This work identifies molecular components of Arabidopsis AP-2 complex, a heterotetramer, which sorts cargo proteins into clathrin-coated endocytic vesicles. Transient formation of punctate structures of AP-2 on the plasma membrane is visualized. AP-2 is required for proper floral organ development and efficient pollination.
Abstract
The adaptor protein-2 (AP-2) complex is a heterotetramer involved in clathrin-mediated endocytosis of cargo proteins from the plasma membrane in animal cells. The homologous genes of AP-2 subunits are present in the genomes of plants; however, their identities and roles in endocytic pathways are not clearly defined in plants. Here, we reveal the molecular composition of the AP-2 complex of Arabidopsis thaliana and its dynamics on the plasma membrane. We identified all of the α-, β-, σ-, and μ-subunits of the AP-2 complex and detected a weak interaction of the AP-2 complex with clathrin heavy chain. The μ-subunit protein fused to green fluorescent protein (AP2M-GFP) was localized to the plasma membrane and to the cytoplasm. Live-cell imaging using a variable-angle epifluorescence microscope revealed that AP2M-GFP transiently forms punctate structures on the plasma membrane. Homozygous ap2m mutant plants exhibited abnormal floral structures, including reduced stamen elongation and delayed anther dehiscence, which led to a failure of pollination and a subsequent reduction of fertility. Our study provides a molecular basis for understanding AP-2–dependent endocytic pathways in plants and their roles in floral organ development and plant reproduction.
INTRODUCTION
Membrane trafficking is vital for developmental and physiological processes in eukaryotic cells. Cargo proteins must be captured by specific adaptor proteins that mediate sorting and uptake into transport vesicles. One of the best-characterized adaptors is the adaptor protein (AP) complex, a heterotetramer containing two large subunits, one medium subunit, and one small subunit, which is found in all eukaryotes including mammals, yeast, nematodes, and flies (Robinson, 2004). There are five types of AP complexes (AP-1 to AP-5), which are involved in different pathways. AP-1 is involved in trafficking between the trans-Golgi network (TGN) and endosomes (Hirst et al., 2012). AP-2 is involved in clathrin-mediated endocytosis from the plasma membrane (McMahon and Boucrot, 2011). AP-3 is involved in protein trafficking from TGN/endosomes to vacuole/lysosomes (Dell’Angelica, 2009). AP-4 is involved in TGN-to-endosome trafficking for specific cargo proteins, such as amyloid precursor protein (Burgos et al., 2010). The recently identified AP-5 complex is localized to late endosomes (Hirst et al., 2011). The Arabidopsis thaliana genome contains all five of the putative AP genes (Bassham et al., 2008; Hirst et al., 2011); however, our knowledge of their roles in membrane trafficking and physiological function is still limited. Arabidopsis AP-2 has the potential to interact with a vacuolar sorting receptor (Happel et al., 2004). AP-3 plays roles in post-Golgi trafficking and is involved in the regulation of vacuolar biogenesis (Niihama et al., 2009; Feraru et al., 2010; Zwiewka et al., 2011). AP-1 is required for trafficking of the cytokinesis-specific soluble N-ethylmaleimide–sensitive attachment protein receptor (SNARE) KNOLLE to the cell plate, which mediates proper cell division (Park et al., 2013; Teh et al., 2013). AP-1 also influences vascular development in the aerial tissues (Wang et al., 2013).
In mammalian cells, AP-2 is essential for the formation of clathrin-coated vesicles (CCVs) and acts as a major hub of interactions between molecular components, including clathrin (McMahon and Boucrot, 2011). Depletion of AP-2 by RNA interference severely reduces CCV accumulation on the plasma membrane (Motley et al., 2003; Boucrot et al., 2010). The subunits of AP-2 interact with the sorting signals of cargo proteins for selective incorporation into CCV. The sorting mechanism is best characterized for the medium (μ) subunit, which is known to recognize the YXXΦ motif (Φ represents Leu, Ile, Phe, Met, or Val) that is present in the cytosolic domains of cargo proteins (Ohno et al., 1995). Mutations of the YXXΦ motif abolish the interaction with μ (Ohno et al., 1995; Boll et al., 1996; Stephens et al., 1997) and alter the subcellular localization of the cargo proteins (Bos et al., 1993; Humphrey et al., 1993). The interaction of μ with cargo proteins through the YXXΦ motif is disrupted by the structural Tyr analog Tyrphostin A23 (TyrA23), which is an efficient inhibitor of clathrin-mediated endocytosis (Banbury et al., 2003). Evidence for the importance of AP-2 during development is suggested by studies of mutants with disrupted subunit genes. Homozygous knockout mice that lack the AP-2 μ-subunit exhibit an embryonic lethal phenotype at an early stage of development (Mitsunari et al., 2005). Drosophila melanogaster embryos lacking the AP-2 α-subunit exhibit the termination of synaptic vesicle recycling, which leads to larval lethality before hatching (González-Gaitán and Jäckle, 1997).
Recent studies suggest that AP-2 is involved in endocytosis for the regulation of signaling and transport events in plants. Treatment with TyrA23 inhibits internalization of the PIN-FORMED auxin transporters and the water channel PLASMA MEMBRANE INTRINSIC PROTEIN2 (Dhonukshe et al., 2007), the iron transporter IRON-REGULATED TRANSPORTER1 (Barberon et al., 2011), the plant-specific endocytic SNARE VESICLE-ASSOCIATED MEMBRANE PROTEIN727 (Ebine et al., 2011), and the ligand-activated brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1 (Irani et al., 2012). Amino acid substitutions of the YXXΦ motif eliminate polar localization of the Arabidopsis boron transporter REQUIRES HIGH BORON1 in the plasma membrane of root tip cells (Takano et al., 2010). The YXXΦ motif is also present in the cytoplasmic domain of the following two leucine-rich repeat proteins involved in the plant immune response to pathogens: Ve2, which is involved in fungal race-specific resistance in tomato (Solanum lycopersicum) (Kawchuk et al., 2001), and EFR, which is a leucine-rich repeat receptor for bacterial elongation factor EF-Tu (Zipfel et al., 2006).
Although evidence for the function of the plant AP-2 complex is accumulating, little is known about its molecular characteristics and physiological roles. In this study, we investigated the molecular composition, intracellular localization, and dynamics of the Arabidopsis AP-2 complex. Mutants lacking the AP-2 μ-subunit exhibited multiple defects in plant development and physiological functions, including fertility and floral organ development. Our results provide valuable insight into the role of AP-2 during plant growth and development.
RESULTS
Arabidopsis AP2M Localizes at the Plasma Membrane in a TyrA23-Dependent Manner
The Arabidopsis genome contains AP-2 subunit homologs: two genes for each of the large subunits (α and β) and single genes for the medium subunit (μ) and the small subunit (σ) (Boehm and Bonifacino, 2001; Bassham et al., 2008). To be consistent with the nomenclature for AP complexes of other organisms, the following nomenclature is used for the Arabidopsis genes: ADAPTOR PROTEIN-2 ALPHA-ADAPTIN1 (AP2A1) and AP2A2 for α-subunits; ADAPTOR PROTEIN-1/2 BETA-ADAPTIN1 (AP1/2B1) and AP1/2B2 for β-subunits; ADAPTOR PROTEIN-2 MU-ADAPTIN (AP2M) for the μ-subunit; and ADAPTOR PROTEIN-2 SIGMA-ADAPTIN (AP2S) for the σ-subunit. AP2M was formerly denoted as μA (Happel et al., 2004).
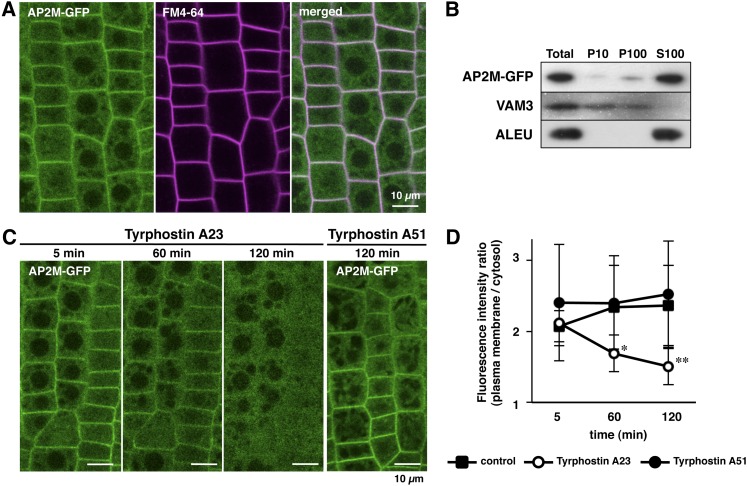
To reveal the subcellular localization of AP2M, AP2M-GFP ap2m transformant plants expressing the AP2M protein fused to green fluorescent protein (GFP) under the control of the endogenous promoter in the ap2m mutant background were generated. In these transformant plants, the ap2m mutant phenotype was complemented (described below), suggesting that the AP2M-GFP fusion protein is functional and behaves similarly to its endogenous counterpart. Confocal laser scanning microscopy revealed that, in the root tip cells of AP2M-GFP ap2m plants, the fluorescence of AP2M-GFP colocalized with FM4-64, a fluorescent lipophilic dye that labels the plasma membrane (Ueda et al., 2001; Dhonukshe et al., 2007), and was dispersed throughout the cytosol (Figure 1A). Colocalization of AP2M-GFP fluorescence with FM4-64 was also observed in the cotyledon epidermal cells of AP2M-GFP ap2m plants (see Supplemental Figure 1 online). Subcellular fractionation of AP2M-GFP ap2m seedlings revealed that a major part of the AP2M-GFP protein was found in the soluble fraction (S100), which accumulated the vacuolar Cys protease aleurain (ALEU) (Rogers et al., 1997; Watanabe et al., 2004), whereas a minor part of the AP2M-GFP protein was detected in the microsome fraction (P100), which accumulated the vacuolar SNARE protein VAM3 (Sato et al., 1997) (Figure 1B). These results suggest that the AP2M protein is predominantly accumulated in the cytosol and is also localized to the plasma membrane.
Figure 1.
AP2M-GFP Is Localized at the Plasma Membrane in a TyrA23-Dependent Manner.
(A) Confocal images of the root tip cells of 4-d-old AP2M-GFP ap2m seedling expressing the AP2M-GFP transgene under the control of the endogenous promoter in the homozygous ap2m mutant background. The plasma membrane is stained with FM4-64.
(B) Immunoblot analysis of subcellular fractions from AP2M-GFP ap2m seedlings. Total lysates (Total), 10,000g pellet (P10), 100,000g pellet (P100, microsome fraction), and 100,000g supernatant (S100, soluble fraction) from the AP2M-GFP ap2m seedlings were immunoblotted with anti-GFP (AP2M-GFP), anti-VAM3 (VAM3), and anti-ALEU (ALEU) antibodies.
(C) Confocal images of the AP2M-GFP fluorescence from the root tip cells of AP2M-GFP ap2m seedlings incubated with TyrA23 and TyrA51. The images were obtained at the indicated time points after incubation with the tyrphostins.
(D) Time course of the ratio of AP2M-GFP fluorescence intensity of plasma membrane to cytosol during incubation of the AP2M-GFP ap2m seedlings with TyrA23 and TyrA51. The means + sd from five root tip cells of three different seedlings (total 15 cells) are shown for each treatment; *P < 10−3 versus control and TyrA51 treatment and **P < 10−4 versus control and TyrA51 treatment, t test.
In mammalian cells, the AP-2 complex is recruited to the plasma membrane to interact with cargo proteins during CCV formation (McMahon and Boucrot, 2011). To examine whether the plasma membrane localization of AP2M-GFP is dependent on the interaction with cargo proteins, the root tip cells of the AP2M-GFP ap2m seedlings were incubated with the clathrin-mediated endocytosis inhibitor TyrA23. Tyrphostin A51 (TyrA51), which has no effect on the interaction between AP-2 and cargo proteins (Banbury et al., 2003), was used as the negative control. TyrA23 significantly reduced the AP2M-GFP fluorescence at the plasma membrane in a time-dependent manner, although the fluorescence at the plasma membrane was not altered by TyrA51 even after the 2-h incubation (Figure 1C). Quantitative analysis demonstrated that the ratio of the AP2M-GFP fluorescence at the plasma membrane to the cytosol was significantly reduced by the TyrA23 treatment compared with the mock and TyrA51 treatments (Figure 1D). These results indicate that the TyrA23 treatment selectively disrupts AP2M localization to the plasma membrane through interrupting the interaction with cargo proteins.
AP2M Transiently Forms Punctate Structures on the Plasma Membrane
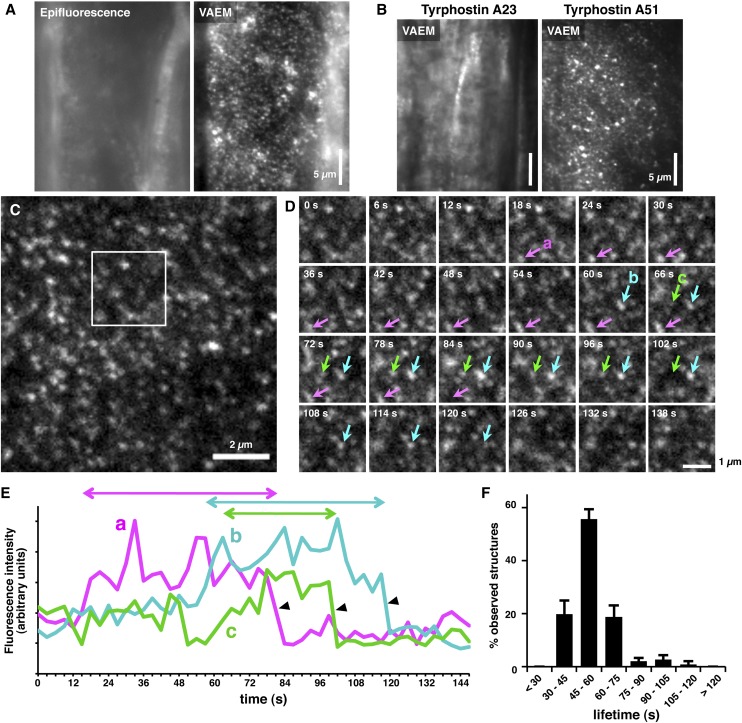
Variable-angle epifluorescence microscopy (VAEM) is a powerful tool to visualize the dynamics of CCV components, including clathrin light chain (CLC) and dynamin-related proteins on the plasma membrane (Fujimoto et al., 2007, 2010; Konopka et al., 2008; Konopka and Bednarek, 2008). To reveal the structure and dynamics of AP2M on the plasma membrane, root cells of AP2M-GFP ap2m plants were analyzed by VAEM. By the perpendicular incidence of the laser to the root cell surface, the AP2M-GFP fluorescence from the cytosol was mainly observed (Figure 2A, epifluorescence). When the incidence angle was changed within the highly oblique yet subcritical angle range against the cell surface, the punctate structures of AP2M-GFP became illuminated (Figure 2A, VAEM). These punctate structures were eliminated after treatment with TyrA23, but not with TyrA51 (Figure 2B), suggesting that the structures are CCV containing the AP2M-GFP protein on the plasma membrane. The punctate structures of AP2M-GFP appeared on the surface of cotyledon epidermal cells at a density of 3.0 ± 0.3 structures per 1 μm2 (n = 9) during 147 s (Figure 2C). Time-lapse imaging demonstrated that the AP2M-GFP structures appeared on the cell surface in a transient manner (Figure 2D; see Supplemental Movie 1 online). Their fluorescence intensities gradually increased and then rapidly decreased (Figures 2D and 2E; see Supplemental Movie 1 online). Quantitative analysis of the fluorescence intensity revealed that the average lifetime of the AP2M-GFP structures was 54.5 s, and ∼75% of the structures disappeared from the cell surface within 1 min (Figure 2F). These observations suggest that AP2M transiently forms the punctate structures of CCV on the plasma membrane.
Figure 2.
AP2M-GFP Forms Transient Punctate Structures at the Plasma Membrane.
(A) Epifluorescence and VAEM images of an identical surface area of the root epidermal cells of AP2M-GFP ap2m seedling.
(B) VAEM images of the surface of root epidermal cells of AP2M-GFP ap2m seedlings incubated with TyrA23 and TyrA51 for 2 h.
(C) VAEM image of the surface of a cotyledon epidermal cell of AP2M-GFP ap2m seedlings.
(D) Time series of the squared area in (C). Each of the frames was photographed at the indicated time points. Arrows indicate the transiently observed AP2M-GFP punctate structures.
(E) Profiles of the fluorescence intensity of the AP2M-GFP punctate structures indicated by the arrows in (D). Two-way arrows indicate the lifetimes of each structure. Arrowheads indicate the rapid decrease of fluorescence intensity observed in (D).
(F) Distribution of the lifetime of the AP2M-GFP punctate structures. The means + sd from >200 structures of three different epidermal cells (total 889 structures) are shown.
Identification of the Arabidopsis AP-2 Complex
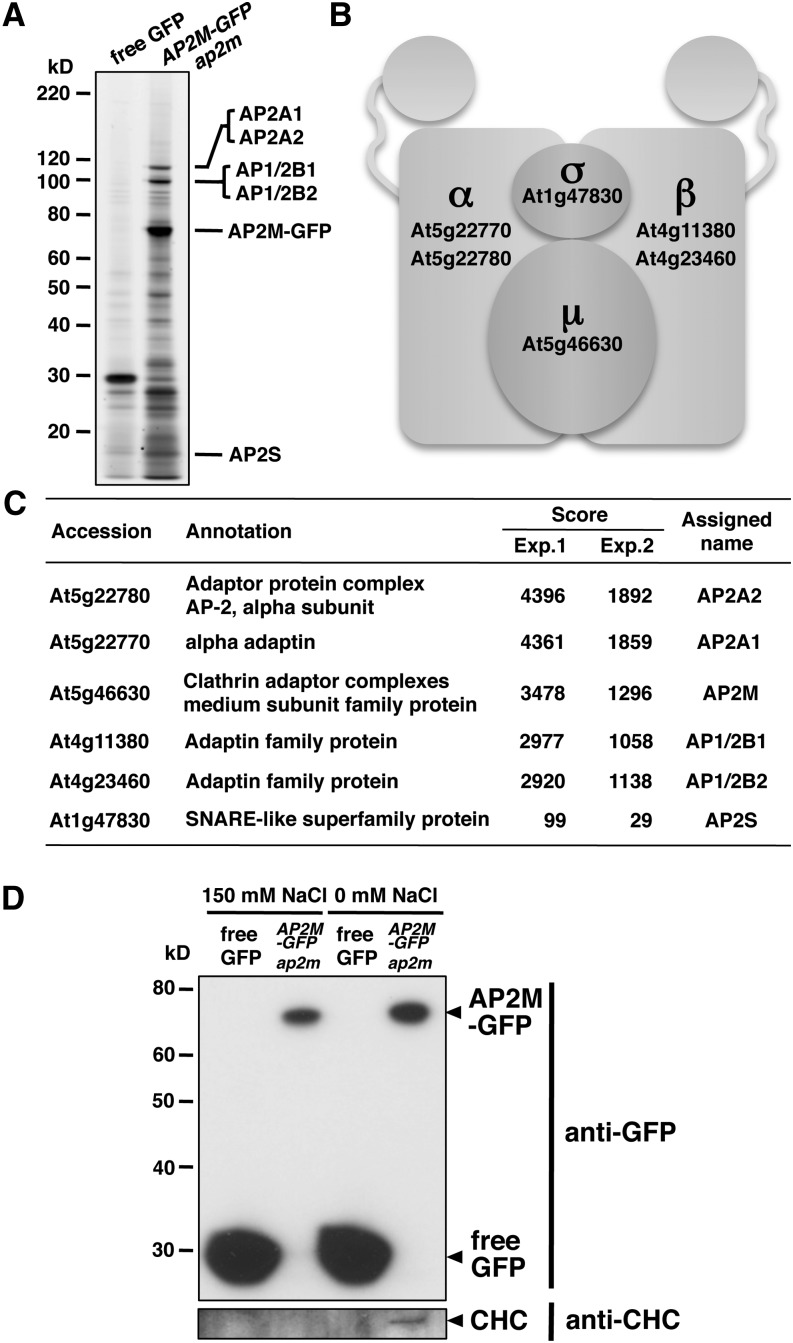
The Arabidopsis genome contains two homologous genes for each of the α and β subunits (Boehm and Bonifacino, 2001; Bassham et al., 2008; Dacks et al., 2008). This suggests that multiple sets of the different gene products are used to assemble the Arabidopsis AP-2 complex. However, its molecular composition has not been revealed experimentally. To identify which gene products are present in the AP-2 complex, anti-GFP antibody was used to immunoprecipitate detergent-soluble proteins from AP2M-GFP ap2m plants. Transgenic Arabidopsis plants expressing GFP were used as a control. The immunoprecipitated proteins were separated by SDS-PAGE and visualized by fluorescent staining. This revealed the isolation of proteins specifically bound to AP2M-GFP, which were not present in the control (Figure 3A). Mass spectrometric analysis of the immunoprecipitated proteins (see Supplemental Tables 1 and 2 online) repeatedly identified AP2A1 (α), AP2A2 (α), AP1/2B1 (β), AP1/2B2 (β), AP2M (μ), and AP2S (σ) (Figure 3C). This suggests that all of the subunits encoded in the Arabidopsis genome are expressed and physically associated with each other to form the AP-2 complex in planta. Simultaneous detection of the two α- and two β-subunits suggests that multiple molecular species of the AP-2 complex composed of different α and β homologous subunits are present in Arabidopsis (Figure 3B).
Figure 3.
Identification of the Arabidopsis AP-2 Complex.
(A) Fluorescent gel image of immunoprecipitates with an anti-GFP antibody from control (free GFP) and AP2M-GFP ap2m seedlings. Predicted molecular sizes of the AP-2 subunits are indicated.
(B) Schematic representation of the Arabidopsis AP-2 complex. Accession numbers of the genes corresponding to the subunits identified in (C) are indicated in boldface.
(C) The Arabidopsis AP-2 complex subunits identified by mass spectrometry. The accession numbers and annotations were obtained from The Arabidopsis Information Resource database (http://www.Arabidopsis.org). Scores from the two independent experiments were calculated by Mascot (Matrix Science).
(D) Immunoblot analysis of immunoprecipitates with an anti-GFP antibody from control (free GFP) and AP2M-GFP ap2m seedlings. The immunoprecipitates were prepared in the presence (150 mM NaCl) and absence of NaCl and immunoblotted with anti-GFP and anti-CHC antibodies.
In mammalian cells, the AP-2 complex interacts with clathrin to form CCV (McMahon and Boucrot, 2011). The clathrin heavy chain (CHC) was detected in the first mass spectrometric analysis of the immunoprecipitated proteins from the AP2M-GFP ap2m plants (see Supplemental Table 1 online). Notably, the AP2M-GFP protein was efficiently coimmunoprecipitated with CHC in the absence of salt, whereas it was not in the presence of 150 mM NaCl (Figure 3D). These results suggest that the AP-2 complex is weakly associated with CHC as a component of CCV.
AP2M Is Involved in Floral Organ Development
To investigate the roles of AP-2 in plant growth and development, homozygous ap2m mutant plants were used for further molecular analysis and morphological dissection. Sequence analysis demonstrated that the ap2m mutant plants harbor a T-DNA insertion in the seventh exon of the AP2M gene (Figure 4A). RT-PCR analysis revealed that homozygous ap2m mutant plants accumulated the 5′ half region of the AP2M transcripts, but neither the full-length nor the 3′half transcripts (Figure 4B). This implies that the ap2m mutant plants might accumulate a truncated from of the AP2M protein lacking the C-terminal region (the red underlined sequence in Supplemental Figure 2 online).
Figure 4.
Homozygous ap2m Mutation Impairs Multiple Aspects of Plant Development.
(A) Schematic representation of the structure of the AP2M gene and its predicted transcript. For the gene structure, closed boxes and solid lines indicate exons and introns, respectively, and a solid line with triangle indicates the position of the inserted T-DNA fragment. For the gene transcript, bold lines indicate the regions of the AP2M gene transcript amplified by RT-PCR shown in (B).
(B) RT-PCR analysis of AP2M gene transcripts isolated from 10-d-old seedlings of wild-type (WT), homozygous ap2m mutant, and AP2M-GFP ap2m plants. The agarose gel images were obtained by ethidium bromide staining.
(C) Petioles are shorter in 4-week-old homozygous ap2m mutant plants than in wild-type or AP2M-GFP ap2m plants.
(D) The homozygous ap2m mutant plant exhibits severely reduced fertility with unfertilized pistils (arrowheads) compared with wild type and AP2M-GFP ap2m plants.
The ap2m mutant plants exhibited smaller and slender rosette leaves with shorter petioles during vegetative growth compared with wild-type plants (Figure 4C). After bolting, the ap2m mutant plants exhibited unfertilized pistils (Figure 4D) and subsequently produced significantly reduced number of seeds (∼200 seed grains per plant) compared with the wild type (∼2500 seed grains per plant), indicating that fertility of the ap2m mutant plants was severely reduced. These mutant phenotypes were complemented in the AP2M-GFP ap2m plants (Figures 4B to 4D). These results suggest an involvement of AP2M in leaf morphogenesis and the reproductive process.
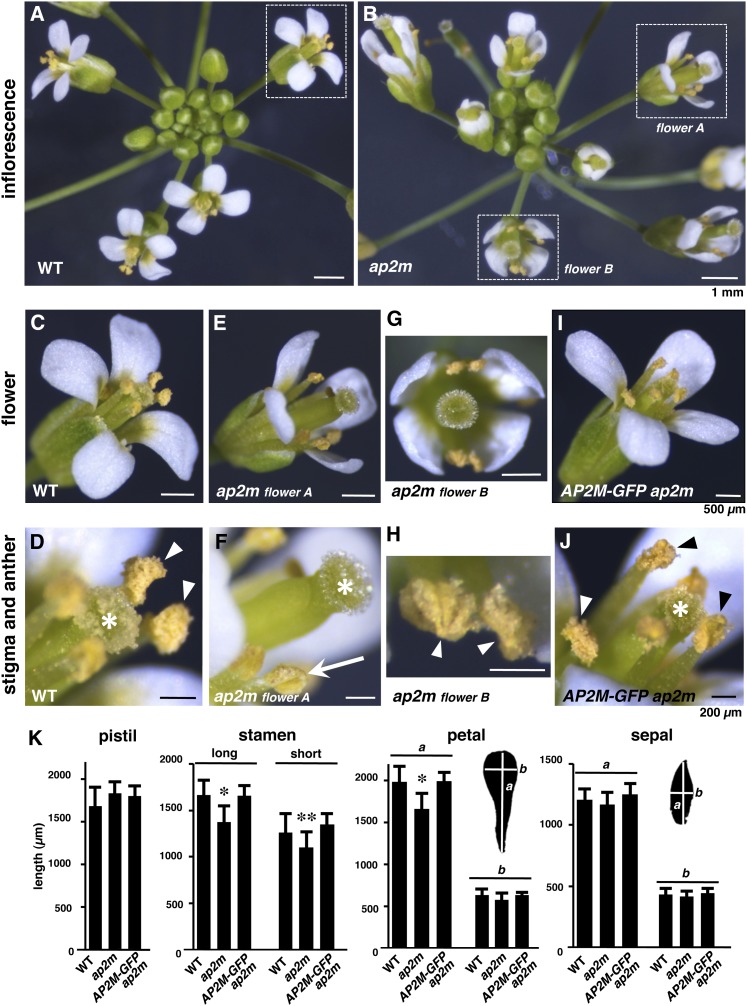
To gain further insight into the role of AP2M in fertility, the floral structures of the ap2m mutant plants were further examined. Compared with the wild type (Figure 5A), the ap2m mutant inflorescences contained morphologically abnormal flowers (Figure 5B). In fully opened wild-type flowers, the stamens were positioned close to the pistils with enough length to reach the stigma (Figure 5C). These organs were tightly surrounded with petals and sepals (Figure 5C). This arrangement of the floral organs leads to efficient pollination (Figure 5D). The ap2m mutant flowers exhibited shorter stamens and incompletely opened petals compared with the wild type (Figure 5E). Dissection of the flowers revealed that the ap2m mutant floral organs exhibited normal numbers and morphology comparable to that of the wild type (see Supplemental Figure 3 online), although their stamens and petals were slightly smaller than those of the wild type (Figure 5K). These defects created the pistil-protruding appearance of the mutant flowers (Figures 5E and 5F). The mutant flowers also exhibited a looser overall structure in which the stamens were positioned away from the pistils (Figure 5G). In addition, the ap2m mutant inflorescences contained flowers with undehisced anthers (Figure 5F) and flowers with dehisced anthers (Figure 5H), suggesting that anther dehiscence was delayed in the mutant plants. These defects led to the nonpollinated stigmas observed in most of the mutant flowers (Figure 5F). These multiple defects in the floral structures and reproductive functions of the ap2m mutant plants were complemented by expression of the AP2M-GFP transgene (Figures 5I to 5K; see Supplemental Figure 3 online).
Figure 5.
Homozygous ap2m Mutant Plants Exhibit Abnormal Floral Organ Development.
(A) to (J) Inflorescence ([A] and [B]), flowers ([C], [E], [G], and [I]), and stigmas and anthers ([D], [F], [H], and [J]) from wild-type (WT; [A], [C], and [D]), homozygous ap2m mutant ([B], [E], to [H]), and AP2M-GFP ap2m plants ([I] and [J]). Higher magnifications of the floral structures with dotted squares in (A) and (B) are shown in (C), (E), and (G). In (D), (F), (H), and (J), asterisks, arrowheads, and arrow indicate stigmas, dehisced anthers, and an undehisced anther, respectively. The sizes of the scale bars in each row are indicated in the panels on the right.
(K) Lengths of pistils, long and short stamens, petals, and sepals from wild-type, homozygous ap2m mutant, and AP2M-GFP ap2m plants. The means + sd from 10 pistils, 40 long stamens, 20 short stamens, 40 petals, and 40 sepals (total 10 flowers) are shown. For petals and sepals, lengths of the long (a) and short (b) axes are shown. *P < 10−3 versus wild-type and AP2M-GFP ap2m plants and **P < 10−2 versus wild-type and AP2M-GFP ap2m plants, t test.
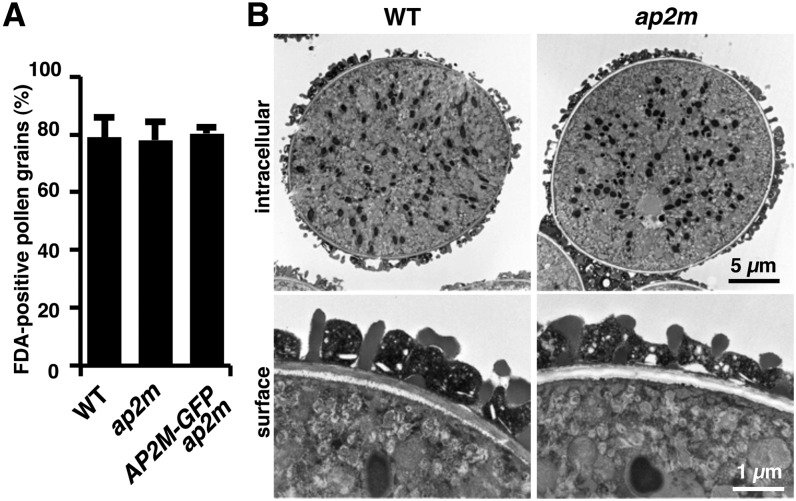
To exclude the possibility that the reduced fertility of the ap2m mutant flowers is due to impaired pollen development, the viability and structure of pollen grains were investigated. Fluorescein diacetate staining indicated that mature ap2m pollen exhibited essentially the same viability as that of the wild type under the plant growth conditions of 22°C and 60% relative humidity (Figure 6A). Transmission electron microscopy revealed that the ultrastructure of mature wild-type pollen grains was indistinguishable from that of the ap2m mutant (Figure 6B). These observations suggest that the ap2m mutation does not strongly affect pollen development. Taken together, these observations suggest that AP2M plays a vital role in floral organ development, which is essential for successful fertilization.
Figure 6.
The ap2m Mutant Pollen Exhibits Normal Viability and Intracellular Structure.
(A) Viability of pollen grains from wild-type (WT), homozygous ap2m mutant, and AP2M-GFP ap2m plants. The means + sd of percentages of pollen grains stained with fluorescein diacetate (FDA) in three independent experiments are shown (n > 300 for each experiment).
(B) Transmission electron micrographs of the intracellular and surface structures of pollen grains from wild-type and homozygous ap2m mutant plants.
DISCUSSION
Arabidopsis AP-2 Complex Is Recruited from the Cytosolic Pool to the Plasma Membrane for Clathrin-Mediated Endocytosis
This study revealed the structure and dynamics of the AP-2 complex on the plasma membrane in Arabidopsis. Immunoprecipitation and subsequent mass spectrometry identified all the predicted AP-2 subunits with high scores when the subunits were isolated in the presence of detergent and salt (Figures 3A to 3C), suggesting that these subunits interact tightly with each other. Detection of the α- and β-subunit isoforms in the mass spectrometry (Figure 3C) implies they are functionally equivalent to each other. Our recent study revealed that both AP1/2B1 and AP1/2B2 also interact with the μ-subunit of AP-1 complex (ADAPTOR PROTEIN-1 MU-ADAPTIN2 [AP1M2]) (Teh et al., 2013), indicating a common use of the β-subunit isoforms by AP-1 and AP-2 as predicted (Boehm and Bonifacino, 2001; Bassham et al., 2008; Dacks et al., 2008). Coimmunoprecipitation assays demonstrated the weak interaction between AP-2 and CHC (Figure 3D), implying that the AP-2 complex is transiently associated with clathrin during CCV formation. Live-cell imaging and subcellular fractionation suggested that the AP-2 complexes are predominantly accumulated in the cytosol, and a part of them is localized to the plasma membrane through interactions between AP2M and cargo proteins (Figures 1A and 1B). The AP2M-GFP fluorescence was remarkably reduced by the TyrA23 treatment (Figures 1C and 1D), suggesting that AP2M interacts with the YXXΦ motif of cargo proteins, and this appears to be the predominant interaction between the AP-2 complex and cargo proteins at the plasma membrane in Arabidopsis. Together, these observations suggest that the AP-2 subunit homologs form a stable protein complex in the cytosol, and their subpopulation is constitutively recruited from the cytosolic pool to the plasma membrane to capture cargo proteins and form CCV for endocytosis.
VAEM Revealed the Dynamics of the AP-2 Complex in Arabidopsis
In studies of animal cells, total internal reflection fluorescence microscopy, which uses evanescent wave fluorescence to visualize intracellular structures near to the plasma membrane with a high signal-to-noise ratio, has been widely used to visualize the formation and internalization of CCV. Theoretically, this technique is not applicable to plant cells because evanescent wave fluorescence would not penetrate cell walls (200 to 400 nm in thickness) to reach the plasma membrane underneath due to its exponential decay within 400-nm depth from the glass cover slip. Instead, VAEM has been used in the studies of plant cells because the excitation laser with a highly oblique yet subcritical angle of incidence is suitable to penetrate the cell wall and illuminate the narrow field adjacent to the plasma membrane with a high signal-to-noise ratio (Fujimoto et al., 2007; Konopka and Bednarek, 2008; Sparkes et al., 2011).
The behavior of the AP2M-GFP structures on the plasma membrane was visualized and quantified using the VAEM technique. Quantitative analysis gave an estimation of formation of one or two punctate structures of AP2M-GFP within 1 min in the 1-μm2 region of the surface of cotyledon epidermal cells. The average lifetime of the structures was ∼1 min (Figure 2), suggesting that the duration of CCV formation and internalization in plant cells is similar to that of mammalian cells (Kirchhausen, 2009). A recent study of CLC and dynamin-related protein (DRP) in Arabidopsis demonstrated that both of the GFP-fused versions of CLC and DRP1C form transient punctate structures on the expanding root epidermal cells at a density of ∼3.5 foci per 1 μm2 with an average lifetime of ∼20 s (Konopka et al., 2008). It also showed that the lifetimes of the structures are different between the expanding and nonexpanding root epidermal cells (Konopka et al., 2008). Two redundant proteins, DRP2B and DRP1A, are colocalized with CLC to form transient punctate structures on the plasma membrane of Arabidopsis cultured cells at a density of ∼1.3 to 1.4 foci per 1 μm2 with average lifetimes of 90 to 100 s (Fujimoto et al., 2010). Simultaneous observations of AP2M-GFP with the other CCV components will give more detailed kinetics of CCV formation and internalization in plant cells. The AP2M-GFP structures exhibited a gradual increase and rapid decrease of fluorescence intensity (Figures 2D and 2E). Similar kinetics of the fluorescent protein–labeled CCV components have been observed in mammalian cells (Merrifield et al., 2002; Ehrlich et al., 2004; Cocucci et al., 2012). The dynamics of the AP2M-GFP fluorescence may reflect the molecular behavior of AP2M on the plasma membrane, including incorporation into CCV and subsequent scission by the activity of DRPs, as is well studied in mammalian cells (Kirchhausen, 2009; McMahon and Boucrot, 2011).
Homozygous ap2m Mutant Plants Are Viable
Homozygous ap2m mutant plants appear to accumulate a truncated version of the AP2M protein lacking the C-terminal half (Figure 4B). This region contains a Trp residue conserved among eukaryotes, which is responsible for efficient interaction with cargo proteins in mammalian cells (see Supplemental Figure 2 online) (Owen and Evans, 1998; Nesterov et al., 1999), implying that the ap2m mutation causes accumulation of a nonfunctional fragment of the AP2M protein. However, homozygous ap2m mutant plants were viable despite abnormal development and reduced fertility (Figures 4C and 4D). These observations may imply that the AP-2 complex is not essential for plant growth and development.
The importance of AP-2 for growth and development varies among different organisms. AP-2 is central to the development of mammals and Drosophila because disruption of the subunit genes leads to lethality (González-Gaitán and Jäckle, 1997; Mitsunari et al., 2005). By contrast, the budding yeast Saccharomyces cerevisiae lacking all three AP β-subunit genes, including the AP-2 homologs, exhibits no deleterious effects on cell growth (Yeung et al., 1999). The only role for the budding yeast AP-2 reported to date is in endocytosis of the killer toxin K28, whereas the internalization of other cargo proteins appears to be independent of AP-2 (Carroll et al., 2009). In the nematode Caenorhabditis elegans, complete loss and C-terminal deletion of the μ-subunit have weak impacts on synaptic vesicle recycling, and the mutants are viable (Gu et al., 2008). The null mutant of the μ-subunit still accumulates the α- and σ-subunits, and the double mutant lacking μ- and α-subunits is embryo lethal (Gu et al., 2013). It has been suggested that the α/σ- and β/μ-subunits have the potential to form large/small subunit heterodimers (Collins et al., 2002; Jackson et al., 2010), which may partially compensate for the loss of AP-2 (Gu et al., 2013). Such mechanisms might complement the lack of AP-2 function in the ap2m mutant plants. Alternatively, it is possible that AP-2 is replaced with unidentified adaptor proteins. A candidate for the unidentified adaptor is the plant-specific protein TPLATE, which has a sequence similarity with AP large subunits at the N terminus where the subunits physically interact with clathrin light and heavy chains (Van Damme et al., 2011). Future investigations on the mechanisms of CCV formation and cargo protein incorporation will provide further information on endocytic systems that may be unique to plants.
AP-2 Plays Roles in Floral Organ Development
In this study, we demonstrated that Arabidopsis AP-2 is required for proper floral organ development. Homozygous ap2m mutant plants exhibited defective flower development, including disrupted stamen elongation, leading to a failure of pollination. This appears to be the major cause of the observed reduction in fertility. The maturation of stamens involves cell elongation of the stamen filament, which synchronizes with development of the other floral organs to maximize the fertilization efficiency (Scott et al., 2004; Wilson et al., 2011). It is possible that AP-2 plays a role in filament cell elongation that requires cell wall biogenesis. Mutations of the genes involved in cell wall biogenesis have been shown to impair multiple aspects of plant development, including floral organ development and petiole elongation (Lane et al., 2001; Williamson et al., 2001). Cell wall biogenesis is likely to require endocytosis because a null mutation of the dynamin-related protein DRP1A reduces cellulose synthesis, leading to impaired cell expansion in various tissues (Collings et al., 2008). A potential target of endocytosis involved in cell wall biogenesis is the cellulose synthase complex, of which internalization under stress conditions has been observed (Crowell et al., 2009). Another reason may be related to phytohormones. The regulation of stamen and anther development is known to require phytohormones, such as jasmonate, gibberellins, and auxin. Mutations of genes involved in the biosynthesis and signaling of these phytohormones affect stamen filament elongation, anther dehiscence, and pollen viability. However, the molecular mechanisms underlying these effects are unknown (Scott et al., 2004; Wilson et al., 2011). AP-2 may play a role in clathrin-mediated endocytosis for the regulation of phytohormone signaling during floral organ development.
METHODS
Plant Materials and Growth Conditions
A mutant Arabidopsis thaliana line in the SAIL collection, SAIL_165_A05 (background Columbia with the homozygous quartet (qrt1) mutation; Preuss et al., 1994), renamed as ap2m, was obtained from the ABRC. The genotype of this line was confirmed by PCR amplification and sequencing of the genomic DNA fragments using the gene-specific primers and the T-DNA–specific primer. Sequences of the primers are as follows: for the gene-specific primers, 5′-GAGGGGAAATTATATAAACTCACTTCC-3′ and 5′-TATTTCCCGCAATAAATAGAGATTATG-3′; for the T-DNA specific primer, 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′. PCR was performed at 96°C for 1 min followed by 35 cycles at 96°C for 15 s, 50°C for 15 s, and 72°C for 1 min, using Ex Taq DNA polymerase (Takara Bio). The homozygous qrt1 mutant plants were used as the wild-type control. Seeds were surface-sterilized and plated onto 0.5% (w/v) gellan gum (Wako) medium containing 1% Suc and Murashige and Skoog salts (Wako), which were supplemented with 7.5 mg/L phosphinothricin as required. The seeds were vernalized and germinated under continuous fluorescent-tube light, and 10-d-old seedlings were transferred onto soil and grown at 22°C with 60% relative humidity under a 16-h-light/8-h-dark regime.
Plasmid Construction and Plant Transformation
The 5-kb sequence including the coding region and the 5′-upstream region of the AP2M gene was amplified using Arabidopsis genomic DNA as template. Sequences of the primer pair are 5′-CACCTGAAACACAAGCATATCGAAGCCAAT-3′ and 5′-GCATCTGATCTCGTAAGATCCCGCTTTCGT-3′. Thirty-five cycles of PCR were performed at 98°C for 10 s, 55°C for 5 s, and 72°C for 25 s using PrimeSTAR Max DNA polymerase (Takara Bio). The amplified fragment was transferred into a binary vector pGWB4 (Nakagawa et al., 2007) using the Gateway cloning system (Invitrogen). The construct was introduced into heterozygous ap2m mutant plants using Agrobacterium tumefaciens–mediated transformation via the floral dip method (Clough and Bent, 1998), and T2 plants homozygous for the T-DNA insertion and the transgene were used for further analysis.
RT-PCR Analysis
Total RNA was isolated from 10-d-old seedlings using an RNeasy plant mini kit (Qiagen) and used for the synthesis of cDNA with Ready-To-Go RT-PCR beads (GE Healthcare) and oligo(dT)12-18 primer (Invitrogen). The cDNA was then used to amplify the full-length coding sequence of AP2M. The partial coding sequence of ACT2, which encodes an actin isoform, was amplified for the loading control. Sequences of the primer pairs are as follows: for full-length AP2M (CDS), 5′-CCGGTGGCTGCTTCCGCCATCTATTTTCTG-3′ and 5′-GCATCTGATCTCGTAAGATCCCGCTTTCGT-3′; for 5′ half of AP2M, 5′-CCGGTGGCTGCTTCCGCCATCTATTTTCTG-3′ and 5′-TCAACCCCAACTTCAAATCAGGCATTCCCG-3′; for 3′ half of AP2M, 5′-ATGATAAAATTGGTCTTGAGAAGGAATCAGAAATG-3′ and 5′-GCATCTGATCTCGTAAGATCCCGCTTTCGT-3′; for ACT2, 5′-AGAGATTCAGATGCCCAGAAGTCTTGTTCC-3′ and 5′-AACGATTCCTGGACCTGCCTCATCA-3′. PCR was performed at 96°C for 1 min followed by 30 cycles at 96°C for 15 s, 50°C for 15 s, and 72°C for 90 and 30 s for AP2M and ACT2, respectively, using Ex Taq DNA polymerase (Takara Bio). The PCR products were separated on 1% agarose gel and visualized by ethidium bromide staining.
Confocal Microscopy
Fluorescence from root tip cells and cotyledon epidermal cells of 4- to 6-d-old AP2M-GFP seedlings stained with 0.5 μM FM4-64 were excited with a 488-nm 25-mW Ar/Kr laser and a 561-nm 20-mW diode-pumped solid-state laser. The images were acquired with a Carl Zeiss LSM780 confocal laser scanning microscope equipped with a ×63 C-Apochromat water immersion objective (numerical aperture = 1.2). Images were processed using LSM image examiner software (Carl Zeiss) and Adobe Photoshop CS4 version 11.0.2 (Adobe Systems).
VAEM
Fluorescence from root tip cells and cotyledon epidermal cells of 4- to 6-d-old AP2M-GFP seedlings were excited with a Coherent Sapphire 488-nm laser. The epifluorescence and VAEM images were acquired with an Olympus IX83 fluorescence microscope equipped with an IX2-RFAEVA total internal reflection fluorescence microscopy system and a ×100 UApoN OTIRF oil immersion objective (numerical aperture = 1.49). Images were processed using MetaMorph version 7.8 (Molecular Devices) and Image J version 1.47q. A macro routine was designed to measure fluorescence intensity and lifetime of the punctate structures: Time series of VAEM image data were corrected for photobleaching and averaged to obtain the fluorescence maxima. Fluorescence intensity of the maxima at each time point was measured and then the time intensity curves of the maxima were subjected to smoothing by the Savitzky-Golay filter (Savitzky and Golay, 1964) and subsequent differentiation followed by resmoothing. The corresponding differential curves were then used to obtain durations between their maximum and minimum, which indicate the lifetimes of the AP2M-GFP punctate structures.
Subcellular Fractionation
Approximately 3 g of 7-d-old seedlings of the AP2M-GFP plants were chopped with a razor blade in a Petri dish on ice in 9 mL of chopping buffer containing 50 mM HEPES-KOH, pH 7.5, 300 mM Suc, and a protease inhibitor cocktail (Roche), filtered through a cell strainer with 70-μm pores (BD Biosciences), and centrifuged at 1000g for 10 min at 4°C. Four milliliters of the supernatant was used as the total fraction, and the second 4 mL was centrifuged at 10,000g for 10 min at 4°C. The pellet (P10) was resuspended in 4 mL of chopping buffer, and the supernatant was further centrifuged at 100,000g for 30 min at 4°C. The pellet (P100, microsome fraction) was resuspended in 4 mL of chopping buffer, and the supernatant was used as the S100 (soluble) fraction. Aliquots of the total, P10, P100, and S100 suspensions were separated by SDS-PAGE and used for immunoblot analysis using anti-VAM3 antibody (Sato et al., 1997) and anti-ALEU antibody (Rogers et al., 1997). The remaining suspensions were mixed with magnetic beads conjugated with anti-GFP antibody (Miltenyi Biotec), incubated on ice for 30 min, and then applied to μ-Columns (Miltenyi Biotec) in a magnetic field. After washing the column with chopping buffer, the immunoprecipitates were eluted with 50 μL of 2 × SDS sample buffer (100 mM Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [w/v] glycerol, and 5% [v/v] 2-mercaptoethanol), separated by SDS-PAGE, and used for immunoblot analysis using a monoclonal anti-GFP antibody (JL-8; Clontech). Full scans of the immunoblots shown in Figure 1B are provided (see Supplemental Figure 4A online).
Mass Spectrometry
Approximately 1 g of 7-d-old seedlings of the AP2M-GFP plants and the transgenic Arabidopsis plants expressing GFP were homogenized on ice in 3 mL of buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% (v/v) Triton X-100, and a protease inhibitor cocktail (Roche) and centrifuged at 20,000g for 15 min at 4°C. The supernatants were immunoprecipitated as described above, separated by SDS-PAGE, and stained with Flamingo Gel Stain (Bio-Rad). The immunoprecipitates were then digested in-gel with 0.01 mg/mL trypsin and examined by mass spectrometry according to the methods described previously (Tamura et al., 2010).
Immunoprecipitation with Clathrin
Approximately 0.5 g of 7-d-old seedlings of the AP2M-GFP plants and the transgenic Arabidopsis plants expressing GFP were homogenized on ice in 1.5 mL of buffer containing 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% (v/v) Triton X-100, and a protease inhibitor cocktail (Roche) in the presence and absence of 150 mM NaCl and then centrifuged at 20,000g for 15 min at 4°C. The supernatants were immunoprecipitated as described above, separated by SDS-PAGE, and used for immunoblot analysis using the monoclonal anti-GFP antibody and a polyclonal anti-CHC antibody (a gift from D.G. Robinson and P. Oliviusson). Full scans of the immunoblots shown in Figure 3D are provided (see Supplemental Figure 4B online).
Transmission Electron Microscopy
Pollen grains were fixed with 4% paraformaldehyde and 2% glutaraldehyde in 50 mM cacodylate buffer, pH 7.4, at 4°C overnight followed by further fixation with 2% osmium tetroxide in 50 mM cacodylate buffer at 4°C for 10 h. After dehydration with a series of aqueous ethanol solutions followed by propylene oxide, they were embedded in resin, ultrathin sectioned using an ultramicrotome (Leica), and stained with 2% uranyl acetate. The images were acquired by a JEM-1200EX transmission electron microscope (JEOL) equipped with an Olympus VELETA charge-coupled device digital camera and processed using Adobe Photoshop CS4.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g22770 (AP2A1), At5g22780 (AP2A2), At4g11380 (AP1/2B1), At4g23460 (AP1/2B2), At5g46630 (AP2M), At1g47830 (AP2S), At1g60780 (AP1M2), At3g18780 (ACT2), NP_004059.2 (Homo sapiens AP-2 μ-subunit), NP_446289.1 (Rattus norvegicus AP-2 μ-subunit), NP_732744.1 (Drosophila melanogaster AP-2 μ-subunit), NP_001024865.1 (Caenorhabditis elegans AP-2 μ-subunit), and NP_592921.1 (Schizosaccharomyces pombe AP-2 μ-subunit).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Images of the Cotyledon Epidermal Cells of AP2M-GFP ap2m Seedling.
Supplemental Figure 2. Sequence Alignment of the AP-2 μ-Subunits from Various Eukaryotes and the Predicted AP2M Sequence Expressed in the ap2m Mutant Plants.
Supplemental Figure 3. Morphology of Floral Organs Dissected from Wild-Type, Homozygous ap2m Mutant, and AP2M-GFP ap2m Plants.
Supplemental Figure 4. Full Scans of the Immunoblots Shown in Figures 1B and 3D.
Supplemental Table 1. List of the Immunoprecipitated Proteins from AP2M-GFP ap2m Seedlings Identified by the First LTQ-Orbitrap Mass Spectrometric Analysis.
Supplemental Table 2. List of the Immunoprecipitated Proteins from AP2M-GFP ap2m Seedlings Identified by the Second LTQ-Orbitrap Mass Spectrometric Analysis.
Supplemental Movie 1. Transient Formation of AP2M-GFP Punctate Structures on the Plasma Membrane of a Cotyledon Epidermal Cell of an AP2M-GFP ap2m Seedling.
Acknowledgments
We thank David G. Robinson and Peter Oliviusson for providing anti-CHC antibody, and Ooi-kock Teh and Tadashi Kunieda for technical assistance and discussions. This work was supported by Grants-in-Aid for Specially Promoted Research to I.H.-N. (22000014), and Grants-in-Aid for Scientific Research to S.Y. (25120715 and 25840108) and K.T. (25650096) from the Japan Society for the Promotion of Science.
AUTHOR CONTRIBUTIONS
T.S., S.Y., M.S., and I.H.-N. conceived the study and designed the experiments. S.Y., Y.S., M.S., T.K., K.T., and T.S. performed live-cell imaging analysis. T.K. and S.Y. developed a macro routine for VAEM imaging analysis. S.Y., Y.S., and Y.F. performed mass spectrometry. S.Y., Y.S., N.H., and T.S. investigated plant development. S.Y., T.S., and I.H.-N. wrote the article.
Glossary
- TGN
trans-Golgi network
- CCV
clathrin-coated vesicle
- VAEM
Variable-angle epifluorescence microscopy
- CHC
clathrin heavy chain
- CLC
clathrin light chain
References
- Banbury D.N., Oakley J.D., Sessions R.B., Banting G. (2003). Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278: 12022–12028 [DOI] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C., Brandizzi F., Otegui M.S., Sanderfoot A.A. (2008). The secretory system of Arabidopsis. In The Arabidopsis Book 6: e0116, /10.1199/tab.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Bonifacino J.S. (2001). Adaptins: The final recount. Mol. Biol. Cell 12: 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W., Ohno H., Songyang Z., Rapoport I., Cantley L.C., Bonifacino J.S., Kirchhausen T. (1996). Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15: 5789–5795 [PMC free article] [PubMed] [Google Scholar]
- Bos K., Wraight C., Stanley K.K. (1993). TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 12: 2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Saffarian S., Zhang R., Kirchhausen T. (2010). Roles of AP-2 in clathrin-mediated endocytosis. PLoS ONE 5: e10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos P.V., Mardones G.A., Rojas A.L., daSilva L.L.P., Prabhu Y., Hurley J.H., Bonifacino J.S. (2010). Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev. Cell 18: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.Y., Stirling P.C., Stimpson H.E.M., Giesselmann E., Schmitt M.J., Drubin D.G. (2009). A yeast killer toxin screen provides insights into a/b toxin entry, trafficking, and killing mechanisms. Dev. Cell 17: 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cocucci E., Aguet F., Boulant S., Kirchhausen T. (2012). The first five seconds in the life of a clathrin-coated pit. Cell 150: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings D.A., Gebbie L.K., Howles P.A., Hurley U.A., Birch R.J., Cork A.H., Hocart C.H., Arioli T., Williamson R.E. (2008). Arabidopsis dynamin-like protein DRP1A: A null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J. Exp. Bot. 59: 361–376 [DOI] [PubMed] [Google Scholar]
- Collins B.M., McCoy A.J., Kent H.M., Evans P.R., Owen D.J. (2002). Molecular architecture and functional model of the endocytic AP2 complex. Cell 109: 523–535 [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.-D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks J.B., Poon P.P., Field M.C. (2008). Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. USA 105: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E.C. (2009). AP-3-dependent trafficking and disease: The first decade. Curr. Opin. Cell Biol. 21: 552–559 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.-D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M.L., Kirchhausen T. (2004). Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118: 591–605 [DOI] [PubMed] [Google Scholar]
- Feraru E., Paciorek T., Feraru M.I., Zwiewka M., De Groodt R., De Rycke R., Kleine-Vehn J., Friml J. (2010). The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Nakazono M., Tsutsumi N. (2007). Imaging of plant dynamin-related proteins and clathrin around the plasma membrane by variable incidence angle fluorescence microscopy. Plant Biotechnol. 24: 449–455 [Google Scholar]
- Fujimoto M., Arimura S., Ueda T., Takanashi H., Hayashi Y., Nakano A., Tsutsumi N. (2010). Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. USA 107: 6094–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Gaitán M., Jäckle H. (1997). Role of Drosophila α-adaptin in presynaptic vesicle recycling. Cell 88: 767–776 [DOI] [PubMed] [Google Scholar]
- Gu, M., Liu, Q., Watanabe, S., Sun, L., Hollopeter, G., Grant, B.D., and Jorgensen, E.M. (2013). AP2 hemicomplexes contribute independently to synaptic vesicle endocytosis. Elife 2: e00190. [DOI] [PMC free article] [PubMed]
- Gu M., Schuske K., Watanabe S., Liu Q., Baum P., Garriga G., Jorgensen E.M. (2008). μ2 adaptin facilitates but is not essential for synaptic vesicle recycling in Caenorhabditis elegans. J. Cell Biol. 183: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N., Höning S., Neuhaus J.-M., Paris N., Robinson D.G., Holstein S.E. (2004). Arabidopsis μ A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Hirst J., Barlow L.D., Francisco G.C., Sahlender D.A., Seaman M.N.J., Dacks J.B., Robinson M.S. (2011). The fifth adaptor protein complex. PLoS Biol. 9: e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Borner G.H.H., Antrobus R., Peden A.A., Hodson N.A., Sahlender D.A., Robinson M.S. (2012). Distinct and overlapping roles for AP-1 and GGAs revealed by the “knocksideways” system. Curr. Biol. 22: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey J.S., Peters P.J., Yuan L.C., Bonifacino J.S. (1993). Localization of TGN38 to the trans-Golgi network: Involvement of a cytoplasmic tyrosine-containing sequence. J. Cell Biol. 120: 1123–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589 [DOI] [PubMed] [Google Scholar]
- Jackson L.P., Kelly B.T., McCoy A.J., Gaffry T., James L.C., Collins B.M., Höning S., Evans P.R., Owen D.J. (2010). A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141: 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk L.M., Hachey J., Lynch D.R., Kulcsar F., van Rooijen G., Waterer D.R., Robertson A., Kokko E., Byers R., Howard R.J., Fischer R., Prüfer D. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. (2009). Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 19: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196 [DOI] [PubMed] [Google Scholar]
- Lane D.R., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Boucrot E. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12: 517–533 [DOI] [PubMed] [Google Scholar]
- Merrifield C.J., Feldman M.E., Wan L., Almers W. (2002). Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4: 691–698 [DOI] [PubMed] [Google Scholar]
- Mitsunari T., Nakatsu F., Shioda N., Love P.E., Grinberg A., Bonifacino J.S., Ohno H. (2005). Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 25: 9318–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Bright N.A., Seaman M.N.J., Robinson M.S. (2003). Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nesterov A., Carter R.E., Sorkina T., Gill G.N., Sorkin A. (1999). Inhibition of the receptor-binding function of clathrin adaptor protein AP-2 by dominant-negative mutant μ2 subunit and its effects on endocytosis. EMBO J. 18: 2489–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihama M., Takemoto N., Hashiguchi Y., Tasaka M., Morita M.T. (2009). ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 50: 2057–2068 [DOI] [PubMed] [Google Scholar]
- Ohno H., Stewart J., Fournier M.-C., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., Bonifacino J.S. (1995). Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269: 1872–1875 [DOI] [PubMed] [Google Scholar]
- Owen D.J., Evans P.R. (1998). A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Song K., Reichardt I., Kim H., Mayer U., Stierhof Y.-D., Hwang I., Jürgens G. (2013). Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc. Natl. Acad. Sci. USA 110: 10318–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Robinson M.S. (2004). Adaptable adaptors for coated vesicles. Trends Cell Biol. 14: 167–174 [DOI] [PubMed] [Google Scholar]
- Rogers S.W., Burks M., Rogers J.C. (1997). Monoclonal antibodies to barley aleurain and homologs from other plants. Plant J. 11: 1359–1368 [DOI] [PubMed] [Google Scholar]
- Sato M.H., Nakamura N., Ohsumi Y., Kouchi H., Kondo M., Hara-Nishimura I., Nishimura M., Wada Y. (1997). The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 272: 24530–24535 [DOI] [PubMed] [Google Scholar]
- Savitzky A., Golay M.J.E. (1964). Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36: 1627–1639 [Google Scholar]
- Scott R.J., Spielman M., Dickinson H.G. (2004). Stamen structure and function. Plant Cell 16 (suppl.): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Graumann K., Martinière A., Schoberer J., Wang P., Osterrieder A. (2011). Bleach it, switch it, bounce it, pull it: Using lasers to reveal plant cell dynamics. J. Exp. Bot. 62: 1–7 [DOI] [PubMed] [Google Scholar]
- Stephens D.J., Crump C.M., Clarke A.R., Banting G. (1997). Serine 331 and tyrosine 333 are both involved in the interaction between the cytosolic domain of TGN38 and the μ2 subunit of the AP2 clathrin adaptor complex. J. Biol. Chem. 272: 14104–14109 [DOI] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh O.-K., Shimono Y., Shirakawa M., Fukao Y., Tamura K., Shimada T., Hara-Nishimura I. (2013). The AP-1 μ adaptin is required for KNOLLE localization at the cell plate to mediate cytokinesis in Arabidopsis. Plant Cell Physiol. 54: 838–847 [DOI] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D., Gadeyne A., Vanstraelen M., Inzé D., Van Montagu M.C.E., De Jaeger G., Russinova E., Geelen D. (2011). Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc. Natl. Acad. Sci. USA 108: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-G., Li S., Zhao X.-Y., Zhou L.-Z., Huang G.-Q., Xie H.T., Feng C., Zhang Y. (2013). HAPLESS13, the Arabidopsis μ1 adaptin, is essential for protein sorting at the trans-Golgi network/early endosome. Plant Physiol. 162: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe E., Shimada T., Tamura K., Matsushima R., Koumoto Y., Nishimura M., Hara-Nishimura I. (2004). An ER-localized form of PV72, a seed-specific vacuolar sorting receptor, interferes the transport of an NPIR-containing proteinase in Arabidopsis leaves. Plant Cell Physiol. 45: 9–17 [DOI] [PubMed] [Google Scholar]
- Williamson R.E., Burn J.E., Birch R., Baskin T.I., Arioli T., Betzner A.S., Cork A. (2001). Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215: 116–127 [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Song J., Taylor B., Yang C. (2011). The final split: The regulation of anther dehiscence. J. Exp. Bot. 62: 1633–1649 [DOI] [PubMed] [Google Scholar]
- Yeung B.G., Phan H.L., Payne G.S. (1999). Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell 10: 3643–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zwiewka M., Feraru E., Möller B., Hwang I., Feraru M.I., Kleine-Vehn J., Weijers D., Friml J. (2011). The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21: 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]