chloroplast DNA
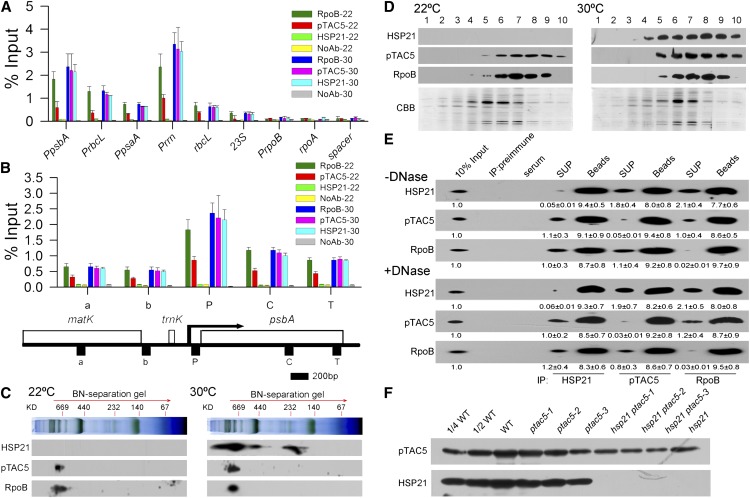
The above results strongly suggest that HSP21 and pTAC5 are involved in PEP-dependent transcription in chloroplasts under heat stress. Thus, we further investigated whether HSP21 and pTAC5 are associated with chloroplast DNA using chloroplast chromatin immunoprecipitation (cpChIP) (Yagi et al., 2012). We examined the association of HSP21, pTAC5, and RpoB (β-subunit of the PEP core) with specific regions of plastid DNA, including PEP promoters (PpsbA, PrbcL, PpsaA, and Prrn), a NEP promoter (PrpoB), coding regions of PEP-dependent genes (rbcL and 23S), the coding region of a NEP-dependent gene (rpoA), and the noncoding region between rps12 and rrn16 (spacer). Our results show that pTAC5 and HSP21 preferentially bound to promoter regions of PEP-dependent genes, but not of NEP-dependent genes. In addition, heat stress enhanced the association of pTAC5 with promoter regions of PEP-dependent genes, indicating that pTAC5 associates with PEP-dependent genes in a temperature-dependent manner (Figure 8A). We further examined the local patterns of spatial association of HSP21 and pTAC5 with the psbA transcription unit (Figure 8B). Our results show that pTAC5 and HSP21 bound not only to the promoter region but also the transcription elongation region during transcription, suggesting that pTAC5 and HSP21 are associated with chloroplast DNA along the psbA transcription unit. Therefore, it is unlikely that HSP21 and pTAC5 bind to specific sequences in the psbA promoter; rather, they may be components of the PEP complex in chloroplasts under heat stress.
Figure 8.
Association of HSP21 and pTAC5 with the PEP Complex.
(A) Association of HSP21, pTAC5, and RpoB with chloroplast DNA in wild-type seedlings grown for 5 d at 22 and 30°C. Association of HSP21, pTAC5, and RpoB with PEP promoter regions (PpsbA, PrbcL, PpsaA, and Prrn), PEP coding sequence regions (rbcL and 23S), a NEP promoter region (PrpoB), a NEP coding region (rpoA), and a noncoding spacer region located between rps12 and rrn16 (spacer) was analyzed by ChIP assay. Chloroplasts were prepared from the cotyledons of wild-type seedlings grown for 5 d at 22 or 30°C, respectively, and then subjected to ChIP assays using antibodies against HSP21, pTAC5, and RpoB. NoAb, no antibody control. The amount of immunoprecipitated DNA in each sample is presented as a percentage of the total input chromatin. Mean values ± sd of three independent experiments are shown.
(B) Spatial association of HSP21, pTAC5, and RpoB along the psbA transcription unit in wild-type seedlings grown for 5 d at 22 and 30°C, respectively. Below is a schematic gene map of the matK-psbA region. Arrow indicates the transcription start site of the psbA gene and the direction of transcription. P, C, T, a, and b indicate the promoter, coding region, terminator, and two units in loci upstream of psbA, respectively. Error bars indicate sd (n = 3).
(C) Immunoblot detection of HSP21, pTAC5, and RpoB on two-dimensional gel electrophoresis. Thylakoid membrane proteins (corresponding to 8 μg chlorophyll) from wild-type seedlings grown for 5 d at 22 or 30°C were fractionated by BN-PAGE in the first dimension and by SDS-PAGE in the second dimension. The approximate molecular masses of the labeled protein complexes are indicated above. Three independent experiments were performed and one representative experiment is presented.
(D) Separation of the PEP complex by glycerol density gradient centrifugation. Total chloroplast proteins from the cotyledons of wild-type seedlings grown for 5 d at 22 or 30°C were loaded onto a 10 to 30% (v/v) glycerol density gradient and separated by centrifugation. Ten fractions were collected from top to bottom and analyzed by immunoblotting with HSP21, pTAC5, or RpoB antibodies. Coomassie blue (CBB) staining is shown below.
(E) Coimmunoprecipitation analysis of the HSP21-pTAC5-RpoB complex. Total chloroplast extracts from wild-type seedlings grown for 5 d at 30°C treated without (top) or with (bottom) DNase were subjected to immunoprecipitation with HSP21, pTAC5, and RpoB antibodies, respectively, and then analyzed by immunoblotting. Three independent experiments were done, and a representative one is shown. The relative protein contents shown below each lane were estimated by normalizing to the content of corresponding 10% input proteins. Values represent means ± sd of three independent experiments. X-ray films were scanned and analyzed using ImageMaster 2D Platinum software. SUP, supernatant.
(F) Immunoblot analysis of HSP21 and pTAC5 in wild-type (WT), ptac5, hsp21 ptac5, and hsp21 seedlings grown for 5 d at 30°C. The lanes on each gel were loaded on the basis of equal total leaf proteins (15 μg).
To further examine whether HSP21 and pTAC5 are the components of the PEP complex, we analyzed thylakoid membranes using blue native (BN) gels and subsequent two-dimensional SDS-PAGE and probed for the presence of HSP21 and pTAC5 by immunoblotting (Figure 8C). Our results showed that both HSP21 and pTAC5 migrated in the PEP complex. In addition, glycerol density gradient centrifugation of chloroplasts showed that HSP21 and pTAC5 cosedimented with the PEP complex (Figure 8D). Furthermore, coimmunoprecipitation assays of intact chloroplasts treated with or without DNase demonstrated the presence of HSP21, pTAC5, and RpoB in the complex immunoprecipitated by either of HSP21, pTAC5, or RpoB antibodies (Figure 8E). Moreover, we found that a decrease in the content of HSP21 resulted in a decrease in the content of pTAC5 and vice versa (Figure 8F). These results suggest that HSP21 and pTAC5 form a complex which is associated mainly with the PEP complex.
Light-Dependent Association of the HSP21-pTAC5 Complex with Chloroplast DNA
PEP transcription activity is greatly stimulated by light in chloroplasts (Yagi et al., 2012). Thus, we examined the light-dependent association of HSP21, pTAC5, and RpoB with promoter regions of several plastid genes in vivo (Figure 9). According to the method of Yagi et al. (2012), chloroplasts were prepared from Arabidopsis seedlings grown for 5 d in the light and then dark adapted for 24 h or seedlings reilluminated for 6 h after the 24-h dark adaptation. Immunoblot analyses demonstrated the constitutive accumulation of HSP21, pTAC5, and RpoB upon illumination, suggesting that the expression level of the PEP complex is not affected by light. The cpChIP assays showed that under heat stress, the relative amounts of HSP21, pTAC5, and RpoB associated with the promoter and coding regions of PEP genes were two- to fivefold higher in the illuminated chloroplasts than in the dark-adapted chloroplasts. The amounts of chloroplast DNA in input samples between dark and light conditions were not significantly different. Thus, the light-dependent cpChIP signals are indicative of light-dependent association of HSP21, pTAC5, and RpoB to chloroplast DNA. Light-dependent association of PEP with NEP-dependent genes, including rpoA and rpoB, and the spacer region was not detected. In addition to promoter regions, we also detected light-dependent association of HSP21, pTAC5, and RpoB with coding and termination regions of psbA. Light accelerated the association of HSP21 and pTAC5 with not only the PEP promoter region but also the coding region in the psbA transcription unit similar to the distribution pattern of RpoB, suggesting that HSP21 and pTAC5 associate with PEP-dependent transcribed regions as well as the components of the PEP complex in a light-dependent manner.
Figure 9.
Light-Dependent Association of HSP21, pTAC5, and RpoB with Chloroplast DNA.
(A) Protein levels of HSP21, pTAC5, and RpoB in dark and light conditions at 22 or 30°C. Chloroplasts were prepared from wild-type Arabidopsis seedlings grown for 5 d in the light and then dark adapted for 24 h (dark) or seedlings reilluminated for 6 h (light) after the 24 h dark adaptation. Protein levels were analyzed by immunoblot on the basis of equal total cotyledon proteins (10 μg) for each lane.
(B) Association of HSP21, pTAC5, and RpoB with chloroplast DNA in response to light in wild-type Arabidopsis seedlings grown at 22°C. ChIP was performed to determine association level of HSP21, pTAC5, and RpoB with PEP promoter regions (PpsbA, PrbcL, PpsaA, and Prrn), PEP coding sequence regions (rbcL and 23S), a NEP promoter region (PrpoB), a NEP coding region (rpoA), and a noncoding spacer region located between rps12 and rrn16 (spacer) using antibodies against HSP21, pTAC5, or RpoB or no antibody (NoAb). The immunoprecipitated DNA was analyzed by quantitative PCR and quantified via standard curves based on a dilution series of input samples. The amount of immunoprecipitated DNA in each sample is presented as a percentage of the total input chromatin. Data are shown as means ± sd of three independent experiments.
(C) Association of HSP21, pTAC5, and RpoB with chloroplast DNA in response to light in wild-type Arabidopsis seedlings grown at 30°C. The procedures and data are presented as in (B).
(D) Relative amounts of chloroplast DNA in dark and light input samples at 22 or 30°C. Amounts of chloroplast DNA in dark and light input samples were analyzed by quantitative real-time PCR with specific primer sets to amplify PpsbA and PrpoB loci. Values are shown as a ratio to that in light input sample at each locus and are represented as means ± sd of three independent experiments.
(E) Association of HSP21, pTAC5, and RpoB along the psbA transcription unit in response to light in wild-type Arabidopsis seedlings grown at 22°C. Data are presented as in (B). Schematic gene map of the matK-psbA region is shown as in Figure 8B.
(F) Association level of HSP21, pTAC5, and RpoB along the psbA transcription unit in response to light in wild-type Arabidopsis seedlings grown at 30°C. Data are presented as in (B). Schematic gene map of the matK-psbA region is shown as in Figure 8B.
pTAC5 Has Protein Zinc-Dependent Disulfide Isomerase Activity
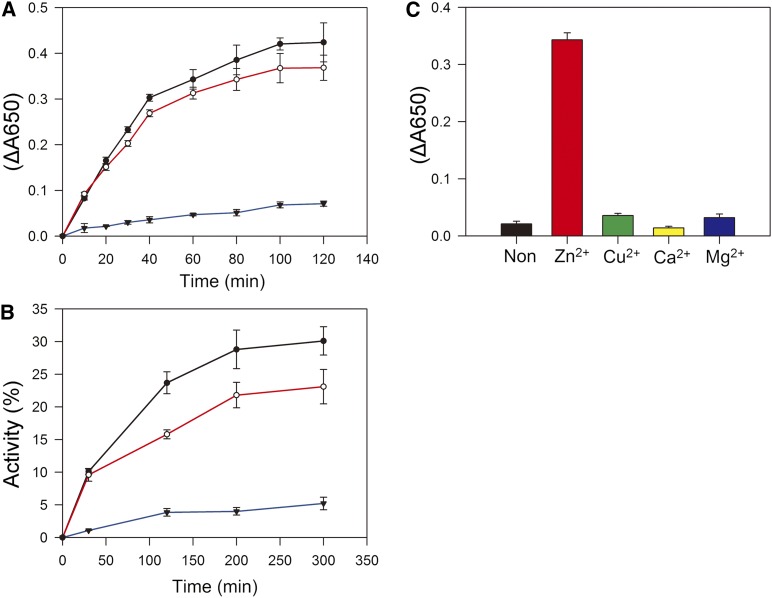
pTAC5 residues 327 to 387 amino acids are similar to the C4-type zinc finger of Escherichia coli DnaJ (see Supplemental Figure 7 online). The zinc finger is important for the enzymatic activity of DnaJ (Tang and Wang, 2001). To analyze the catalytic properties of pTAC5, we expressed the C-terminal portion of pTAC5 including residues 253 to 387 amino acids and used it in rapid spectrophotometric assays of disulfide isomerase activity based on the reduction of insulin (Holmgren, 1979). Mature insulin contains two polypeptide chains, A and B, linked by disulfide bonds. When these bonds are broken, the free B chain is insoluble and precipitates, increasing absorbance at 650 nm. E. coli DnaJ catalyzes the DTT-dependent reduction of insulin (Tang and Wang, 2001; Shi et al., 2005). We measured the reduction of insulin by DTT in the presence of pTAC5 or E. coli DnaJ (Figure 10A). Insulin B chain precipitation was observed after 10 min in the presence of pTAC5 or E. coli DnaJ.
Figure 10.
pTAC5 Has Reductase and Oxidase Activity.
(A) Purified truncated pTAC5 (253 to 387 amino acids) catalyzes the reduction of insulin. The reaction was initiated by adding DTT into 0.1 M potassium phosphate, pH 6.6, containing 0.13 mM insulin in the absence (blue) or presence of 1.0 μM pTAC5 (red) or 1.0 μM E. coli DnaJ (black). The resulting precipitation of the B chain was monitored by following the optical density at 650 nm. Values represent means ± sd of three independent experiments.
(B) Effect of truncated pTAC5 (253 to 387 amino acids) on refolding of reduced and denatured RNase A. Refolding of denatured and reduced RNase A (40 μM) was initiated in 50 mM Tris-HCl, pH 8.0, containing 0.1 M NaCl and 0.3 mM DTT in the absence (blue) or presence of 1.0 μM pTAC5 (red) or 1.0 μM E. coli DnaJ (black). At the indicated time points, an aliquot containing 40 μM RNase A was withdrawn from the reaction to assay RNase A activity. Activity is expressed as a percentage of native RNase A activity. Values are means ± sd of three independent experiments.
(C) Effect of metal on pTAC5 activity. Purified truncated pTAC5 (253 to 387 amino acids) was denatured with 6 M guanidine hydrochloride, and then the denatured proteins were renatured with dialyzing buffer containing 5.0 mM ZnCl2, CuCl2, CaCl2, MgCl2, or no divalent metal ion. The dialyzed proteins were used for enzymatic assays. The reduction of insulin and the resulting precipitation of the B chain were monitored by following the optical density at 650 mm. Data represent mean ± sd of three independent assays.
Renaturation of reduced and denatured RNase A containing eight sulfhydryl groups involves the oxidation of its thiol groups followed by rearrangement of the disulfides to the native conformation (with four disulfide bridges) (Anfinsen and Scheraga, 1975). Addition of pTAC5 or E. coli DnaJ to the reaction stimulated RNase A renaturation, suggesting that pTAC5 has protein disulfide isomerase (PDI) activity (Figure 10B). To determine whether pTAC5 requires Zn2+ for its enzymatic activity, as DnaJ does, purified C-terminal pTAC5 was denatured and renatured in a dialyzing buffer containing different divalent metal ions. C-terminal pTAC5 catalyzed the reduction of insulin when renatured in the presence of ZnCl2 but lacked activity when renatured in the presence of any of the other buffers (Figure 10C), indicating that Zn2+ is required for the enzymatic activity of pTAC5.
In this study, our results showed that there was a significant decrease in the levels of plastid-encoded proteins in hsp21 and ptac5 under heat stress (Figures 2B and 7B). In order to investigate whether the decreased plastid-encoded protein levels are due to increased levels of heat-induced oxidative stress, we made a comparison in the production of reactive oxygen species (ROS) by detecting 2,7-dichlorodihydrofluorescein diacetate (H2DCFDA) fluorescence of the cotyledons in wild-type, hsp21, and RNAi pTAC5 lines according to Tang et al. (2012). Our results showed that there was no significant accumulation of ROS in wild-type, hsp21, and RNAi pTAC5 lines (see Supplemental Figure 8 online). Therefore, it seems that the decrease in the levels of plastid-encoded proteins in the mutants may be not due to increased levels of heat-induced oxidative stress. The results in this study suggest that the decrease in the levels of plastid-encoded proteins in the mutants can be explained by the decrease in the PEP activity in the hsp21 and ptac5 mutants. On the other hand, we also observed that there was a decrease in the levels of nuclear-encoded proteins (e.g., PsbO and LHCII) in the mutants (Figures 2B and 7B), which may possibly be an indirect result of the failed formation of the thylakoid membranes in the mutants.
DISCUSSION
sHSPs are of significant interest in many areas of biology and biochemistry due to their protein dynamics, diverse evolutionary history, and connection to human disease and stress acclimation in plants and other organisms (Basha et al., 2012). In order to further understand the molecular mechanism of sHSPs, much more work is needed to clarify how sHSPs function in vivo and to identify in vivo target proteins of sHSPs (Basha et al., 2012). Although many studies have demonstrated that HSP21, a nuclear-encoded chloroplast-localized sHSP, plays an important role in protecting PSII against heat stress and oxidative stress, the molecular mechanism by which HSP21 is involved in cell protection remains unknown, and its target protein(s) has also not been identified. Here, using genetic and biochemical approaches, we found that HSP21, cooperating with its in vivo target pTAC5, is essential for proper chloroplast development in Arabidopsis under heat stress. Such a function is different from those reported for other sHSPs in prokaryotic and eukaryotic cells.
Our results show that loss of HSP21 function in Arabidopsis leads to an ivory phenotype and a significant decrease in the contents of the chloroplast proteins under heat stress (Figure 2), indicating that HSP21 is required for chloroplast development under heat stress (Pfalz and Pfannschmidt, 2013). PEP is responsible for the transcription of photosynthesis genes and is essential for proper chloroplast development. Any factor affecting PEP activity may block chloroplast development (Pfalz and Pfannschmidt, 2013). PEP activity was decreased in the hsp21 mutant under heat stress (Figure 4; see Supplemental Figure 2 online), similar to findings in Δrpo mutants that do not accumulate PEP (Serino and Maliga, 1998; Krause et al., 2000), ptac mutants (Pfalz et al., 2006; Gao et al., 2011; Yagi et al., 2012), and mutants with lesions in PEP function (Hess et al., 1993; Allison et al., 1996; Chateigner-Boutin et al., 2008; Chi et al., 2008). In addition, HSP21 is localized in plastid nucleoids (Figure 3) and is associated with the PEP complex (Figures 8C to 8E). Thus, our results suggest that HSP21 plays an important role in maintaining the proper function of the PEP transcription machinery under heat stress.
To reveal the molecular mechanism of HSP21 in PEP function, we attempted to identify HSP21 target proteins using protein affinity chromatography and were able to identify pTAC5 as putative HSP21 target (see Supplemental Figure 3 and Supplemental Table 1 online). pTAC5 is localized to chloroplast nucleoids (see Supplemental Figure 4 online) and has been shown to be a component of TACs (Pfalz et al., 2006). BiFC and coimmunoprecipitation analyses further confirmed the interaction of HSP21 and pTAC5 (Figures 5 and 6). Moreover, the in planta analysis of pTAC5 provides further evidence for its functional association with HSP21. Transgenic plants with reduction of pTAC5 exhibited a similar phenotype to that of loss of HSP21 function and had reduced expression of PEP-dependent genes under heat stress (Figure 7). In addition, BN gel and subsequent two-dimensional SDS-PAGE, density gradient centrifugation, and coimmunoprecipitation assays show that HSP21 and pTAC5 form a complex in vivo as part of a functional PEP complex (Figures 8C to 8E). Taken together, our results suggest that HSP21 and pTAC5 form a complex that is associated with the PEP complex, involved in maintaining PEP function, and required for chloroplast development in Arabidopsis under heat stress.
How do HSP21 and pTAC5 maintain the PEP activity under heat stress? It has been reported that the knockout mutants with severe lesions in plastid transcription lead to almost identical phenotypes in Δrpo mutants and mutants impaired in PEP function (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-MacIossek et al., 1999; Krause et al., 2000; Legen et al., 2002). In this study, we observed that the transcript levels of PEP-dependent genes were decreased, while the transcript levels of NEP-dependent genes were upregulated, which is similar to the expression profiles in Δrpo mutants and mutants impaired in PEP function (Hess et al., 1993; Allison et al., 1996; Hajdukiewicz et al., 1997; Silhavy and Maliga, 1998; De Santis-MacIossek et al., 1999; Krause et al., 2000; Legen et al., 2002). Moreover, the transcription rates of PEP-dependent genes were decreased in hsp21 and ptac5 (Figure 4). Defects in mRNA processing are observed in ptac2, 6, and 12, with larger transcripts accumulating for many genes (e.g., psaAB, rbcL, accD, and ndhB). Thus, it is suggested that pTAC2, 6, and 12 may be involved in plastid transcription and RNA processing (Pfalz et al., 2006). However, we observed no accumulation of larger transcripts for representative genes in hsp21 and ptac5 under heat stress (see Supplemental Figure 2A online). It thus seems unlikely that HSP21 is involved in RNA processing. In addition, polysome association analyses further showed that HSP21 and pTAC5 may also not be involved in translation (see Supplemental Figure 2B online). Thus, our results suggest that HSP21 and pTAC5 are required preferentially for transcription by PEP rather than mRNA processing and translation. However, whether they are involved in other processes (e.g., replication/DNA inheritance and RNA stability) remains to be further investigated (Pfalz and Pfannschmidt, 2013).
In order to examine how the HSP21-pTAC5 complex regulates PEP-dependent transcription, we searched for its binding regions on plastid DNA using cpChIP assays. We found that HSP21 and pTAC5 exclusively bound to the promoter regions of PEP-dependent psaA, rbcL, and rrn genes. For psbA, HSP21 and pTAC5 showed binding to both the promoter and transcribed regions. Moreover, we observed that the binding of HSP21 and pTAC5 on the promoter regions of the PEP-dependent genes exhibited temperature dependence (Figure 8). Several PEP promoters, including psbA, rbcL, psaA, and rrn, are regulated by unique cis-elements that are located upstream of or within the core promoter and recognized by promoter-specific transcription factors (Cheng et al., 1997; Suzuki et al., 2003). Therefore, our results suggest that the HSP21-pTAC5 complex is a PEP-associated general, rather than sequence-specific, factor. In contrast with PEP-dependent transcribed loci, HSP21 and pTAC5 were associated weakly with NEP-dependent transcribed loci, suggesting that the HSP21-pTAC5 complex is not associated with the NEP transcription complex.
It has been demonstrated recently that light-dependent chloroplast transcription is mediated by light-induced association of the PEP-pTAC3 complex with promoters possibly through the light-dependent expression of σ-factors (Yagi et al., 2012). Thus, we investigated the molecular mechanism of light-dependent transcription by the HSP21-pTAC5 complex in chloroplasts under heat stress. If PEP is trapped at promoter regions in the dark, ChIP signals at PEP promoters would not decrease in dark-adapted leaves. However, our results show that ChIP signals at both promoters and coding regions of PEP-dependent photosynthesis and rRNA genes were reduced in dark-adapted seedlings under heat stress (Figure 9), suggesting that PEP dissociates from chloroplast genomic DNA in the dark. Therefore, it is likely that light regulates the association of the HSP21-pTAC5-PEP complex with the promoter region under heat stress possibly through the light-dependent expression of σ-factors.
pTAC5 residues 327 to 387 amino acids are very similar to the C4-type zinc finger of E. coli DnaJ (see Supplemental Figure 7 online). Moreover, pTAC5 has PDI activity (Figure 10). DnaJ proteins are molecular chaperones that specifically regulate ATP-dependent DnaK-like chaperones involved in protein folding, such as HSP70. Because pTAC5 lacks the J domain responsible for stimulating DnaK ATPase activity, it may function without HSP70. Plastid DnaJ domain proteins play diverse roles in chloroplast biogenesis, including plastid division and protein assembly. Well-studied plastid DnaJ domain proteins include ribulose-1,5-bis-phosphate carboxylase/oxygenase assembly factor BSD2 (Brutnell et al., 1999), cotyledon-specific chloroplast biogenesis factor CYO1 (Shimada et al., 2007), chloroplast division factor PARC6 (Glynn et al., 2009), and thylakoid and photosystem assembly factor SCO2 (Tanz et al., 2012). It should be noted that none of these plastid DnaJ domain proteins appears to be localized to chloroplast nucleoids. Thus, our results suggest that pTAC5 is a plastid DnaJ domain protein that acts as a chaperone-like factor for the PEP complex under heat stress.
What might be the function of pTAC5 in the PEP complex under heat stress? Recent studies have shown that the PEP complex is composed of the subunits of the PEP core and the components of TAC (Pfalz et al., 2006; Steiner et al., 2011; Majeran et al., 2012). As pTAC5 has PDI activity, we searched for the number of Cys residues of the subunits of the PEP core and some components of TAC (Pfalz et al., 2006). Interestingly, we found that the PEP core subunits and many TAC components have multiple or many Cys residues (see Supplemental Table 2 online). For example, there are 5, 11, 17, 17, 21, 16, and 15 Cys residues in RpoA, RpoB, RpoC1, RpoC2, polA, gyrA, and pTAC2, respectively. It is well known that heat stress often results in protein denaturation (Vierling, 1991) and sHSPs bind to partially folded or denatured proteins and prevent their aggregation under heat stress (Sun et al., 2002; Basha et al., 2012). Based on the results in this study, we tentatively propose that HSP21 may stabilize pTAC5 so that pTAC5 can function directly in disulfide bond formation and/or accelerating the folding of Cys-rich proteins in the PEP complex in order to maintain PEP-dependent plastid transcription under heat stress. However, this hypothesis needs further investigation.
It should be noted that heat stress resulted in enhanced association of pTAC5 with promoter regions of PEP-dependent genes in wild-type plants (Figures 8A and 8B), whereas heat stress did not significantly affect transcription rates of PEP-dependent genes (Figures 4B and 4C). Our results show that the transcript levels and the transcription rates of PEP-dependent genes were significantly decreased in RNAi pTAC5 lines under heat stress (Figures 7C to 7E). Our results and previous studies have demonstrated that pTAC5 is a component of plastid transcriptionally active chromosome proteins (see Supplemental Figure 4 online; Pfalz et al., 2006). These results indicate that pTAC5 plays an important role in maintaining PEP function under heat stress through enhancing association with promoter regions of PEP-dependent genes and stabilizing the proper structure of the PEP complex. Otherwise, there would be a decrease in the transcription rates of PEP-dependent genes under heat stress if there were no enhanced association of pTAC5 with promoter regions of PEP-dependent genes. This would explain why we observed enhanced association of pTAC5 with promoter regions of PEP-dependent genes in wild-type plants but with maintained transcription rates of PEP-dependent genes under heat stress.
In this study, we observed that there were no phenotypic differences between the wild type and the hsp21 and ptac5 mutants under normal temperature. Instead, as discussed above, HSP21 and pTAC5 are required for maintaining PEP function and chloroplast development only under heat stress. In addition, our results show that HSP21 was expressed only under heat stress and pTAC5 was induced significantly under heat stress, though it is also expressed under normal temperature (Figure 1C; see Supplemental Figures 1, 5, and 6 online). Inactivation of a fundamental component of the PEP transcription machinery should lead to a total block of PEP activity (Pfalz and Pfannschmidt, 2013). However, we did not observe a total block of PEP activity in hsp21 and ptac5. Therefore, it seems that HSP21 and pTAC5 are not fundamental components of the PEP complex. Instead, they are regulators of plastid transcription that maintain PEP function under heat stress.
Plastid transcription is mediated by bacterial-type PEP and phage-type NEP. Recent genomic and proteomic studies reveal that land plants have lost most prokaryotic nucleoid proteins involved in DNA packaging, replication, transcription, and translation but have acquired eukaryotic-type chloroplast nucleoid proteins during plant evolution (Sato, 2001; Pfalz et al., 2006; Steiner et al., 2011). These nucleoid proteins are involved in diverse processes of chloroplast biogenesis, including plastid gene expression (Pfalz et al., 2006; Gao et al., 2011; Yagi et al., 2012), the redox regulation of PEP function (Arsova et al., 2010), and protecting chloroplast nucleoids against oxidative stress (Myouga et al., 2008). Plants are often exposed to environmental stresses such as heat stress. Understandably, some pTAC proteins and PEP-associated proteins must have been acquired during plant evolution to protect the PEP-dependent transcription and maintain proper chloroplast development and plant growth under environmental stress conditions. Our results show that HSP21 and pTAC5 are conserved among land plants, but not in cyanobacteria and algae (see Supplemental Figures 9 and 10 online). In addition, with the exception of TrxZ, no orthologs of PAPs or of many TAC components have been found in Chlamydomonas reinhardtii (Pfalz and Pfannschmidt, 2013). Thus, our results suggest that HSP21 and pTAC5 are likely to be an evolutionary acquisition of terrestrial plants in order to maintain proper chloroplast development by the regulation of the PEP-dependent transcription under the environment with heat stress. Obviously, understanding the molecular functions of HSP21 and pTAC5 will provide insight into the mechanisms enabling environmental regulation of plastid transcription and also offer a unique way of revealing plant evolution.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana (ecotype Columbia-0) ethyl methanesulfonate mutation line CS85472 was obtained from the ABRC. This line was found to have three mutation sites: a DNA rearrangement in At2g26330 that encodes a putative receptor kinase ERECTA, a point mutation (414 bp relative to the ATG codon; G to A) in At4g27670 that encodes HSP21, and a synonymous mutation (678 bp relative to the ATG codon; G to C) in At2g46080 that encodes a protein related to BYPASS1. These three mutation sites were confirmed by PCR and sequencing of the full-length genomic DNA fragments of the three genes from the mutant CS85472 and wild-type plants (for primers used, see Supplemental Table 3 online). In order to obtain the HSP21 single mutation line, CS85472 was backcrossed to the wild type three times. The HSP21 single mutation line was verified by PCR, sequencing, immunoblot, complementation, and phenotype analyses (Figures 1 and 2). Single knockout fsd3 mutant (SALK_103228) was obtained from the ABRC (Myouga et al., 2008).
Arabidopsis seeds were surface-sterilized in 50% bleach for 15 min and washed five times with sterile double distilled water before plating on a Murashige and Skoog (MS) plate with 2% Suc and 0.8% agar. The seeds were stratified in the dark at 4°C for 2 d. After dark germinated for 2 d at 22°C, the seedlings were grown on MS medium plates in growth chambers at 30°C with a 16-h photoperiod (100 µmol photons m−2 s−1) and 50% humidity.
In order to investigate the effects of heat stress on the expression of pTAC5 gene and pTAC5 protein, the seedlings grown for 5 d at 22°C and 100 µmol photons m−2 s−1 were exposed to 30°C for different times (0 to 24 h) at 100 µmol photons m−2 s−1. To observe the response of pTAC5/pTAC5 to light during greening of etiolated seedlings, wild-type Arabidopsis plants were grown in continuous darkness for 5 d either at 22 or 30°C, and then the etiolated plants were exposed to light (100 µmol photons m−2 s−1) for 1, 3, and 6 h at their respective temperatures.
Isolation of Chloroplasts, Thylakoid Membranes, and Stroma Proteins
For each isolation procedure of chloroplasts, 30 Petri seedlings (5 d old) were used, which is equivalent to 4000 individuals. All the chloroplast isolation procedures were performed at 4°C. Cotyledons were homogenized for 3 to 4 s using a polytron (Kinematica PT10-35GT) with a small rotor (13-mm diameter, 40% max speed) in 20 mL isolation buffer (20 mM HEPES/KOH, pH 8.0, 0.3 M sorbitol, 5 mM MgCl2, 5 mM EGTA, 5 mM EDTA, and 10 mM NaHCO3) in a 50-mL beaker. The homogenate was filtered through a double layer of Miracloth and then centrifuged at 3000g for 3 min, and the pellet was resuspended in 1 mL isolation buffer. The resuspended chloroplasts were loaded onto a 20/40/80% v/v three-step Percoll gradient and centrifuged in a swing-out rotor at 3500g for 30 min. The intact chloroplasts appeared in the phase between 40 and 80% Percoll. The intact chloroplasts were recovered and washed by isolation buffer and then centrifuged at 3000g for 3 min. The pellet was isolated chloroplasts. Thylakoid membranes and stroma proteins were prepared from isolated intact chloroplasts according to Stöckel and Oelmüller (2004).
Immunoblot, SDS-PAGE, and BN-PAGE Analyses
Immunoblot, SDS-PAGE, and BN-PAGE analyses were performed according to our previous studies (Peng et al., 2006; Liu et al., 2012). For immunoblot analysis, total proteins were prepared and quantified as previously described (Ouyang et al., 2011). The isolated thylakoid pellets were suspended in resuspension buffer (25 mM BisTris-HCl, pH 7.0, 1% n-Dodecyl β-D-maltoside, and 20% glycerol [w/v]) at 1.0 mg chlorophyll mL−1. After incubation at 4°C for 5 min and centrifugation at 12,000g for 10 min, the supernatant was added with one-tenth volume of loading buffer (100 mM BisTris-HCl, pH 7.0, 0.5 M 6-amino-n-caproic acid, 5% Serva blue G, and 30% [w/v] glycerol) and applied to 0.75-mm-thick 4 to 12% acrylamide gradient gels in a Hoefer Mighty Small vertical electrophoresis unit connecting with a cooling circulator. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer for 30 min and layered onto 1-mm-thick 15% SDS polyacrylamide gels containing 6 M urea. After electrophoresis, the proteins were transferred to nitrocellulose membranes, probed with specific antibodies, and visualized by the enhanced chemiluminescence method. The PsaA and PsaN antibodies were purchased from Agrisera, and all other antibodies were produced in our laboratory (Peng et al., 2006).
Fluorescence Imaging of ROS
Fluorescence imaging of ROS was performed as described by Tang et al. (2012). The seedlings were incubated with 10 μM H2DCFDA in 10 mM Tris-HCl, pH 7.2, for 10 min. H2DCFDA and chlorophyll fluorescence images were captured by DMI-4500 fluorescence microscope equipped with a charge-coupled device camera (Leica).
Subcellular Localization of GFP and RFP Proteins
For subcellular localization of GFP protein, the full length of HSP21 or pTAC5 was subcloned into the pBI221-P35S-GFP vector with the GFP at C terminus. For subcellular localization of RFP protein, the full length of pTAC2 was subcloned into the pBI221-P35S-RFP vector with the RFP at C terminus (for primers used, see Supplemental Table 3 online). The constructs for nuclear, chloroplast, and mitochondria localization were constructed according to our previous study (Cai et al., 2009). The resulting fusion constructs and the empty vector were transformed into the protoplasts of Arabidopsis. GFP and RFP fluorescence of transiently transformed Arabidopsis protoplasts was observed by confocal scanning microscopy (LSM 510 Meta; Zeiss). For GFP, we used 488 and 509 nm for excitation and emission, respectively. For RFP, we used 585 and 608 nm for excitation and emission, respectively. For chlorophyll autofluorescence, we used 488 and 650 to 750 nm for excitation and emission, respectively. For double-labeled protoplasts, GFP and RFP tracks were switched line by line during scanning, while chlorophyll fluorescence track was set as an additional track and activated independently.
BiFC
BiFC assay was performed according to Walter et al. (2004). Full-length cDNAs or specific fragments of cDNAs were subcloned into pUC-SPYNE and pUC-SPYCE, and plasmids were cotransformed into protoplasts (for primers used for fusion constructs, see Supplemental Table 3 online). YFP fluorescence was imaged using a confocal laser scanning microscope (LSM 510 Meta).
Pull-Down and Coimmunoprecipitation Assays
Coimmunoprecipitation and pull down assays were performed basically according to our previous studies (Sun et al., 2007; Ouyang et al., 2011). The intact chloroplasts were solubilized with 50 mM HEPES, pH 7.5, 150 mM KCl, 5 mM MgCl2, 10 μM ZnSO4, and 1% (v/v) Triton X-100, and the supernatant was further used for coimmunoprecipitation assays. To exclude the possibility that the protein association identified by coimmunoprecipitation may result from DNA tethering with plastid nucleoids, isolated intact chloroplast was treated with DNase (10 units RQ1 DNase at 37°C for 30 min) prior to coimmunoprecipitation assays (Prikryl et al., 2008).
RNA Gel Blot and Polysome Association Analyses
RNA gel blot and polysome association analyses (for primers used, see Supplemental Table 3 online) were performed according to our previous study (Liu et al., 2012).
Chloroplast Run-on Transcription and Chloroplast ChIP Assays
Chloroplast run-on transcription was performed essentially as described in our previous study (Chi et al., 2010). Chloroplast ChIP was performed principally following the protocol described by Yagi et al. (2012). Isolated chloroplast pellets were suspended in 1 mL of chloroplast isolation buffer containing 1% (v/v) formaldehyde and incubated at 25°C for 10 min with rotation to cross-link protein-DNA. After incubation, 150 μL 1 M Gly was added to the chloroplasts and further incubated at 25°C for 5 min with rotation to stop the cross-linking reaction. Cross-linked chloroplasts were pelleted by centrifugation and washed with chloroplast isolation buffer. The cross-linked chloroplast pellets were suspended in lysis buffer (50 mM Tris-HCl, pH 7.6, 0.15 M NaCl, 1 mM EDTA, 1% [v/v] Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, and protease inhibitor mixture [Roche]). Subsequently, chloroplast DNA was sheared to 0.2 to 1 kb by sonication (30% output power, 15 times, 30 s with 30 s interval). The supernatant was collected by centrifugation at 20,000g, 4°C, for 10 min and incubated at 4°C with rotation for 1 h, followed by dilution with 2 mL of lysis buffer containing 2 μg mL−1 RNase. Diluted extracts (200 μL) were incubated with or without 5 μL antibodies for 2 h at 4°C with rotation, and then 20 μL protein A/G microbeads was added to the extracts. After 1 h incubation at 4°C with rotation, the microbeads were washed three times with lysis buffer, and the DNA-protein complex was recovered by 200 μL ice cold Gly elution buffer (0.1 M Gly, 0.5 M NaCl, 0.05% Tween 20, pH 2.8). For reverse cross-linking, 8 μL of 5 M NaCl and 2 μL 10 mg mL−1 Proteinase K were added to the elution fraction and 200 μL input sample and incubated at 65°C overnight. Immunoprecipitated DNA was purified with a PCR purification kit (Qiagen) according to the manufacturer’s instructions.
Glycerol Density Gradient Centrifugation
The analysis of chloroplast protein complex by glycerol density gradient centrifugation was performed according to Yagi et al. (2012). Isolated intact chloroplasts were solubilized at 2 mg chlorophyll mL−1 in lysis buffer (20 mM Tris-HCl, pH 7.4, 0.1% Triton X-100, 50 mM NaCl, 0.1 mM EDTA, 10% glycerol [v/v], and 1 mM PMSF) for 20 min on ice and were centrifuged at 20,000g for 15 min. The supernatant was layered onto a linear glycerol gradient (10 to 30% v/v) in 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 50 mM 6-aminohexanoic acid, 0.1 mM EDTA, 0.1% Triton X-100, and complete protease inhibitor cocktail (Roche) and separated by centrifugation at 320,000g (SW55Ti; Beckman) at 4°C for 16 h. Ten fractions were collected from top to bottom. After trichloroacetic acid precipitation, samples were separated by SDS-PAGE and detected with specific antibodies.
Assays of Insulin Disulfide Reduction and PDI Activity
The reduction of insulin (Sigma-Aldrich) was assayed by measuring the increase in absorbance at 650 nm (Holmgren, 1979). The reduced RNase A (Sigma-Aldrich) was prepared as described previously (Pigiet and Schuster, 1986). PDI activity was determined by measurement of the reactivation of reduced RNase A (Hasegawa et al., 2003).
RNA Isolation, cDNA Synthesis, RT-PCR, and Quantitative Real-Time RT-PCR
Procedures for the purification of total RNAs for cDNA synthesis, RT-PCR, and quantitative real-time RT-PCR (for primers used, see Supplemental Table 3 online) were performed according to our previous study (Chi et al., 2008). The amplification of 18S rrn was used as an internal control for normalization.
Affinity Chromatography of Proteins Associated with HSP21-His
Affinity chromatography of proteins associated with HSP21 was performed according to Peng et al. (2012). Total cotyledon proteins from wild-type and HSP21-His plants were generally mixed with 50 μL anti-His MicroBeads (Miltenyi Biotec). After incubation of the mixture for 30 min at 4°C, the beads were transferred to columns placed in a magnetic field. Columns were rinsed four times with 200 μL washing buffer I (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, and 0.1% SDS). After final washing with 200 μL washing buffer II (20 mM Tris-HCl, pH 7.5), total proteins were eluted with elution buffer (50 mM Tris-HCl, pH 6.8, 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromophenol blue, and 10% glycerol). The proteins were separated on 12.5% SDS-PAGE gels and stained with Coomassie Brilliant Blue. SDS-PAGE lanes were cut into several slices and analyzed by LC-MS/MS.
Peptide Preparation, Mass Spectrometry Analysis, and Database Search
Peptide preparation, LC-MS/MS analyses, and database search were performed as described previously (Peng et al., 2012). LC-MS/MS analyses were performed on a LTQ-Orbitrap XL-HTC-PAL system. MS/MS spectra were compared using the Mascot server (version 2.3.2) against TAIR8 (The Arabidopsis Information Resource), with the following search parameters: set-off threshold at 0.05 in the ion score cutoff; peptide tolerance, 10 ppm; MS/MS tolerance, 0.8 D; peptide charge, 2+ or 3+; trypsin as enzyme allowing up to one missed cleavage.
Antiserum Production
For the production of polyclonal antibodies against HSP21 and pTAC5, the nucleotide sequences encoding the soluble part of HSP21 (amino acids 61 to 227) and the soluble part of pTAC5 (amino acids 151 to 313) were amplified from cDNA (for primers used, see Supplemental Table 3 online). The resulting DNA fragments were fused in frame with the N-terminal His affinity tag of pET28a, and the resulting plasmids were transformed into Escherichia coli strain BL21 (DE3). The fusion proteins were purified on a nickel-nitrilotriacetic acid agarose resin matrix and raised in rabbit with purified antigen. The dilution ratios for both antibodies against HSP21 and pTAC5 in immunoblot analyses were 1:1000.
HSP21 Promoter Construction and GUS Staining
The PHSP21:GUS was made by amplifying the 2-kb sequence upstream of the HSP21 translation start sites and subcloning the fragment into pCAMBIA 1301 binary vector (for primers used, see Supplemental Table 3 online). The different tissues of the transgenic lines were harvested and incubated in staining solution (50 mM sodium phosphate buffer, pH 7.2, 0.2% Triton X-100, 10 mM potassium ferrocyanide, 10 mM potassium ferricyanide, and 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid, cyclohexylammonium salt) at 37°C overnight. Samples were then washed in 70% ethanol before photographs were taken.
RNAi and Complementation of the hsp21 Mutant
For RNAi vector construction, short sequences of Arabidopsis pTAC5 were cloned into the pHANNIBAL vector (Wesley et al., 2001) between the XbaI-BamHI sites in sense orientation and between the XhoI-KpnI sites in antisense orientation (for primers used, see Supplemental Table 3 online). The expression cassette was excised with NotI and cloned into the NotI site of the binary vector pART27. To complement hsp21, the full-length At4g27670 coding sequence was subcloned into the pCAMBIA1301 vector under the control of P35S. The resultant construct was transformed into Agrobacterium tumefaciens GV3101 strain and introduced into hsp21 plants. Individual transgenic plants were selected on the basis of resistance to 50 mg L−1 hygromycin in half-strength MS medium and 0.8% agar. The resistant ones were transferred to soil and grown in the growth chamber to produce seeds. The success of complementation was confirmed by phenotypic analyses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: HSP21 (AT4G27670), pTAC2 (AT1G74850), pTAC5 (AT4G13670), pTAC12 (AT2G34640), FSD3 (AT5G23310), RpoA (ATCG00740), RpoB (ATCG00190), AccD (ATCG00500), ClpP (ATCG00670), NdhB (ATCG00890), PsaA (ATCG00350), PsaE (AT2G20260), PsaH (AT1G52230), PsaN (AT5G64040), LHCII (AT1G29920), PsbA/D1 (ATCG00020), D2 (ATCG00270), CP43 (ATCG00280), CP47 (ATCG00680), PsbO (AT5G66570), Cytf (ATCG00540), CF1β (ATCG00480), RbcL (ATCG00490), ERECTA (AT2G26330), Tubulin (AT1G04820), and FNR (AT1G20020).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Patterns of HSP21.
Supplemental Figure 2. RNA Gel Blot and Polysome Association Analyses in the Wild Type and the Mutants.
Supplemental Figure 3. Affinity Chromatography of Proteins Associated with HSP21.
Supplemental Figure 4. Subcellular Localization of pTAC5.
Supplemental Figure 5. Identification and Phenotypes of Transgenic Plants (ptac5-1, ptac5-2, and ptac5-3) with Reduction of pTAC5.
Supplemental Figure 6. Expression Patterns of pTAC5.
Supplemental Figure 7. Putative Topology of the Zinc Finger Motif in pTAC5.
Supplemental Figure 8. Accumulation of ROS in Wild Type, ptac5, and hsp21.
Supplemental Figure 9. Alignment of the Amino Acid Sequences of HSP21 and Its Homolog Proteins.
Supplemental Figure 10. Alignment of the Amino Acid Sequences of pTAC5 and Its Homolog Proteins.
Supplemental Table 1. LC-MS/MS Based Identification of pTAC5 in the Copurified Protein from His-Tagged HSP21.
Supplemental Table 2. Number of Cys Residues in the Subunits of TAC.
Supplemental Table 3. List of Primers Used in This Study.
Acknowledgments
We thank for the ABRC and RIKEN for the seed stocks. This work was supported by the State Key Basic Research and Development Plan of China (2009CB118503) and the Solar Energy Initiative of the Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
L.Z. and C.L. designed the study. L.Z. and W.Z. performed the research. L.Z., H.W., S.D., Q.L., X.W., L.P., L.X.Z., and C.L. analyzed the data. L.Z. and C.L. wrote the article.
Glossary
- sHSP
small heat shock protein
- PSII
photosystem II
- PEP
plastid-encoded RNA polymerase
- NEP
nuclear-encoded RNA polymerase
- pTAC
plastid transcriptionally active chromosome
- GUS
β-glucuronidase
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- BiFC
bimolecular fluorescence complementation
- RNAi
RNA interference
- cpChIP
chloroplast chromatin immunoprecipitation
- BN
blue native
- PDI
protein disulfide isomerase
- ROS
reactive oxygen species
- H2DCFDA
2,7-dichlorodihydrofluorescein diacetate
- MS
Murashige and Skoog
References
- Allison L.A. (2000). The role of sigma factors in plastid transcription. Biochimie 82: 537–548 [DOI] [PubMed] [Google Scholar]
- Allison L.A., Simon L.D., Maliga P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Anfinsen C.B., Scheraga H.A. (1975). Experimental and theoretical aspects of protein folding. Adv. Protein Chem. 29: 205–300 [DOI] [PubMed] [Google Scholar]
- Arsova B., Hoja U., Wimmelbacher M., Greiner E., Ustün S., Melzer M., Petersen K., Lein W., Börnke F. (2010). Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A.J., Hilton G.R., Lioe H., Bagnéris C., Benesch J.L., Kay L.E. (2011). Quaternary dynamics of αB-crystallin as a direct consequence of localised tertiary fluctuations in the C-terminus. J. Mol. Biol. 413: 310–320 [DOI] [PubMed] [Google Scholar]
- Basha E., O’Neill H., Vierling E. (2012). Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 37: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce Cahoon A., Stern D.B. (2001). Plastid transcription: A menage à trois? Trends Plant Sci. 6: 45–46 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Sawers R.J., Mant A., Langdale J.A. (1999). BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Ji D., Peng L., Guo J., Ma J., Zou M., Lu C., Zhang L. (2009). LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A.L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L.F., Lurin C., Small I., León P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Chauhan H., Khurana N., Nijhavan A., Khurana J.P., Khurana P. (2012). The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 35: 1912–1931 [DOI] [PubMed] [Google Scholar]
- Chen Q., Vierling E. (1991). Analysis of conserved domains identifies a unique structural feature of a chloroplast heat shock protein. Mol. Gen. Genet. 226: 425–431 [DOI] [PubMed] [Google Scholar]
- Cheng M.C., Wu S.P., Chen L.F., Chen S.C. (1997). Identification and purification of a spinach chloroplast DNA-binding protein that interacts specifically with the plastid psaA-psaB-rps14 promoter region. Planta 203: 373–380 [DOI] [PubMed] [Google Scholar]
- Chi W., Ma J., Zhang D., Guo J., Chen F., Lu C., Zhang L. (2008). The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Mao J., Li Q., Ji D., Zou M., Lu C., Zhang L. (2010). Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 64: 14–25 [DOI] [PubMed] [Google Scholar]
- De Santis-MacIossek G., Kofer W., Bock A., Schoch S., Maier R.M., Wanner G., Rüdiger W., Koop H.U., Herrmann R.G. (1999). Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: Molecular biology, biochemistry and ultrastructure. Plant J. 18: 477–489 [DOI] [PubMed] [Google Scholar]
- Gao Z.P., Yu Q.B., Zhao T.T., Ma Q., Chen G.X., Yang Z.N. (2011). A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.Y., Osteryoung K.W. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59: 700–711 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P.T., Allison L.A., Maliga P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härndahl U., Hall R.B., Osteryoung K.W., Vierling E., Bornman J.F., Sundby C. (1999). The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones 4: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa G., Suwa M., Ichikawa Y., Ohtsuka T., Kumagai S., Kikuchi M., Sato Y., Saito Y. (2003). A novel function of tissue-type transglutaminase: Protein disulphide isomerase. Biochem. J. 373: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005). Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Heckathorn S.A., Downs C.A., Sharkey T.D., Coleman J.S. (1998). The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol. 116: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W.R., Prombona A., Fieder B., Subramanian A.R., Börner T. (1993). Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (1979). Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254: 9627–9632 [PubMed] [Google Scholar]
- Kim K.H., Alam I., Kim Y.G., Sharmin S.A., Lee K.W., Lee S.H., Lee B.H. (2012). Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol. Lett. 34: 371–377 [DOI] [PubMed] [Google Scholar]
- Krause K., Maier R.M., Kofer W., Krupinska K., Herrmann R.G. (2000). Disruption of plastid-encoded RNA polymerase genes in tobacco: Expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol. Gen. Genet. 263: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Kuromori T., Hirayama T., Kiyosue Y., Takabe H., Mizukado S., Sakurai T., Akiyama K., Kamiya A., Ito T., Shinozaki K. (2004). A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Lambert W., Koeck P.J., Ahrman E., Purhonen P., Cheng K., Elmlund D., Hebert H., Emanuelsson C. (2011). Subunit arrangement in the dodecameric chloroplast small heat shock protein Hsp21. Protein Sci. 20: 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.J., Roseman A.M., Saibil H.R., Vierling E. (1997). A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J., Kemp S., Krause K., Profanter B., Herrmann R.G., Maier R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011). Function of plastid sigma factors in higher plants: Regulation of gene expression or just preservation of constitutive transcription? Plant Mol. Biol. 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., Lu C. (2012). PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24: 4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. (1998). Two plastid RNA polymerases of higher plants: An evolving story. Trends Plant Sci. 3: 4–6 [Google Scholar]
- McHaourab H.S., Godar J.A., Stewart P.L. (2009). Structure and mechanism of protein stability sensors: Chaperone activity of small heat shock proteins. Biochemistry 48: 3828–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F., Hosoda C., Umezawa T., Iizumi H., Kuromori T., Motohashi R., Shono Y., Nagata N., Ikeuchi M., Shinozaki K. (2008). A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neta-Sharir I., Isaacson T., Lurie S., Weiss D. (2005). Dual role for tomato heat shock protein 21: Protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 17: 1829–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M., Li X., Ma J., Chi W., Xiao J., Zou M., Chen F., Lu C., Zhang L. (2011). LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat. Commun. 2: 277. [DOI] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Shikanai T. (2012). Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24: 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Liere K., Kandlbinder A., Dietz K.J., Oelmüller R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Pfannschmidt T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18: 186–194 [DOI] [PubMed] [Google Scholar]
- Pigiet V.P., Schuster B.J. (1986). Thioredoxin-catalyzed refolding of disulfide-containing proteins. Proc. Natl. Acad. Sci. USA 83: 7643–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2008). A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N. (2001). Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 6: 151–155 [DOI] [PubMed] [Google Scholar]
- Serino G., Maliga P. (1998). RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 117: 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeel S., Haq N.U., Heckathorn S.A., Hamilton E.W., Luthe D.S. (2011). Ecotypic variation in chloroplast small heat-shock proteins and related thermotolerance in Chenopodium album. Plant Physiol. Biochem. 49: 898–908 [DOI] [PubMed] [Google Scholar]
- Shi Y.Y., Tang W., Hao S.F., Wang C.C. (2005). Contribution of cysteine residues in Zn2 to zinc fingers and thiol-disulfide oxidoreductase activities of chaperone Dna. J. Biochem. 44: 1683–1689 [DOI] [PubMed] [Google Scholar]
- Shimada H., Mochizuki M., Ogura K., Froehlich J.E., Osteryoung K.W., Shirano Y., Shibata D., Masuda S., Mori K., Takamiya K. (2007). Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19: 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M., Gernhard S., von Koskull-Döring P., Vierling E., Scharf K.D. (2008). The plant sHSP superfamily: Five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D., Maliga P. (1998). Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet. 33: 340–344 [DOI] [PubMed] [Google Scholar]
- Steiner S., Schröter Y., Pfalz J., Pfannschmidt T. (2011). Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel F., Baldwin A.J., Painter A.J., Jaya N., Basha E., Kay L.E., Vierling E., Robinson C.V., Benesch J.L. (2010). Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. USA 107: 2007–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J., Oelmüller R. (2004). A novel protein for photosystem I biogenesis. J. Biol. Chem. 279: 10243–10251 [DOI] [PubMed] [Google Scholar]
- Sun W., Van Montagu M., Verbruggen N. (2002). Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. (2007). Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., MacRae T.H. (2005). Small heat shock proteins: Molecular structure and chaperone function. Cell. Mol. Life Sci. 62: 2460–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J.Y., Sriraman P., Svab Z., Maliga P. (2003). Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell 15: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J.Y., Ytterberg A.J., Beardslee T.A., Allison L.A., Wijk K.J., Maliga P. (2004). Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 40: 164–172 [DOI] [PubMed] [Google Scholar]
- Tang W., Wang C.C. (2001). Zinc fingers and thiol-disulfide oxidoreductase activities of chaperone Dna. J. Biochem. 40: 14985–14994 [DOI] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., Lin R. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz S.K., Kilian J., Johnsson C., Apel K., Small I., Harter K., Wanke D., Pogson B., Albrecht V. (2012). The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J. 69: 743–754 [DOI] [PubMed] [Google Scholar]
- Vierling E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 579–620 [Google Scholar]
- Volkov R.A., Panchuk I.I., Schöffl F. (2005). Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol. Biol. 57: 487–502 [DOI] [PubMed] [Google Scholar]
- Wadhwa R., Ryu J., Gao R., Choi I.K., Morrow G., Kaur K., Kim I., Kaul S.C., Yun C.O., Tanguay R.M. (2010). Proproliferative functions of Drosophila small mitochondrial heat shock protein 22 in human cells. J. Biol. Chem. 285: 3833–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang D., Luthe D.S. (2003). Heat sensitivity in a bentgrass variant. Failure to accumulate a chloroplast heat shock protein isoform implicated in heat tolerance. Plant Physiol. 133: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E.R., Aevermann B.D., Sanders-Reed Z. (2008). Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones 13: 127–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yagi Y., Ishizaki Y., Nakahira Y., Tozawa Y., Shiina T. (2012). Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. USA 109: 7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]